Abstract
Genome‐wide association studies (GWAS) have identified several risk loci for nonalcoholic fatty liver disease (NAFLD). Previous studies have largely relied on small sample sizes and have assessed quantitative traits. We performed a case‐control GWAS in the UK Biobank using recorded diagnosis of NAFLD based on diagnostic codes recommended in recent consensus guidelines. We performed a GWAS of 4,761 cases of NAFLD and 373,227 healthy controls without evidence of NAFLD. Sensitivity analyses were performed excluding other co‐existing hepatic pathology, adjusting for body mass index (BMI) and adjusting for alcohol intake. A total of 9,723,654 variants were assessed by logistic regression adjusted for age, sex, genetic principal components, and genotyping batch. We performed a GWAS meta‐analysis using available summary association statistics. Six risk loci were identified (P < 5*10−8) (apolipoprotein E [APOE], patatin‐like phospholipase domain containing 3 [PNPLA3, transmembrane 6 superfamily member 2 [TM6SF2], glucokinase regulator [GCKR], mitochondrial amidoxime reducing component 1 [MARC1], and tribbles pseudokinase 1 [TRIB1]). All loci retained significance in sensitivity analyses without co‐existent hepatic pathology and after adjustment for BMI. PNPLA3 and TM6SF2 remained significant after adjustment for alcohol (alcohol intake was known in only 158,388 individuals), with others demonstrating consistent direction and magnitude of effect. All six loci were significant on meta‐analysis. Rs429358 (P = 2.17*10−11) is a missense variant within the APOE gene determining ϵ4 versus ϵ2/ϵ3 alleles. The ϵ4 allele of APOE offered protection against NAFLD (odds ratio for heterozygotes 0.84 [95% confidence interval 0.78‐0.90] and homozygotes 0.64 [0.50‐0.79]). Conclusion: This GWAS replicates six known NAFLD‐susceptibility loci and confirms that the ϵ4 allele of APOE is associated with protection against NAFLD. The results are consistent with published GWAS using histological and radiological measures of NAFLD, confirming that NAFLD identified through diagnostic codes from consensus guidelines is a valid alternative to more invasive and costly approaches.

Abbreviations
- ALT
alanine aminotransferase
- APOE
apolipoprotein E
- BMI
body mass index
- CI
confidence interval
- EHR
electronic health record
- GCKR
glucokinase regulator
- GS‐SFHS
Generation Scotland: Scottish Family Health Study
- GWAS
genome‐wide association study
- HbA1c
glycated hemoglobin
- HER
electronic health record
- HSD17B13
hydroxysteroid 17‐beta dehydrogenase 13
- LD
linkage disequilibrium
- MARC1
mitochondrial amidoxime reducing component 1
- NAFLD
nonalcoholic fatty liver disease
- OR
odds ratio
- PNPLA3
patatin‐like phospholipase domain containing 3
- SNP
single nucleotide polymorphism
- TM6SF2
transmembrane 6 superfamily member 2
- TRIB1
tribbles pseudokinase 1
- UKB
UK Biobank
- VLDL
very‐low‐density lipoprotein
Nonalcoholic fatty liver disease (NAFLD) is the most common form of metabolic disease worldwide with an estimated prevalence of 24%, which appears to be increasing in all populations.( 1 ) NAFLD has an even higher prevalence in those with other forms of metabolic disease including obesity, type 2 diabetes mellitus and hyperlipidemia, which are rising in prevalence. Therefore, the prevalence of NAFLD is also anticipated to rise.( 1 )
NAFLD covers a spectrum of disease severity including elevated hepatic triglyceride content (isolated steatosis), inflammation (nonalcoholic steatohepatitis), fibrosis and cirrhosis, and is associated with elevated risk of further morbidity. It has risen to the second‐most‐common indication for liver transplantation in the United States and carries a significantly elevated risk of hepatocellular carcinoma, cardiovascular disease, diabetes mellitus, and all‐cause mortality.( 1 ) However, the pathogenesis of NAFLD is complex, and disease progression is highly variable. Despite contributing to significant morbidity and mortality, there are no licensed pharmacological therapies for NAFLD, and several agents under investigation have thus far shown limited effectiveness.( 2 )
Development of NAFLD is influenced by both environmental and genetic factors, its heritability estimated at 22%‐50%.( 3 ) Several genome‐wide association studies (GWASs) have been published examining various phenotype definitions for NAFLD or NAFLD severity encompassing radiological evidence of hepatic steatosis, visceral fat content, lean NAFLD (NAFLD in a nonobese individual), and histological severity of fibrosis. These studies have highlighted several loci and candidate mechanisms underlying NAFLD pathogenesis.( 4 , 5 , 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 )
Studies using a case‐control GWAS methodology have identified five loci associated with NAFLD at genome‐wide significance (P < 5*10−8; mitochondrial amidoxime reducing component 1 (MARC1), glucokinase regulator (GCKR), hydroxysteroid 17‐beta dehydrogenase 13 (HSD17B13), transmembrane 6 superfamily member 2 (TM6SF2), and patatin‐like phospholipase domain containing 3 [PNPLA3]).( 7 , 8 , 9 , 14 , 15 , 16 , 17 , 20 ) Known loci are shown in Supporting Tables S1 and S2. Most case‐control analyses have relied on histological and radiological confirmation of NAFLD limiting the sample size in these studies. Use of routinely collected administrative data allows for larger cohorts and reduces the need for invasive or expensive investigations such as biopsy or magnetic resonance imaging (MRI).( 25 ) One published GWAS relied on natural language processing to identify NAFLD cases from electronic health record (EHR) results and found significant associations at loci previously identified in cohorts with complete histological classification.( 15 ) Studies such as the UK Biobank (UKB) have extensive genotypic data linked to hospital and primary care discharge codes.
A subgroup of 14,440 participants in the UKB has been analyzed in a published GWAS focusing on MRI‐based measures of steatohepatitis and fibrosis.( 18 ) Separately a case control for all‐cause cirrhosis using several sources including UKB has been published with an additional single variant analysis for NAFLD at rs2642438 using only limited International Classification of Diseases, 10th Revision diagnostic codes for NAFLD.( 17 ) Finally, a recent exome‐wide association study analyzed association of missense and nonsense mutations with serum alanine aminotransferase (ALT) before analyzing significant variants for association with hepatic fat content in a subset of 8,930 UKB participants.( 22 ) The analysis identified several known NAFLD‐susceptibility variants as well as three NAFLD‐susceptibility loci (apolipoprotein E [APOE], glycerol‐3‐phosphate acyltransferase 1 mitochondrial [GPAM], and olfactory receptor family 12 subfamily D member 2 [OR12D2]), although APOE was identified in an earlier subanalysis.( 18 ) There are no published case‐control GWAS using administrative data based on recent consensus recommendations.( 25 ) We therefore undertook a GWAS of participants with recorded diagnostic codes attributed to NAFLD compared with healthy controls.
Materials and Methods
Diagnostic codes used for the identification of NAFLD are shown in Supporting Tables S3 and S4. Methods for identification of participants, determination of NAFLD status, and genotyping for the UKB and Generation Scotland–Scottish Family Health Study (GS‐SFHS) are provided in Supporting Materials 2. The UKB received ethical approval (research ethical committee reference 11/NW/0382). UKB data access was approved under projects 30439 (phenotype data) and 19655 (genotype data). Ethical approval for GS‐SFHS was granted by National Health Service Tayside Research Ethics Committee (REC reference number 05/S1401/89).
GWAS
Initial Analysis
Genotyped and imputed single nucleotide polymorphisms (SNPs) were analyzed. Participants of self‐reported European ancestry were considered eligible for inclusion. Outliers for heterozygosity and unexpected runs of homozygosity were excluded. One participant from each pair of related individuals in the UKB (Kinship > 0.0884( 26 )) was excluded.
Association with NAFLD was analyzed using logistic regression adjusted for age, sex, the first 20 genetic principal components, and batch, with batch included as a random effect. Imputation dosage was used for imputed SNPs.
Quality control to exclude SNPs with low minor allele frequency (), low imputation quality score (), and deviation from Hardy‐Weinberg Equilibrium (HWE: P < 5*10−6) were applied. Genome‐wide significance was determined as P < 5*10−8 in the UKB and P < 0.0083 in the GS‐SFHS replication cohort (P value of 0.05 subjected to Bonferroni correction for each locus tested). Replication was undertaken by analyzing the lead SNP within each locus for the GS‐SFHS study.
Linkage Disequilibrium Clumping and Conditional Analyses
SNPs showing genome‐wide significance (P < 5*10−8) were considered significant. Linkage disequilibrium (LD) clumping was performed using the functional mapping and annotation of GWAS (FUMA) web application v1.3.5d (https://fuma.ctglab.nl/). ( 27 ) Loci were established for lead SNPs with a minimum distance of 250 Kb between loci and using an r 2 < 0.25 to indicate independent SNPs within the same locus. Full details of the parameters passed to FUMA are available in Supporting Materials3A.
Each locus was re‐analyzed while conditioning on the lead SNP, and further signals with genome‐wide significance were identified. This process was repeated until no remaining SNPs reached genome‐wide significance.
Population Stratification
The genomic inflation factor was calculated using the summary association statistics. Evidence of test statistic inflation in was investigated with LD score regression.( 28 )
Sensitivity Analyses
Sensitivity analyses were conducted to ensure the robustness of the case definition. First, the analysis was conducted at each of the NAFLD‐susceptibility loci after exclusion of individuals with alternative hepatic pathology as described previously. Second, each of the analyses was conducted adjusting for body mass index (BMI) as a covariate. Third, the analysis was conducted adjusting for estimated consumption of alcohol (units per day). The alcohol estimate was derived from a 24‐hour dietary recall questionnaire during online follow‐up and was available for about 40% of participants. The logistic regression methodology for the sensitivity analyses was identical to the main GWAS.
An additional analysis in the UKB cohort was undertaken to assess the relationship between APOE genotype and NAFLD based on the GWAS results (one lead SNP was a missense mutation within APOE). See Supporting Materials 2 for methods explaining the determination of APOE alleles and statistical analysis of association with NAFLD.
Association of NAFLD‐Susceptibility Loci With Phenotypic Traits
NAFLD is strongly associated with obesity, hyperlipidemia, hyperglycemia, inflammation, and deranged liver function tests, in particular ALT.( 29 , 30 ) Associations with serum biochemistry values for each identified variant were assessed in an age‐adjusted and sex‐adjusted linear regression model. Serum ALT, aspartate aminotransferase, gamma‐glutamyltransferase, alkaline phosphatase, total cholesterol (TC), low‐density lipoprotein cholesterol (LDL‐C), high‐density lipoprotein cholesterol, triglycerides, apolipoprotein A and B, lipoprotein A, glucose, glycated hemoglobin (HbA1c), C‐reactive protein, BMI, waist circumference, hip circumference, and waist‐to‐hip ratio were assessed. Each trait was assessed visually through histogram and log‐transformed in the case of skewed distribution. To evaluate potential amplification of steatogenic effects in the context of obesity, insulin resistance and alcohol intake,( 21 , 31 ) a gene‐environment interaction was assessed for each NAFLD‐susceptibility variant with BMI, HbA1c, and alcohol intake.
A linear regression adjusted for age, sex, the first 20 genetic principal components, and genotyping batch was undertaken for the association of NAFLD‐susceptibility variants with the Fibrosis‐4 Index (FIB‐4) score and the NAFLD fibrosis score; both validated noninvasive measures associated with NAFLD‐related fibrosis.( 32 , 33 )
Genome‐wide Association Meta‐analysis
Two NAFLD case‐control GWASs with available summary association statistics (Namjou et al,( 15 ) 1,106 cases and 8,571 controls; and Anstee et al,( 16 ) 1,483 cases and 17,781 controls) from independent populations of European ancestry were also assessed. Namjou et al. reported on participants from American health centers and relied on natural language processing to ascertain cases of NAFLD from the EHR, and therefore was more closely related to the methodology used in this study. Namjou et al. performed a case‐control logistic regression adjusted for age, sex, BMI, the medical center, and the first three genetic principal components. Anstee et al. recruited participants from European tertiary liver centers with biopsy‐proven NAFLD and compared them to population controls with a linear mixed model adjusting for sex and the first five genetic principal components before repeating the analysis as a logistic regression. Identified lead SNPs from the UKB GWAS were inspected in these studies for direction and magnitude of effect.
Meta‐analysis was conducted using METAL( 34 ) with an inverse‐variance fixed‐effects meta‐analysis. Although between‐study heterogeneity was expected, the fixed‐effects model was used in preference to the random effects model due to the low number of studies and anticipated deviation from the Gaussian distribution required for a random effects model.( 35 )
Plotting and Statistical Analysis of Results
Genomic analyses were performed using SNPtest (version 2.5.2),( 36 ) QCTOOLS (version 2.0.6), and METAL (2018 version) on the University of Edinburgh Linux high‐performance compute cluster. Post‐GWAS analysis, regression analyses, and plotting were performed using R version 3.6.3.( 37 ) Methods and results are reported in accordance with the STREGA (STrengthening the REporting of Genetic Association Studies) guidelines( 38 ) (Supporting Materials 4).
Results
NAFLD Cohorts
A total of 502,616 participants entered the UKB study with 377,998 taken forward for GWAS. A history of NAFLD was present in 4,761 with 373,227 controls. After exclusion of alternative hepatic pathology there were 3,954 NAFLD cases and 355,942 controls. The median follow‐up was 9.0 years (range 7.3‐11.9). Compared with controls, cases were more likely to be male (51.0% vs. 46.3%), older (mean 57.4 vs. 56.9 years), heavier (mean 89.2 vs. 78.2 kg), and diabetic (32.9% vs. 7.7%). The baseline characteristics are shown in Supporting Materials 1 (Supporting Table S5).
A total of 24,096 participants entered GS‐SFHS with 6,317 taken forward for GWAS. A history of NAFLD was present in 67 with 6,250 controls (both without alternative pathologies). The mean follow‐up was 11.2 years (range 9.6‐14.7 years) (Fig. 1).
FIG. 1.
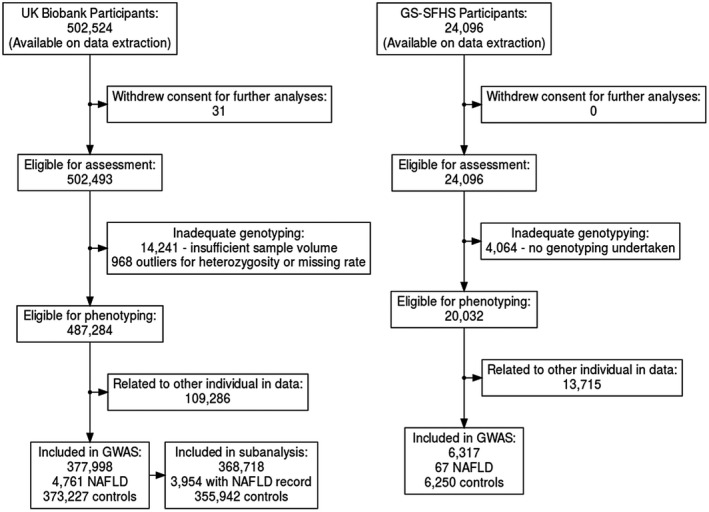
Flowchart describing participant recruitment in UKB and GS‐SFHS.
GWAS
Initial Analysis
A total of 460 SNPs were identified with genome‐wide significant P values after removal of low‐quality SNPs in the UKB. A total of 1,313 SNPs demonstrated borderline significance (P < 5*10−5). LD clumping revealed six loci with at least one significant NAFLD‐associated signal. Significant associations were seen with rs2642442 (MARC1 intron), rs1260326 (GCKR exon), rs17321515 (tribbles pseudokinase 1 [TRIB1] intergenic), rs73001065 (maternal‐effect uncoordinated sister chromatid cohesion factor [MAU2] intron), rs429358 (APOE exon), and rs3747207 (PNPLA3 intron). Rs73001065 is in strong linkage with the previously identified missense variant (rs58542926) within TM6SF2 (R 2 = 0.82),( 14 , 16 ) which is thought to be the causal variant.( 39 ) Rs3747207 is in perfect linkage (R 2 = 1) with the established protein‐altering variant rs738409. In both cases, P values of the lead SNP and the putative causal variants were almost identical. Loci boundaries of the loci are shown in Supporting Materials 3B and the results at the 460 significant SNPs in Supporting Materials 5.
Of the six loci, all six are known from previous case‐control GWASs of NAFLD or related traits such as hepatic steatosis, including the recently identified APOE variant (rs429358( 18 , 21 , 22 , 23 )) (Table 1). One locus (rs1260326) was replicated within the GS‐SFHS cohort. Of the five loci established by previous case‐control format GWASs of NAFLD, we replicated all five with four reaching genome‐wide significance (MARC1, GCKR, TM6SF2, and PNPLA3) and one (HSD17B13, P = 7.41−03) reaching the Bonferroni‐corrected threshold (see Table 2, Supporting Materials 1, and Supporting Table S2). We replicated three of the four significant loci reported by Anstee et al. and the only significant locus identified by Namjou et al. in their case‐control GWAS (P < 0.0083; Supporting Materials 1 and Supporting Table S2). Of the two loci identified previously by quantitative trait GWAS but not case‐control GWAS, TRIB1 reached the replication threshold in both Anstee et al. and Namjou et al., whereas APOE reached the replication threshold in Anstee et al. and nominal significance in Namjou et al. Both TRIB1 and APOE have been identified in other GWASs.
TABLE 1.
Loci Associated With NAFLD in the UKB Cohort
| RSID | Chr:Pos | Reference Allele/Effect Allele | EAF | OR (95% CI) | P Value | Consequence (Gene) | Hypothesized Functional Gene |
|---|---|---|---|---|---|---|---|
| rs2642442 | 1:220973563 | C/T | 0.317 | 1.15 (1.10‐1.20) | 7.67−10 | Intron (MARC1) | MARC1 |
| rs1260326 | 2:27730940 | T/C | 0.392 | 0.87 (0.84‐0.91) | 2.54−11 | Missense (GCKR) | GCKR |
| rs17321515 | 8:126486409 | A/G | 0.476 | 0.86 (0.82‐0.89) | 1.81−13 | Intergenic (TRIB1) | TRIB1 |
| rs73001065 | 19:19460541 | G/C | 0.071 | 1.41 (1.32‐1.51) | 1.08−24 | Intron (MAU2) | TM6SF2 |
| rs429358 | 19:45411941 | T/C | 0.156 | 0.82 (0.77‐0.87) | 2.17−11 | Missense (APOE) | APOE |
| rs3747207 | 22:44324855 | G/A | 0.215 | 1.45 (1.38‐1.51) | 6.74−60 | Intron (PNPLA3) | PNPLA3 |
Functional role is based on assessment of published literature. Chromosome and position based on Genome Reference Consortium Human Build 37. Effect allele is the minor allele.
Abbreviations: Chr:Pos, chromosome:position; EAF, effect allele frequency; P Value, P value using allelic model.
TABLE 2.
Association of Identified Loci with NAFLD in the Replication Cohort
| RSID | Chr:Pos | Reference Allele/Effect Allele | GS‐SFHS | Namjou et al. | Anstee et al. | Meta‐analysis | Gene | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| EAF | OR (95% CI) | P Value | OR* | P Value | OR (95% CI) | P Value | P Value | ||||
| rs2642442 | 1:220973563 | C/T | 0.311 | 1.06 (0.73‐1.54) | 0.551 | 1.19 | 5.96−03 | 1.16 (1.06‐1.26) | 9.70−04 | 5.83−12 | MARC1 |
| rs1260326 | 2:27730940 | T/C | 0.387 | 0.73 (0.52‐1.03) | 0.00737 | 0.90 | 7.34−02 | 0.78 (0.73‐0.84) | 1.06−10 | 3.08−15 | GCKR |
| rs17321515 | 8:126486409 | A/G | 0.478 | 0.94 (0.67‐1.32) | 0.913 | 0.80 | 1.08−04 | 0.86 (0.79‐0.93) | 1.99−04 | 1.24−16 | TRIB1 |
| rs73001065 | 19:19460541 | G/C | 0.065 | 0.79 (0.37‐1.69) | 0.591 | 1.30 | 1.19−02 | 1.58 (1.37‐1.82) | 1.59−10 | 7.51−30 | TM6SF2 |
| rs429358 | 19:45411941 | T/C | 0.162 | 0.70 (0.41‐1.18) | 0.126 | 0.81 | 9.57−03 | 0.85 (0.77‐0.95) | 4.16−03 | 3.42−13 | APOE |
| rs3747207 | 22:44324855 | G/A | 0.194 | 1.37 (0.92‐2.03) | 0.142 | 1.78 | 2.63−20 | 1.83 (1.68‐1.98) | 2.58−49 | 1.67−87 | PNPLA3 |
Chromosome and position based on Genome Reference Consortium Human Build 37.
The summary association statistics for Namjou et al. did not provide a CI or standard error for the odds ratio.
Abbreviations: Chr:Pos, chromosome:position; EAF, effect allele frequency; P Value, P value using allelic model.
GWAS meta‐analysis resulted in broadly similar results with no additional loci reaching genome‐wide significance. Direction and magnitude of effect were similar for all six loci other than rs73001065‐C, for which GS‐SFHS demonstrated a nonsignificant reduction in NAFLD risk but with substantially wider confidence intervals (CIs).
Forest plots showing effect size in the four studies (UKB GWAS cohort, GS‐SFHS replication cohort, Namjou et al. summary association statistics, and Anstee et al. summary association statistics) along with a Manhattan plot from the GWAS meta‐analysis are shown in Supporting Materials 1 (Supporting Figs. S1 and S2).
A Manhattan plot for association of variants with NAFLD in the UKB is shown in Fig. 2.
FIG. 2.
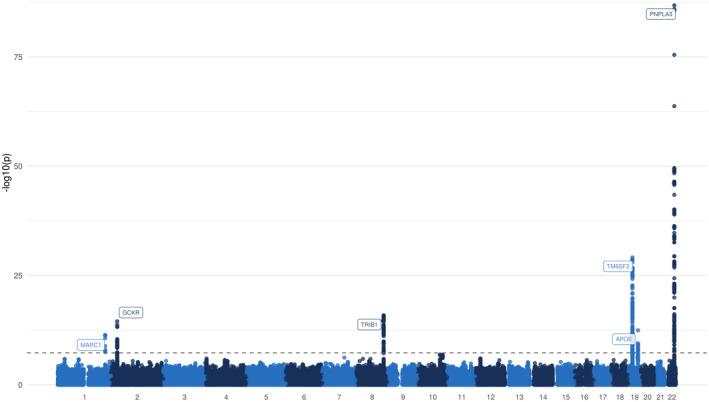
Manhattan plot for the association with NAFLD (4,761 cases and 373,227 controls). Each variant is plotted based on chromosome and position on the x‐axis and ‐log10 P values on the y‐axis. The horizontal dotted line represents genome‐wide significance (P = 5*10−8).
Conditional Analyses
After conditional analyses, one locus was found to have a further independent signal. Rs182611493 within the TM6SF2 locus showed significant association (P = 9.30*10−13) after conditioning on rs73001065 (see Supporting Materials 5). The remaining five loci did not have any SNPs reaching genome‐wide significance (P < 5*10−8) after conditioning on the lead SNP.
Population Stratification
After adjusting for the first 20 genetic principal components, there was evidence of test statistic inflation (). Inflation may be due to polygenicity rather than unmeasured population substructure.( 40 ) The LD score regression intercept was 1.006, and the proportion of test statistic inflation ascribed to causes other than polygenicity was estimated to be 7.52%, confirming that polygenicity is the main driver of test statistic inflation. The quantile‐quantile plot is shown in Supporting Materials 1 (Supporting Fig. S3).
Sensitivity Analyses
Across all sensitivity analyses, the estimated genetic effects at each lead SNP had the same direction and broadly similar magnitude. All SNPs demonstrated a significant effect when alternative hepatic pathologies were excluded and when adjusting for BMI. Two SNPs retained significance after additionally adjusting for alcohol intake (rs73001065, rs3747207), with one retaining suggestive significance (rs17321515, P = 1.39*10−7). The other three SNPs no longer retained suggestive significance, although this is likely to be due to the greatly reduced sample size in those individuals who had completed the alcohol intake questionnaire (158,388 vs. 377,998), with all six SNPs showing greatly attenuated significance. Visual inspection of the lattice plots showed that odds ratio (OR) estimates at each SNP were broadly similar with wider CIs in the alcohol‐adjusted analysis (see Supporting Materials 1 and Supporting Fig. S4).
A total of 377,998 individuals had sufficient data available to calculate the APOE genotype. As expected, the most common genotype was (219,869), whereas there were a total of 19 individuals who were either or who were excluded (there were no homozygotes). The distribution of individuals by genotype is shown in Supporting Materials 1 (Supporting Table S6). The APOE genotype was significantly associated with NAFLD (). The allele was strongly associated with reduced risk. The OR for NAFLD risk for heterozygotes was 0.84 (95% CI 0.78‐0.90) and for homozygotes 0.64 (0.50‐0.79). The allele was not associated with any significant change in NAFLD risk. The OR for NAFLD risk for heterozygotes was 1.02 (95% CI 0.93‐1.11) and for homozygotes 0.76 (0.50‐1.12). All ORs relate to the homozygote reference group. The results of the logistic regression are shown in Figure 3.
FIG. 3.
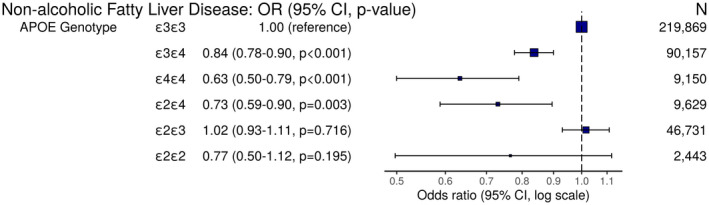
OR plot demonstrating odds of NAFLD by APOE genotype. Each APOE genotype is compared with the ε3 homozygotes reference group (model adjusted for age, sex, genotyping batch, and the first 20 genetic principal components).
Association of NAFLD‐susceptibility Loci with Phenotypic Traits
Assessment of serum lipids was undertaken for each lead SNP in all 377,998 individuals taken forward for GWAS. The NAFLD‐susceptibility alleles were heterogeneous in their influence on serum lipids with most demonstrating reduced levels of TC and LDL other than rs17321515 (see Fig. 4). Four variants were associated with elevated ALT (rs17321515, rs73001065, rs429358, rs3747207). Figures for the other serum biochemistry markers, anthropometric features and disease associations are available in Supporting Materials 1 (Supporting Figs. S5‐S10).
FIG. 4.
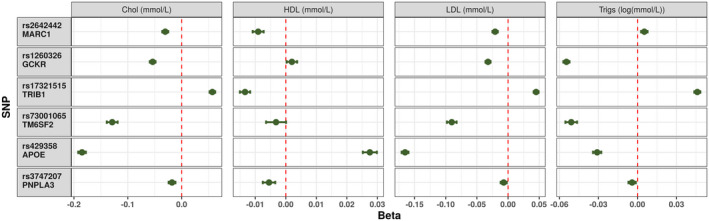
Impact of each NAFLD‐susceptibility allele on the measured serum cholesterol fractions. Each point represents the beta‐coefficient from an age‐adjusted and sex‐adjusted linear regression, and the error bar represents the 95% CI. Triglycerides were log‐transformed before the analysis. Abbreviations: Chol, total cholesterol; trigs, triglycerides.
The 6 variants were each tested for 3 gene‐environment interactions and a Bonferroni‐corrected P value of 0.0028 was considered significant. All 6 variants demonstrated a significant gene‐environment interaction with HbA1c and all but rs1260326 demonstrated significant interaction with BMI. None of the 6 variants demonstrated an interaction with alcohol intake.
In patients with NAFLD, only the locus with the strongest signal was associated with a change in FIB‐4 score on linear regression (rs3747207: Beta 0.08 [95% CI 0.03‐0.14]; P = 0.001). The NAFLD fibrosis score was not significantly influenced by any of the NAFLD‐susceptibility loci.
Numeric results for the GWAS, sensitivity analyses, and additional analyses are shown in Supporting Materials 5 and 6.
Discussion
We performed a GWAS of NAFLD using 4,761 cases and 373,227 controls from the UKB study. We identified six NAFLD‐susceptibility variants previously identified in GWAS of quantitative NAFLD traits such as hepatic steatosis, including GCKR, ( 6 ) TM6SF2 ( 6 , 10 ) and MARC1,( 17 ) and confirm the recently identified NAFLD‐susceptibility variant of rs429358‐C within APOE, a protein‐altering variant, which is protective against NAFLD. We replicate all five previously established loci associated with NAFLD in case‐control GWASs including HSD17B13.( 13 , 16 )
Rs429358 is a recently identified( 18 , 21 , 22 , 23 ) missense variant within APOE, which in combination with rs7412 defines the three main alleles of APOE, namely, , , and . APOE plays various roles in peripheral lipid and lipoprotein metabolism, and the three common alleles influence metabolic and cardiovascular disease and Alzheimer’s disease.( 41 ) The role of APOE in NAFLD has been examined previously in candidate‐gene studies and has recently been detected by GWAS. While this manuscript was under preparation, three independent analyses were published confirming an association at the APOE locus,( 21 , 22 , 23 ) although it was first identified in a subgroup analysis of an earlier paper.( 18 ) An exome‐wide array meta‐analysis was published, in which rs2075650 in TOMM40 was identified; the authors suggested, based on conditional analysis, that rs429358 in APOE is the causal variant with the C allele conferring protection.( 23 ) Candidate genes studies have previously shown a decreased risk with the allele( 42 , 43 , 44 ) and the allele,( 43 , 45 ) although some studies reported no difference in the risk of NAFLD.( 46 , 47 ) Elevated serum levels of APOE appear to correlate with higher fatty liver index, regardless of genotype.( 47 ) Perhaps surprisingly, the allele may be associated with greater NAFLD fibrosis severity, although this finding was made in a very small sample.( 48 ) The existence of additional populations demonstrating a similar association confirms that the APOE finding is likely to be a genuine association. The apparent lack of effect in some of the earlier studies is likely related to smaller sample sizes.
The mechanisms by which APOE influences NAFLD development remain unclear. There is a linear increase in both cardiovascular risk and serum LDL and total cholesterol, with transition from to and from to .( 41 ) The association between NAFLD and cardiovascular and metabolic disease( 1 ) is unlikely to be explained by APOE activity, given that the allele simultaneously offers protection against NAFLD and increases the risk of cardiovascular and metabolic disease. APOE influences hepatic very‐low‐density lipoprotein (VLDL) secretion.( 49 ) Apoe‐deficient mice demonstrate reduced VLDL secretion and greater steatohepatitis severity. This is not corrected by APOE‐producing bone marrow transplants with hepatic VLDL secretion remaining low, confirming that APOE plays an important role in liver autonomous VLDL secretion.( 50 ) The allele is associated with enhanced hepatic VLDL secretion, and this may explain the association with hypertriglyceridemia and cardiovascular disease as well as protection against hepatic steatosis.( 51 )
In our study, the overall rate of NAFLD detected using the EHR is lower than would be expected for NAFLD diagnosed by histological or radiological approaches. This is an expected limitation of the approach, as not all individuals with NAFLD undergo histological or radiological evaluations and may live in the community unaware of their disease. Despite the difference between the detected and anticipated rates of NAFLD in this study cohort, there are several elements of the analysis that suggest that the phenotype is valid for NAFLD research. First, all six loci have been identified by previous GWASs with strength of signal commensurate to that seen in earlier studies. Detection of such a unique genetic architecture strongly supports the validity and specificity of the EHR‐based phenotype. Second, we base our case definition on recent, independently authored expert consensus guidelines,( 25 ) and based on these guidelines conduct sensitivity analyses after exclusion of alternative hepatic pathology, in which our results are consistent. Third, the baseline characteristics of the NAFLD cohort compared with controls is similar to that which is expected, as shown in Supporting Materials 1 (Supporting Table S5). Fourth, the UKB is affected by healthy volunteer bias, suggesting that the overall rate of NAFLD may be lower than an age‐matched population.( 52 ) Finally, the results of our analysis are consistent with a histological analysis of a subgroup of UKB participants, in which several variants including the APOE variant were identified.( 18 , 21 , 22 , 23 )
The strengths of this study include using a larger sample size than previous studies, with greater power to detect association. The study also used a second cohort for external replication as well as available GWAS summary association statistics from other populations, resulting in replication of all identified loci. The use of administrative records for identification of cases also demonstrates that this technique can be used to study NAFLD without the requirement for invasive procedures or radiological assessment and, as discussed, the results are consistent with published literature. Our study has confirmed four loci identified in the same cohort using radiological assessment of NAFLD in a smaller subset (up to 14,400 participants; APOE, GCKR, TM6SF2, and PNPLA3)( 18 ) and two other SNPs identified in other population. Furthermore, the strongest known signal at PNPLA3 was verified in this study with a highly significant association, and relative effect sizes at each locus were highly similar to results from histologically or radiologically characterized cohorts. The study also benefits from demonstrating the robustness of the associations within sensitivity analyses using diagnostic codes based on published recommendations( 25 ) and provides mechanistic evidence by using validated biomarkers to determine the potential influence of each locus on NAFLD development. Notably, using consensus recommendations to define NAFLD has resulted in 4,761 eligible cases, whereas previous studies have classified only 704 individuals from within the UKB as NAFLD cases, with the remainder misclassified as controls.( 17 )
The limitations of this study include misclassification of individuals with alternative hepatic pathology on administrative records and low detection rate of NAFLD due to limited documentation in the EHR. The definition of cases based on administrative records also prevents any assessment of NAFLD severity; thus, variants that contribute solely to the progression of steatohepatitis, fibrosis, or cirrhosis without promoting initial occurrence of steatosis may not be identified. The binary case definition offers lower power to detect associations than continuous traits, although this is partly overcome by the very large sample size. The results of our study do, however, support published literature. The GS‐SFHS replication cohort only demonstrated significant association at the GCKR locus, but none of the other loci. This is likely to be due to the underpowered sample size with only 67 NAFLD cases compared with almost 5,000 analyzed in the UKB cohort. The overall rate of NAFLD ascertained using the EHR‐based approach was similar between the cohorts (1.2% in the UKB and 1.1% in the GS‐SFHS cohorts), suggesting that the lack of replication may be determined by sample size rather than differences in clinical coding. Despite this, all six identified loci have been detected by earlier GWASs. Although the definition of NAFLD is based on consensus recommendations,( 25 ) it is possible that regional variation in recording before these recommendations has influenced documentation of NAFLD.
This paper supports the use of administrative data as a means to conduct research into NAFLD. Such approaches are likely to require large sample sizes, but do overcome the need for invasive and costly recruitment and investigations.
In summary, we have performed a GWAS of NAFLD using UKB data and identified six loci associated with NAFLD. We have also demonstrated the feasibility of EHR‐based NAFLD research without reliance on invasive investigations, validating the consensus guidelines.
Supporting information
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgment
We thank Dr. Colin Fischbacher for the support in selection of ICD10, ICD9, and read codes.
Supported by a Medical Research Council Clinical Research Training Fellowship (MR/T008008/1); Chief Scientist Office of the Scottish Government Health Directorates (CZD/16/6); Scottish Funding Council (HR03006); and Wellcome Trust (216767/Z/19/Z); Wellcome Ph.D. training fellowship for clinicians (204979/Z/16/Z); Edinburgh Clinical Academic Track (ECAT) programme; Wellcome Trust Senior Research Fellowship in Clinical Science (ref. 219542/Z/19/Z); MRC Human Genetics Unit programme grant, “Quantitative traits in health and disease” (U. MC_UU_00007/10); British Heart Foundation through a Senior Clinical Research Fellowship (FS/16/14/32023); Research Excellence Award (RE/18/5/34216); and Wellcome Trust (grant 210752/Z/18/Z). Genotyping of the GS:SFHS samples was carried out by the Genetics Core Laboratory at the Edinburgh Clinical Research Facility, University of Edinburgh, Scotland, and was funded by the Medical Research Council UK and the Wellcome Trust (Wellcome Trust Strategic Award “STratifying Resilience and Depression Longitudinally” (Reference 104036/Z/14/Z).
Potential conflict of interest: Nothing to report.
References
Authors names in bold designate shared co‐first authorship.
- 1. Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol 2018;15:11‐20. [DOI] [PubMed] [Google Scholar]
- 2. Dufour J‐F, Caussy C, Loomba R. Combination therapy for non‐alcoholic steatohepatitis: rationale, opportunities and challenges. Gut 2020;69:1877‐1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sookoian S, Pirola CJ. Genetic predisposition in nonalcoholic fatty liver disease. Clin Mol Hepatol 2017;23:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2008;40:1461‐1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chalasani N, Guo X, Loomba R, Goodarzi MO, Haritunians T, Kwon S, et al. Genome‐wide association study identifies variants associated with histologic features of nonalcoholic fatty liver disease. Gastroenterology 2010;139:1567‐1576.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Speliotes EK, Yerges‐Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, et al. Genome‐wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet 2011;7:e1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kawaguchi T, Sumida Y, Umemura A, Matsuo K, Takahashi M, Takamura T, et al. Genetic polymorphisms of the human PNPLA3 gene are strongly associated with severity of non‐alcoholic fatty liver disease in Japanese. PLoS One 2012;7:e38322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adams LA, White SW, Marsh JA, Lye SJ, Connor KL, Maganga R, et al. Association between liver‐specific gene polymorphisms and their expression levels with nonalcoholic fatty liver disease. Hepatology 2013;57:590‐600. [DOI] [PubMed] [Google Scholar]
- 9. Kitamoto T, Kitamoto A, Yoneda M, Hyogo H, Ochi H, Nakamura T, et al. Genome‐wide scan revealed that polymorphisms in the PNPLA3, SAMM50, and PARVB genes are associated with development and progression of nonalcoholic fatty liver disease in Japan. Hum Genet 2013;132:783‐792. [DOI] [PubMed] [Google Scholar]
- 10. Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjærg‐Hansen A, et al. Exome‐wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2014;46:352‐356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wood KL, Miller MH, Dillon JF. Systematic review of genetic association studies involving histologically confirmed non‐alcoholic fatty liver disease. Gastroenterology 2015;2:e000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wattacheril J, Lavine JE, Chalasani NP, Guo X, Kwon S, Schwimmer J, et al. Genome wide associations related to hepatic histology in nonalcoholic fatty liver disease in hispanic boys. J Pediatr 2017;190:100‐107.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abul‐Husn N, Cheng X, Li A, Xin Y, Schurmann C, Stevis P, et al. A protein‐truncating HSD17B13 variant and protection from chronic liver disease. N Eng J Med 2018;378:1096‐1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chung GE, Lee Y, Yim JY, Choe EK, Kwak M‐S, Yang JI, et al. Genetic polymorphisms of PNPLA3 and SAMM50 are associated with nonalcoholic fatty liver disease in a Korean population. Gut Liver 2018;12:316‐323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Namjou B, Lingren T, Huang Y, Parameswaran S, Cobb BL, Stanaway IB, et al. GWAS and enrichment analyses of non‐alcoholic fatty liver disease identify new trait‐associated genes and pathways across eMERGE Network. BMC Med 2019;17:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anstee QM, Darlay R, Cockell S, Meroni M, Govaere O, Tiniakos D, et al. Genome‐wide association study of non‐alcoholic fatty liver and steatohepatitis in a histologically characterised cohort. J Hepatol 2020;73:505‐515. [DOI] [PubMed] [Google Scholar]
- 17. Emdin CA, Haas ME, Khera AV, Aragam K, Chaffin M, Klarin D, et al. A missense variant in mitochondrial amidoxime reducing component 1 gene and protection against liver disease. PLoS Genet 2020;16:e1008629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parisinos CA, Wilman HR, Thomas EL, Kelly M, Nicholls RC, McGonigle J, et al. Genome‐wide and Mendelian randomisation studies of liver MRI yield insights into the pathogenesis of steatohepatitis. J Hepatol 2020;73:241‐251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park SL, Li Y, Sheng X, Hom V, Xia L, Zhao K, et al. Genome‐wide association study of liver fat: the multiethnic cohort adiposity phenotype study. Hepatol Commun 2020;4:1112‐1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yoshida K, Yokota K, Kutsuwada Y, Nakayama K, Watanabe K, Matsumoto A, et al. Genome‐wide association study of lean nonalcoholic fatty liver disease suggests human leukocyte antigen as a novel candidate locus. Hepatol Commun 2020;4:1124‐1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Emdin CA, Haas M, Ajmera V, Simon TG, Homburger J, Neben C, et al. Association of genetic variation with cirrhosis: a multi‐trait genome‐wide association and gene environment interaction study. Gastroenterology 2021;160:1620‐1633.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jamialahmadi O, Mancina RM, Ciociola E, Tavaglione F, Luukkonen PK, Baselli G, et al. Exome‐wide association study on alanine aminotransferase identifies sequence variants in the GPAM and APOE associated with fatty liver disease. Gastroenterology 2021;160:1634‐1646.e7. [DOI] [PubMed] [Google Scholar]
- 23. Palmer ND, Kahali B, Kuppa A, Chen Y, Du X, Feitosa MF, et al. Allele specific variation at apoe increases non‐alcoholic fatty liver disease and obesity but decreases risk of Alzheimer’s disease and myocardial infarction. Hum Mol Genet 2021;30:1443‐1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Teo K, Abeysekera KW, Adams L, Aigner E, Anstee QM, Banales JM, et al. rs641738C T near MBOAT7 is associated with liver fat, ALT and fibrosis in NAFLD: a meta‐analysis. J Hepatol 2021;74:20‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hagström H, Adams LA, Allen AM, Byrne CD, Chang Y, Grønbæk H, et al. Administrative coding in electronic health care record‐based research of NAFLD: an expert panel consensus statement. Hepatology 2021;74:474‐482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen W‐M. Robust relationship inference in genome‐wide association studies. Bioinformatics 2010;26:2867‐2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017;8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bulik‐Sullivan BK, Loh P‐R, Finucane HK, Ripke S, Yang J, Patterson N, et al. LD score regression distinguishes confounding from polygenicity in genome‐wide association studies. Nat Genet 2015;47:291‐295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yoneda M, Mawatari H, Fujita K, Iida H, Yonemitsu K, Kato S, et al. High‐sensitivity C‐reactive protein is an independent clinical feature of nonalcoholic steatohepatitis (NASH) and also of the severity of fibrosis in NASH. J Gastroenterol 2007;42:573‐582. [DOI] [PubMed] [Google Scholar]
- 30. Verma S, Jensen D, Hart J, Mohanty SR. Predictive value of ALT levels for non‐alcoholic steatohepatitis (NASH) and advanced fibrosis in non‐alcoholic fatty liver disease (NAFLD). Liver Int 2013;33:1398‐1405. [DOI] [PubMed] [Google Scholar]
- 31. Stender S, Kozlitina J, Nordestgaard BG, Tybjærg‐Hansen A, Hobbs HH, Cohen JC. Adiposity amplifies the genetic risk of fatty liver disease conferred by multiple loci. Nat Genet 2017;49:842‐847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Angulo P, Hui JM, Marchesini G, Bugianesi E, George J, Farrell GC, et al. The NAFLD fibrosis score: a noninvasive system that identifies liver fibrosis in patients with NAFLD. Hepatology 2007;45:846‐854. [DOI] [PubMed] [Google Scholar]
- 33. Vallet‐Pichard A, Mallet V, Nalpas B, Verkarre V, Nalpas A, Dhalluin‐Venier V, et al. FIB‐4: an inexpensive and accurate marker of fibrosis in HCV infection. Comparison with liver biopsy and fibrotest. Hepatology 2007;46:32‐36. [DOI] [PubMed] [Google Scholar]
- 34. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta‐analysis of genomewide association scans. Bioinformatics 2010;26:2190‐2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Begum F, Ghosh D, Tseng GC, Feingold E. Comprehensive literature review and statistical considerations for GWAS meta‐analysis. Nucleic Acids Res 2012;40:3777‐3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marchini J, Howie B. Genotype imputation for genome‐wide association studies. Nat Rev Genet 2010;11:499‐511. [DOI] [PubMed] [Google Scholar]
- 37. R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 38. Little J, Higgins JPT, Ioannidis JPA, Moher D, Gagnon F, von Elm E, et al. STrengthening the REporting of Genetic Association Studies (STREGA) of the STROBE Statement. PLoS Med 2009;6:e1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu Y‐L, Reeves HL, Burt AD, Tiniakos D, McPherson S, Leathart JBS, et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non‐alcoholic fatty liver disease. Nat Commun 2014;5:4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang J, Weedon MN, Purcell S, Lettre G, Estrada K, Willer CJ, et al. Genomic inflation factors under polygenic inheritance. Eur J Hum Genet 2011;19:807‐812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bennet AM, Di Angelantonio E, Ye Z, Wensley F, Dahlin A, Ahlbom A, et al. Association of apolipoprotein E genotypes with lipid levels and coronary risk. JAMA 2007;298:1300‐1311. [DOI] [PubMed] [Google Scholar]
- 42. Yang MH, Son HJ, Sung JD, Choi YH, Koh KC, Yoo BC, et al. The relationship between apolipoprotein E polymorphism, lipoprotein (a) and fatty liver disease. Hepatogastroenterology 2005;52:1832‐1835. [PubMed] [Google Scholar]
- 43. Sazci A, Akpinar G, Aygun C, Ergul E, Senturk O, Hulagu S. Association of apolipoprotein E polymorphisms in patients with non‐alcoholic steatohepatitis. Dig Dis Sci 2008;53:3218‐3224. [DOI] [PubMed] [Google Scholar]
- 44. De Feo E, Cefalo C, Arzani D, Amore R, Landolfi R, Grieco A, et al. A case‐control study on the effects of the apolipoprotein E genotypes in nonalcoholic fatty liver disease. Mol Biol Rep 2012;39:7381‐7388. [DOI] [PubMed] [Google Scholar]
- 45. Demirag MD, Onen HI, Karaoguz MY, Dogan I, Karakan T, Ekmekci A, et al. Apolipoprotein E gene polymorphism in nonalcoholic fatty liver disease. Dig Dis Sci 2007;52:3399‐3403. [DOI] [PubMed] [Google Scholar]
- 46. Lee D‐M, Lee S‐O, Mun B‐S, Ahn H‐S, Park H‐Y, Lee H‐S, et al. Relation of apolipoprotein E polymorphism to clinically diagnosed fatty liver disease. Taehan Kan Hakhoe Chi 2002;8:355‐362. [PubMed] [Google Scholar]
- 47. van den Berg EH, Corsetti JP, Bakker SJL, Dullaart RPF. Plasma ApoE elevations are associated with NAFLD: the PREVEND study. PLOS ONE 2019;14:e0220659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stachowska E, Maciejewska D, Ossowski P, Drozd A, Ryterska K, Banaszczak M, et al. Apolipoprotein E4 allele is associated with substantial changes in the plasma lipids and hyaluronic acid content in patients with nonalcoholic fatty liver disease. J Physiol Pharm 2013;64:711‐717. [PubMed] [Google Scholar]
- 49. Riches FM, Watts GF, van Bockxmeer FM, Hua J, Song S, Humphries SE, et al. Apolipoprotein B signal peptide and apolipoprotein E genotypes as determinants of the hepatic secretion of VLDL apoB in obese men. J Lipid Res 1998;39:1752‐1758. [PubMed] [Google Scholar]
- 50. Schierwagen R, Maybüchen L, Zimmer S, Hittatiya K, Bäck C, Klein S, et al. Seven weeks of Western diet in apolipoprotein‐E‐deficient mice induce metabolic syndrome and non‐alcoholic steatohepatitis with liver fibrosis. Sci Rep 2015;5:12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kypreos KE, van Dijk KW, van der Zee A, Havekes LM, Zannis VI. Domains of apolipoprotein E contributing to triglyceride and cholesterol homeostasis in vivo: carboxyl‐terminal region 203299 promotes hepatic very‐low‐density‐lipoprotein‐triglyceride secretion. J Biol Chem 2001;276:19778‐19786. [DOI] [PubMed] [Google Scholar]
- 52. Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, Sprosen T, et al. Comparison of sociodemographic and health‐related characteristics of UK biobank participants with those of the general population. Am J Epidemiol 2017;186:1026‐1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material


