Abstract
Because human immunodeficiency virus type 1 (HIV-1) subtypes and circulating recombinant forms (CRFs) are spreading rapidly worldwide and are becoming less confined to a geographical area, RNA assays that can detect and quantify all HIV-1 isolates reliably are in demand. We have developed a fast, real-time monitored RNA assay based on an isothermal nucleic acid sequence-based amplification technology that amplifies a part of the long terminal repeat region of the HIV-1 genome. Real-time detection was possible due to the addition of molecular beacons to the amplification reaction that was monitored in a fluorimeter with a thermostat. The lower level of detection of the assay was 10 HIV-1 RNA molecules per reaction, and the lower level of quantification was 100 copies of HIV-1 RNA with a dynamic range of linear quantification between 102 and 107 RNA molecules. All HIV-1 groups, subtypes, and CRFs could be detected and quantified with equal efficiency, including the group N isolate YBF30 and the group O isolate ANT70. To test the clinical utility of the assay, a series of 62 serum samples containing viruses that encompassed subtypes A through G and CRFs AE and AG of HIV-1 group M were analyzed, and these results were compared to the results of a commercially available assay. This comparison showed that the quantification results correlated highly (R2 = 0.735) for those subtypes that could be well quantified by both assays (subtypes B, C, D, and F), whereas improved quantification was obtained for subtypes A and G and CRFs AE and AG. A retrospective study with six individuals infected with either a subtype A, B, C, or D or an AG isolate of HIV-1 group M, who were treated with highly active antiretroviral therapy, revealed that the assay was well suited to the monitoring of therapy effects. In conclusion, the newly developed real-time monitored HIV-1 assay is a fast and sensitive assay with a large dynamic range of quantification and is suitable for quantification of most if not all subtypes and groups of HIV-1.
The viral RNA level in the plasma or serum of human immunodeficiency virus type 1 (HIV-1) infected individuals has become the most important marker for several aspects of an HIV-1 infection. The HIV-1 RNA level is predictive for disease progression in untreated infected individuals (7, 13, 17, 18, 25), and it is the best marker for monitoring therapy effects (14, 20, 31). In addition, the presence of viral RNA in the serum of newborn children is used as evidence for mother-to-child transmission, since maternal antibodies present in infant serum hamper antibody-screening assays. In this decade the number of newborn infants screened will increase as soon as cheaper and easier assays become available for use in developing countries, where they are needed most. The presence of viral RNA will also be one of the most important markers for monitoring breakthroughs in vaccine studies due to the presence of viral antibodies in sera as a result of vaccination. In addition, vaccines that do not have a strict preventive effect but evoke immune responses against the virus that keep viral RNA levels low increase the necessity for viral RNA assays that can quantify the levels of any HIV-1 subtype RNA reliably. Such vaccines might delay or even block the progression to AIDS, analogous to the effects of antiretroviral therapy.
To assess the amount of genomic RNA of any HIV-1 isolate in either serum or plasma, we developed a quantitative nucleic acid sequence-based amplification (NASBA) (10, 29) assay based on a highly conserved region of the HIV-1 genome, the long terminal repeat (LTR) region. In previous studies, we have shown that RNA assays based on the LTR region of HIV-1 can detect all HIV-1 isolates of group M and group O and are preferred over assays that amplify another region of the viral genome (4, 6). To increase throughput as well as to have a closed-tube format to minimize the risk of contamination, we have developed a fast, real-time monitored assay by using molecular beacons in the NASBA reaction (16). Molecular beacons are stem-and-loop-structured oligonucleotides with a fluorescent label at the 5′ end and a universal quencher at the 3′ end (26). If the molecular beacon has a closed stem-and-loop structure, the fluorophore and quencher are in close proximity and fluorescent energy is transferred to the quencher. When the loop of the molecular beacon hybridizes to its target, the molecular beacon undergoes a conformational change resulting in a physical separation of the fluorophore and quencher and emission of photons at the wavelength that is specific for the fluorophore (26). Molecular beacons are highly specific for their target. When present in a NASBA amplification reaction, the beacons hybridize with their amplified target RNA to form a stable hybrid. The intensity of the fluorescence upon hybridization is a direct measure of the amplicon concentration. An additional advantage of the use of molecular beacons to measure the amplicon concentration is a decreased contamination risk, because less handling is required, and the reactions take place in closed tubes that are not opened after amplification.
We describe the development and the performance of a new HIV-1 viral RNA assay that was validated in a clinical setting on a panel of serum and plasma samples from individuals infected with any of the group M HIV-1 subtypes. In addition, we show for five treated HIV-1–infected individuals that this assay is highly suited for monitoring the effectiveness of therapy, independent of the genetic subtype of the viral isolates.
MATERIALS AND METHODS
LTR-based real-time NASBA HIV-1 RNA assay (Retina HIV-1).
Three regions with a highly conserved sequence, located in the 5′ LTR region of the HIV-1 genome, were used to develop the LTR-based assay. The assay was based on standard NASBA technology (30), whereas for real-time detection, molecular beacons were added to the reaction (50 nM). Viral RNA from a total volume of 200 μl of plasma or serum sample was isolated using a silica-based method (2). Five microliters of the 50 μl of isolated viral RNA was used in the NASBA reaction, using a 5′ sense primer (5′-CTCAATAAAGCTTGCCTTGA) (HIVHXB2CG [GenBank accession no. K03455]; nucleotides [nt] 508 to 523) and a 3′ antisense primer elongated with a T7 sequence (in italics) (5′-aattctaatacgactcactatagggagagGGGCGCCACTGCTAGAGA) (nt 643 to 628) to amplify the 135-nt fragment (6). We developed a molecular beacon that could hybridize with all known HIV-1 isolates from all groups. The molecular beacon consists of a stem-and-loop-structured oligonucleotide with a fluorescein (FAM) label at the 5′ end and a universal quencher (DABCYL) at the 3′ end (5′-FAM-cgtacg agtagtgtgtgcccgtctgt cgtacg-DABCYL [the bases that form the stem are in lowercase italics]; the hybridizing loop encompasses nt 532 to 551). The hybridization reaction was monitored every 40 s in a 96-well fluorimeter equipped with a thermostat. A calibration curve with 2 × 102, 2 × 103, 2 × 104, and 2 × 105 molecules of HIV-1 was included in each experiment. For real-time NASBA amplification, the time-to-positivity (TTP) principle is applicable, similar to real-time PCR (11). The number of wild-type RNA copies per ml of serum could be extrapolated from the standard calibration curve.
Samples.
Sixty-two serum samples taken from individuals with known HIV-1 subtypes, based on both LTR and gag sequences, were tested for validation of the assay. Selection and description of these individuals has been described elsewhere (4, 6). Briefly, most of these individuals were non-European and non-U.S. immigrants to The Netherlands, who probably were infected in their home country or were individuals known or suspected to be infected with an HIV-1 strain of non-European and non-U.S. origin. They were identified by a thorough epidemiological investigation that is part of the routine evaluation of every newly diagnosed HIV case at the outpatient clinic of the Academic Medical Center. In all the serum samples, viral RNA levels were determined by both the Retina HIV-1 and NucliSens HIV-1 QT assays (Organon-Teknika, Boxtel, The Netherlands). The latter is a commercially available NASBA assay that amplifies a part of the gag gene that was performed according to the manufacturer's instructions.
In addition, serum samples from five HIV-1-infected individuals, who were treated with antiretroviral therapy and from whom a serum sample was drawn and stored at −80°C every 6 months, were analyzed with both the Retina HIV-1 and NucliSens assays.
Several well-characterized and calibrated standards were tested in threefold serial dilutions; they were obtained from Virology Networks (Utrecht, The Netherlands; ENVAS panel) and the National Institute for Biological Standards and Calibration (NIBSC, Hertsfordshire, United Kingdom). The latter is an international World Health Organization supported standard for nucleic acid diagnostics. Additionally, a well-characterized stock of cultured subtype B virus (HXB3) (15) was tested in these experiments. An RNA calibration molecule that encompassed a fragment of HXB3 (nt 465 to 1642) was synthesized in vitro using T7 RNA polymerase. The concentration of the RNA synthesized in vitro was determined on ethidium bromide-stained gel using RNA standards and, independently, by UV spectrophotometry. This RNA synthesized in vitro was used as the calibration standard in all experiments.
To investigate the linearity and sensitivity of the Retina HIV-1 assay for the various subtypes and groups of HIV-1, threefold serial dilutions were made of a panel of 10 cultured virus stocks that encompassed HIV-1 subtypes A through G and the circulating recombinant forms (CRFs) AE and AG. These series of viruses were tested and compared to published results, which were obtained after extensive characterization and calibration (19). Viral culture supernatants of group O ANT-70 and group N YBF30 isolates also were included in the experiment.
Statistical analysis.
For statistical analysis, Pearson's correlation coefficient procedure and Spearman's rank order correlation procedure, as implemented in the SPSS v10.0.5 software package (SPSS Inc., Chicago, Ill.), were used. The results from the NucliSens assays that were below 400 copies of RNA per ml were replaced with a value of 400 copies per ml to facilitate analysis.
RESULTS
Performance of Retina HIV-1.
The newly developed Retina HIV-1 assay could be used as both a qualitative and a quantitative assay by testing a standard curve within the same experiment. The isothermal (41°C) amplification process resulted in the synthesis of large amounts of single-stranded RNA (10, 29) to which the molecular beacon could hybridize immediately upon synthesis, enabling the emission of fluorescence. This resulted in an assay that combines amplification and detection within the same step. In other assays like NucliSens and Amplicor HIV-1 RNA v1.5 (Roche Molecular Diagnostics, Branchburg, N.J.), the detection step is separate and time consuming. Compared to the NucliSens assay, the saving of time for each 10 samples is approximately 1.5 h.
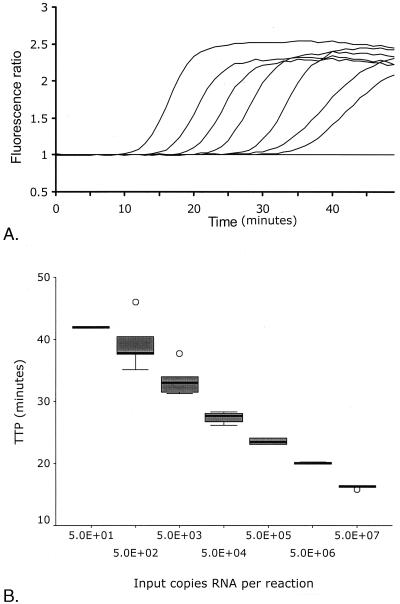
Typical amplification curves could be plotted in which an increase in fluorescence was observed, until most of the molecular beacon had hybridized with the synthesized amplicons and the fluorescence reached a maximum level (Fig. 1A). The time point at which the fluorescence signal became detectable over the background was linear over a range of at least five orders of magnitude of input RNA molecules, as shown by a dilution series of an RNA molecule synthesized in vitro (Fig. 1B). Based on five such serial dilutions, the linear quantification of the assay was determined to be between 102 and 107 copies of RNA per reaction (R2 = 0.94, P < 0.001). Precision and accuracy of the assay were within 0.2 and 0.1 log10, respectively, over the complete linear quantification range. The analytical sensitivity of the assay (lower detection level) at which amplification occurred in 50% of the reactions, was 10 RNA copies per reaction. In our case, HIV-1 RNA was isolated from 0.2 ml of serum and eluted in 50 μl, of which only one-tenth was used in a reaction. Thus, the equivalent of HIV-1 RNA isolated from 20 μl of serum or plasma was added to the reaction, which resulted in a sensitivity of 500 copies of HIV RNA per ml. The sensitivity could be improved if a precipitation protocol was applied to use all of the isolated RNA in the amplification, or if more serum input was used (32).
FIG. 1.
(A) Amplification plots of a tenfold serial dilution series of calibrator RNA synthesized in vitro. The amount of input RNA was 5 × 107, 5 × 106, 5 × 105, 5 × 104, 5 × 103, 5 × 102, 5 × 101, or 0 molecules. (B) Box plot depicting calculated TTP versus amount of input RNA copies from five replications of 10-fold serial dilutions of calibrator series, as described for panel A (5.0E+02 = 5.0 × 102, etc.). The box represents the interquartile range. The “whiskers” that extend from the box indicate the highest and lowest values, excluding the outliers, which are separately plotted (○). The line across each box indicates the median.
To evaluate whether the assay was quantitative, we tested several independent standard panels. Two homemade standard panels, one from RNA synthesized in vitro and one from a well-characterized subtype B isolate (HXB3) (15), were compared to ENVAS and NIBSC standards. Analysis of the dilution series indicated similar quantification efficiencies for all of the panels (data not shown). Thus, all standards tested were able to function as standards for calibration curves in combination with our Retina HIV-1 assay.
Linear quantification of the Retina HIV-1 assay on various HIV-1 subtypes.
The linear quantification capabilities of the Retina HIV-1 assay were determined by serial dilutions of viral culture supernatant for each subtype (A to G) of the HIV-1 M group in human serum. These samples were tested in several commercially available assays, and the results have been reported for the complete group (12) and for each sample separately, as determined by the Amplicor assay (19). A linear increase in the calculated TTP values was observed with increasing dilution for all tested samples, indicating that all subtypes were quantified similarly and with similar efficiencies (data not shown). The calculated viral RNA levels were compared to the reported results of the Amplicor assay (Pearson's correlation r = 0.407). The Retina HIV-1 assay assessed the viral RNA levels at mean values that were 0.35 log10 copies of RNA per ml higher than the mean values reported by the Amplicor assay.
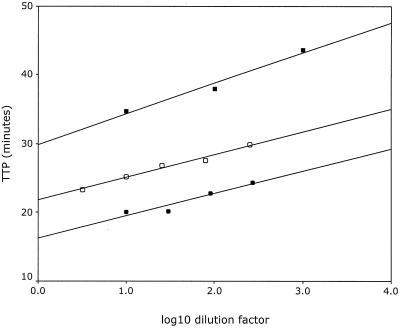
In addition, we tested serial dilutions of isolated RNA from culture supernatants of ANT70, a group O virus, and YBF30, a group N virus. Figure 2 depicts the relationship between the TTP value and the dilution factor before isolation of the nucleic acid for the group O and group N isolates, as well as for the calibration curve of the in vitro-synthesized standard RNA as a reference. The dilution series of both the group O and the group N samples demonstrated a linear relationship between the TTP values and the dilution factor, paralleling that of the calibration curve and indicating that these group O and group N isolates could be reliably quantified.
FIG. 2.
Measured TTP in minutes versus log10 of the dilution factor for the group O isolate ANT70 (●), the group N isolate YBF30 (■), and as a reference, the calibration curve (○).
RNA quantification in serum samples from 62 individuals infected with HIV-1 group M, subtypes A through G and CRFs AE and AG.
To evaluate the clinical utility of the Retina HIV-1 assay, the viral RNA levels in a set of 62 serum samples were assessed and compared to the values obtained by the commercially available NucliSens assay. Based on gag sequences and phylogeny, our sample set encompassed 12 subtype A, 11 subtype B, 7 subtype C, 9 subtype D, 1 subtype F, 6 subtype G, 1 subtype H, 5 CRF AE, and 10 CRF AG isolates. Subtype assignment based on the LTR sequences uncovered 4 recombinant isolates from the 59 isolates for which we had obtained the complete LTR sequence as well. Those four recombinant isolates had an LTR/gag subtype pattern of A/C, A/D, D/A, and F/D.
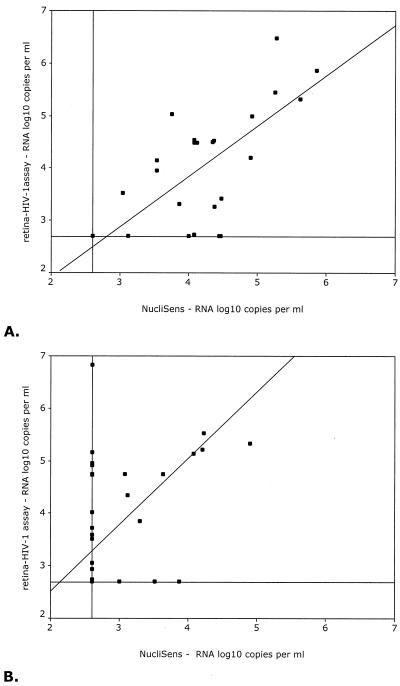
To facilitate analysis, the serum samples were divided into two groups, based on their subtype, since the NucliSens assay did not reliably quantify the subtype A and G and CRF AE and AG isolates (1, 6, 24). The first group (G1; n = 29) included all samples with subtypes B, C, D, F, and H (Fig. 3A), whereas the second group (G2; n = 33) contained the subtypes A and G and CRFs AE and AG (Fig. 3B). The calculated correlation coefficients (Pearson's correlation) between the Retina HIV-1 and NucliSens assays were 0.485 (P < 0.001) for the complete group of samples and 0.735 (P < 0.001) and 0.388 (P = 0.026) for groups G1 and G2, respectively. Since comparison of data from two different assays can lead to biased results when the correlation between the two sets of data is determined, because of extreme discrepancies in just a few samples, we also used the Spearman rank order test to calculate correlation coefficients (data not shown). The Pearson and Spearman correlation coefficients did not differ substantially, which indicates that the influence on the correlation coefficient of substantially deviating samples was limited. In a separate analysis we considered only those samples that yielded discrete values in both assays, and we found higher correlation coefficients (0.611, 0.747, and 0.621 for the complete group, G1, and G2, respectively).
FIG. 3.
Scatter diagrams of log10 RNA levels (copies/milliliter) as assessed by the Retina HIV-1 assay versus the log10 RNA levels (copies/ml) as assessed by the NucliSens assay. Depicted are the results from individuals infected with HIV-1 subtypes B (n = 11), C (n = 7), D (n = 9), F (n = 1), and H (n = 1) (A) and subtypes A (n = 12) and G (n = 6) and CRFs AE (n = 5) and AG (n = 10) (B). The horizontal and vertical lines depict the lower detection level of each assay, whereas the diagonal line represents the regression line through the origin.
In total, 13 samples containing viruses of various subtypes of HIV-1 group M could not be detected by the NucliSens assay but were positive in the Retina HIV-1 assay. All 13 of these serum samples were categorized in the group that contained subtypes A and G and CRFs AE and AG. Of these, two samples were subtype A, five were CRF AG, one was CRF AE, and five were subtype G. Previously, we have reported that due to mismatches in primer and probe regions, the NucliSens assay either cannot detect or underestimates this group of subtypes and CRFs (6).
In six serum samples no viral RNA was detected by the Retina HIV-1 assay, but the NucliSens assay showed the presence of viral RNA. Of these six samples, two samples contained viruses from subtype A, one from subtype B, two from subtype D, and one from subtype F. Of 62 serum samples tested, 14 samples had no viral RNA that was detectable by either assay, probably due to the effects of therapy or to naturally low viral RNA levels.
Analysis of mismatches in primer and molecular-beacon hybridization sequences.
Since our new Retina HIV-1 assay uses molecular beacons as a detection method, the influence of mismatches with the hybridizing region could be more pronounced than with linear probes under the same conditions. Therefore, we analyzed the sequences of the molecular-beacon-hybridizing regions as well as the primer-hybridizing regions from those samples that were negative in the Retina HIV-1 assay but were positive in the NucliSens assay in order to explain the discrepancies. None of the samples showed a mismatch in any of the two primer-hybridizing regions. The subtype B sample that was negative in the Retina HIV-1 assay showed a deletion mutation in the hybridizing region of the molecular beacon. For the subtype A, D, and F samples, no mismatches could be found for the molecular-beacon-hybridizing region.
Retina HIV-1 assay is suitable for monitoring therapy effects.
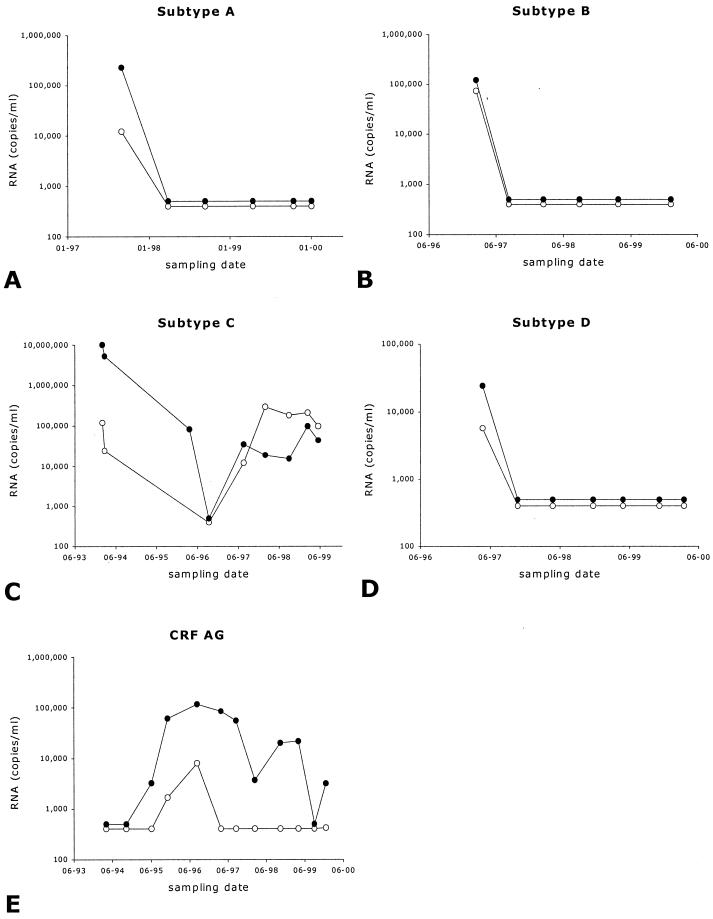
The effects of antiretroviral therapy are evaluated by the changes in the viral RNA level in serum of HIV-1-infected individuals. A fast decrease after initiation of antiretroviral therapy is usually seen if therapy is effective. During the course of therapy, the RNA levels are assessed every 6 months in order to detect whether therapy is still effective or whether virological drug failure has occurred. We selected five individuals, each infected with a different group M subtype of HIV-1, who were on antiretroviral therapy for more than 4 years. Approximately every 6 months a blood sample was drawn, and the serum was stored at −80°C. Each of these serum samples was analyzed for the viral RNA level using both the Retina HIV-1 and the NucliSens assays. The results are depicted in Fig. 4. We observed that, after initiation of antiretroviral therapy, both assays detected a rapid drop in viral RNA levels, which remained low or even below the detection level of each assay until a virological drug failure was observed as determined by increasing viral RNA levels. Independently of which subtype was analyzed, therapy effects were efficiently monitored by both assays, with no significant differences.
FIG. 4.
Viral RNA levels in sera from five longitudinally followed HIV-1-infected individuals who were treated with antiretroviral therapy. Depicted are the results from both Retina HIV-1 (●) and NucliSens (○) assays. Each panel depicts the results from an individual infected with the indicated subtype of virus.
DISCUSSION
We have developed a real-time NASBA HIV-1 viral RNA assay in which a 135-nt conserved region of the LTR of the HIV-1 genome was amplified. In a previous study we have shown that the primer and probe locations within the amplified region of the LTR are highly conserved among the various subtypes of HIV-1 group M, as well as for the N and O groups (6). To detect the synthesized amplicons, molecular-beacon probes were added to the reaction, which enabled real-time detection and monitoring of the amplification reactions. By using real-time monitoring, reaction and detection could occur simultaneously, omitting a time-consuming detection step. An additional advantage was that the tubes stayed closed after initiation of the amplification, minimizing the risk of contamination by the relatively large amounts of amplified product (10). Since NASBA is an isothermal amplification reaction, a fluorimeter with a thermostat was sufficient to monitor the continuous amplification reaction. Such fluorimeters are relatively inexpensive compared to the instruments required to monitor real-time PCRs. The amplification time of the Retina HIV-1 assay was 60 min for 96 samples, making it a very fast, high-throughput assay.
The lower detection level of the Retina HIV-1 assay was approximately 10 copies of HIV-1 RNA per reaction. The lower quantification level of the Retina HIV-1 assay was approximately 100 copies per reaction. Quantification was achieved within a linear range of at least five orders of magnitude (102 to 107 molecules per reaction; Fig. 1B.). The accuracy and precision could vary within the limits of 0.1 and 0.2 log10 copies of RNA per ml. These characteristics of the Retina HIV-1 assay compare well to commercially available assays like NucliSens HIV-1 RNA QT (Organon-Teknika), Amplicor HIV-1 Monitor v1.5 (Roche Molecular Diagnostics), and Quantiplex HIV-1 RNA v3.0 (Bayer Diagnostics, Mannheim, Germany) (3, 8, 22, 23, 27, 28). Since our assay measures RNA that was isolated from an equivalent of 20 μl, the sensitivity was approximately 500 copies per ml. Different isolation protocols could lead to higher sensitivity, similar to that obtained by the ultrasensitive version of the Amplicor assay, in which a lower limit of 50 copies per ml is reached due to ultracentrifugation of the samples.
The quantification by the Retina HIV-1 was tested on a number of available HIV-1 standards like those of ENVAS and NIBSC (all subtype B), and it was shown that the assay could reliably quantify these standards. The Retina HIV-1 assay was able to quantify all group M subtypes of HIV-1 with a similar efficiency. The result of the quantification of a panel of subtype isolates showed that Retina HIV-1 and Amplicor v1.5 assay gave concordant values. In addition, the Retina HIV-1 assay could also linearly quantify isolates from HIV-1 groups N and O (Fig. 2). The broad specificity of the Retina HIV-1 assay is an asset, since it is known that the various subtypes and groups of HIV-1 are rapidly spreading worldwide.
The group N and O viruses were as efficiently amplified as group M viruses, but the signals were lower than for group M isolates. This difference in signals was most likely due to the mismatch found in the hybridizing loop of the molecular beacon, based on the reported LTR sequence of both viral isolates (5; B. Korber, C. Kuiken, B. Foley, B. Hahn, F. McCutchan, J. Mellors, and J. Sodroski, http://hiv-web.lanl.gov/).
The clinical utility of the Retina HIV-1 assay was tested on 62 serum samples. The results were compared to those of the NucliSens assay. Five of 62 isolates were not detected by the Retina HIV-1 but were detected by the NucliSens assay, whereas 13 of 62 were detected by the Retina HIV-1 assay and not by the NucliSens assay. The sequence of one of the five isolates not detected by the Retina HIV-1 assay showed a deletion mismatch in the hybridizing region of the molecular beacon, a likely cause of the negative result. For the remaining four samples, no obvious explanation for the detection failure could be found. The Retina HIV-1 assay was evaluated in a set of longitudinally drawn serum samples of patients on antiretroviral therapy. When compared to the results of the NucliSens assay, the results from the Retina HIV-1 assay showed comparable profiles of viral RNA levels in response to antiretroviral therapy, with RNA values dropping below the detection limit when effective therapy was administered and RNA levels rebounding when therapy failed.
In the perspective of the current direction of HIV-1 research, the Retina HIV-1 assay can prove its value in different fields of HIV-1 diagnostics. First, the Retina HIV-1 assay could be of use in screening for vertical transmission. Mother-to-child transmission is mostly present in developing countries, where subtypes of HIV-1 group M other than subtype B are most prevalent. Improved detection of the subtypes, using the LTR region as an amplification region, will probably lead to an increase in the number of infants diagnosed with HIV-1 infections, especially if the infants are infected with a subtype A, G, CRF AE or AG, or group N or O virus. An additional advantage of the Retina HIV-1 over the Quantiplex v3.0 and the ultrasensitive Amplicor v1.5 assay formats is the smaller serum volume (200 versus 500 μl) that is necessary to detect HIV-1 RNA with similar sensitivity, provided that a precipitation protocol is included to concentrate the RNA that serves as input for the Retina HIV-1 assay (data not shown). Related, among other factors, to mother-to-child transmission is the development of vaccines that evoke an immune response that can keep viral RNA levels low, thereby prohibiting vertical transmission (9, 21). Viral RNA levels in this context reflect the efficacy of the vaccine used and should be monitored.
Second, the Retina HIV-1 assay could prove its value in monitoring prophylactic vaccine breakthroughs because the assay is relatively cheap, can be used as a high-throughput application, and can detect isolates of any of the HIV-1 subtypes. These features will be important because phase III trials on the efficacy of vaccines will include large cohorts of HIV-1 virus-negative individuals who have detectable HIV-1 antibodies because of the immune responses induced by the vaccine. These large cohorts will result in high numbers of samples to be tested from all over the world for the presence or absence of HIV-1 RNA of any subtype or group.
In summary, our LTR-based Retina HIV-1 assay can quantify viral isolates from HIV-1 group M, N, or O. Due to the real-time detection, the throughput of the assay is high, with 96 samples amplified and detected within 60 min. The reactions are monitored in a relatively cheap thermostat-equipped fluorimeter, which makes the assay feasible for laboratories all over the world. Since the amplification as well as the detection takes place in one closed tube, the risk of contamination is reduced to a minimum. The characteristics of the assay make it highly suitable for application in HIV diagnostics and HIV-1 therapy management, as well as in the diagnosis of HIV-1-infected infants, vaccine efficacy, and vaccine breakthrough.
ACKNOWLEDGMENTS
We thank Eveline Timmermans and Irene Bosboom for technical assistance.
The YBF30 group N isolate isolated by F. Simon, Hopital Bichat, Paris, France, was obtained from the NIBSC Centralised Facility for AIDS Reagents supported by EU Programme EVA (contract BMH 97/2515) and the United Kingdom Medical Research Council.
REFERENCES
- 1.Alaeus A, Lidman K, Sonnerborg A, Albert J. Subtype-specific problems with quantification of plasma HIV-1 RNA. AIDS. 1997;11:859–865. doi: 10.1097/00002030-199707000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Boom R, Sol C J, Salimans M M, Jansen C L, Wertheim-van Dillen P M, Van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coste J, Montes B, Reynes J, Peeters M, Segarra C, Vendrell J P, Delaporte E, Segondy M. Comparative evaluation of three assays for the quantitation of human immunodeficiency virus type 1 RNA in plasma. J Med Virol. 1996;50:293–302. doi: 10.1002/(SICI)1096-9071(199612)50:4<293::AID-JMV3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 4.de Baar M P, De Ronde A, Berkhout B, Cornelissen M, van der Horn K H M, van der Schoot A M, De Wolf F, Lukashov V V, Goudsmit J. Subtype-specific sequence variation of the HIV type 1 long terminal repeat and primer-binding site. AIDS Res Hum Retroviruses. 2000;16:499–504. doi: 10.1089/088922200309160. [DOI] [PubMed] [Google Scholar]
- 5.de Baar M P, Janssens W, De Ronde A, Fransen K, Colebunders R, Kestens L, van der Groen G, Goudsmit J. Natural residues versus antiretroviral drug-selected mutations in HIV type 1 group O reverse transcriptase and protease related to virological drug failure in vivo. AIDS Res Hum Retroviruses. 2000;16:1385–1394. doi: 10.1089/08892220050140937. [DOI] [PubMed] [Google Scholar]
- 6.de Baar M P, van der Schoot A M, Goudsmit J, Jacobs F, Ehren R, van der Horn K H, Oudshoorn P, De Wolf F, De Ronde A. Design and evaluation of a human immunodeficiency virus type 1 RNA assay using nucleic acid sequence-based amplification technology able to quantify both group M and O viruses by using the long terminal repeat as target. J Clin Microbiol. 1999;37:1813–1818. doi: 10.1128/jcm.37.6.1813-1818.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Wolf F, Spijkerman I, Schellekens P T, Langendam M, Kuiken C, Bakker M, Roos M, Coutinho R, Miedema F, Goudsmit J. AIDS prognosis based on HIV-1 RNA, CD4+ T-cell count and function: markers with reciprocal predictive value over time after seroconversion. AIDS. 1997;11:1799–1806. doi: 10.1097/00002030-199715000-00003. [DOI] [PubMed] [Google Scholar]
- 8.Dyer J R, Gilliam B L, Eron J J, Jr, Grosso L, Cohen M S, Fiscus S A. Quantitation of human immunodeficiency virus type 1 RNA in cell free seminal plasma: comparison of NASBA with Amplicor reverse transcription-PCR amplification and correlation with quantitative culture. J Virol Methods. 1996;60:161–170. doi: 10.1016/0166-0934(96)02063-0. [DOI] [PubMed] [Google Scholar]
- 9.Garcia P M, Kalish L A, Pitt J, Minkoff H, Quinn T C, Burchett S K, Kornegay J, Jackson B, Moye J, Hanson C, Zorrilla C, Lew J F. Maternal levels of plasma human immunodeficiency virus type 1 RNA and the risk of perinatal transmission. Women and Infants Transmission Study Group. N Engl J Med. 1999;341:394–402. doi: 10.1056/NEJM199908053410602. [DOI] [PubMed] [Google Scholar]
- 10.Guatelli J C, Whitfield K M, Kwoh D Y, Barringer K J, Richman D D, Gingeras T R. Isothermal, in vitro amplification of nucleic acids by a multienzyme reaction modeled after retroviral replication. Proc Natl Acad Sci USA. 1990;87:1874–1878. doi: 10.1073/pnas.87.5.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology. 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 12.Jagodzinski L L, Wiggins D L, McManis J L, Emery S, Overbaugh J, Robb M, Bodrug S, Michael N L. Use of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 to assess the performance of viral RNA quantitation tests. J Clin Microbiol. 2000;38:1247–1249. doi: 10.1128/jcm.38.3.1247-1249.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jurriaans S, Van Gemen B, Weverling G J, Van Strijp D, Nara P, Coutinho R, Koot M, Schuitemaker H, Goudsmit J. The natural history of HIV-1 infection: virus load and virus phenotype independent determinants of clinical course? Virology. 1994;204:223–233. doi: 10.1006/viro.1994.1526. [DOI] [PubMed] [Google Scholar]
- 14.Kappes J C, Saag M S, Shaw G M, Hahn B H, Chopra P, Chen S, Emini E A, McFarland R, Yang L C, Piatak M, Jr, Lifson J D. Assessment of antiretroviral therapy by plasma viral load testing: standard and ICD HIV-1 p24 antigen and viral RNA (QC-PCR) assays compared. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;10:139–149. doi: 10.1097/00042560-199510020-00005. [DOI] [PubMed] [Google Scholar]
- 15.Layne S P, Merges M J, Dembo M, Spouge J L, Conley S R, Moore J P, Raina J L, Renz H, Gelderblom H R, Nara P L. Factors underlying spontaneous inactivation and susceptibility to neutralization of human immunodeficiency virus. Virology. 1992;189:695–714. doi: 10.1016/0042-6822(92)90593-e. [DOI] [PubMed] [Google Scholar]
- 16.Leone G, van Schijndel H, Van Gemen B, Kramer F R, Schoen C D. Molecular beacon probes combined with amplification by NASBA enable homogeneous, real-time detection of RNA. Nucleic Acids Res. 1998;26:2150–2155. doi: 10.1093/nar/26.9.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mellors J W, Kingsley L A, Rinaldo C R, Jr, Todd J A, Hoo B S, Kokka R P, Gupta P. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122:573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 18.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 19.Michael N L, Herman S A, Kwok S, Dreyer K, Wang J, Christopherson C, Spadoro J P, Young K K, Polonis V, McCutchan F E, Carr J, Mascola J R, Jagodzinski L L, Robb M L. Development of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 and performance of an improved AMPLICOR HIV-1 MONITOR test with isolates of diverse subtypes. J Clin Microbiol. 1999;37:2557–2563. doi: 10.1128/jcm.37.8.2557-2563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Brien W A, Hartigan P M, Martin D, Esinhart J, Hill A, Benoit S, Rubin M, Simberkoff M S, Hamilton J D. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. Veterans Affairs Cooperative Study Group on AIDS. N Engl J Med. 1996;334:426–431. doi: 10.1056/NEJM199602153340703. [DOI] [PubMed] [Google Scholar]
- 21.Quinn T C, Wawer M J, Sewankambo N, Serwadda D, Li C, Wabwire-Mangen F, Meehan M O, Lutalo T, Gray R H. Viral load and heterosexual transmission of human immunodeficiency virus type 1. Rakai Project Study Group. N Engl J Med. 2000;342:921–929. doi: 10.1056/NEJM200003303421303. [DOI] [PubMed] [Google Scholar]
- 22.Revets H, Marissens D, De Wit S, Lacor P, Clumeck N, Lauwers S, Zissis G. Comparative evaluation of NASBA HIV-1 RNA QT, AMPLICOR-HIV monitor, and QUANTIPLEX HIV RNA assay, three methods for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:1058–1064. doi: 10.1128/jcm.34.5.1058-1064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuurman R, Descamps D, Weverling G J, Kaye S, Tijnagel J, Williams I, van Leeuwen R, Tedder R, Boucher C A, Brun-Vezinet F, Loveday C. Multicenter comparison of three commercial methods for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:3016–3022. doi: 10.1128/jcm.34.12.3016-3022.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segondy M, Ly T D, Lapeyre M, Montes B. Evaluation of the Nuclisens HIV-1 QT assay for quantitation of human immunodeficiency virus type 1 RNA levels in plasma. J Clin Microbiol. 1998;36:3372–3374. doi: 10.1128/jcm.36.11.3372-3374.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spijkerman I J, Prins M, Goudsmit J, Veugelers P J, Coutinho R A, Miedema F, De Wolf F. Early and late HIV-1 RNA level and its association with other markers and disease progression in long-term AIDS-free homosexual men. AIDS. 1997;11:1383–1388. doi: 10.1097/00002030-199711000-00013. [DOI] [PubMed] [Google Scholar]
- 26.Tyagi S, Kramer F R. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 27.Vandamme A M, Schmit J C, Van Dooren S, Van Laethem K, Gobbers E, Kok W, Goubau P, Witvrouw M, Peetermans W, De Clercq E, Desmyter J. Quantification of HIV-1 RNA in plasma: comparable results with the NASBA HIV-1 RNA QT and the AMPLICOR HIV monitor test. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13:127–139. doi: 10.1097/00042560-199610010-00003. [DOI] [PubMed] [Google Scholar]
- 28.Vandamme A M, Van Dooren S, Kok W, Goubau P, Fransen K, Kievits T, Schmit J C, De Clercq E, Desmyter J. Detection of HIV-1 RNA in plasma and serum samples using the NASBA amplification system compared to RNA-PCR. J Virol Methods. 1995;52:121–132. doi: 10.1016/0166-0934(94)00151-6. [DOI] [PubMed] [Google Scholar]
- 29.Van Gemen B, Kievits T, Nara P, Huisman H G, Jurriaans S, Goudsmit J, Lens P. Qualitative and quantitative detection of HIV-1 RNA by nucleic acid sequence-based amplification. AIDS. 1993;7(Suppl.):S107–S110. doi: 10.1097/00002030-199311002-00020. [DOI] [PubMed] [Google Scholar]
- 30.Van Gemen B, van Beuningen R, Nabbe A, Van Strijp D, Jurriaans S, Lens P, Kievits T. A one-tube quantitative HIV-1 RNA NASBA nucleic acid amplification assay using electrochemiluminescent (ECL) labelled probes. J Virol Methods. 1994;49:157–167. doi: 10.1016/0166-0934(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 31.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 32.Weverling G J, Lange J M A, Jurriaans S, Prins J M, Lukashov V V, Notermans D W, Roos M, Schuitemaker H, Hoetelmans R M W, Danner S A, Goudsmit J, De Wolf F. Alternative multidrug regimen provides improved suppression of HIV-1 replication over triple therapy. AIDS. 1998;12:F117–F122. doi: 10.1097/00002030-199811000-00003. [DOI] [PubMed] [Google Scholar]