Abstract
To improve the reliability of the serodiagnosis of Chlamydia trachomatis infections, an immunoblot analysis, a microimmunofluorescence titration, and different immunoassays using synthetic peptides derived from species-specific epitopes in variable domain IV of the major outer membrane protein or recombinant antigens (heat shock protein 70 [hsp70], hsp60, hsp10, polypeptide encoded by open reading frame 3 of the plasmid [pgp3], macrophage infectivity potentiator, and a fragment of the total lipopolysaccharide) were evaluated. Because cross-reactions between chlamydial species have been reported, the microimmunofluorescence tests were also performed with Chlamydia pneumoniae and Chlamydia psittaci used as antigens, and C. pneumoniae-specific antibodies were also determined by immunoassays. Since the presence of antimicrobial antibodies must be interpreted in light of their prevalence in the general population, responses obtained with serum samples from patients with well-defined infection (i.e., with positive urethral or endocervical C. trachomatis DNA amplification) were compared to those obtained with samples from healthy blood donors. The best sensitivity (86%) with a specificity of 81% was obtained for immunoblotting results, when the number of individuals with ≥10 immunoglobulin G (IgG) and/or ≥2 IgM responses to the different C. trachomatis antigens was considered. A 13-kDa antigen was recognized by most of the samples (86% for IgG) from patients with acute urogenital infection but rarely (3%) by those from healthy blood donors (P < 0.0001). The sensitivity and specificity results obtained for serum antibodies to peptides or recombinant antigens were slightly lower than those results obtained for the number of responses to whole C. trachomatis antigens, which were 76 and 77%, respectively, when IgG responses to both recombinant hsp60 and pgp3 were considered.
Although serology can never replace methods aiming at the direct detection of Chlamydia trachomatis, there are situations in which reliable serological tests can be helpful. Indeed, urogenital infections with these bacteria are frequently inapparent (18, 30, 34, 40). Therefore, determination of antibodies to C. trachomatis antigens may be useful in determining whether a patient has had a previous infectious encounter. For example, in chronically infected patients in whom the bacteria are no longer detectable locally, a positive serological test may be the only indication of chlamydial involvement.
Different tests have been used for chlamydial serology. Early studies were performed with a complement fixation test, but this test could not differentiate between chlamydial species, and it lacked sensitivity. The microimmunofluorescent (MIF) test is still considered the serologic “gold standard.” Although it is claimed to be species specific, cross-reactions between chlamydial species have been reported (37, 38). Recently, several enzyme-linked immunosorbent assays (ELISAs) have been commercially developed with Chlamydia recombinant antigens, some of them known to be C. trachomatis specific.
We therefore used different approaches to investigate whether a test or a combination of tests could be sensitive and specific enough to be used for the serodiagnosis of C. trachomatis infection. We first performed immunoblot assays of C. trachomatis antigens, since this technique is widely used in the serodiagnosis of Lyme borreliosis (36) but is not currently used in Chlamydia serology. However, because antibodies directed to conformational epitopes can be missed by immunoblot analysis, we also developed an ELISA using, as antigens, five different Chlamydia recombinant proteins, most of which were purified in native conditions. The selected proteins were C. trachomatis heat shock protein 70 (hsp70), hsp60, hsp10, a polypeptide encoded by open reading frame 3 of the plasmid (pgp3), and a macrophage infectivity potentiator (MIP). hsp70 (5, 13, 28) and MIP (24, 27) have been identified as in vitro targets of neutralizing antibodies. hsp60, which is supposed to play an important role in the host immune response (31), is coexpressed with hsp10 (29), but hsp10 has been reported to be an independent marker (4, 22). pgp3, which is predominantly found in chlamydial outer membrane complex preparations (10), has been found to be a major immunogen in chlamydial infections (11). Antigens were prepared and tested under exactly the same conditions in order to compare the respective sensitivity and specificity of the tests. Two commercially available ELISA tests were also evaluated. One uses synthetic peptides derived from species-specific epitopes in variable domain IV of the major outer membrane protein (MOMP) of C. trachomatis, which is not homologous to C. pneumoniae MOMP sequence (Labsystems Research Laboratory, Helsinki, Finland). The other ELISA was based on an exclusively Chlamydia-specific recombinant fragment of the total lipopolysaccharide (LPS) 3-deoxy-d-manno-2-octulopyranosonic acid (3-Kdo) (Medac GmbH, Hamburg, Germany). Finally, the MIF tests were performed with C. trachomatis, C. pneumoniae, and C. psittaci as antigens, and anti-C. pneumoniae-specific antibodies were determined with commercially available ELISA tests (Labsystems Research Laboratory) in order to investigate possible cross-reactions.
Since the presence of antimicrobial antibodies must be interpreted in light of their prevalence in the general population, we compared the prevalence of anti-Chlamydia antibodies in samples of patients with well-defined disease (i.e., with positive urethral or endocervical C. trachomatis DNA amplification) with those in samples from healthy blood donors with a similar age and sex ratio.
MATERIALS AND METHODS
Patients.
Serum samples were stored at −70°C until processed. The study subjects were categorized into one of the two following groups: group 1 patients (n = 45) had acute C. trachomatis urogenital infection with positive findings on urethral or endocervical C. trachomatis DNA amplification with the Amplicor test (Roche Diagnostic Systems, Branchburg, N.J.); group 2 subjects (n = 31) were healthy blood donors.
The median ages (in years), age range, and percentages of female subjects are given in Table 1.
TABLE 1.
Serum antibody responses to whole C. trachomatis antigens determined by immunoblot analysis
| Subjects and analysis | Median age in years (range) | % Female | Response (mean no. detectable bands ± SD) to antigen by antibody:
|
||
|---|---|---|---|---|---|
| IgG | IgM | IgA | |||
| Patients with acute urogenital C. trachomatis infection | 32 (17–65) | 27 | 18.4 ± 9.3 | 2 ± 2 | 11.1 ± 8.0 |
| Healthy blood donors | 28 (20–39) | 29 | 5.6 ± 5.2 | 0.5 ± 0.7 | 3.3 ± 3.7 |
| Statistical analysis (P)a | <0.0001 | 0.0004 | 0.0001 | ||
Results (P values) from Student's t test analysis of indicated antibody responses of both groups to C. trachomatis antigens.
Recombinant protein preparation. (i) Template DNA.
Template DNA for the PCR was obtained from C. trachomatis serovar D, strain UW-3/Cx, purchased from the American Type Culture Collection (No. VR-885), or from purified recombinant plasmid clones pUC18 for hsp70 and pCVB2 for hsp60, which were kindly provided by I. Maclean (University of Manitoba, Winnipeg, Canada).
(ii) Primers and DNA amplification.
The different C. trachomatis sequences amplified (8, 12, 13, 26), with their restriction endonuclease sites and GenBank accession numbers, are indicated in Table 2. Oligonucleotides used as primers were synthesized by Microsynth, Balgach, Switzerland.
TABLE 2.
Sequences of primers used for DNA amplification and size and localization of the cloned genes
| Genes | Location and PCR primer sequencesa | Restriction enzyme site | Size of DNA fragment (bp) | Localization of DNA fragment (bp)b | GenBank accession no. | Reference |
|---|---|---|---|---|---|---|
| hsp70 | 5′ end, 5′-CATGccatggcGCGAAAAAAGAAAGTCTAACAAAAT-3′ | NcoI | 1,959 | 151–2109 | M27580 | 13 |
| 3′ end, 5′-CGggatccCTCAGGTTTATCAACAATTTCAACA-3′ | BamHI | |||||
| hsp60 | 5′ end, 5′-CATGccatggTCGCTAAAAACATTAAATACAACGA-3′ | NcoI | 1,635 | 383–2017 | M58027 | 8 |
| 3′ end, 5′-CATGagatctATAGTCCATTCCTGCGCCAGGCATTGC-3′ | BglII | |||||
| pgp3 | 5′ end, 5′-CATGccatggGAAATTCTGGTTTTTATTTGTATAA-3′ | NcoI | 795 | 4054–4848 | J03321 | 12 |
| 3′ end, 5′-CATGagatctAGCGTTTGTTTGAGGTATTACCTCT-3′ | BglII | |||||
| MIP | 5′ end, 5′-CATGtcatgaAGAATATATTAAGTTGGATGCTTAT-3′ | BspHId | 732 | 100–831 | X66126, S47522, X53481 | 26 |
| 3′ end, 5′-CGggatccTTCTGTAACAGATACATTATCGTCG-3′ | BamHI | |||||
| hsp10 | 5′ end, 5′-CATGccatggeCAGATCAAGCAACGACCCTCAAGAT-3′ | NcoI | 309 | 37–345 | M58027 | 8 |
| 3′ end, 5′-CATGagatctTTGCAGAACTGCGATAACTTCGCTC-3′ | BgIII |
The lowercase characters correspond to restriction enzyme sites.
Nucleotides are numbered according to coordinates of the sequences and refer to their position on the GenBank entries.
G will be replaced by A with the QuickChange site-directed mutagenesis kit of Stratagene, La Jolla, Calif.
The BspHI restriction site is TCATGA which is able to replace the NcoI (CCATGG), since CATG is conserved.
G will be replaced by T with the Quick Change site-directed mutagenesis kit of Stratagene.
The amplification was performed in a reaction volume of 50 μl containing 0.2 mM concentrations of each deoxynucleoside triphosphate, 0.25 μM concentrations of each oligonucleotide primer, and 20 mM Tris-HCl (pH 8.8), 2 mM MgSO4, 10 mM KCl, 10 mM (NH4)2SO4, 0.1% Triton X-100, 0.1 mg of nuclease-free bovine serum albumin (BSA) per ml, and 2.5 U of recombinant Pfu DNA polymerase from Pyrococcus furiosus (Stratagene, La Jolla, Calif.). Samples were subjected to 33 cycles of amplification in a DNA Thermal Cycler (Perkin-Elmer Cetus, Norwalk, Conn.) as follows: 1 min (except for the first cycle, which was 3 min) of denaturation at 95°C, 1 min of primer annealing to template at 55°C, and 4 min of primer extension at 72°C. After the last cycle, all samples were incubated for 10 min at 72°C to ensure that the final extension step was complete.
(iii) Cloning of PCR-amplified products in the pQE-60 vector.
The pQE-60 vector (Qiagen, Chatsworth, Calif.) was chosen to append a tag of six histidines (6xHis) to the C termini of recombinant proteins for large-scale purification via nickel chelate affinity chromatography. The C-terminal 6xHis tag position allows purification of only full-length proteins. To perform this purification step, the native stop codon was replaced in frame with the BamHI or the BglII sites (Table 2).
Amplified products were separated on an 0.8% agarose gel, and bands of the expected sizes (Table 2) were excised and purified with the QIAquick spin column purification protocol (Qiagen). Both the amplified insert and the pQE-60 vector were cut with appropriate restriction enzymes (Table 2), ligated with T4 DNA ligase (New England Biolabs) and transformed into M15(pREP4)-competent Escherichia coli cells (Qiagen) using standard protocols (CaCl2 treatment and heat shock).
(iv) Identification of bacterial colonies containing recombinant plasmids.
The transformed cells were plated onto 1.5% agar containing kanamycin (Sigma) (25 μg/ml) and ampicillin (Boehringer) (100 μg/ml). Colonies were screened for the presence of an insert of the predicted size (Table 2) by DNA amplification. Oligonucleotides used were the primer-promoter region and the primer-reverse sequencing for pQE vectors (Qiagen).
Bacteria from each tested colony were grown overnight in 1.5 ml of Luria broth (LB) medium containing kanamycin (25 μg/ml) and ampicillin (100 μg/ml). When recombinant clones were identified, glycerol stocks were prepared.
(v) Sequence analysis.
The identity of positive clones was confirmed by nucleotide sequencing in both orientations of the entire DNA inserts. Nucleotide sequences were analyzed with the basic local alignment search tool (BLAST) software. They were translated into protein sequences and compared with known sequences in ExPASy.
(vi) In vitro site-directed mutagenesis (for hsp70 and hsp10).
In order to change an incorrect base introduced at the cloning step of hsp70 and hsp10, in vitro site-directed mutagenesis was performed with the QuickChange site-directed mutagenesis kit from Stratagene. For the amplification reaction, oligonucleotide primers containing the desired mutations were the hsp70 5′ end, 5′-AGAAATTAACCATGAGCGAAAAAAGAAAGTCTAA-3′; hsp70 3′ end, 5′-TTAGACTTTCTTTTTTCGCTCATGGTTAATTTCT-3′; hsp10 5′ end, 5′-AGAAATTAACCATGTCAGATCAAGCAACGACCCT-3′; and hsp10 3′ end, 5′-AGGGTCGTTGCTTGATCTGACATGGTTAATTTCT-3′.
(vii) Small expression cultures and purification of 6xHis tagged proteins.
In order to check the protein molecular weight and the presence of the C-terminal 6xHis tag, 10 ml of culture medium was inoculated with overnight cultures of recombinant clones. Expression was induced by adding 1 mM of isopropyl-β-d-thiogalacto-pyranoside (Life Technologies). Purification was performed under denaturing conditions to isolate any tagged proteins, independently of their solubility within the cell, according to Qiagen protocol. Nickel-nitrilotriacetic acid (Ni-NTA) spin columns from Qiagen were used, and the eluted proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Coomassie blue R-250 was used for staining.
(viii) Large-scale expression and purification under native conditions of 6xHis-tagged chlamydial recombinant proteins.
A small inoculate of glycerol stock was added to 25 ml of LB containing kanamycin (25 μg/ml) and ampicillin (100 μg/ml) and was incubated overnight at 37°C. This seeding culture was added to 500 ml of LB that also contained kanamycin and ampicillin, incubated at 37°C for about 1 h, and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (except for MIP, for which a concentration of 0.4 mM was used) for about 3.5 h (2.5 for MIP) with vigorous shaking. Cultures of E. coli were pelleted by using an RC 5 B Sorvall centrifuge with a GS-3 rotor at 4,000 × g for 20 min at 4°C. Pellets were washed in phosphate-buffered saline (PBS) and frozen at −20°C until used. Pellets were thawed and resuspended in lysis buffer (50 mM Na2HPO4/NaH2PO4 buffer [pH 8.0], 300 mM NaCl, 10 mM imidazole, 1 mM phenylmethylsulfonyl fluoride, 20 mM 2-mercaptoethanol, and 1 mg of lysozyme per ml [Sigma]). They were sonicated for 6 × 10 s on ice to ensure complete resuspension. Lysate was then centrifuged at 10,000 × g for 30 min at 4°C to pellet cell debris.
Fast protein liquid chromatography (FPLC) equipment (Pharmacia) was used to purify proteins under native conditions by nickel chelate affinity chromatography (Qiagen Ni-NTA Superflow resin).
For MIP purification a different experimental procedure was used. The recombinant protein was extracted by Triton X-114. A cushion of 6% (wt/vol) sucrose–10 mM Tris-HCl (pH 7.4)–150 mM NaCl–0.06% Triton X-114 was placed at the bottom of a centrifuge tube. The lysis supernatant was overlaid on this sucrose cushion, and the tube was incubated for 10 min at 30°C. Clouding of the solution occurred. The tube was centrifuged for 10 min at 300 × g at room temperature with a swinging bucket rotor. The upper aqueous phase was removed from the tube and received 0.5% fresh Triton X-114. After dissolution of the surfactant at 0°C, the mixture was again overlaid on the sucrose cushion used previously, incubated 10 min at 30°C, and centrifuged (6). The Triton X-114 phase was then diluted in lysis buffer and passed through the column. After extensive washing with wash buffer containing 0.5% 3[(3-cholamidopropyl)-dimethylammonio]-1-propane-sulfonate (CHAPS), MIP was eluted by a 20 to 250 mM imidazole gradient containing 50 mM Na2HPO4/NaH2PO4 buffer (pH 8.0), 300 mM NaCl, and 0.5% CHAPS. Fractions containing MIP were pooled and dialyzed against PBS with 0.5% CHAPS (25).
The protein concentration was determined with the Bradford assay (Bio-Rad Laboratories, Richmond, Calif.). Purified proteins were subsequently characterized by SDS-PAGE and immunoblotting, separated into aliquots, and frozen at −80°C.
(ix) Characterization of expressed recombinant proteins: control of apparent molecular weight, in-frame translation of the 6xHis tag, and binding by IgG from C. trachomatis-infected patients.
Precast Tris-glycine polyacrylamide gels (4 to 20% with a 4% stacking gel; 10 wells) were purchased from Novex (San Diego, Calif.). Protein preparations were reduced by heating at 95°C for 5 min in the presence of 0.0625 M Tris (pH 6.8)–10% (vol/vol) glycerol–3% (wt/vol) SDS–0.003% (wt/vol) bromophenol blue–3% (vol/vol) 2-mercaptoethanol. Ten microliters of protein preparation or 3 to 5 μl of protein standard (Mark12 wide range protein standard from Novex for the Coomassie blue staining gel, 6xHis protein ladder from Qiagen for the immunoblot assay with anti-His antibody, and prestained, multicolored protein standard from Novex for the immunoblot assay with a pool of sera from C. trachomatis-infected patients) was loaded in each well. Electrophoresis was performed for 1 h and 50 min at a constant voltage of 125 V in an Xcell II vertical slab gel system (Novex). The running buffer consisted of 0.025 M Tris, 0.192 M glycine, and 0.1% (wt/vol) SDS (pH 8.3).
Three identical gels were prepared, one of which was stained for protein visualization with Coomassie blue and the other two of which were transferred to a Hybond C-extra nitrocellulose membrane purchased from Amersham, Little Chalfont, England. The transfer was performed for 1 h and 30 min at a constant voltage of 40 V by semidry blotting in an Xcell II blot module (Novex) at 4°C. The transfer buffer consisted of 0.012 M Tris, 0.096 M glycine, and 20% (vol/vol) methanol (pH 8.3). The blotted nitrocellulose membranes were incubated in blocking buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 3% [wt/vol] BSA), in order to block unspecific binding sites. The membrane was then washed twice for 10 min with Tris-buffered saline (TBS)-Tween-Triton buffer (20 mM Tris [pH 7.5], 500 mM NaCl, 0.05% [vol/vol] Tween 20, and 0.2% [vol/vol] Triton X-100) and 10 min with TBS buffer (10 mM Tris [pH 7.5], 150 mM NaCl).
For the 6xHis tag detection, a mouse monoclonal penta His antibody (Qiagen) was diluted 1/1,000 in blocking buffer and incubated with the membrane for 1 h at room temperature. The membrane was then washed twice for 10 min with TBS-Tween-Triton buffer and 10 min with TBS buffer and incubated for 1 h, at room temperature, with the second antibody (alkaline phosphatase-conjugated F(ab′)2 fragment of polyclonal sheep anti-mouse IgG) (Cappel, Embrach, Switzerland) at a dilution of 1:1,000 in blocking buffer.
For the binding by IgG-specific antibodies, a pool of 22 sera from patients with positive urethral or endocervical C. trachomatis DNA amplification was diluted 1:100 and incubated for 2 h at 37°C. The second antibody, an alkaline phosphatase-conjugated F(ab′)2 fragment of polyclonal goat IgG anti-human IgG (Fc specific) (Cappel) was diluted 1:1,600 and incubated for 2 h at 37°C.
The membranes were then washed four times with TBS-Tween-Triton buffer. The color was developed by adding a buffer containing 100 mM Tris (pH 9.5), 100 mM NaCl, 5 mM MgCl2, 0.33 mg of nitroblue tetrazolium per ml, and 0.165 mg of 5-bromo-4-chloro-3-indoyl phosphate per ml. Color development was stopped by washing the membrane with water after 15 to 20 min.
Measurement of IgG and IgA antibodies against the 6xHis-tagged native recombinant proteins by ELISA.
The solid phase was Ni-NTA HisSorb strips (Qiagen), which were used to allow the oriented binding of the 6xHis-tagged native proteins. These flat-well strips were coated with 100 μl of each of the five recombinant protein solutions, prepared at the same concentration of 100 pmol per ml of PBS with 0.25% (wt/vol) BSA and incubated overnight at 4°C. The strips were washed four times in PBS-Tween 20. Serum samples were tested in quadruplicate; they were diluted 1:100 for IgG and 1:50 for IgA determination in PBS with 0.25% BSA and incubated overnight at 4°C. After four washes in PBS-Tween, 100 μl of an alkaline phosphatase-conjugated F(ab′)2 fragment of polyclonal goat IgG, anti-human IgG (Fc specific), or IgA (α chain specific) (Cappel), diluted with PBS-Tween 20 containing 0.25% BSA, was added to each well and incubated for 2 h at 37°C. After four washes in PBS-Tween, 100 μl of substrate (1 mg of p-nitrophenyl phosphate per ml in 10% diethanolamine) was added for 1 h at 37°C. Color development was stopped by the addition of 3 M NaOH (25 μl/well). Optical density (OD) at 410 nm was measured by a Dynatech MR 5000 Microplate Reader (Dynatech, Alexandria, Va.). The reactivity of each serum was also tested in four noncoated wells. The net OD values are presented (mean OD value obtained for the four coated wells − nonspecific OD [mean OD value obtained for the four noncoated wells]).
The determination of absorbance cutoff values was of 14 sera from patients with acute urogenital C. trachomatis infection and 31 from healthy blood donors was performed by using the prediction limit calculated from child serum values, used as negative controls, and a 99% confidence level. For IgG and IgA antibodies to all of the proteins, the absorbance cutoff values were found to be 0.1 (14). To evaluate the reproducibility of the immunoassay results, one positive serum was tested 11 times. The reproducibilities of the ODs from run to run were ±10%.
Immunoblot analysis of sera.
Immunoblot analysis of 14 sera from patients with acute urogenital C. trachomatis infection and 31 from healthy blood donors was performed as previously described (2). Briefly, elementary body antigens of C. trachomatis LGV2 strain 434 were purchased from Biodesign International (Kennebunk, Maine). The antigens were suspended in sample buffer, heated to 100°C for 5 min, and centrifuged at 10,000 × g for 5 min. Two hundred microliters (145 μg of protein) was loaded in a two-dimensional well of precast Tris-glycine polyacrylamide gels (10 to 27% with a 4% stacking gel; two-dimensional well) purchased from Novex. After electrophoresis, C. trachomatis antigens were transferred to a Hybond C-extra nitrocellulose membrane purchased from Amersham. The blotted nitrocellulose membrane was cut into strips 4 mm in width. These strips were incubated overnight at 4°C in blocking buffer, incubated for 2 h at 37°C in sera diluted 1:100 in PBS-Tween, and incubated for 2 h at 37°C with alkaline phosphatase-conjugated F(ab′)2 fragment of polyclonal goat IgG anti-human IgG (Fc-specific), IgM (μ chain specific) or IgA (α chain specific) (Cappel) diluted with PBS-Tween containing 5% (wt/vol) BSA. After color development, the numbers of bands and their intensities on nitrocellulose paper were evaluated both visually (blots were assessed blindly, always by the same person) and with a GS-700 imaging densitometer and with Molecular Analyst software version 2.1 (Bio-Rad Laboratories, Hercules, Calif.) from weak (+) to very strong (++++) intensity. In order to avoid biases due to limitations of reproducibility, at least two immunoblots were done using each serum sample and were evaluated independently. The percentage of patients with serum antibodies to each antigen was calculated. The total response corresponded to all bands visually detectable, and the high-intensity responses corresponded only to bands with +++ and ++++ intensities.
Commercially available ELISA.
The ELISA kits used to detect the IgG and IgA anti-C. trachomatis MOMP antibodies and those used to detect IgG, IgM, and IgA anti-C. pneumoniae were purchased from Labsystems Research Laboratory (Helsinki, Finland). The ELISA kits used to detect the IgG, IgM, and IgA anti-Chlamydia LPS antibodies were purchased from Medac GmbH (Hamburg, Germany). The assays and calculations were performed according to the manufacturers' instructions. However, equivocal results for the anti-MOMP and anti-C. pneumoniae assays or gray-zone results for anti-LPS assays were considered negative. Results were considered positive when the signal/cutoff value was ≥1.4 for the anti-MOMP antibodies or >1.1 for the IgM anti-C. pneumoniae antibodies, when enzyme immunounits were >45 for IgG or >12 for IgA anti-C. pneumoniae antibodies and when OD values were greater than or equal to cutoff plus 10% (+15% for IgM) for the anti-LPS antibodies.
MIF tests.
Specific IgG, IgM, and IgA antibodies to C. trachomatis, C. pneumoniae, and C. psittaci antigens were determined with slides purchased from Labsystems. The assays were performed according to the manufacturer's instructions. Results were considered positive when titers were ≥1:16 for IgG and IgA and ≥1:10 for IgM.
Calculations.
Sensitivity, specificity, positive predictive value, negative predictive value, and false-positive and false-negative rates were calculated as described previously (1).
Statistical analysis.
Where appropriate, results were analyzed by the Student's t test and chi-square test.
RESULTS
Analysis of cloned genes.
The nucleotide sequences of cloned genes were analyzed with BLAST, and those giving the best alignment showed a high similarity to the previously published C. trachomatis sequences. When the protein sequences were compared, the hsp70 sequence was found to have 100% identity to the protein sequence obtained from the AE001312 (GenBank accession number) nucleotide sequence, hsp60 to have 99.4% identity to M31739, pgp3 to have 100% identity to M19487, MIP to have 100% identity to AE001324, and hsp10 to have 100% identity to AE001285.
Analysis of purified recombinant proteins.
Protein solubility was determined according to Qiagen protocol; hsp70, hsp60, pgp3, and hsp10 were found to be present mainly in soluble fractions. MIP was found to be present mainly with insoluble matter. The five recombinant proteins were, however, prepared in native conditions. Mild nonionic (Triton X-114) and amphoteric (CHAPS) detergents were used to purify MIP.
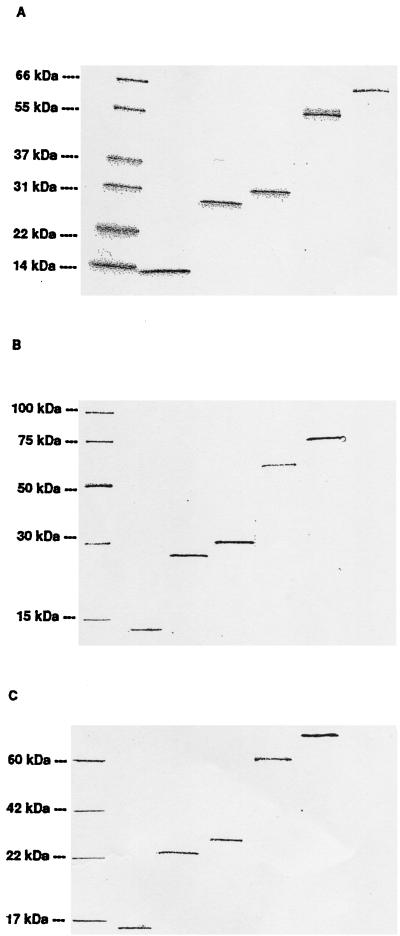
After migration on SDS-PAGE and Coomassie blue staining (Fig. 1A), immunodetection with the penta-His antibody (Fig. 1B), or immunodetection with a pool of sera from C. trachomatis-infected patients (Fig. 1C), purified proteins showed a band representing the expected apparent molecular weight and confirming correct insertion of the coding fragments and in-frame translation of the 6xHis tag, and these showed a recognition by IgG from infected patients.
FIG. 1.
Characterization of expressed recombinant proteins. The separation of proteins by SDS-PAGE was performed in three identical gels, one of which was stained for protein visualization with Coomassie blue (A). The two other gels were transferred to a Hybond C-extra nitrocellulose membrane and immunodetected with either anti-His antibody (penta-His antibody) according to Qiagen instructions for 6xHis-tagged protein detection (B) or a pool of 22 sera from patients with positive urethral or endocervical C. trachomatis DNA amplification for IgG-specific antibody binding (C). Lanes (left to right) MW standard, hsp10, MIP, pgp3, hsp60, and hsp70.
Antibody responses to whole C. trachomatis antigens determined by immunoblot analysis of serum samples from healthy blood donors and C. trachomatis-infected patients.
Immunoblot analysis showed that the IgM response seemed to be limited to a small number of antigens (maximum, 6), whereas IgA, and particularly IgG, recognized a high number of antigens (maximum, 23 and 31, respectively).
A highly significant (P ≤ 0.0004) increase (three- to fourfold) in the number of IgG, IgM, and IgA responses to whole Chlamydia antigens obtained after immunoblotting was observed for serum from C. trachomatis-infected patients compared to serum from healthy blood donors (Table 1).
Antibody responses to recombinant or synthetic Chlamydia antigens determined by ELISA.
When the percentage of individuals with serum antibodies to synthetic peptides or recombinant antigens was examined, C. trachomatis-infected patients had significantly more IgG to LPS, MOMP, hsp60, and pgp3 and more IgA to LPS and MOMP than healthy blood donors. The highest significant difference was observed for IgG responses to hsp60 (Table 3).
TABLE 3.
Percentages of individuals with serum antibodies to recombinant or synthetic Chlamydia antigens as determined by ELISA
| Antigen (source) | Serum antibody response | % of patients with acute C. trachomatis urogenital infection (n = 45) | % of healthy blood donors (n = 31)a | Statistical analysis (chi-square test) between both groupsb |
|---|---|---|---|---|
| LPS (Medac) | IgG | 84 | 52 | 9.62; P = 0.0019 |
| IgM | 9 | 3 | NS | |
| IgA | 62 | 23 | 11.61; P = 0.0007 | |
| MOMP (Labsystems) | IgG | 58 | 32 | 4.80; P = 0.029 |
| IgA | 47 | 16 | 7.61; P = 0.0058 | |
| Recombinant proteins prepared in our laboratory | ||||
| hsp70 | IgG | 29 | 17 | NS |
| IgA | 2 | 0 | NS | |
| hsp60 | IgG | 62 | 20 | 13.64; P = 0.0002 |
| IgA | 24 | 13 | NS | |
| pgp3 | IgG | 53 | 20 | 8.87; P = 0.0029 |
| IgA | 9 | 3 | NS | |
| MIP | IgG | 20 | 7 | NS |
| IgA | 0 | 0 | NS | |
| hsp10 | IgG | 2 | 3 | NS |
| IgA | 2 | 0 | NS |
Only 30 healthy blood donors were tested for serum antibodies to recombinant proteins.
NS, not significant.
The patterns of the IgG and IgA responses to the different peptides or recombinant antigens obtained for C. trachomatis-infected patients were different for each individual. Detailed IgG responses to different individual antigens in the seven immunoassays as well as the number of IgG responses to whole C. trachomatis antigens determined after immunoblotting are presented in Fig. 2. Except for one donor (number 11), an IgG response to pgp3 was observed only for samples in which >10 antigens of C. trachomatis were recognized after immunoblotting.
FIG. 2.
Sera from healthy blood donors or C. trachomatis-infected patients tested in immunoassays have different patterns of reactivity with synthetic peptides or recombinant antigens. MOMP antigens are synthetic peptides derived from species-specific epitopes in the variable domain IV of the MOMP of C. trachomatis and are not homologous to C. pneumoniae MOMP (Labsystems Research Laboratory). LPS is an exclusively Chlamydia-specific recombinant fragment of the total LPS (3-deoxy-d-manno-2-octulopyranosonic acid) (Medac GmbH). hsp70, hsp60, MIP, pgp3, and hsp10 are native purified recombinant antigens prepared in our laboratory. Two mild detergents were used to purify MIP, which otherwise was prepared under the same conditions as the other recombinant proteins (cf., Materials and Methods). Black boxes, high reactivity (≥cutoff value with OD 1.0); gray boxes, lower reactivity (≥cutoff value with OD < 1.0); white boxes, reactivity below the cutoff values. The number of positive IgG responses obtained for each sample tested in the seven immunoassays is compared with the number of IgG responses to LGV2 antigens observed in immunoblots.
The number of IgG responses to Chlamydia antigens determined after immunoblotting and the total number of positive IgG responses in the seven immunoassays were highly correlated (0.87; 95% confidence interval, 0.78 to 0.93; P < 0.0001 [Z test]).
Antibody responses to C. trachomatis, C. pneumoniae, or C. psittaci antigens determined by MIF test.
When the percentage of individuals with serum antibodies to the three different species of Chlamydia was examined, C. trachomatis-infected patients had significantly more IgG to C. trachomatis and C. pneumoniae than did healthy blood donors. However, the difference observed for C. pneumoniae was at the limit of a nonsignificant result (P = 0.046) (Table 4).
TABLE 4.
Percentages of individuals with serum antibodies to C. trachomatis, pneumoniae, or psittaci antigens as determined by microimmunofluorescence test
| Bacteria | Serum antibody | % of patients with acute C. trachomatis urogenital infection (n = 25) | % of healthy blood donors (n = 19) | Statistical analysis (chi-square test) between both groupsa |
|---|---|---|---|---|
| C. trachomatis | IgG | 44 | 11 | 5.81; P = 0.016 |
| IgM | 4 | 0 | NS | |
| IgA | 0 | 0 | NS | |
| C. pneumoniae | IgG | 72 | 42 | 3.99; P = 0.046 |
| IgM | 0 | 0 | NS | |
| IgA | 0 | 0 | NS | |
| C. psittaci | IgG | 0 | 0 | NS |
| IgM | 0 | 0 | NS | |
| IgA | 0 | 0 | NS |
NS, not significant.
Antibody responses to C. pneumoniae antigens determined by ELISA.
No significant difference was observed between the percentages of C. trachomatis-infected patients with IgG, IgM, or IgA antibodies and those percentages obtained for healthy blood donors (Table 5).
TABLE 5.
Percentages of individuals with serum antibodies to C. pneumoniae antigens determined by ELISA
| Serum antibody | % of patients with acute C. trachomatis urogenital infection (n = 32) | % of healthy blood donors (n = 26) | Statistical analysis (chi-square test) between both groupsa |
|---|---|---|---|
| IgG | 50 | 73 | NS |
| IgM | 0 | 0 | NS |
| IgA | 25 | 54 | NS |
NS, not significant.
Comparison of the results obtained for C. pneumoniae antibodies by MIF test and ELISA.
The ELISA appeared to have significantly higher sensitivity than the MIF test, particularly for the IgA determination. For the IgG measurement in healthy blood donor sera, 73% of donors were found to be positive with ELISA, and 42% were found to be positive with the MIF test (P = 0.036). For IgA, 25% of C. trachomatis-infected patients were found to be positive with ELISA, but none were found with the MIF test (P = 0.007), and 54% of healthy blood donors were found to be positive with ELISA, but none were found with the MIF test (P = 0.0001).
Comparison of C. trachomatis- and C. pneumoniae-specific assays with anti-Chlamydia LPS ELISA.
The presence of IgG anti-Chlamydia LPS is associated with the presence of IgG anti-C. pneumoniae alone (donors 13 and 18), with IgG anti-C. trachomatis alone (patients 31, 37, and 38), or with both in a high number of cases (Fig. 3).
FIG. 3.
Sera from healthy blood donors or C. trachomatis-infected patients tested for IgG anti-C. pneumoniae antibodies (MIF test and ELISA from Labsystems), anti-Chlamydia LPS antibodies (ELISA kit from Medac) and anti-C. trachomatis antibodies (MIF test and ELISA kit for anti-MOMP antibodies from Labsystems; anti-hsp60 and pgp3 antibody tests developed in our laboratory). For anti-C. pneumoniae MIF results, no titration was performed and positive results (≥1:16) are depicted as black boxes; for anti-C. trachomatis MIF results, high reactivity (≥1/128) is depicted as black boxes, and lower reactivity (1:16 to 1:64) is depicted as gray boxes. For IgG anti-C. pneumoniae antibodies determined with ELISA, high reactivity (>100) is depicted as black boxes, and lower reactivity (45 to 100) is depicted as gray boxes. For anti-LPS, MOMP, hsp60, and pgp3 antibodies, high reactivity (≥cutoff value with OD of >1.0) is depicted as black boxes; lower reactivity (≥cutoff value with OD of <1.0) is depicted as gray boxes. For all tests, reactivity below the cutoff values is depicted by white boxes.
Diagnostic value of the different antibody determinations.
From immunoblot results, when the numbers of individuals with ≥10 IgG, ≥2 IgM, or ≥6 IgA responses to C. trachomatis antigens were considered, the best results were obtained for IgG, with a sensitivity of 79% and a specificity of 84%. When the numbers of individuals with ≥10 IgG and/or ≥2 IgM responses were considered, the best sensitivity (86%) was obtained with a slight decrease of specificity (81%). With these criteria, a negative predictive value (likelihood that a negative result means the patient does not have disease) of 93% was obtained.
When the numbers of individuals with serum antibodies to peptides or recombinant antigens were considered, the diagnostic values of the tests were always lower. The highest sensitivity was obtained for IgG anti-LPS (84%) but with a low specificity (48%). The best results were obtained for IgG anti-hsp60 (sensitivity, 62%; specificity, 80%). When the numbers of individuals with IgG responses to hsp60 and/or pgp3 were considered, a higher sensitivity was obtained (76%) for a slightly lower specificity (77%).
The MIF test presented good specificity (89%) but poor sensitivity (44%) (Table 6).
TABLE 6.
Best results obtained for the different anti-Chlamydia antibody assays
| Assay | Sensitivity (%) | Specificity (%) | Predictive value (+) (%) | Predictive value (−) (%) | False-positive rate (%) | False-negative rate (%) |
|---|---|---|---|---|---|---|
| IgG anti-C. trachomatis (blot) (≥10 bands)a | 79 | 84 | 69 | 90 | 16 | 21 |
| IgM anti-C. trachomatis (blot) (≥2 bands)a | 50 | 94 | 78 | 81 | 6 | 50 |
| IgA anti-C. trachomatis (blot) (≥6 bands)a | 71 | 87 | 71 | 87 | 13 | 29 |
| IgG anti-C. trachomatis (blot) (≥10 bands) + | ||||||
| IgM anti-C. trachomatis (blot) (≥2 bands)a | 86 | 81 | 67 | 93 | 19 | 14 |
| IgA anti-C. trachomatis (blot) (≥6 bands) + | ||||||
| IgM anti-C. trachomatis (blot) (≥2 bands)a | 79 | 81 | 65 | 89 | 19 | 21 |
| IgG anti-LPSb | 84 | 48 | 70 | 68 | 52 | 16 |
| IgG anti-MOMPb | 58 | 68 | 72 | 53 | 32 | 42 |
| IgG anti-hsp60c | 62 | 80 | 82 | 59 | 20 | 38 |
| IgG anti-pgp3c | 53 | 80 | 80 | 53 | 20 | 47 |
| IgG anti-MOMP + pgp3c | 71 | 67 | 76 | 61 | 33 | 29 |
| IgG anti-hsp60 + pgp3c | 76 | 77 | 83 | 68 | 23 | 24 |
| IgG anti-C. trachomatis (MIF test)d | 44 | 89 | 85 | 55 | 11 | 56 |
Positive patients were 14 patients with C. trachomatis urogenital infection, and negative individuals were 31 healthy blood donors.
Positive patients were 45 patients with C. trachomatis urogenital infection, and negative individuals were 31 healthy blood donors.
Positive patients were 45 patients with C. trachomatis urogenital infection, and negative individuals were 30 healthy blood donors.
Positive patients were 25 patients with C. trachomatis urogenital infection, and negative individuals were 19 healthy blood donors.
From immunoblot results, when the percentages of individuals with serum antibodies to different C. trachomatis antigens were analyzed, the nine bands giving the most significant differences between patients with acute urogenital infection and healthy blood donors were of about 77, 71, 64, 61, 52, 36, 21, 18, and 13 kDa. The last band was particularly discriminant since P ≤ 0.0001 for IgG, IgM, and IgA when all of the responses were considered, and P ≤ 0.0001 for IgG and IgA when only the responses of high intensity were considered (Tables 7 and 8).
TABLE 7.
C. trachomatis antigens of diagnostic value
| Antigens (kDa) | % of individuals with serum antibodies to C. trachomatis antigens (total responses)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| IgG
|
IgM
|
IgA
|
|||||||
| Patients with urogenital infection (n = 42) | Healthy blood donors (n = 31) | P | Patients with urogenital infection (n = 42) | Healthy blood donors (n = 31) | P | Patients with urogenital infection (n = 42) | Healthy blood donors (n = 31) | P | |
| 77 | 79 | 6 | <0.0001 | 5 | 0 | NSa | 42 | 3 | <0.0001 |
| 71 | 65 | 0 | <0.0001 | 28 | 0 | 0.0011 | 56 | 6 | <0.0001 |
| 64 | 74 | 13 | <0.0001 | 33 | 0 | 0.0003 | 56 | 10 | <0.0001 |
| 61 | 84 | 26 | <0.0001 | 35 | 10 | 0.011 | 88 | 35 | <0.0001 |
| 52 | 81 | 26 | <0.0001 | 35 | 0 | 0.0002 | 70 | 26 | 0.0003 |
| 36 | 79 | 3 | <0.0001 | 12 | 0 | 0.047 | 65 | 10 | <0.0001 |
| 21 | 81 | 26 | <0.0001 | 28 | 3 | 0.0051 | 63 | 0 | <0.0001 |
| 18 | 79 | 48 | 0.0072 | 63 | 10 | <0.0001 | 74 | 10 | <0.0001 |
| 13 | 86 | 3 | <0.0001 | 51 | 0 | <0.0001 | 77 | 3 | <0.0001 |
NS, not significant.
TABLE 8.
Chlamydia trachomatis antigens of diagnostic value
| Antigen size (kDa) | % of individuals with serum antibodies to Chlamydia trachomatis antigens (high-intensity responses only)a
|
|||||
|---|---|---|---|---|---|---|
| IgG
|
IgA
|
|||||
| Patients with urogenital infection (n = 42) | Healthy blood donors (n = 31) | P | Patients with urogenital infection (n = 42) | Healthy blood donors (n = 31) | P | |
| 77 | 19 | 0 | 0.01 | 2 | 0 | NS |
| 71 | 30 | 0 | 0.0006 | 12 | 0 | 0.047 |
| 64 | 44 | 6 | 0.0006 | 16 | 0 | 0.017 |
| 61 | 47 | 10 | 0.0006 | 12 | 3 | NS |
| 52 | 35 | 0 | 0.0002 | 21 | 3 | 0.025 |
| 36 | 33 | 3 | 0.0017 | 21 | 0 | 0.0059 |
| 21 | 14 | 3 | NS | 9 | 0 | NS |
| 18 | 28 | 13 | NS | 37 | 0 | 0.0001 |
| 13 | 40 | 0 | <0.0001 | 37 | 0 | 0.0001 |
NS, not significant.
DISCUSSION
Reliable serological tests could be of assistance in the diagnosis of C. trachomatis infection when a test for direct detection of bacteria is negative or difficult to perform, as in cases of upper genital tract infections. In this study the diagnostic value of two complementary approaches, a global and a detailed one, were compared. A global view of the humoral response was obtained by immunoblot analysis and was quantified by counting the number of immunoreactive bands obtained. The molecular specificity of the response was studied in two different ways: its ability to recognize 27 different Chlamydia antigens after immunoblotting and its reactivity to seven Chlamydia peptides or recombinant antigens in immunoassays. To retain an antigenic structure capable of detecting antibodies directed to conformational epitopes, which could be missed by immunoblot analysis, five of these tests were performed with recombinant proteins prepared in their native conditions. These five antigens were tested by ELISA in exactly the same way in order to determine the key immunodominant antigens.
The immunoblot analysis was performed with a lymphogranuloma venereum (LGV) serovar, although it is an infrequent cause of sexually transmitted infections. However, human strains of C. trachomatis are closely related, and the LGV2 preparation seems to have sufficient cross- or group-reactive antigens to detect antibodies against most infecting strains. Antibodies directed against antigens common to all serovars of C. trachomatis have been detected but not antibodies directed against a particular serovar. In any case, it was not possible to test all trachoma serovars by this method. When immunoblots were analyzed, some C. trachomatis antigens appeared to have high reactivities with healthy blood donor samples. These bindings could correspond to cross-binding reactions with antigenic proteins from other microorganisms. Chlamydial proteins having homologues in other bacteria have been reported (33). However, the total number of immunoreactive bands appeared to be useful for diagnosis. Indeed, the best sensitivity, specificity, and negative predictive values for the serodiagnosis of C. trachomatis infection were obtained when considering serum specimens with at least 10 IgG bands and/or 2 IgM bands in immunoblot results. Thus, this study shows that immunoblotting is significantly more sensitive and specific for diagnosing C. trachomatis infection than the ELISAs. It is also more sensitive than the MIF test. A similar conclusion has already been obtained for serodiagnosis of early Lyme disease (15, 16) in which an immunoblot test has been recommended by the Centers for Disease Control and Prevention (7).
A C. trachomatis antigen of about 13 kDa was recognized by most of the samples (86% for IgG) from patients with acute urogenital infection and rarely (3%) by those from healthy blood donors (P < 0.0001) and could therefore be of great diagnostic value, but its identity was not determined. It is unlikely that this antigen is an LPS fraction or hsp10, since no similar results were obtained with immunoassays using these recombinant antigens. Another possible antigen is the ribosomal protein L7/12 (33).
Concerning the antigens tested by ELISA, the highest sensitivity (84%) was observed for the IgG reactivity to LPS but this determination had a low specificity (45%), confirming our previous observations (3) which attributed these results to the high prevalence of anti-C. pneumoniae antibodies in the population (19, 21), since LPS is a genus-specific antigen. This high prevalence was also observed in the present study, in which as high as 73% of healthy blood donors were found to have IgG anti-C. pneumoniae. The highest specificity (80%) was obtained with the IgG response to both hsp60, another conserved antigen, and pgp3, a C. trachomatis-specific antigen. When results obtained for IgG anti-hsp60 were associated with those obtained for pgp3, the sensitivity was increased (76%) with only a slight decrease of specificity (77%). This highest specificity observed with hsp60 was not expected, since it is one of the most conserved proteins in evolution and C. trachomatis and C. pneumoniae hsp60 are immunologically similar (20, 39). However, in this study, we found no evidence that anti-hsp60 antibodies might be elicited by other microorganisms, as was suggested by Sziller et al. (35), since the anti-hsp60 antibodies detected in our blood donor samples were always present with other Chlamydia-specific antibodies. Indeed, one blood donor sample also had IgG anti-LPS, and five samples had IgG targeting C. trachomatis-specific antigens (MOMP and pgp3). Moreover, none of the six blood donors with evidence of IgG anti-C. pneumoniae but without evidence of IgG anti-C. trachomatis (donors 3, 5, 7, 8, 13, and 18) had IgG anti-hsp60. Our results agree rather with those of Persson et al. (32), which suggest that anti-hsp60 antibodies are related to C. trachomatis but not to C. pneumoniae, and with those of Chernesky et al. (9), who obtained a high specificity with an ELISA using this protein. However, it could be useful to evaluate the specificity of anti-C. trachomatis hsp60 antibody determination in serum samples of patients with various bacterial infections.
Concerning the MIF test for C. trachomatis antibody detection, a high specificity was obtained for IgG, but this test had poor sensitivity. No IgA was detected when 47% of C. trachomatis-infected patients had IgA anti-MOMP antibodies and 71% recognized more than six antigens of C. trachomatis in immunoblots. We did not observe cross-reactions between chlamydial species, since no antibody was found to recognize C. psittaci.
As already reported (17, 33), a considerable heterogeneity was observed in the antigens recognized by sera from C. trachomatis-infected patients after immunoblotting as well as by ELISA. When the patterns were compared between the 45 infected patients, no diagnostic profile of antibodies to C. trachomatis antigens was found but a great diversity was observed. These individual variations in humoral response could be due to differences in the host ability to develop an immune response to a given antigen; it may be related to HLA class II alleles, since Zhong and Brunham (41) have reported that the antibody responses to C. trachomatis hsp60 and hsp70 are major histocompatibility complex-linked in mice. This finding also underlines the usefulness of testing different Chlamydia antigens in order to avoid false-negative results. Thus, four C. trachomatis-infected patients (9%) were found to have no IgG, IgA, or IgM response to the seven different Chlamydia antigens tested by immunoassays. However, in one of them a high number of Chlamydia antigens was recognized after immunoblotting. Therefore, in terms of diagnostic value, increasing the number of tested antigens decreases the number of patients negative results. The time course of the antibody appearance could also differ between the different antigens or between LPS and protein antigens. Immunoaccessibility is also certainly an important factor, and the different reactivities observed for the five recombinant proteins are probably a function of their surface exposure, since bacterial infection usually induces serum antibody against surface-exposed components. Thus, the poor reactivity to MIP can be explained by its limited immunoaccessibility (28). The different reactivities observed in the present study between hsp70, hsp60, and hsp10, and already reported for hsp60 and hsp10 (4, 22) could be explained either by a greater surface exposure of hsp60, different genetic regulation of antibody responses to the different hsp types, or lack of epitopes due to the absence of posttranslational modification, as suggested by LaVerda and Byrne for hsp10 (23).
In conclusion, although the MIF test is still considered as the serologic “gold standard,” we did not find it to be the best test. No cross-reactions between chlamydial species were observed, but a lower sensitivity (44%) was obtained than with other tests (86%). Because this test has also been criticized for its requirement of considerable expertise, its subjective interpretation, and its unsuitability for testing large numbers of specimens, we think that immunoblotting or ELISA with appropriate antigen(s) is more useful. Thus, by comparing the humoral immune responses of patients with proven C. trachomatis infection with those of healthy blood donors of similar ages and sex ratios, the best balance between sensitivity and specificity was found with the total number of IgG and IgM reactive bands obtained by immunoblotting. As a complementary method, the association of two recombinant proteins (hsp60 and pgp3), prepared under native conditions and used as antigens in ELISA, improved both specificity and sensitivity compared to individual recombinant antigens.
The anti-LPS antibody test from Medac appears to be of low usefulness due to its nonspecificity. Concerning anti-C. pneumoniae assays, we observed no good agreement between results obtained from MIF and ELISA tests.
ACKNOWLEDGMENTS
The technical assistance of Carol Brugger, Yvette Froment, Ursula Spenato, and Monique Stucker is gratefully acknowledged.
This work was supported by grant 32-47299.96 from the Fonds National Suisse de la Recherche Scientifique.
REFERENCES
- 1.Bas S, Cunningham T, Kvien T K, Glennas A, Melby K, Vischer T L. The value of isotype determination of serum antibodies against Chlamydia for the diagnosis of Chlamydia reactive arthritis. Br J Rheumatol. 1996;35:542–547. doi: 10.1093/rheumatology/35.6.542. [DOI] [PubMed] [Google Scholar]
- 2.Bas, S., C. Scieux, and T. L. Vischer. Male gender predominance in Chlamydia trachomatis sexually acquired reactive arthritis: are women more protected by anti-Chlamydia antibodies? Ann. Rheum. Dis., in press. [DOI] [PMC free article] [PubMed]
- 3.Bas S, Vischer T L. Chlamydia trachomatis antibody detection and diagnosis of reactive arthritis. Br J Rheumatol. 1998;37:1054–1059. doi: 10.1093/rheumatology/37.10.1054. [DOI] [PubMed] [Google Scholar]
- 4.Betsou F, Sueur J M, Orfila J. Serological investigation of Chlamydia trachomatis heat shock protein 10. Infect Immun. 1999;67:5243–5246. doi: 10.1128/iai.67.10.5243-5246.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birkelund S, Larsen B, Holm A, Lundemose A G, Christiansen G. Characterization of a linear epitope on Chlamydia trachomatis serovar L2 DnaK-like protein. Infect Immun. 1994;62:2051–2057. doi: 10.1128/iai.62.5.2051-2057.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Recommendations for test performance and interpretation from the Second National Conference on Serologic Diagnosis of Lyme Disease. Morbid Mortal Weekly Rep. 1995;44:590–591. [PubMed] [Google Scholar]
- 8.Cerrone M C, Ma J J, Stephens R S. Cloning and sequence of the gene for heat shock protein 60 from Chlamydia trachomatis and immunological reactivity of the protein. Infect Immun. 1991;59:79–90. doi: 10.1128/iai.59.1.79-90.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chernesky M, Luinstra K, Sellors J, Schachter J, Moncada J, Caul O, Paul I, Mikaelian L, Toye B, Paavonen J, Mahony J. Can serology diagnose upper genital tract Chlamydia trachomatis infections? Studies on women with pelvic pain, with or without chlamydial plasmid DNA in endometrial biopsy tissue. Sex Transm Dis. 1998;25:14–19. doi: 10.1097/00007435-199801000-00004. [DOI] [PubMed] [Google Scholar]
- 10.Comanducci M, Cevenini R, Moroni A, Giuliani M M, Ricci S, Scarlato V, Ratti G. Expression of a plasmid gene of Chlamydia trachomatis encoding a novel 28 kDa antigen. J Gen Microbiol. 1993;139:1083–1092. doi: 10.1099/00221287-139-5-1083. [DOI] [PubMed] [Google Scholar]
- 11.Comanducci M, Manetti R, Bini L, Santucci A, Pallini V, Cevenini R, Sueur J M, Orfila J, Ratti G. Humoral immune response to plasmid protein pgp3 in patients with Chlamydia trachomatis infection. Infect Immun. 1994;62:5491–5497. doi: 10.1128/iai.62.12.5491-5497.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comanducci M, Ricci S, Cevenini R, Ratti G. Diversity of the Chlamydia trachomatis common plasmid in biovars with different pathogenicity. Plasmid. 1990;23:149–154. doi: 10.1016/0147-619x(90)90034-a. [DOI] [PubMed] [Google Scholar]
- 13.Danilition S L, Maclean I W, Peeling R, Winston S, Brunham R C. The 75-kilodalton protein of Chlamydia trachomatis: a member of the heat shock protein 70 family? Infect Immun. 1990;58:189–196. doi: 10.1128/iai.58.1.189-196.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frey A, Di Canzio J, Zurakowski D. A statistically defined endpoint titer determination method for immunoassays. J Immunol Methods. 1998;221:35–41. doi: 10.1016/s0022-1759(98)00170-7. [DOI] [PubMed] [Google Scholar]
- 15.Fung B P, McHugh G L, Leong J M, Steere A C. Humoral immune response to outer surface protein C of Borrelia burgdorferi in Lyme disease: role of the immunoglobulin M response in the serodiagnosis of early infection. Infect Immun. 1994;62:3213–3221. doi: 10.1128/iai.62.8.3213-3221.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grodzicki R L, Steere A C. Comparison of immunoblotting and indirect enzyme-linked immunosorbent assay using different antigen preparations for diagnosing early Lyme disease. J Infect Dis. 1988;157:790–797. doi: 10.1093/infdis/157.4.790. [DOI] [PubMed] [Google Scholar]
- 17.Inman R D, Johnston M E, Chiu B, Falk J, Petric M. Immunochemical analysis of immune response to Chlamydia trachomatis in Reiter's syndrome and nonspecific urethritis. Clin Exp Immunol. 1987;69:246–254. [PMC free article] [PubMed] [Google Scholar]
- 18.Karam G H, Martin D H, Flotte T R, Bonnarens F O, Joseph J R, Mroczkowski T F, Johnson W D. Asymptomatic Chlamydia trachomatis infections among sexually active men. J Infect Dis. 1986;154:900–903. doi: 10.1093/infdis/154.5.900. [DOI] [PubMed] [Google Scholar]
- 19.Karvonen M, Tuomilehto J, Pitkaniemi J, Naukkarinen A, Saikku P. Chlamydia pneumoniae IgG antibody prevalence in south-western and eastern Finland in 1982 and 1987. Int J Epidemiol. 1994;23:176–184. doi: 10.1093/ije/23.1.176. [DOI] [PubMed] [Google Scholar]
- 20.Kikuta L C, Puolakkainen M, Kuo C C, Campbell L A. Isolation and sequence analysis of the Chlamydia pneumoniae GroE operon. Infect Immun. 1991;59:4665–4669. doi: 10.1128/iai.59.12.4665-4669.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo C C, Jackson L A, Campbell L A, Grayston J T. Chlamydia pneumoniae (TWAR) Clin Microbiol Rev. 1995;8:451–461. doi: 10.1128/cmr.8.4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LaVerda D, Albanese L N, Ruther P E, Morrison S G, Morrison R P, Ault K A, Byrne G I. Seroreactivity to Chlamydia trachomatis Hsp10 correlates with severity of human genital tract disease. Infect Immun. 2000;68:303–309. doi: 10.1128/iai.68.1.303-309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaVerda D, Byrne G I. Use of monoclonal antibodies to facilitate identification, cloning, and purification of Chlamydia trachomatis hsp10. J Clin Microbiol. 1997;35:1209–1215. doi: 10.1128/jcm.35.5.1209-1215.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lundemose A G, Birkelund S, Fey S J, Larsen P M, Christiansen G. Chlamydia trachomatis contains a protein similar to the Legionella pneumophila mip gene product. Mol Microbiol. 1991;5:109–115. doi: 10.1111/j.1365-2958.1991.tb01831.x. [DOI] [PubMed] [Google Scholar]
- 25.Lundemose A G, Kay J E, Pearce J H. Chlamydia trachomatis Mip-like protein has peptidyl-prolyl cis/trans isomerase activity that is inhibited by FK506 and rapamycin and is implicated in initiation of chlamydial infection. Mol Microbiol. 1993;7:777–783. doi: 10.1111/j.1365-2958.1993.tb01168.x. [DOI] [PubMed] [Google Scholar]
- 26.Lundemose A G, Rouch D A, Birkelund S, Christiansen G, Pearce J H. Chlamydia trachomatis Mip-like protein. Mol Microbiol. 1992;6:2539–2548. doi: 10.1111/j.1365-2958.1992.tb01430.x. [DOI] [PubMed] [Google Scholar]
- 27.Lundemose A G, Rouch D A, Penn C W, Pearce J H. The Chlamydia trachomatis Mip-like protein is a lipoprotein. J Bacteriol. 1993;175:3669–3671. doi: 10.1128/jb.175.11.3669-3671.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maclean I W, Peeling R W, Brunham R C. Characterization of Chlamydia trachomatis antigens with monoclonal and polyclonal antibodies. Can J Microbiol. 1988;34:141–147. doi: 10.1139/m88-028. [DOI] [PubMed] [Google Scholar]
- 29.Morrison R P, Su H, Lyng K, Yuan Y. The Chlamydia trachomatis hyp operon is homologous to the groE stress response operon of Escherichia coli. Infect Immun. 1990;58:2701–2705. doi: 10.1128/iai.58.8.2701-2705.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh M K, Cloud G A, Baker S L, Pass M A, Mulchahey K, Pass R F. Chlamydial infection and sexual behavior in young pregnant teenagers. Sex Transm Dis. 1993;20:45–50. doi: 10.1097/00007435-199301000-00009. [DOI] [PubMed] [Google Scholar]
- 31.Peeling R W, Kimani J, Plummer F, Maclean I, Cheang M, Bwayo J, Brunham R C. Antibody to chlamydial hsp60 predicts an increased risk for chlamydial pelvic inflammatory disease. J Infect Dis. 1997;175:1153–1158. doi: 10.1086/516454. [DOI] [PubMed] [Google Scholar]
- 32.Persson K, Osser S, Birkelund S, Christiansen G, Brade H. Antibodies to Chlamydia trachomatis heat shock proteins in women with tubal factor infertility are associated with prior infection by C. trachomatis but not by C. pneumoniae. Hum Reprod. 1999;14:1969–1973. doi: 10.1093/humrep/14.8.1969. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez-Campillo M, Bini L, Comanducci M, Raggiaschi R, Marzocchi B, Pallini V, Ratti G. Identification of immunoreactive proteins of Chlamydia trachomatis by Western blot analysis of a two-dimensional electrophoresis map with patient sera. Electrophoresis. 1999;20:2269–2279. doi: 10.1002/(SICI)1522-2683(19990801)20:11<2269::AID-ELPS2269>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 34.Shafer M A, Schachter J, Moncada J, Keogh J, Pantell R, Gourlay L, Eyre S, Boyer C B. Evaluation of urine-based screening strategies to detect Chlamydia trachomatis among sexually active asymptomatic young males. JAMA. 1993;270:2065–2070. [PubMed] [Google Scholar]
- 35.Sziller I, Witkin S S, Ziegert M, Csapo Z, Ujhazy A, Papp Z. Serological responses of patients with ectopic pregnancy to epitopes of the Chlamydia trachomatis 60 kDa heat shock protein. Hum Reprod. 1998;13:1088–1093. doi: 10.1093/humrep/13.4.1088. [DOI] [PubMed] [Google Scholar]
- 36.Tugwell P, Dennis D T, Weinstein A, Wells G, Shea B, Nichol G, Hayward R, Lightfoot R, Baker P, Steere A C. Laboratory evaluation in the diagnosis of Lyme disease. Ann Intern Med. 1997;127:1109–1123. doi: 10.7326/0003-4819-127-12-199712150-00011. [DOI] [PubMed] [Google Scholar]
- 37.Wagenvoort J H, Koumans D, van de Cruijs M. How useful is the Chlamydia micro-immunofluorescence (MIF) test for the gynaecologist? Eur J Obstet Gynecol Reprod Biol. 1999;84:13–15. doi: 10.1016/s0301-2115(98)00303-0. [DOI] [PubMed] [Google Scholar]
- 38.Wong Y K, Sueur J M, Fall C H, Orfila J, Ward M E. The species specificity of the microimmunofluorescence antibody test and comparisons with a time resolved fluoroscopic immunoassay for measuring IgG antibodies against Chlamydia pneumoniae. J Clin Pathol. 1999;52:99–102. doi: 10.1136/jcp.52.2.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan Y, Lyng K, Zhang Y X, Rockey D D, Morrison R P. Monoclonal antibodies define genus-specific, species-specific, and cross-reactive epitopes of the chlamydial 60-kilodalton heat shock protein (hsp60): specific immunodetection and purification of chlamydial hsp60. Infect Immun. 1992;60:2288–2296. doi: 10.1128/iai.60.6.2288-2296.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zelin J M, Robinson A J, Ridgway G L, Allason-Jones E, Williams P. Chlamydial urethritis in heterosexual men attending a genitourinary medicine clinic: prevalence, symptoms, condom usage and partner change. Int J STD AIDS. 1995;6:27–30. doi: 10.1177/095646249500600106. [DOI] [PubMed] [Google Scholar]
- 41.Zhong G, Brunham R C. Antibody responses to the chlamydial heat shock proteins hsp60 and hsp70 are H-2 linked. Infect Immun. 1992;60:3143–3149. doi: 10.1128/iai.60.8.3143-3149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]