Abstract
Bioinformatics analysis is a key element in the development of in-house next-generation sequencing assays for tumor genetic profiling that can include both tumor DNA and RNA with comparisons to matched-normal DNA in select cases. Bioinformatics analysis encompasses a computationally heavy component that requires a high-performance computing component and an assay-dependent quality assessment, aggregation, and data cleaning component. Although there are free, open-source solutions and fee-for-use commercial services for the computationally heavy component, these solutions and services can lack the options commonly utilized in increasingly complex genomic assays. Additionally, the cost to purchase commercial solutions or implement and maintain open-source solutions can be out of reach for many small clinical laboratories. Here, we present Software for Clinical Health in Oncology for Omics Laboratories (SCHOOL), a collection of genomics analysis workflows that (i) can be easily installed on any platform; (ii) run on the cloud with a user-friendly interface; and (iii) include the detection of single nucleotide variants, insertions/deletions, copy number variants (CNVs), and translocations from RNA and DNA sequencing. These workflows contain elements for customization based on target panel and assay design, including somatic mutational analysis with a matched-normal, microsatellite stability analysis, and CNV analysis with a single nucleotide polymorphism backbone. All of the features of SCHOOL have been designed to run on any computer system, where software dependencies have been containerized. SCHOOL has been built into apps with workflows that can be run on a cloud platform such as DNANexus using their point-and-click graphical interface, which could be automated for high-throughput laboratories.
Keywords: Bioinformatics, cancer, NGS
1. Introduction
The rapid expansion and decreasing cost of next-generation sequencing (NGS) technology present an opportunity to improve the diagnosis and treatment of cancer through identifying tumor-specific mutations and enabling physicians to adapt treatment plans that suit the unique molecular profile of each patient.1,2 To address the evolving list of clinically actionable and prognostic biomarkers in the treatment of cancer, academic clinical laboratories have developed sequencing assays with varying size gene panels (100–1600 genes), with consistent quality to detect relevant genetic variants.
Bioinformatics analysis of sequence data includes two phases: (i) primary analysis, which converts the raw sequencing reads into predicted genetic variants and read abundances, and (ii) secondary analysis, which is customized for each clinical assay to maximize sensitivity and specificity by identifying artifact and poor-quality variant predictions. For a quick turn-around-time, the primary analysis is computationally demanding and requires computational resources with high memory (>32 GB) and multiple processors using a local high-performance cluster or cloud-computing resources. Furthermore, the primary analysis requires multiple elements for complete somatic variant detection including single nucleotide variants (SNVs), insertions/deletions (indels), copy number variants (CNVs), and structural variants such as translocations, large deletions, internal tandem duplications, and differences in microsatellite length. Because each assay might include different elements for detecting these different variant types, the primary analysis should be customizable for a variety of assays. Secondary analysis is much less resource-intensive, can be run on a desktop computer, and should be tailored to the needs of the specific assay.
Both commercial and open-source solutions have been introduced to address primary analysis needs in cancer genomics. Commercially developed bioinformatics pipelines are proprietary, often have limited options for customization, and require licensing, which can increase computational costs. By contrast, open-source solutions are usually customizable and lack licensing costs.3,4 However, common best-practice tools for variant detection, such as BWA and GATK4, require computational programming expertise to run in a Linux command line environment. Some commonly used open-source tools for more complex variant detection lack thorough documentation, continued support for development, or the flexibility to process varied data types (tumor-only samples versus matched tumor/normal control). Additionally, many existing software tools are difficult to install and maintain, due to the sometimes difficult installation of software dependencies, which make them sensitive to updates and changes to default programs. Finally, these tools are not natively packaged as an end-to-end analysis pipeline, which starts with raw sequencing reads and results in predicted variants. A user-friendly interface is critical in a customizable, open-source bioinformatics pipeline that is easy to install and run without specialized computational training.
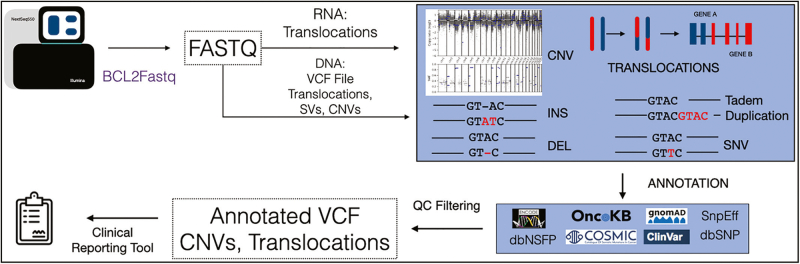
In order to address the need for an end-to-end customizable bioinformatics pipeline for the primary analysis of sequence data, we have developed a collection of analysis workflows for NGS data and the detection of genetic alterations in cancer called Software for Clinical Health in Oncology for Omics Laboratories (SCHOOL) [Fig. 1]. SCHOOL: (i) can detect SNVs, indels, CNVs, and translocations from RNA and DNA sequencing, (ii) has tools for mutational profiling and omics integration, and (iii) is designed to be easy to run on local computing resources or the cloud, with all software packaged alongside dependencies, so that they can work on any system where singularity or docker programming packages are available. We have optimized each step to use a minimal amount of RAM and processors to reduce computation costs on the cloud. These workflows contain the steps necessary to complete primary NGS analysis, including variant detection and annotation. Furthermore, these workflows can execute on a local cluster using Nextflow,5 a command-line workflow manager, or on the cloud, https://platform.dnanexus.com/panx/projects/FvPKK200Y9g81KqkKjJ9X818/data/, using the DNANexus applets and workflows code.
Fig. 1.
Overview of SCHOOL workflow from sequencing through reporting. In SCHOOL, data flow from the sequencer into the primary analysis pipeline, which includes quality control, alignment, and variant calling appropriate for the sample type. Then, in secondary analysis, the variants can be annotated for eventual clinical reports.
2. Technical background
2.1. NGS analysis
The primary analysis of sequence data for the detection of somatic variants in tumor samples requires five main steps including (i) alignment of the raw sequencing data to a reference genome, (ii) identification of SNVs and indels in DNA, (iii) identification of CNVs in DNA, (iv) identification of copy number and structural variants in RNA and DNA, and (v) annotation and the prediction of effect.6 For each step, there are many considerations for the bioinformatics workflow that can affect accuracy.6
For alignments, the user should consider the genome reference, removal of duplicate reads, and the alignment program. There are currently two available versions of the reference human genome: GRCh37 (hg19), released in 2009, and GRCh38 (hg38), released in 2013. GRCh38 has been shown to produce more accurate alignments.7 Sequence duplicates are caused when polymerase chain reaction (PCR) errors are amplified. Duplicates can be marked or removed, so that they are ignored in downstream steps, Picard (http://broadinstitute.github.io/picard/) can be used to mark duplicates, and Samtools can be used to remove duplicates. When unique molecular barcodes are included in the sequencing adapter, added during sample preparation, FGBio (see URLs) can be used to create consensus sequences of duplicates. BWA-MEM8 is the most used tool for sequence alignment of sequencing reads to the human reference genome. The output of BWA must be converted to BAM, sorted, and indexed using Samtools9 in order to be used in variant detection tools.
For SNVs and indels, there are many different open-source tools that can be used for the detection of these variant types. Somatic variants can be detected in somatic or tumor-only mode using the following tools: Strelka2,10 Freebayes,11 MuTect2,12 Pindel,13 BCFtools call,14 LoFreq,15 VarScan,16 Platypus,17 GATK4,18 and Scapel.19 Some tools such as GATK and BCFtools are designed to identify germline variants, whereas other tools, such as LoFreq and MuTect2, are designed to identify low-frequency variants common in tumor samples. A comparison of nine somatic variant calling programs found that Mutect2, Strelka2, and Virmid were among the most accurate.20
There are also many different methods for the detection of copy number and structural variation. Some methods use sequence depth of coverage, or the average number of reads overlapping each region of the genome, to determine copy number changes. Because biases in coverage differ in each region of the genome, this coverage is not uniform, but can be normalized with healthy control samples. Additionally, some methods can take into account the allele frequency of common polymorphisms, called b-allele frequency, to correct these biases in the depth of coverage. A comparison of four CNV detection tools found that CNVKit had high sensitivity but a lower specificity relative to other programs like Control-FREEC.21 Although there are also many methods for the detection of structural variants, the accuracy of detection of structural variants is low compared with SNV and indels for short-read data. The strategy that most laboratories employ is the validation of known clinically actionable structural variants such as the FLT3 internal tandem duplication or known gene fusion events.
2.2. Computation interoperability and graphical interfaces
A software container is a freestanding unit that comprises the software of interest and all of its dependencies. Containerization allows for better maintenance and stability of computer software because it supports: (i) the deployment of the same code on the same running environment on any computer system (on premises or cloud); (ii) the separation of software packages to their own environment to satisfy specific running conditions; and (iii) dependencies and easier implementation of new packages or tools. Two popular containerization tools are docker, which was designed to work in a cloud environment, and singularity, which was designed to work in a local high-performance computing environment. Fortunately, containers that are created using docker can be converted to run using singularity on a local cluster. Additionally, containers can be versioned and easily shared, making them ideal to distribute code for bioinformatics pipelines.
Whether it is running in a high-performance computing environment locally or on the cloud, a bioinformatics pipeline has several important elements including automated advancement from step to step and parallelization. Pipelines are designed to take the input of each step as it becomes available and run blocks of code to convert it into predetermined output files. Some steps are serialized, meaning that they are dependent on the successful completion of earlier steps, such as alignment being dependent on the completion of read trimming. Other processes, such as variant calling using different callers, can be run concurrently to save time in a process called parallelization. Pipelines can be controlled on local computing resources using languages such as Nextflow, WDL, and Snakemate. In the cloud, commercial cloud frameworks exist to allow biologists with limited computational expertise to run pipelines in a point-and-click environment, using platforms such as DNANexus, Seven Bridges, and Illumina Connected Analytics.
3. Approach
To implement our pipelines, we (i) tested several software packages for analysis accuracy, (ii) created docker containers for each self-contained step in our workflow, (iii) implemented input options for customizing each step, and (iv) designed an end-end workflow for primary analysis that could be run on an internal high-performance computing cluster or run on the cloud using a point-and-click graphical interface.
3.1. Tool testing
We tested 10 variant calling methods, including Strelka2,10 Freebayes,11 MuTect2,12 Pindel,13 BCFtools call,14 LoFreq,15 VarScan,16 Platypus,17 GATK4,18 and Scapel,19 using data generated from an engineered cell line that was designed with variants with low allele frequency and validated by quantitative PCR (Table S1). Consistent with previous studies, we found high sensitivity with MuTect2 and Strelka2. Of the 21 SNVs and small indels (<50 bp) present, Freebayes detected all 21 variants, MuTect2 detected 20 and LoFreq detected 16; each caller was within 10% of the expected variants allele frequency (VAF). Callers such as Strelka2, GATK2, Platypus, and VarScan, using default parameters, detected the variants with >20% VAF, as designed (Table S1). Pindel detected the 300 bp internal tandem duplicate (ITD) in the FLT3 gene, along with two other indels at 35%–40% VAF.
3.2. Tools implemented in docker and customization
Our workflow allows users to choose for SNV and indel detection Strelka2,10 Freebayes,11 MuTect2,12 and Pindel13 or a combination of the four. For MuTect2,12 alignments are first recalibrated using GATK418 with BaseRecalibrator and ApplyBQSR. If matched tumor/ normal pairs are sequenced, users can ensure that these samples originate from the same patient using BCFtools14 and NGS Checkmate.22
We implemented several tools for the detection of copy number and structural variants. For CNVs, we implemented CNVKit.23 Because CNVKit works best with a panel of healthy normal control samples, we have also implemented a container and tool to generate this healthy control reference. To detect ITDs, users can use Pindel and ITDSeeker.13,24 When microsatellite-specific baits are included, microsatellite stability can be estimated with an MSI-Sensor pro.25 Gene fusions (translocations) can be detected using DNA- and RNA- specific tools. Star-Fusion is implemented for RNASeq data, and DELLY and SVABA are implemented for DNA26,27 sequencing data.
Because expression can also be assessed with the RNASeq data, we have implemented the steps necessary for assessing gene expression. Reads can be aligned with HiSAT2,28 and expression values are determined using FeatureCount and StringTie.29,30 Variants in the RNA can be determined using Freebayes11 and BamReadCt (see URLs).
Variants can be annotated using gnomAD31 for the detection of common mutations and snpEff for gene effect.32 Other sources of annotations include the database of oncoKB hotspots,33 Encode repeat regions,34 the database of non-synonymous of functional predictions (dbNSFP),35 and variant databases dbSNP,36 Clinvar,37 and COSMIC.38
3.3. End-to-end workflows
The bioinformatics workflow contains three elements: (i) a software container created using Docker, which contains all software dependencies for each step, (ii) scripts written in bash that contain software commands necessary to complete each step of the workflow, and (iii) the workflow script and configuration that defines the inputs and outputs of each step, the compute requirements, the bash script parameters, and the container used for each step. The workflow script and configuration were implemented for execution on a local high-performance computing cluster, using the workflow management program Nextflow, and on the cloud, using the DNANexus Toolkit.
For users with bioinformatics and computing expertise, Nextflow can be configured to run on a variety of platforms locally and on the cloud. Nextflow readily submits jobs to commonly used compute cluster scheduling software such as SGE, PBS, and SLURM but also can be configured to submit jobs to cloud systems on Amazon Web Services (AWS) and Google Cloud. Users can create a Nextflow configuration file to customize the workflow for their hardware. Additionally, Nextflow allows for users to resume failed jobs and has extensive logging of each step, making troubleshooting easy to document. Finally, these Nextflow workflows can be configured to run on individual tumor samples or tumor/normal sample pairs or in a batch mode for processing the data for an entire sequencing run.
In order to make these workflows accessible to users with limited computational expertise, we transformed the workflows to run on AWS resources on the DNANexus platform. DNANexus has a point-and-click user interface for running data analysis. Each step of the workflow was transformed into a DNANexus App, and apps were combined into a DNANexus Workflow. Users can run these pipelines with their raw sequence files in FastQ, a DNA reference tar gzip file and a gene panel reference tar gzip file. To reduce the price to run each step, every DNANexus app was run on test files to determine the minimal resources necessary, largely through monitoring memory and processor consumption and increasing resources incrementally when tools reached memory or disk usage limits (Table S2). We then set these resource requirements as the default settings for each app. Users can alter these settings to decrease computing time. The cost of analysis is highly dependent on the size of the data set and the machines chosen to do the analysis, where the user will want to balance cost and computational time.
4. Conclusion
We have developed SCHOOL, a set of bioinformatics analysis tools and pipelines for the analysis of NGS data in an academic clinical oncology laboratory (Fig. S1), which has been in use at the CAP/CLIA laboratory at UT Southwestern Medical Center for four years. Additionally, SCHOOL pipelines have been used in over 20 research studies ranging from basic science to case reports.39, 40, 41, 42, 43 SCHOOL includes tools and methods for primary analysis of sequencing data from raw reads to finished variant calls, accommodating germline and somatic DNA and tumor RNA. We further include tools for panel-specific customization, including extensions for: copy number analysis, microsatellite stability, integration between DNA and RNA data, and structural variant calling to detect gene fusions in DNA and ITDs.
The SCHOOL pipeline can be optimized for the panel used in the assay. For example, when a panel of normal samples is included in assay development, a panel customization pipeline will align each sample and generate panel reference samples and the input BED files for copy number analysis. The aggregate normal sample VCF file will also be created to use when running MuTect2 to remove artifacts and rare variant sequences. Lastly, the normal samples can be used as a microsatellite reference for predicting microsatellite stability in the absence of a matched-normal sample.
For groups running an RNA Sequencing assay, there are several opportunities for data integrations, including comparison of RNA and DNA breakpoints in gene fusion events, comparison of splice site alteration using RNA data, and independent confirmation of variants by concurrent expression in RNA. The presence of variants in both the tumor RNA and tumor DNA provides enhanced confidence that a variant is not an artifact of the assay. Additionally, the presence of variants in RNA could indicate that the aberrant gene mutation is expressed in the tumor tissue. This could provide additional support of the importance of the variant, particularly for a suspected gain of function variants in oncogenic drivers or potential splice site variants that result in exon exclusion or intron inclusion. However, it is still important to note that variants in regulatory regions or gene deletions could prevent RNA expression.
These workflows represent a user-friendly, inexpensive, and flexible way to implement NGS bioinformatics for mutation detection and annotation in CAP/CLIA laboratories. SCHOOL can be easily installed on a local computing cluster that uses a queueing system such as SLURM or SGE with Nextflow with minimal dependency on pre-installed packages or runs on the cloud for laboratories that lack local resources and expertise with a user-friendly point-and-click graphic interface. We estimate the costs for analysis using cloud resources is a fraction of the costs of data generation and for some labs will reduce the need for additional computational expertise and resources on site.
These pipelines implemented in SCHOOL perform computationally heavy analysis in variant detection, which we consider to be the primary analysis. Users should, in the course of their validation studies, determine the filtering parameters of these results to maximize sensitivity and specificity for their assay using a set of quality metrics for the variants based on variant type, including a number of alternate reads, percent of alternate reads, strand bias, and other quality scores. In this secondary analysis step, in addition to filtering variants and removing artifacts, the user can determine mutational profiling metrics like tumor mutational burden and distributions of SNV by codon change. These secondary analysis steps often need tuning based on the assay and are less computationally intensive, meaning they can be done locally on a PC or laptop computer.
URLs
bcl2fastq (https://support.illumina.com/sequencing/ sequencing_software/bcl2fastq-conversion-software.html).
fgbio (https://github.com/fulcrumgenomics/fgbio).
picard (http://broadinstitute.github.io/picard/).
COSMIC (cancer.sanger.ac.uk).
bamreadct (https://github.com/genome/bam-readcount).
Financial support and sponsorship
Nil.
Declaration of Competing Interest
There are no conflicts of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.4103/jpi.jpi_20_21.
Appendix A. Supplementary data
Supplementary material
Data availability
All the code used in SCHOOL is available at the UTSW Clinical Laboratory github wiki site: https://medforomics.github.io/schoolwiki/ using the repos: school for pipelines, process_scripts used in each docker container, and dnanexus_applets for running on DNANexus.
References
- 1.Kamps R., Brandão R.D., BJVD Bosch, et al. Next-generation sequencing in oncology: Genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci. 2017;18:308. doi: 10.3390/ijms18020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Surrey L.F., Luo M., Chang F., Li M.M. The genomic era of clinical oncology: Integrated genomic analysis for precision cancer care. Cytogenet Genome Res. 2016;150:162–175. doi: 10.1159/000454655. [DOI] [PubMed] [Google Scholar]
- 3.Chang L., BChir MB, Chang M, Chang HM, Chang F. Microsatellite instability: A predictive biomarker for cancer immunotherapy. Appl Immunohistochem Mol Morphol. 2017;26:e15–e21. doi: 10.1097/PAI.0000000000000575. [DOI] [PubMed] [Google Scholar]
- 4.Salipante S.J., Scroggins S.M., Hampel H.L., Turner E.H., Pritchard C.C. Microsatellite instability detection by next generation sequencing. Clin Chem. 2014;60:1192–1199. doi: 10.1373/clinchem.2014.223677. [DOI] [PubMed] [Google Scholar]
- 5.Stenzinger A., Allen J.D., Maas J., et al. Tumor mutational burden standardization initiatives: Recommendations for consistent tumor mutational burden assessment in clinical samples to guide immunotherapy treatment decisions. Genes Chromosomes Cancer. 2019;58:578–588. doi: 10.1002/gcc.22733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.SoRelle J.A., Wachsmann M., Cantarel B.L. Assembling and validating bioinformatic pipelines for next-generation sequencing clinical assays. Arch Pathol Lab Med. 2020;144:1118–1130. doi: 10.5858/arpa.2019-0476-RA. [DOI] [PubMed] [Google Scholar]
- 7.Ellrott K., Bailey M.H., Saksena G., et al. MC3 Working Group; Cancer Genome Atlas Research Network. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 2018;6:271–281.e7. doi: 10.1016/j.cels.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen P.A., Ni Y., Bao F., et al. Houston methodist variant viewer: An application to support clinical laboratory interpretation of next-generation sequencing data for cancer. J Pathol Inform. 2017;8:44. doi: 10.4103/jpi.jpi_48_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Tommaso P., Chatzou M., Floden E.W., Barja P.P., Palumbo E., Notredame C. Nextflow enables reproducible computational workflows. Nat Biotechnol. 2017;35:316–319. doi: 10.1038/nbt.3820. [DOI] [PubMed] [Google Scholar]
- 10.Zook J.M., Catoe D., McDaniel J., et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci Data. 2016;3 doi: 10.1038/sdata.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013:1–3. [Google Scholar]
- 12.Li H., Handsaker B., Wysoker A., et al. 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and Samtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan B., Kusko R., Xiao W., et al. Correction to: Similarities and differences between variants called with human reference genome HG19 or HG38. BMC Bioinform. 2019;20:252. doi: 10.1186/s12859-019-2776-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benjamin D., Sato T., Cibulskis K., Getz G., Stewart C., Lichtenstein L. Calling somatic SNVs and Indels with Mutect2. bioRxiv. 2019:1–8. doi: 10.1101/861054. [DOI] [Google Scholar]
- 15.DePristo M.A., Banks E., Poplin R., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S., Scheffler K., Halpern A.L., et al. Strelka2: Fast and accurate calling of germline and somatic variants. Nat Methods. 2018;15:591–594. doi: 10.1038/s41592-018-0051-x. [DOI] [PubMed] [Google Scholar]
- 17.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. arXiv. 2012:1–9. [Google Scholar]
- 18.Ye K., Schulz M.H., Long Q., Apweiler R., Ning Z. Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilm A., Aw P.P., Bertrand D., et al. Lofreq: A sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koboldt D.C., Chen K., Wylie T., et al. Varscan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rimmer A., Phan H., Mathieson I., et al. WGS500 Consortium. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet. 2014;46:912–918. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang H., Bergmann E.A., Arora K., et al. Indel variant analysis of short-read sequencing data with scalpel. Nat Protoc. 2016;11:2529–2548. doi: 10.1038/nprot.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cingolani P., Platts A., Wang le L., et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SNPeff: SNPs in the genome of drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karczewski K.J., Francioli L.C., Tiao G., et al. Genome Aggregation Database Consortium. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tate J.G., Bamford S., Jubb H.C., et al. COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rausch T., Zichner T., Schlattl A., Stütz A.M., Benes V., Korbel J.O. DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wala J.A., Bandopadhayay P., Greenwald N.F., et al. Svaba: Genome-wide detection of structural variants and indels by local assembly. Genome Res. 2018;28:581–591. doi: 10.1101/gr.221028.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talevich E., Shain A.H., Botton T., Bastian B.C. Cnvkit: Genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12 doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Au C.H., Wa A., Ho D.N., Chan T.L., Ma E.S. Clinical evaluation of panel testing by next-generation sequencing (NGS) for gene mutations in myeloid neoplasms. Diagn Pathol. 2016;11:11. doi: 10.1186/s13000-016-0456-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S., Lee J., Chae S., et al. Multidimensional histone methylations for coordinated regulation of gene expression under hypoxia. Nucleic Acids Res. 2017;45:11643–11657. doi: 10.1093/nar/gkx747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia P., Yang X., Guo L., et al. Msisensor-pro: Fast, accurate, and matched-normal-sample-free detection of microsatellite instability. Genomics Proteomics Bioinformatics. 2020;18:65–71. doi: 10.1016/j.gpb.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao Y., Smyth G.K., Shi W. Featurecounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 35.Kovaka S., Zimin A.V., Pertea G.M., Razaghi R., Salzberg S.L., Pertea M. Transcriptome assembly from long-read RNA-seq alignments with stringtie2. Genome Biol. 2019;20:278. doi: 10.1186/s13059-019-1910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng Y.-Y., Cotto K.C., Ramu A., et al. RegTools: Integrated analysis of genomic and transcriptomic data for discovery of splicing variants in cancer. bioRxiv. 2018:1–21. doi: 10.1101/436634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mose L.E., Perou C.M., Parker J.S. Improved indel detection in DNA and RNA via realignment with ABRA2. Bioinformatics. 2019;35:2966–2973. doi: 10.1093/bioinformatics/btz033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lagunas-Rangel F.A., Chávez-Valencia V. FLT3-ITD and its current role in acute myeloid leukaemia. Med Oncol. 2017;34:114. doi: 10.1007/s12032-017-0970-x. [DOI] [PubMed] [Google Scholar]
- 39.Zaritsky A., Jamieson A.R., Welf E.S., et al. Interpretable deep learning uncovers cellular properties in label-free live cell images that are predictive of highly metastatic melanoma. Cell Syst. 2021;12 doi: 10.1016/j.cels.2021.05.003. 733–47.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang W., Williams T.A., Bhagwath A.S., et al. GEAMP, a novel gastroesophageal junction carcinoma cell line derived from a malignant pleural effusion. Lab Investig. 2020;100:16–26. doi: 10.1038/s41374-019-0278-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bishop J.A., Gagan J., Krane J.F., Jo V.Y. Low-grade apocrine intraductal carcinoma: Expanding the morphologic and molecular spectrum of an enigmatic salivary gland tumor. Head Neck Pathol. 2020;14:869–875. doi: 10.1007/s12105-020-01128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rooper L.M., Agaimy A., Dickson B.C., et al. DEK-AFF2 carcinoma of the sinonasal region and skull base: Detailed clinicopathologic characterization of a distinctive entity. Am J Surg Pathol. 2021;45:1682–1693. doi: 10.1097/PAS.0000000000001741. [DOI] [PubMed] [Google Scholar]
- 43.Argani P., Palsgrove D.N., Anders R.A., et al. A novel NIPBL-NACC1 gene fusion is characteristic of the cholangioblastic variant of intrahepatic cholangiocarcinoma. Am J Surg Pathol. 2021;45:1550–1560. doi: 10.1097/PAS.0000000000001729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
All the code used in SCHOOL is available at the UTSW Clinical Laboratory github wiki site: https://medforomics.github.io/schoolwiki/ using the repos: school for pipelines, process_scripts used in each docker container, and dnanexus_applets for running on DNANexus.