Abstract
A transframe region within HIV-1 Gag-Pol (referred to as p6* or p6pol), directly linked to the protease (PR) N-terminus, plays a pivotal role in modulating PR activation. To identify specific p6* residues involved in PR activation, we created a series of p6* mutants by making substitutions for conserved p6* residues. Our results indicate that some p6* mutants were defective in terms of virus infectivity, despite displaying a wild-type virus particle processing pattern. Mutations at p6* F8 reduced virus infectivity associated with insufficient virus processing, due in part to impaired PR maturation and RT packaging. Our data strongly suggest that conserved Phe (F) residues at position 8 of p6* are involved in the PR maturation process.
Introduction
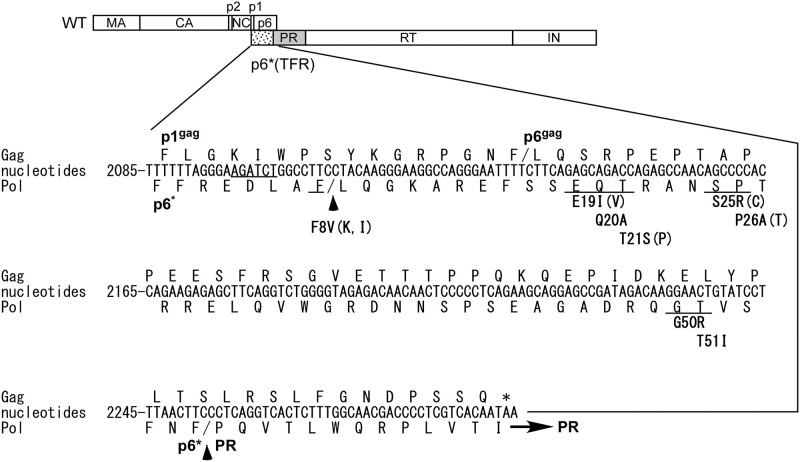
Essential retroviral proteases (PRs) are encoded by part of the pol gene [1]. Due to a partial overlap between gag and pol reading frames, HIV-1 Pol is translated as a Gag-Pol fusion protein via a -1 ribosomal frameshift mechanism that occurs at a frequency of 5–10% during Gag translation [2]. Gag-Pol dimerization triggers the activation of embedded PR, and the activated PR becomes a fully functional PR dimer following autocleavage from Gag-Pol [3]. Cleavage of the viral structural Gag precursor Pr55gag by PR yields four major products: matrix (MA; p17), capsid (CA; p24), nucleocapsid (NC; p7), and C-terminal p6gag [4]. Two spacer peptides, p2 and p1, separate NC from CA and p6gag, respectively. The proteolytic processing of Gag-Pol produces PR, reverse transcriptase, and integrase (IN) in addition to Gag cleavage products. Within Gag-Pol, p1gag-p6gag is truncated and replaced with a transframe region (TFR) referred to as p6* or p6pol (Fig 1). PR-mediated Gag and Gag-Pol processing (referred to as virus maturation) is essential for viral infectivity [5, 6].
Fig 1. Schematic representation of HIV-1 Gag and Gag-Pol constructs.
Indicated are HIV-1 Gag protein domains MA (matrix; p17), CA (capsid; p24), NC (nucleocapsid; p7), p1, p6, pol-encoded transframe region (TFR) p6*, PR, RT and IN. An overlapping reading frame sequence between p1gag-p6gag and p6* and encoded amino acid residues are shown. Positions of mutated amino acid residues are underlined. Alternative substitution residues are in parentheses. Arrowheads indicate cleavages at the p6*/PR junction and the F8/L9 putative internal cleavage site.
Since premature PR activation triggers premature Gag cleavage prior to Gag multimerization and virus particle formation (resulting in markedly reduced virus yields), temporal and spatial PR activation regulation is critical to virus assembly [7–9]. Several lines of evidence suggest that p6*, located adjacent to the PR N-terminus, plays a modulating role in PR activation. First, p6* removal from PR precursor is required for PR activity to either fully undergo autocleavage or mediate virus particle maturation [10–13]. Second, p6*-derived peptides are capable of blocking PR activity in vitro [14, 15]. Molecular model studies further suggest that C-terminal p6* residues may contribute to delayed PR activation by destabilizing PR dimer interface interactions [16–19]. We previously reported that PR dimerization enhancement following p6* replacement with a leucine zipper dimerization motif leads to markedly reduced virus yields due to improved Gag cleavage by PR. However, the presence of a C-terminal p6* tetrapeptide can counteract the Gag cleavage enhancement incurred by a leucine zipper insertion in the deleted p6* region [20–22].
Single amino acid substitutions of the four C-terminal p6* residues can significantly impair virus maturation, strongly suggesting that the C-terminal p6* tetrapeptide is important for PR activation regulation [10, 21]. Data from a study involving stepwise cluster substitutions for p6 codons (3–13 residues) suggest that both C- and N-terminal p6* regions are important, with the middle p6* part being largely dispensable in terms of PR activation [13]. Consistent with these data, a deletion of approximately 63% of p6* amino acid residues, or partial substitution of p6* with a heterologous peptide, does not significantly affect HIV-1 infectivity [23]. Results from our previous study, in which we performed single-cycle-infection assays of HIV-1 virions produced from Gag and Gag-Pol co-transfectants, indicate a significant reduction (20–40% of wt level) in virus infectivity following a major deletion of the p6* coding sequence [21, 24]. The remaining C-terminal p6* tetrapeptide in the p6*-deleted mutant is insufficient for mitigating the reduced infectivity resulting from deficient virus processing [24]. The presence or absence of undefined p6* residues contributing to PR activation regulation is unknown. PR inhibitors in current use largely target mature PR, but this is frequently accompanied by the emergence of PR-resistant HIV-1 variants. Understanding how p6* (especially certain specifically identified p6* residues) contributes to PR activation may support the development of an alternative HIV/AIDS treatment strategy involving p6* or p6*-PR targeting. To identify specific p6* residues involved in PR activation, we created a series of p6* mutants with site-directed mutagenesis of conserved p6* residues without affecting encoded p6gag residues. Mutation effects on virus and PR maturation were analyzed by immunoblot and single-round-infection assays.
Materials and methods
Plasmid construction
The parental HIV-1 proviral sequence in this study is HXB2 [25]. All constructs expressing Gag and Gag-Pol were derived from a replication-defective HIV-1 expression vector, HIVgpt [26]. All constructs including the p6* residue substitution mutants (see Table 1) were engineered by PCR-based overlap extension mutagenesis using HIVgpt as a template. We used a downstream reverse primer 5’-GGTACAGTCTCAATAGGGCTAATG-3’ (nt. 2577–51) or an upstream forward primer 5’-AATGATGCAGAGAGGCAAT-3’ (nt.1916-34). The amplified fragments were digested with BglII and BclI, and subcloned into a plasmid cassette pBRCla-Sal that contains HIV-1 coding sequence (from ClaI-nt.831 to SalI-nt.5786). Each mutation-containing pBRCla-Sal cassette was then digested with SpeI and SalI, and ligated into HIVgpt. The resultant constructs were confirmed by DNA sequencing.
Table 1. Primers used for creating p6* residue substitution mutations.
| No. | Mutation clones | Forward / Reverse primer(5’-3’) |
|---|---|---|
| 1 | F8V | F: 5’-GAA GAT CTG GCC GTC CTA CAA GGG AAG-3’ |
| R: 2578–52 5’-ACT GGT ACA GTC TCA ATA GGG CTA ATG-3’ | ||
| 2 | F8K | F: 5’-GAA GAT CTG GCC AAG CTA CAA GGG AAG-3’ |
| R: 2578–52 5’-ACT GGT ACA GTC TCA ATA GGG CTA ATG-3’ | ||
| 3 | F8I | F: 5’-GAA GAT CTG GCC ATC CTA CAA GG-3’ |
| R: 2578–52 5’-ACT GGT ACA GTC TCA ATA GGG CTA ATG-3’ | ||
| 4 | E19V | F: 5’-G GAA TTT TCT TCA GTC CAG ACC AGA GCC AAC AG-3’ |
| R: 5’-CT GTT GGC TCT GGT CTG GAC TGA AGA AAA TTC C-3’ | ||
| 5 | Q20S | F: 5’-G GAA TTT TCT TCA GAG TCG ACC AGA GCC AAC AG-3’ |
| R: 5’- CT GTT GGC TCT GGT CGA CTC TGA AGA AAA TTC C-3’ | ||
| 6 | E19I/Q20A | F: 5’-G GAA TTT TCT TCA ATC GCG ACC AGA GCC AAC AG-3’ |
| R: 5’-CT GTT GGC TCT GGT CGC GAT TGA AGA AAA TTC C-3’ | ||
| 7 | E19I/Q20A/T21S | F: 5’-G GAA TTT TCT TCA ATC GCG TCC AGA GCC AAC AG-3’ |
| R: 5’-CT GTT GGC TCT GGA CGC GAT TGA AGA AAA TTC C-3’ | ||
| 8 | E19V/Q20A/T21P | F: 5’-G GAA TTT TCT TCA GTC GCG CCC AGA GCC AAC AG-3’ |
| R:5’-CT GTT GGC TCT GGG CGC GAC TGA AGA AAA TTC C-3’ | ||
| 9 | S25R | F: 5’-CC AGA GCC AAC CGC CCC ACC AGA AGA G-3’ |
| R: 5’-C TCT TCT GGT GGG GCG GTT GGC TCT GG-3’ | ||
| 10 | P26A | F: 5’-CC AGA GCC AAC AGC GCC ACC AGA AGA G-3’ |
| R: 5’-C TCT TCT GGT GGC GCT GTT GGC TCT GG-3’ | ||
| 11 | S25C/P26T | F: 5’-CC AGA GCC AAC TGC ACC ACC AGA AGA G-3’ |
| R: 5’-C TCT TCT GGT GGT GCA GTT GGC TCT GG-3’ | ||
| 12 | G50R | F: 5’-CC GAT AGA CAA AGA ACT GTA TCC TTT A-3’ |
| R: 5’-T AAA GGA TAC AGT TCT TTG TCT ATC GG-3’ | ||
| 13 | T51I | F: 5’-CC GAT AGA CAA GGA ATT GTA TCC TTT A-3’ |
| R: 5’-T AAA GGA TAC AAT TCC TTG TCT ATC GG-3’ | ||
| 14 | G50R/T51I | F: 5’-CC GAT AGA CAA AGA ATT GTA TCC TTT A-3’ |
| R: 5’-T AAA GGA TAC AAT TCT TTG TCT ATC GG-3’ |
† Bold-faced nucleotides denote the mutated codons. BglII sites are underlined. Cloning procedures are described in Materials and Methods.
Cell culture and transfection
HEK-293T or HeLa cells were maintained in DMEM supplemented with 10% fetal calf serum. Confluent HEK-293T cells were trypsinized, split 1:10 and seeded onto 10-cm plates 24 hours before transfections. For each construct, HEK-293T cells were transfected with 20 μg of plasmid DNA by the calcium phosphate precipitation method [27], with the addition of 50 μm chloroquine to enhance transfection efficiency. Culture media and cells are harvested for protein analysis at 48–72 h post-transfection.
Immunoblot
Culture media from transfected HEK-293T cells were filtered (0.45-μm pores) and centrifuged through 2 ml 20% sucrose. Viral pellets and cell lysates mixed with sample buffer were subjected to 10% SDS-PAGE as described previously [20]. For separation of HIV-1 PR-associated products, samples were subjected to 4 to 12% bis-Tris gradient gels (NuPage bis-Tris minigels; Thermo Fisher Scientific) and electroblotted onto nitrocellulose membranes as described [28]. HIV-1 Gag proteins were probed with an anti-p24gag monoclonal antibody (mouse hybridoma clone 183-H12-5C). For HIV-1 RT detection, the primary antibody is rabbit antiserum or a mouse anti-RT monoclonal antibody [29, 30]. Membrane bound HIV-1 PR was detected with a mouse anti-PR monoclonal antibody (Abcam; ab8327) or a sheep antiserum. Cellular β-actin was detected using a mouse anti-β-actin monoclonal antibody (Sigma). The secondary antibody is a sheep anti-mouse or a donkey anti-rabbit (HRP)-conjugated antibody and the procedures used for HRP activity detection followed the manufacturer’s protocol (Thermo Fisher Scientific).
Single-cycle infection assays
For infections, 10 μg of wt or each of p6* mutant plasmid plus 5 μg of the VSV- G protein expression plasmid pHCMV-G [31] were co-transfected into HEK-293T cells. Forty-eight hours after transfection, virus-containing supernatant were collected, filtered, and aliquots of the filtrate were diluted to infect HeLa cells in the presence of 4 μg/ml polybrene. The remaining supernatant and cell samples were prepared and subjected to 10% SDS-PAGE. Twenty-four hours after infection, cells were trypsinized, split into dishes and refed with medium containing drug selection cocktail [32]. Selected mycophenolic acid resistant colonies were fixed and stained with 50% methanol containing 0.5% methylene blue. Numbers of drug-resistant colonies are converted into titers (cfu/ml). Infectivity was expressed as the ratio of the mutant titers to wt titers in parallel experiments.
Results
To identify specific p6* residues involved in PR activation, highly conserved residues believed to be replaceable without affecting encoded p6gag residues were altered using PCR-mediated site-directed mutagenesis. p6* sequences in published HIV-1 strains were aligned, with non-polymorphic residues treated as highly conserved residues (https://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/2006_7/5.pdf). Primers used to generate mutants are shown in Table 1. Of the 14 p6* residue substitution mutants created, 9 contained single amino acid residue substitutions, 3 contained double substitutions, and 2 contained triple substitutions. A four-residue substitution mutant designated E19I/Q20A/T21S/S25G was generated during the PCR-mediated cloning of the E19I/Q20A/T21S mutant. This unintentionally created mutation did not affect encoded p6gag residues.
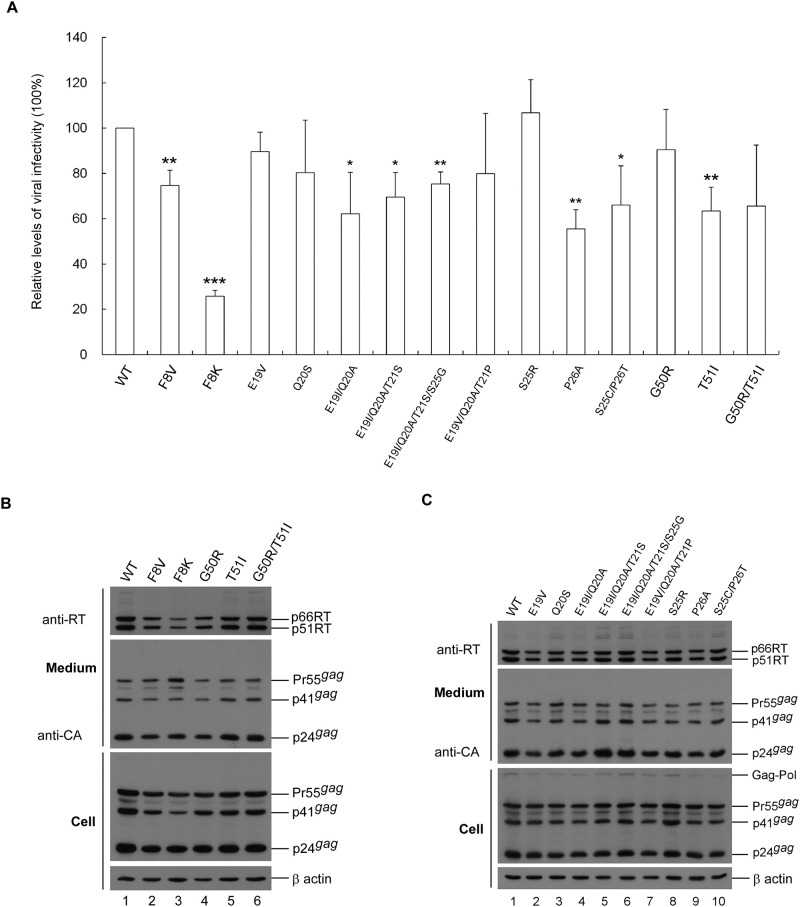
Impacts of p6* mutations on virus maturation were assessed using single-cycle infection assays and immunoblot of released virions following the transient expression of each mutant with a VSV-G expression vector in HEK-293T cells. According to assay results, some single amino acid substitution mutations exerted small or moderate impacts on virus infectivity, and simultaneous substitutions for two, three or four residues led to noticeable reductions in virus infectivity. For example, E19V or Q20S mutations did not significantly impair virus infectivity, but E19I/Q20A, E19I/Q20A/T21S and E19I/Q20A/T21S/S25G did. Reduced E19V/Q20A/T21P infectivity was statistically non-significant (Fig 2A). Likewise, reduced T51I infectivity became statistically non-significant following the additional replacement of Arg for G50 (G50R/T51I). The individual and multiple substitutions of the p6* residues (such as E19V, E19I/Q20A and E19I/Q20A/T21S, etc.) were selected from databases excluding the polymorphic p6* codons. It remains to be determined if an individual single mutation such as E19I, Q20A, T21P, T21S or S25G has any impact on virus infectivity.
Fig 2. Effects of p6* mutations on virus particle processing and infectivity.
HEK293T cells were transfected with a wt HIVgpt or designated p6* mutant construct plus a VSV-G expression vector. Cells and supernatants were collected 48 h post-transfection. Collected and filtered supernatant aliquots were used to infect HeLa cells. Remaining supernatants were used for immunoblot. (A) Relative virus infectivity. Drug-resistant colony infection and selection was performed as described in Materials and Methods section. Mutant infectivity was determined as the ratio of mutant titers to wt titers, normalized to Gag protein levels in parallel experiments. *p < 0.05; **p < 0.05; ***p < 0.001. All results are from three independent experiments. (B-C) Shown is a representative immunoblot. Membrane-bound proteins were initially probed with anti-RT serum prior to stripping and probing with an anti-p24CA monoclonal antibody. HIV-1 Gag-Pol, Pol, 66/51RT, Pr55gag, p41gag and p24gag positions are shown.
Regarding mutations at the putative internal cleavage site F8/L9, we observed that a Val (V) or Lys (K) replacement at F8 significantly reduced virus infectivity. Specifically, F8K exhibited a five-fold decrease in virus infectivity compared to the wt in parallel experiments. Western blot analysis data indicate that despite defective infectivity, most mutants displayed a virus particle processing profile similar to that of the wt (Fig 2B and 2C). However, some mutants (including F8K) exhibited higher virus-associated Pr55gag levels compared to the wt (Fig 2B, lane 3 vs. lane 1), suggesting a virus maturation defect.
Mutations at a putative internal p6* cleavage site trigger defects in PR maturation and RT incorporation
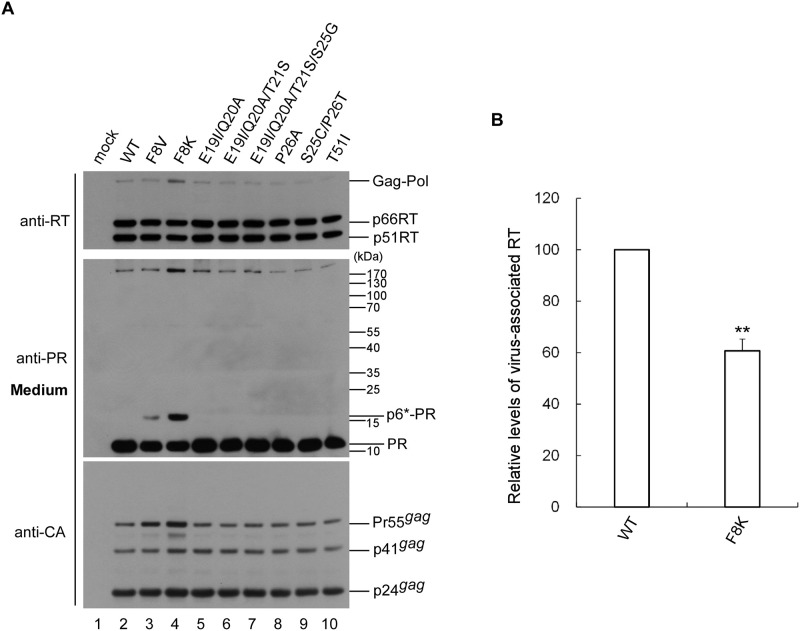
Given the pivotal role of p6* in modulating the PR activation process, it is likely that the reduced infectivity observed in some p6* mutants may have been due to a defect in the PR activation process, leading to subsequent impairments in virus maturation. To test this possibility, all infectivity-defective mutants were subjected to immunoblot with an HIV-1 PR antiserum. Our data readily detected mature PR in all mutant particles. In addition to mature PR, bands corresponding to the PR precursor (p6*-PR) were observed for F8V and F8K, with substantial amounts of F8K PR present in precursor form (Fig 3A, lanes 3 and 4). Since F8V and F8K exhibited higher levels of virus-associated Pr55gag compared to the wt, impaired PR maturation may have contributed, at least in part, to insufficient virus processing (Fig 3A middle panel, lanes 3 and 4 vs. lane 2). In addition, we found substitution mutations at F8, in particular, F8K markedly reduced virus-associated RT levels (Fig 3B). These data suggest that a native Phe (F) at position 8 of p6* is important for PR maturation and RT incorporation.
Fig 3. Mutations at the internal p6* cleavage site impaired PR maturation and RT incorporation.
HEK293T cells were transfected with designated constructs. Cells and supernatants were collected 48 h post-transfection and subjected to immunoblot. To detect PR-associated products, supernatant samples were separated by 4–12% Bis-Tris gradient gels. Membrane-bound proteins were initially probed with anti-PR serum, stripped, and probed with anti-RT serum, followed by probing with anti-p24CA monoclonal antibodies. Molecular weight size markers (in kDa) are indicated on right side (second upper panel). HIV-1 Gag-Pol, Pol, 66/51RT, p6*-PR, PR, Pr55gag, p41gag and p24gag positions are shown. Asterisks indicate 14 kDa PR-associated intermediate precursor positions. (B) Levels of HIV-1 Gag proteins Pr55, p41 and p24, and RT-associated Gag-Pol and p66/51 in each sample were quantified by scanning band densities on immunoblots. Ratios of total Pol versus Gag proteins levels were calculated, and normalized to wt in parallel experiments. Data were obtained from three independent experiments. **p < 0.05.
Since F8/L9 has been proposed as an internal p6* cleavage site [15, 33, 34], changes in F8 residues may block internal p6* cleavage, thus contributing to impaired PR maturation. If it exists, a cleavage at F8/L9 would supposedly produce a PR intermediate (designated delTFP-p6*-PR) with an 8-amino acid transframe peptide (TFP) removed from the p6*-PR. Since the delTFP-p6*-PR migrates slightly faster than the p6*-PR, the two may be detected by immunoblot as a double band [35]. Accordingly, it is likely that inhibiting p6*-PR autocleavage by reducing PR activity might allow for the detection of p6*-PR and delTFP-p6*-PR in wt virions. To test this possibility, wt and F8K transfectants were mock-treated or treated with low doses of an HIV-1 PR inhibitor. An F8I mutation (referred to as F440I in the Pettit group’s reports) is capable of blocking the F8/L9 internal p6* cleavage site in synthesized Gag-Pol in vitro [36, 37]. We used an F8I mutant to serve as a control.
Our results indicate that both F8I and F8K exhibited higher levels of virus-associated Pr55gag compared to the wt (Fig 4, lanes 4 and 6 vs. lane 2), suggesting a virus particle processing defect. Unexpectedly, we did not observe a double band of PR precursors; instead, two distinct immature PR products (17 kDa and 14 kDa) were readily detected in wt samples when PR activity was partially inhibited (Fig 4, lane 3). This suggests that F8/L9 cleavage may occur very rapidly, or that the degree of partial PR activity inhibition completely blocks F8/L9 cleavage. Unlike the wt, both F8K and F8I had readily detected bands corresponding to p6*-PR, regardless of PR inhibition treatment or no treatment (Fig 4, lanes 4–7). The PR-associated 14 kDa products were likely derived from a cleavage at a cryptic site within p6*-PR.
Fig 4. Effects of partial PR activity inhibition on PR maturation.
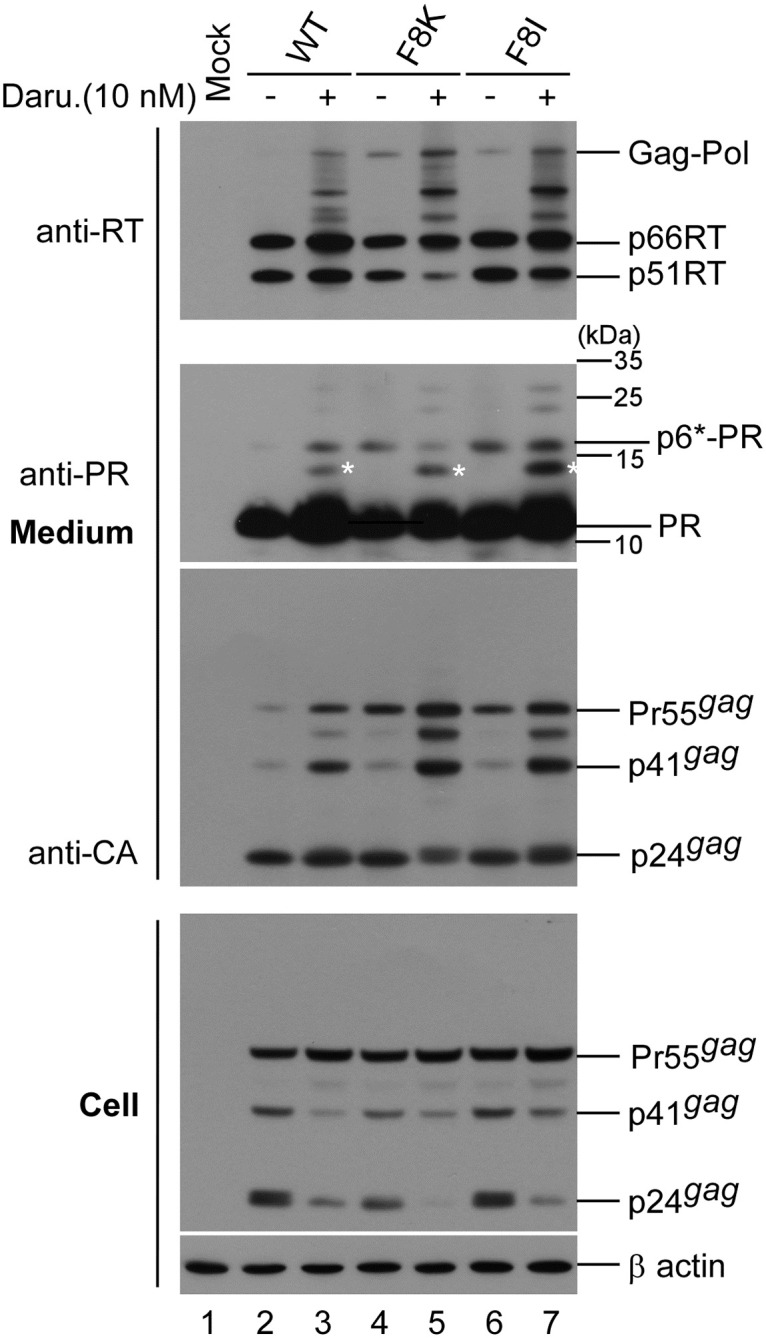
HEK293T cells were transfected with designated construct. At 4 h post-transfection, equal amounts of cells were plated on two dishes and either left untreated or treated with darunavir (an HIV-1 protease inhibitor) at a concentration of 10 nM. Supernatants and cells were collected 48 h post-transfection, prepared, and subjected to immunoblot as described in the Fig 3 caption. Asterisks indicate 14 kDa PR intermediate positions. Shown is a representative immunoblot from three independent experiments.
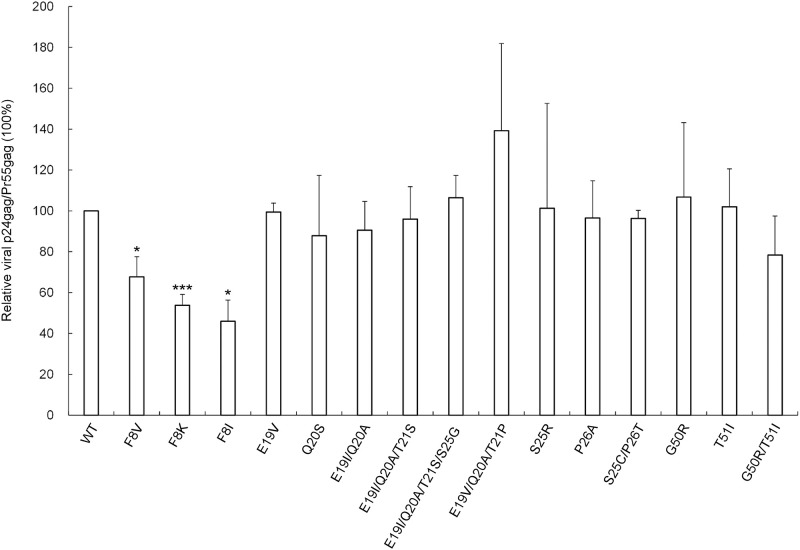
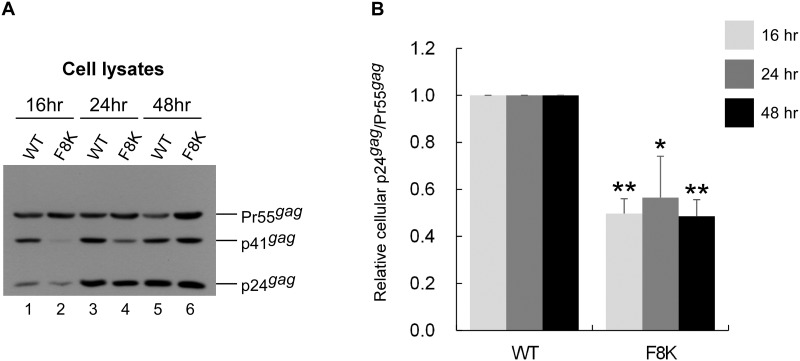
Quantification results for virus particle processing efficiency indicate that both F8V and F8K had significantly lower p24gag/Pr55gag ratios compared to the wt, suggesting a defect in virus maturation (Fig 5). A kinetic analysis of intracellular Gag cleavage indicates F8K had a slower cleavage rate than wt (Fig 6), supporting that mutations at F8 can affect PR-mediated Gag cleavage efficiency. The defects in virus maturation and RT package may partly account for marked reductions in infectivity following mutation substitutions at F8. Since most of the infectivity-impaired mutants we observed exhibited Gag processing efficiency levels comparable to that of the wt, immunoblot assays may be insufficient for detecting subtle but damaging impacts on virus maturation resulting in reduced infectivity. We cannot exclude the possibility that the mutations impaired infectivity during other stages of the virus replication cycle. Combined, our data suggest that mutations at the internal p6* cleavage site can impair virus and PR maturation.
Fig 5. Virus particle processing efficiency data for p6* mutants.
Virus-associated Pr55gag and p24gag levels were quantified as immunoblot band densities. Ratios of p24gag to p55gag were determined for each mutant and normalized to those for a wt in parallel experiments. Data were obtained from three independent experiments. Error bars indicate standard deviations. *p<0.05; ***p<0.001.
Fig 6. Effects of F8K mutation on Gag cleavage.
HEK293T cells were transfected with indicated constructs. At 4 h post-transfection, equal amounts of cells were plated on three dish plates. Cells were collected at 16, 24 and 48 h, and subjected to immunoblot. Panel A is a representative immunoblot from three independent experiments. Cellular Pr55gag and p24gag levels were quantified by scanning immunoblot band densities. Ratios of p24gag to Pr55gag were determined and normalized to those of wt in parallel experiments. Bars indicate standard deviations. *p<0.05;**p<0.01 (panel B).
Discussion
Previous studies have suggested that conserved C-terminal p6* tetrapeptide mutations trigger significant PR maturation impairment [10, 20, 21]. For the present study we focused on analyzing the effects of other conserved p6* residue mutations on virus processing and PR maturation. Since changes in the conserved p6* residues used in this research did not affect p6gag residues, it remains unknown whether the other conserved p6* residues contributed to PR maturation. Although immunoblot data reveal lower virus particle processing efficiency for some p6* mutants (Fig 5), virus-associated RT and PR were readily detected in most mutants. This agrees with a previous report indicating that p6* point mutations did not exert major impacts on the quantitative cleavage of Pol components, despite the possibility that the mutations may have affected Gag cleavage [10]. F8V, F8K and F8I particles contained readily detected p6*-PR (Figs 3 and 4), suggesting that mutations at the internal p6* cleavage site may inhibit the PR maturation process. It is likely that insufficient PR maturation contributes to lower virus processing efficiency, which in turn leads to markedly reduced virus infectivity.
Inhibited PR maturation due to reduced PR activity or an internal p6* cleavage defect may lead to cleavages at cryptic substrate sites [38]. In support of this hypothesis, we found that a 14 kDa PR intermediate was readily detected in wt, F8K and F8I particles following the partial inhibition of PR activity (Fig 4).
Internal p6* cleavages at F8/L9 were determined by observing the autocleaving of synthesized HIV-1 Gag-Pol or p6*-PR expressed and purified from bacterial cells in vitro [19, 36, 37, 39–41]. However, to our knowledge the PR intermediate delTFP-p6*-PR derived from a cleavage at F8/L9 is not present in mature HIV-1 virions. A similar product was found within HIV-1 particles following the partial inhibition of PR activity [42]. Results from an analysis of p6* mutation effects on PR activation also failed to detect delTFP-p6*-PR in virus particles [10]. A mutation at F8V has been described as having no major effects on virus infectivity or PR maturation, with mature PR identified as predominant, and with p6*-PR barely detectable [10]. In a separate study, p6*-PR was the predominant PR species, and mature PR was barely detected in virions released from HIV-1-infected human cells [43]. These discrepancies may be due, at least in part, to the employment of different systems. Ludwig et al. concluded that the F8V mutation exerted no major impacts on virus and PR maturation after transiently expressing p6* mutants in an H1299 human lung cancer cell line (2008). During virus assembly and processing, viral polyprotein folding and conformational change may affect target sequence accessibility [44]. Embedded PR release from Gag-Pol entails an initial intramolecular cleavage at the PR N-terminus. The second step, intermolecular cleavage at the PR C-terminus, releases mature PR [39, 41, 45]. Immature PR precursor possesses substantial enzymatic activity [43, 46] and is less sensitive than mature PR to protease inhibition [39]. In our study, p6*-PR precursors were readily detected in F8 mutant particles (Figs 3 and 4), strongly suggesting that mutations at p6* F8 affect PR maturation. It is likely that F8 mutations incur a conformational change that impairs the PR maturation process by destabilizing PR precursor dimer interactions.
Conclusion
Most of our p6* mutations did not exert major impacts on virus particle processing, but some were found to significantly impair virus infectivity. In particular, we found that mutations at a putative internal p6* cleavage site markedly impaired virus processing and infectivity, likely due in part to defects in PR maturation and RT package. We do not have evidence indicating that a cleavage occurs at F8/L9, but our data strongly suggest that the Phe conserved residue at p6* position 8 is important for PR maturation.
Supporting information
(PDF)
Acknowledgments
The authors thank the following from the National Institutes of Health AIDS Research and Reference Reagent Program for their assistance in obtaining the following reagents: anti-RT monoclonal antibody (MAb21) (Stephen Hughes); antiserum to HIV-1 PR (D. Bailey and Mark Page) and antiserum to HIV-1 RT (Stuart Le Grice).
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
CW received grants from Taipei Veterans General Hospital (V106C-047, V107C-049, V108C-034) and the Taiwan Ministry of Science and Technology (106-2320-B-010-017-MY2, 108-2320-B-010-030). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Petropoulos C. Retroviral Taxonomy, Protein Structures, Sequences, and Genetic Maps. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 2.Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331(6153):280–3. Epub 1988/01/21. doi: 10.1038/331280a0 . [DOI] [PubMed] [Google Scholar]
- 3.Freed EO. HIV-1 assembly, release and maturation. Nature Reviews Microbiology. 2015;13:484. doi: 10.1038/nrmicro3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swanstrom R, Wills JW. Synthesis, Assembly, and Processing of Viral Proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 5.Pettit SC, Moody MD, Wehbie RS, Kaplan AH, Nantermet PV, Klein CA, et al. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68(12):8017–27. Epub 1994/12/01. doi: 10.1128/JVI.68.12.8017-8027.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng C, Ho BK, Chang TW, Chang NT. Role of human immunodeficiency virus type 1-specific protease in core protein maturation and viral infectivity. J Virol. 1989;63(6):2550–6. Epub 1989/06/01. doi: 10.1128/JVI.63.6.2550-2556.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figueiredo A, Moore KL, Mak J, Sluis-Cremer N, de Bethune MP, Tachedjian G. Potent nonnucleoside reverse transcriptase inhibitors target HIV-1 Gag-Pol. PLoS Pathog. 2006;2(11):e119. Epub 2006/11/14. doi: 10.1371/journal.ppat.0020119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tachedjian G, Orlova M, Sarafianos SG, Arnold E, Goff SP. Nonnucleoside reverse transcriptase inhibitors are chemical enhancers of dimerization of the HIV type 1 reverse transcriptase. Proc Natl Acad Sci U S A. 2001;98(13):7188–93. Epub 2001/06/21. doi: 10.1073/pnas.121055998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan YY, Wang SM, Huang KJ, Chiang CC, Wang CT. Placement of leucine zipper motifs at the carboxyl terminus of HIV-1 protease significantly reduces virion production. PLoS One. 2012;7(3):e32845. Epub 2012/03/08. doi: 10.1371/journal.pone.0032845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludwig C, Leiherer A, Wagner R. Importance of protease cleavage sites within and flanking human immunodeficiency virus type 1 transframe protein p6* for spatiotemporal regulation of protease activation. J Virol. 2008;82(9):4573–84. Epub 2008/03/07. doi: 10.1128/JVI.02353-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Partin K, Zybarth G, Ehrlich L, DeCrombrugghe M, Wimmer E, Carter C. Deletion of sequences upstream of the proteinase improves the proteolytic processing of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1991;88(11):4776–80. Epub 1991/06/01. doi: 10.1073/pnas.88.11.4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tessmer U, Krausslich HG. Cleavage of human immunodeficiency virus type 1 proteinase from the N-terminally adjacent p6* protein is essential for efficient Gag polyprotein processing and viral infectivity. J Virol. 1998;72(4):3459–63. Epub 1998/04/03. doi: 10.1128/JVI.72.4.3459-3463.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paulus C, Ludwig C, Wagner R. Contribution of the Gag-Pol transframe domain p6* and its coding sequence to morphogenesis and replication of human immunodeficiency virus type 1. Virology. 2004;330(1):271–83. Epub 2004/11/06. doi: 10.1016/j.virol.2004.09.013 . [DOI] [PubMed] [Google Scholar]
- 14.Paulus C, Hellebrand S, Tessmer U, Wolf H, Krausslich HG, Wagner R. Competitive inhibition of human immunodeficiency virus type-1 protease by the Gag-Pol transframe protein. The Journal of biological chemistry. 1999;274(31):21539–43. Epub 1999/07/27. doi: 10.1074/jbc.274.31.21539 . [DOI] [PubMed] [Google Scholar]
- 15.Louis JM, Dyda F, Nashed NT, Kimmel AR, Davies DR. Hydrophilic peptides derived from the transframe region of Gag-Pol inhibit the HIV-1 protease. Biochemistry. 1998;37(8):2105–10. Epub 1998/03/28. doi: 10.1021/bi972059x . [DOI] [PubMed] [Google Scholar]
- 16.Yu F-H, Chou T-A, Liao W-H, Huang K-J, Wang C-T. Gag-Pol Transframe Domain p6* Is Essential for HIV-1 Protease-Mediated Virus Maturation. PLoS ONE. 2015;10(6):e0127974. doi: 10.1371/journal.pone.0127974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatterjee A, Mridula P, Mishra RK, Mittal R, Hosur RV. Folding regulates autoprocessing of HIV-1 protease precursor. The Journal of biological chemistry. 2005;280(12):11369–78. Epub 2005/01/06. doi: 10.1074/jbc.M412603200 . [DOI] [PubMed] [Google Scholar]
- 18.Ishima R, Torchia DA, Louis JM. Mutational and structural studies aimed at characterizing the monomer of HIV-1 protease and its precursor. The Journal of biological chemistry. 2007;282(23):17190–9. Epub 2007/04/07. doi: 10.1074/jbc.M701304200 . [DOI] [PubMed] [Google Scholar]
- 19.Louis JM, Clore GM, Gronenborn AM. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat Struct Biol. 1999;6(9):868–75. Epub 1999/08/31. doi: 10.1038/12327 . [DOI] [PubMed] [Google Scholar]
- 20.Yu F-H, Huang K-J, Wang C-T. C-Terminal HIV-1 Transframe p6* Tetrapeptide Blocks Enhanced Gag Cleavage Incurred by Leucine Zipper Replacement of a Deleted p6* Domain. Journal of Virology. 2017;91(10). doi: 10.1128/jvi.00103-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu F-H, Wang C-T. HIV-1 protease with leucine zipper fused at N-terminus exhibits enhanced linker amino acid-dependent activity. Retrovirology. 2018;15(1):32. doi: 10.1186/s12977-018-0413-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu FH, Huang KJ, Wang CT. Conditional activation of an HIV-1 protease attenuated mutant by a leucine zipper dimerization motif. Virus Res. 2021;295:198258. Epub 2020/12/15. doi: 10.1016/j.virusres.2020.198258 . [DOI] [PubMed] [Google Scholar]
- 23.Leiherer A, Ludwig C, Wagner R. Uncoupling human immunodeficiency virus type 1 Gag and Pol reading frames: role of the transframe protein p6* in viral replication. J Virol. 2009;83(14):7210–20. Epub 2009/05/01. doi: 10.1128/JVI.02603-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiu H-C, Wang F-D, Chen Y-MA, Wang C-T. Effects of human immunodeficiency virus type 1 transframe protein p6* mutations on viral protease-mediated Gag processing. Journal of General Virology. 2006;87(7):2041–6. doi: 10.1099/vir.0.81601-0 [DOI] [PubMed] [Google Scholar]
- 25.Ratner L, Haseltine W, Patarca R, Livak KJ, Starcich B, Josephs SF, et al. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313(6000):277–84. doi: 10.1038/313277a0 . [DOI] [PubMed] [Google Scholar]
- 26.Page KA, Landau NR, Littman DR. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J Virol. 1990;64(11):5270–6. Epub 1990/11/01. doi: 10.1128/JVI.64.11.5270-5276.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graham R, van der Eb A. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 1973;52:456–67. doi: 10.1016/0042-6822(73)90341-3 [DOI] [PubMed] [Google Scholar]
- 28.Chiang C-C, Wang S-M, Tseng Y-T, Huang K-J, Wang C-T. Mutations at human immunodeficiency virus type 1 reverse transcriptase tryptophan repeat motif attenuate the inhibitory effect of efavirenz on virus production. Virology. 2009;383(2):261–70. doi: 10.1016/j.virol.2008.10.027 [DOI] [PubMed] [Google Scholar]
- 29.Ferris AL, Hizi A, Showalter SD, Pichuantes S, Babe L, Craik CS, et al. Immunologic and proteolytic analysis of HIV-1 reverse transcriptase structure. Virology. 1990;175(2):456–64. doi: 10.1016/0042-6822(90)90430-y [DOI] [PubMed] [Google Scholar]
- 30.Hizi A, McGill C, Hughes SH. Expression of Soluble, Enzymatically Active, Human Immunodeficiency Virus Reverse Transcriptase in Escherichia coli and Analysis of Mutants. Proceedings of the National Academy of Sciences. 1988;85(4):1218–22. doi: 10.1073/pnas.85.4.1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yee JK, Friedmann T, Burns JC. Generation of high-titer pseudotyped retrovirus with very broad host range. Methods in Cell Biology 1994;43:99–112. doi: 10.1016/s0091-679x(08)60600-7 [DOI] [PubMed] [Google Scholar]
- 32.Chen YL, Ts’ai PW, Yang CC, Wang CT. Generation of infectious virus particles by transient co-expression of human immunodeficiency virus type 1 gag mutants. J Gen Virol. 1997;78(10):2497–501. doi: 10.1099/0022-1317-78-10-2497 [DOI] [PubMed] [Google Scholar]
- 33.Phylip LH, Griffiths JT, Mills JS, Graves MC, Dunn BM, Kay J. Activities of precursor and tethered dimer forms of HIV proteinase. Advances in experimental medicine and biology. 1995;362:467–72. Epub 1995/01/01. doi: 10.1007/978-1-4615-1871-6_61 . [DOI] [PubMed] [Google Scholar]
- 34.Chen N, Morag A, Almog N, Blumenzweig I, Dreazin O, Kotler M. Extended nucleocapsid protein is cleaved from the Gag-Pol precursor of human immunodeficiency virus type 1. The Journal of general virology. 2001;82(Pt 3):581–90. Epub 2001/02/15. doi: 10.1099/0022-1317-82-3-581 . [DOI] [PubMed] [Google Scholar]
- 35.Abbink TEM, Ooms M, Haasnoot PCJ, Berkhout B. The HIV-1 Leader RNA Conformational Switch Regulates RNA Dimerization but Does Not Regulate mRNA Translation. Biochemistry. 2005;44(25):9058–66. doi: 10.1021/bi0502588 [DOI] [PubMed] [Google Scholar]
- 36.Pettit SC, Gulnik S, Everitt L, Kaplan AH. The dimer interfaces of protease and extra-protease domains influence the activation of protease and the specificity of GagPol cleavage. J Virol. 2003;77(1):366–74. Epub 2002/12/13. doi: 10.1128/jvi.77.1.366-374.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettit SC, Lindquist JN, Kaplan AH, Swanstrom R. Processing sites in the human immunodeficiency virus type 1 (HIV-1) Gag-Pro-Pol precursor are cleaved by the viral protease at different rates. Retrovirology. 2005;2(1):66. doi: 10.1186/1742-4690-2-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loeb DD, Hutchison CA, Edgell MH, Farmerie WG, Swanstrom R. Mutational analysis of human immunodeficiency virus type 1 protease suggests functional homology with aspartic proteinases. Journal of Virology. 1989;63(1):111–21. doi: 10.1128/JVI.63.1.111-121.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettit SC, Everitt LE, Choudhury S, Dunn BM, Kaplan AH. Initial cleavage of the human immunodeficiency virus type 1 GagPol precursor by its activated protease occurs by an intramolecular mechanism. Journal of virology. 2004;78(16):8477–85. doi: 10.1128/JVI.78.16.8477-8485.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zybarth G, Kräusslich HG, Partin K, Carter C. Proteolytic activity of novel human immunodeficiency virus type 1 proteinase proteins from a precursor with a blocking mutation at the N terminus of the PR domain. Journal of Virology. 1994;68(1):240–50. doi: 10.1128/JVI.68.1.240-250.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettit SC, Clemente JC, Jeung JA, Dunn BM, Kaplan AH. Ordered processing of the human immunodeficiency virus type 1 GagPol precursor is influenced by the context of the embedded viral protease. J Virol. 2005;79(16):10601–7. Epub 2005/07/30. doi: 10.1128/JVI.79.16.10601-10607.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tien C, Huang L, Watanabe SM, Speidel JT, Carter CA, Chen C. Context-dependent autoprocessing of human immunodeficiency virus type 1 protease precursors. PLOS ONE. 2018;13(1):e0191372. doi: 10.1371/journal.pone.0191372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Almog N, Roller R, Arad G, Passi-Even L, Wainberg MA, Kotler M. A p6Pol-protease fusion protein is present in mature particles of human immunodeficiency virus type 1. Journal of Virology. 1996;70(10):7228–32. doi: 10.1128/JVI.70.10.7228-7232.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strop P, Konvalinka J, Stys D, Pavlickova L, Blaha I, Velek J, et al. Specificity studies on retroviral proteinase from myeloblastosis-associated virus. Biochemistry. 1991;30(14):3437–43. Epub 1991/04/09. doi: 10.1021/bi00228a013 . [DOI] [PubMed] [Google Scholar]
- 45.Wondrak EM, Nashed NT, Haber MT, Jerina DM, Louis JM. A transient precursor of the HIV-1 protease. Isolation, characterization, and kinetics of maturation. The Journal of biological chemistry. 1996;271(8):4477–81. Epub 1996/02/23. doi: 10.1074/jbc.271.8.4477 . [DOI] [PubMed] [Google Scholar]
- 46.Lindhofer H, von der Helm K, Nitschko H. In vivo processing of Pr160gag-pol from human immunodeficiency virus type 1 (HIV) in acutely infected, cultured human T-lymphocytes. Virology. 1995;214(2):624–7. Epub 1995/12/20. doi: 10.1006/viro.1995.0074 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting information files.