Abstract
A genomics-based PCR method was developed and used to test specimens from patients involved in a large outbreak of Mycoplasma pneumoniae in a closed religious community in New York State. New P1 adhesin gene primers were designed to bind to 9 of 10 target sequences in the repetitive-element sequences obtained from the whole genome sequence of M. pneumoniae. This PCR method had a sensitivity of 0.006 CFU and a specificity of 100% for M. pneumoniae. The PCR was validated by testing a subset of patient samples by culture and comparing the results to those obtained by PCR. Of the initial 280 samples tested, 73 were positive by PCR and 22 were positive by culture. All samples positive by culture were also positive by PCR. Follow-up testing of selected patients 3 to 6 weeks after antibiotic treatment revealed that eight samples remained positive by PCR and that three samples remained positive by culture. Additionally, no nonspecific PCR inhibition was detected as a result of the specimen type, transport medium, or sample preparation methodology. The study demonstrates that the PCR described here is a rapid, sensitive, and specific method for the identification of M. pneumoniae and was helpful for the detection and monitoring of the outbreak.
Mycoplasma pneumoniae is primarily a respiratory pathogen that is responsible for approximately 15 to 20% of all community-acquired pneumonias (8). The clinical presentation of patients with M. pneumoniae infection is not significantly different from that of patients with infections caused by other respiratory pathogens such as Chlamydia pneumoniae (22), so diagnosis of infection relies primarily on laboratory testing. M. pneumoniae is, however, a fastidious organism, requiring a laborious effort and 21 days or more for growth in culture. Likewise, serology is lacking in sensitivity, has questionable specificity (22), and is dependent on specific collection times relative to the onset of illness (5). DNA probes have also been used, but cross-reactivity between M. pneumoniae and Mycoplasma genitalium has been observed, and methods that use these probes also lack sensitivity (4).
Numerous PCR approaches have been developed to provide a rapid, sensitive method for the detection of M. pneumoniae. PCR targets have included the P1 adhesin gene (3, 4, 5, 12, 17, 21), the ATPase operon (11), genomic clones (2, 7), or a combination of these (1). The P1 adhesin gene is responsible for cytadherence, which is necessary for colonization and infection of host cells by M. pneumoniae (22). This gene is also an intriguing target for PCR because of its repetitive nature within the M. pneumoniae genome. Approximately 8% of this genome is composed of repetitive DNA elements with regions homologous to the P1 adhesin gene (10). Use of the entire genomic DNA sequence enabled us to design primers that would theoretically amplify the maximum number of target molecules, resulting in an improved sensitivity of the PCR for M. pneumoniae.
A large outbreak of respiratory illness occurred in a closed religious community in rural New York State in the summer of 1998, providing the opportunity to evaluate improved molecular biology-based methods for M. pneumoniae detection. A study was designed to evaluate new, whole genome-based PCR primers and compare the detection capability of PCR to that of culture of throat swab specimens from community members. Additionally, the outbreak allowed us to determine whether PCR or culture of follow-up specimens could detect M. pneumoniae after antibiotic treatment (azithromycin) of the infected individuals.
MATERIALS AND METHODS
Patients.
Specimens were obtained from patients who were associated with an outbreak of respiratory illness and whose clinical presentation was consistent with disease caused by M. pneumoniae. The outbreak occurred between 1 May 1998 and 11 August 1998 in a closed religious community in New York State. Throat swab specimens were initially obtained from 280 persons, consisting of symptomatic and asymptomatic persons. Five-day antimicrobial therapy with azithromycin (kindly donated by Pfizer Pharmaceuticals) was established for symptomatic individuals, and a 3-day prophylactic treatment was given to asymptomatic individuals following the collection of specimens. The total cumulative dose of azithromycin was identical (1,500 mg) for all treated individuals. After a 3- to 6-week period, follow-up throat swab specimens were collected from 69 of the initially tested individuals.
Specimen collection.
Throat swabs were collected with a Dacron applicator swab. The swabs were expressed in tubes containing 3 ml of Mycoplasma transport medium (Trypticase soy broth containing 0.25% glucose, 0.5% bovine serum albumin, 0.01% thallium acetate, 1,000 U of penicillin per ml), and the swabs were discarded. The specimen tubes were then transported on ice to the laboratory on the same day of collection and processed for culture and PCR or stored at 4°C for 24 to 48 h. If culture was not performed at the time of PCR, the samples were stored at −70°C until culture was initiated.
Culture.
The culturing of M. pneumoniae was accomplished by a method adapted from that of Tully et al. (20) and presented recently (C. J. Carlyn, A. L. Waring, C. K. Csiza, T. A. Halse, S. J. Wong, and R. J. Limberger, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. G-16, 1999). Briefly, the specimen tubes were vortexed, a volume of 100 μl was added to 3 ml of spiroplasma diphasic medium (SP4 agar; 2 ml of broth and 1 ml of solid media), and two serial 10-fold dilutions were made to dilute out potential inhibitors of Mycoplasma growth. These samples were then incubated at 37°C and examined twice weekly. When a color change occurred, 25 μl was subcultured onto SP4 agar. These subcultured samples were incubated in 5% CO2 in a 37°C incubator and were then examined twice weekly for the presence of typical M. pneumoniae colonies. Typical colonies were identified by guinea pig red blood cell hemadsorption and confirmed by a growth inhibition test (20).
PCR.
A total of 500 μl of the original specimen was aliquoted into a microcentrifuge tube for PCR analysis. This aliquot was concentrated by centrifugation at 13,000 × g for 10 min at room temperature, 480 μl of the medium was removed, and 30 μl of sterile water was added to the remaining 20 μl of medium and the pellet. The samples were then vortexed and heated to 95°C for 15 min. A 10-μl aliquot of the sample lysate was then used directly for PCR amplification. An M. pneumoniae-specific PCR was performed with a total reaction mixture volume of 100 μl. Each primer at a final concentration of 1 μM (primer MP-F [5′-CCCTCGACCAAGCCAACCTC-3′] and primer MP-R [5′-TGCGCGTTGTTCTTGTTGGTG-3′]), each deoxynucleoside triphosphate at a final concentration of 200 μM, and final concentrations of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, and 2.5 U of AmpliTaq Gold (Perkin-Elmer, Foster City, Calif.) were used for PCR in a Perkin-Elmer 9600 thermocycler. Thermocycler conditions consisted of an initial incubation of 95°C for 9 min, followed by 40 cycles of 94°C for 30 s, 62°C for 30 s, and 72°C for 30 s. An additional incubation at 72°C for 7 min was added to complete the elongation. Negative controls with no template were included, as were controls known to be positive. To assess the overall effect of potential nonspecific inhibition of the amplification, an additional PCR analysis was performed with 41 randomly selected PCR-negative samples. Each specimen was spiked with 150 ng of HeLa cell DNA (laboratory stock), and a PCR specific for the human β-globin gene was used to evaluate the patient samples. The PCR mixtures were spiked with this DNA to maintain a precise, consistent amount of template for analysis of nonspecific inhibition. The amplified products were analyzed on 2% GTG agarose gels containing ethidium bromide and were visualized at maximal levels of intensity with a Gel Doc 1000 gel analysis system (Bio-Rad Laboratories, Hercules, Calif.). All reagents and conditions for the PCR assay were optimized before the assays for sensitivity and specificity were performed. The oligonucleotides were synthesized in the Wadsworth Center Molecular Genetics Core Facility.
Dot blot hybridization of PCR products.
In order to specifically confirm amplification of M. pneumoniae DNA, a 26-bp probe specific for the internal region of the PCR product, MPP2 (5′-AATCCCGACTCGTTAAAGCAGGATAA-3′), was used in all hybridization assays. A 30-μl aliquot of PCR product was denatured at 37°C for 5 min with 0.1 volume of 1 N NaOH. Neutralization was obtained by adding 1 volume of 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and sterile water was then added to give a total volume of 400 μl. The total volume was transferred onto a Hybond-N nylon membrane (Amersham Pharmacia Biotech, Piscataway, N.J.) with a dot blot apparatus (Schleicher & Schuell, Keene, N.H.). The membrane was UV cross-linked, prehybridized overnight at 42°C, and then hybridized at 42°C for 2 h with labeled probe (the hybridization solutions were specified by the manufacturer). The internal oligonucleotide probe was labeled according to the manufacturer's protocol by using the ECL 3′ oligolabeling and detection kit (Amersham Pharmacia Biotech). DNA hybridization was visualized by detection of a chemiluminescent signal when the blot was exposed to X-Omat AR film (Kodak, Rochester, N.Y.). Positive results by both PCR and hybridization assays were interpreted as a positive result for the specimen.
PCR sensitivity and specificity.
The sensitivity of the PCR was determined by using a viable suspension of M. pneumoniae ATCC 29342. Tenfold serial dilutions were made in SP4 culture medium to 10−10. Identical volumes of each of these dilutions were run in PCR assays and were plated onto SP4 agar. The concentration of M. pneumoniae was determined by plate counts, performed in triplicate.
A number of additional Mycoplasma spp., respiratory organisms, and other common bacteria were tested to determine the specificity of the PCR assay. Concentrations of either 3 × 106 CFU or 0.1 ng of DNA (equivalent to ∼1 × 106 copies of the genome) were used. The following organisms were tested by both PCR and probe hybridization assays to determine the specificities of the assays: M. pneumoniae ATCC 29342, ATCC 15531, and NIH 1428; Mycoplasma arthritidis ATCC 14152; Mycoplasma hominis ATCC 23114; M. genitalium ATCC 33530; Mycoplasma salivarium ATCC 23064D; Mycoplasma orales ATCC 23714D; Mycoplasma fermentans ATCC 19989D; Ureaplasma urealyticum; Bordetella pertussis; Legionella pneumophila; group A streptococcus; Streptococcus pneumoniae; Haemophilus influenzae; Staphylococcus aureus; Neisseria meningitidis; Streptococcus sanguis; Branhamella catarrhalis; Corynebacterium aquaticum; Klebsiella pneumoniae; Chlamydia pneumoniae; Chlamydia trachomatis; Treponema pallidum; Escherichia coli; and Haemophilus ducreyi.
RESULTS
Primer and probe design.
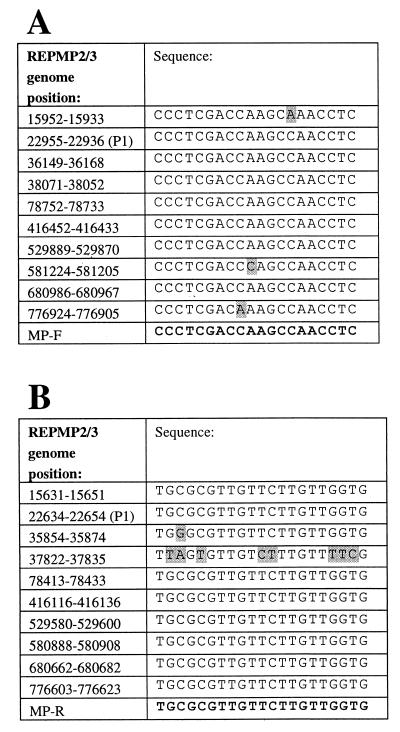
Analysis of the complete sequence of the M. pneumoniae genome (found at the website http://www . zmbh . uniheidelberg . de / M_pneumoniae / MP_Home . html ) (10) enabled the selection of two new PCR primers. This sequence was manipulated by using the programs available in the Wisconsin Package (Wisconsin Package, version 10.1; Genetics Computer Group, Madison, Wis.), followed by manual sequencing analysis. Sequences within the repetitive element REPMP2/3 and the P1 adhesin gene (16, 19) were aligned, and the most highly conserved regions were selected to design primers for PCR. As shown in Fig. 1, nine amplicons could theoretically be synthesized in the first round of PCR with these primers, and thus, a significant increase in sensitivity over that of a single-target PCR assay would be achieved. The sizes of the expected products ranged from 309 to 339 bp and were visualized as one broad band upon gel electrophoresis (Fig. 2 and 3).
FIG. 1.
Primer design strategy for the forward and reverse PCR primers (primers MP-F and MP-R, respectively) indicating the binding positions within the M. pneumoniae genome (10). The primer sequences are shown at the bottom of each panel. Shaded areas indicate mismatches between the primer and the genome sequences within REPMP2/3 regions. The P1 adhesin gene sequence (P1) is indicated by the appropriate genome sequence. (A) Alignment of forward primer MP-F; (B) alignment of reverse primer MP-R.
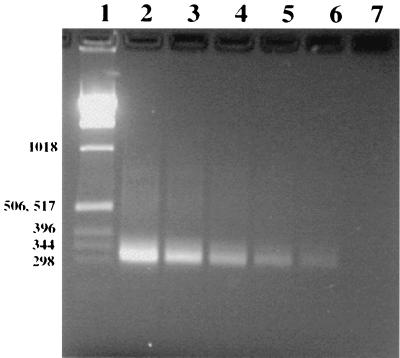
FIG. 2.
Ethidium bromide-stained 2% agarose gel showing sensitivity of PCR after dilution of viable M. pneumoniae. Lane 1, 1-kb DNA ladder (numbers on the left are in base pairs); lanes 2, 3, and 4, amplified M. pneumoniae culture extracts containing 60, 6, and 0.6 CFU, respectively, in the total PCR mixture; lanes 5 and 6, extracts that did not produce any CFU (extrapolated to dilutions of 0.06 and 0.006 CFU, respectively); lane 7, negative control.
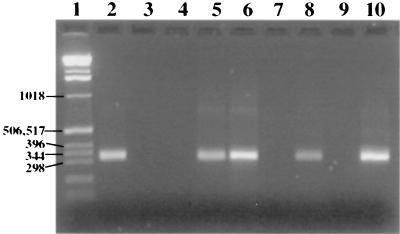
FIG. 3.
PCR amplification of repetitive element REPMP2/3 within the P1 operon for direct detection of M. pneumoniae in patient samples. Samples were centrifuged, lysed, amplified by PCR, electrophoresed, and stained with ethidium bromide. Lane 1, 1-kb ladder (numbers on the left are in base pairs); lanes 2 to 8, patient specimens; lane 9, negative control; lane 10, positive control.
The internal probe, MPP2, was speculated to bind efficiently to seven of the amplified PCR products (sequence identities, 92 to 100%). The two remaining amplified PCR products had sequence identities to the probe of 61 and 69%, respectively, but were also capable of binding to the probe under our hybridization conditions (data not shown). The dot blot hybridization procedure was designed to provide specificity for the assay to ensure that the PCR products were indeed M. pneumoniae.
Sensitivity and specificity of PCR method.
Under the conditions used in this study, PCR produced an amplification product in the range of 309 to 339 bp, as expected by the design of the assay. The sensitivity of detection was 0.006 CFU, as determined by comparison of the PCR results with the culture results after plating of serial dilutions (Fig. 2). Negative controls with no template produced no PCR products, and positive controls for M. pneumoniae always produced a product of 309 to 339 bp, as shown in Fig. 3.
The non-M. pneumoniae species and the unrelated respiratory species tested as well as the additional bacterial species tested produced no detectable PCR products upon analysis by the PCR assay and no hybridization products by the hybridization assay. Thus, the combined results of PCR and probing demonstrated 100% specificity.
PCR inhibition analysis.
Forty-one of the specimens that were negative by both culture and PCR were tested for nonspecific PCR inhibition. After spiking of the samples with HeLa cell DNA, a region of the human β-globin gene was amplified from all specimens by a PCR assay that is routinely used by our laboratory for inhibition testing. Because all of these specimens produced a band after the β-globin PCR, they were determined to be devoid of any significant PCR inhibitory factors (data not shown). However, this does not preclude the potential presence of inhibitors in some of the patient samples that were not assayed by the β-globin PCR.
Analysis of patient specimens.
A total of 349 specimens were tested by PCR. Of those, 280 specimens were from the original outbreak and 69 were received as follow-up specimens. Of the 280 specimens tested initially, 73 were positive and 207 were negative after PCR and hybridization. A typical ethidium bromide-stained 2% agarose gel, containing 11 patient specimens along with both positive and negative controls, is shown in Fig. 3. A total of 108 initial specimens were also tested by culture. Of those initial specimens tested, 22 were culture positive and 86 were culture negative.
PCR and culture results on posttherapeutic follow-up.
The second follow-up specimen collected from 69 individuals was taken 3 to 6 weeks after antibiotic therapy. It was of interest to determine the effect that antibiotic treatment would have on the detection of M. pneumoniae by PCR and culture. Most of the individuals retested (53 of 69) were originally positive. Of those individuals retested, 8 remained positive by PCR and 61 were negative by PCR. All of those who were originally PCR negative remained PCR negative. The eight secondary specimens that remained positive by PCR were also tested by culture. Of these, three specimens were positive by culture and had initially been positive by culture. The other five specimens were negative by culture. Two of these were initially culture negative, one was initially culture positive, and two had not been previously tested by culture.
Dot blot hybridization analysis.
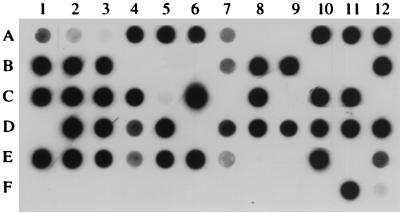
Hybridization analysis was carried out to confirm the specificity of the PCR. Figure 4 shows a typical blot with positive and negative controls, organisms used for testing of specificity (Treponema pallidum and Chlamydia pneumoniae), and 65 patient specimens that were analyzed by PCR. A total of 131 patient specimens (initial and follow-up specimens) were tested by hybridization. Of those tested, 81 (73 initial specimens and 8 follow-up specimens) were positive by DNA hybridization and 50 were negative. These results correlated with the PCR results; thus, no PCR-negative specimens were determined to be positive by hybridization and no PCR-positive samples were determined to be negative by hybridization. Thus, as expected, the hybridization assay provided sensitivity equivalent to those of PCR and Gel Doc gel analysis system for the 131 samples tested, and none of the observed PCR products from patient specimens had false-positive results.
FIG. 4.
Dot blot hybridization assay of amplified DNA from clinical specimens. Dots A1 to A3 represent hybridization to an M. pneumoniae PCR product diluted 1:10, 1:40, and 1:160, respectively. Dot F1 is a negative control. Dots F2 and F3 indicate Treponema pallidum and Chlamydia pneumoniae DNA samples, respectively, that were used as part of the specificity assessment. The remaining dots are patient specimens. Negative samples are dots A8, A9, B4 to B6, B10, B11, C7, C9, C12, D1, D6, E8, E9, E11, and F1 to F10; the rest of the samples are positive.
Comparison of culture and PCR.
A total of 116 patient specimens (108 primary specimens and 8 follow-up specimens) were tested by both PCR and culture. A comparison of the results obtained by these methods with patient specimens is summarized in Table 1. All samples that were culture positive were also PCR positive. Of the 91 specimens found to be culture negative, 28 were PCR positive. The data in Table 1 demonstrate that PCR analysis was capable of markedly increasing the level of detection of M. pneumoniae. It should be noted that the first 87 outbreak-related specimens were tested by both culture and PCR; thereafter, the remaining specimens were tested only by culture if they were PCR positive. As a consequence of using PCR to prescreen samples for culture, the culture positivity rate was slightly higher than it would have been had all samples been cultured.
TABLE 1.
Summary of results for paired outbreak-related specimens tested by both culture and PCR
| Result | No. of specimens
|
||
|---|---|---|---|
| PCR negative | PCR positive | Total culture results | |
| Culture negative | 63 | 28 | 91 |
| Culture positive | 0 | 25 | 25 |
| Total PCR results | 63 | 53 | 116 |
DISCUSSION
Pneumonia caused by M. pneumoniae has long been a difficult disease to diagnose because there are both clinical and laboratory diagnostic problems associated with its identification. It has been realized for quite some time that the detection of M. pneumoniae is greatly enhanced by the use of the PCR methodology. PCR methods have provided an advantage because they are fast, specific, and sensitive: in this case 1 day is required for amplification and 1 day is required for the dot blot assay, whereas 1 to 3 weeks is required for traditional culture methods. We have designed a genomics-based PCR primer pair that targets multiple sites and that, we believe, on the basis of published CFU, provides an improvement over the current published methods.
An outbreak of M. pneumoniae in New York State provided the opportunity to both validate our procedure and compare its sensitivity and specificity to those of culture. This outbreak was also the largest outbreak in New York State and one of the largest in the country to be evaluated by testing of specimens by PCR. Before development of this PCR assay, our New York State reference laboratory had gained significant experience with and expertise in Mycoplasma culture. The results of the present study indicate that our PCR method is at least twice as sensitive as the current culture methodology. Our PCR method was highly sensitive (0.006 CFU) and specific and displayed no nonspecific inhibition due to the transport or processing of throat swab specimens.
The present PCR method was designed to amplify the P1 adhesin gene of M. pneumoniae. Numerous investigators have recognized the benefit of using the P1 adhesin gene as a target for PCR identification of M. pneumoniae. A number of investigators have published descriptions of PCR methods that target sequences within this gene (3, 4, 5, 12, 17, 21). The P1 adhesin gene is an ideal amplification target for PCR because primers can be designed to target conserved regions within known repeats of the M. pneumoniae genome. Portions of this gene are repeated up to 10 times within the genome, thus allowing an increase in the sensitivity of a PCR assay above that of a typical one-target PCR method. Our efforts were focused on taking full advantage of the repetitive elements in the genome by creating an assay that would be capable of amplifying the genome in the maximum number of regions. The newly designed PCR had the theoretical advantage of amplifying nine regions of the M. pneumoniae genome (Fig. 1). The method was demonstrated to be further improved by including the addition of a reverse primer to bind to the 10th region (data not shown). Indeed, this method was highly sensitive (0.006 CFU). This detection limit is the equivalent to 3 serial dilutions below the 6 CFU concentration of Mycoplasma determined by culture. In addition to the multiple-target advantage of this PCR, it is also possible that nonviable bacterial cells and extracellular DNA contribute to the discrepancy between the high level of sensitivity of PCR compared to that of culture.
Other groups have also attempted to capitalize on this repetitive aspect of the genome, but analysis suggests that those methods can potentially amplify only six or seven regions of the genome (3, 12, 21). From our sensitivity experiments, on the basis of serial dilution of M. pneumoniae (ATCC 29342) and comparison of plate counts to PCR results, we found the level of detection of PCR to be 0.006 CFU. This was considerably more sensitive than the sensitivity reported for most M. pneumoniae PCR assays. One group (1) found a similar sensitivity (0.019 CFU), but a nested PCR was used to achieve its PCR sensitivity. Because nested PCR methods can be troubled by contamination problems (1, 6, 15), the simple, nonnested PCR assay described here, which has a sensitivity comparable to that of nested PCR, should provide an improvement over the current diagnostic tools used for laboratory detection of M. pneumoniae by PCR.
Hybridization by a dot blot method and with an internal probe specific for our PCR product was used to add specificity to our level of detection. On occasion, after specimen amplification we found secondary PCR products in addition to the expected 309- to 339-bp product. Additionally, we occasionally found nonspecific PCR products in the PCR-negative specimens. Both of these events usually occurred when specimens were tested on the same day that they were received or when recently obtained specimens were tested, and spurious bands often disappeared upon retesting. For these reasons, it was important to probe the PCR products with a specific oligonucleotide internal to the PCR primer set to ensure that any positive result was truly a positive result and any negative result was truly a negative result. This added test allowed confirmation that the specimen was indeed M. pneumoniae.
Amplicon contamination is an important issue that must be addressed with any PCR assay, especially one that is used frequently in a setting such as a large outbreak. Appropriate precautions were taken to maintain standard laboratory practices to avoid contamination. At least one negative control, which contained no template, was run in every PCR, and this control was always negative. The hybridization reactions also contained multiple negative controls that were never positive. Finally, a second PCR was used to confirm the results for a number of specimens with PCR-positive, culture-negative results. The PCR assay amplified a different portion of the P1 adhesin gene that was included in the REPMP4 portions of the M. pneumoniae genome (21).
As noted earlier in the report, there were twice as many PCR-confirmed cases of M. pneumoniae infection as there were culture-confirmed cases. We believe that these PCR-positive, culture-negative cases were true cases of M. pneumoniae infection for several reasons. There have been studies that suggest that PCR can detect mild cases of infection (8, 18), infection in patients who have previously been treated with antibiotics (8, 14), and infection in patients believed to be late in the course of disease (24). Moreover, PCR can detect nonviable bacteria, and in each of these instances culture would be negative. Epidemiological analysis suggested that the community members who were PCR positive had reported contact with patients in this closed community confirmed to be positive by culture (data not shown). Conceivably, the significance of M. pneumoniae as a cause of infection is underestimated because of the difficulties in the past of obtaining samples from patients with laboratory-confirmed cases of M. pneumoniae infection.
Of the 172 primary specimens tested only by PCR and hybridization, a positivity rate of 30% was established within the community during the outbreak period. By comparison, a 20% positivity rate was demonstrated by culture. However, this number is probably the maximum culture positivity rate because many of these specimens tested by culture were tested only after they were determined to be PCR positive to decrease the amount of laborious culture manipulation during the outbreak. Because the sensitivity of this PCR assay is quite good, the likelihood of finding a culture-positive, PCR-negative specimen was extremely unlikely. Most likely, the use of culture methods on all of the samples would have resulted in a lower culture positivity rate.
In the present study PCR provided a marked improvement over culture, as was expected. Table 1 illustrates the results for those paired samples that were tested by both PCR and culture. These data demonstrate that, of the specimens tested, no PCR-negative specimen was found to be culture positive. This indicates the increased sensitivity of PCR over that of culture. Additionally, when those specimens that were PCR positive were observed for their rate of culture positivity, it was determined that PCR was approximately twice as sensitive as culture. Additionally, our sensitivity studies revealed that the level of detection by PCR was 2 log units above the level of detection by culture and above the levels of detection by many of the published PCR assays.
An additional question addressed in the present outbreak study was what effect treatment with the antibiotic azithromycin would have on the ability of members of the community to harbor M. pneumoniae. To address this question, secondary specimens were taken from selected members of the community 3 to 6 weeks following the initial specimen collection from a group of 53 patient specimens that were initially positive by PCR, 8 specimens remained positive by PCR (including three specimens that remained culture positive). Because this group of specimens contained specimens from both symptomatic and asymptomatic patients, no clear link between the duration of therapy or the presentation of the infected individual and the persistence of the organism can be concluded from these results. However, a marked decrease in the number of affected individuals was found after the 3- to 6-week period. Azithromycin therapy has previously been found to be effective at preventing outbreaks of infection with M. pneumoniae (9). Additionally, the combined use of the 1,500-mg cumulative dose of azithromycin and standard epidemic control measures has been associated with a significant reduction in the rate of transmission of M. pneumoniae (13).
The results of the follow-up testing could be interpreted in several ways. First, the result can be interpreted as a 15% rate of carriage of M. pneumoniae within the community following an outbreak, which would be consistent with past studies (23). One study (8) examined families in Seattle and found that a carrier state that lasted up to several months did exist, regardless of symptoms or treatment with a broad-spectrum antibiotic. That group also concluded that the rate of carriage of M. pneumoniae may increase during epidemic periods within communities. Second, the result can be interpreted as a failure of the antibiotic treatment to completely clear the infection because of either the therapy or patient compliance. Third, the result can be interpreted as a possible reinfection of the patient due in part to the closeness of the community. Fourth, it is also possible that PCR testing is detecting nonviable organisms or residual cellular DNA that has not been cleared. However, it is possible that the very high sensitivity of PCR achieved the detection of infections that were not clinically relevant. This is a question relevant to a number of diseases that has not yet been fully addressed.
In conclusion, this report describes a genomics-based PCR that provided rapid detection of M. pneumoniae in a closed community during a large outbreak. This PCR assay was developed for the testing of throat swab specimens suspected to harbor M. pneumoniae and is highly sensitive (0.006 CFU) without the addition of a nested PCR component. This PCR is also 100% specific. In our hands this method was found to be twice as sensitive as culture, and as expected for a PCR procedure, it is extremely rapid, especially when compared to the speed of culture.
ACKNOWLEDGMENTS
We thank Susan Wong, Wendy Archinal, Joel Ackelsberg, Stan Kondracki, Christopher Maendel, the Wadsworth Center Molecular Genetics Core Facility, and the Wadsworth Center Photography and Illustrations Department for valuable assistance.
REFERENCES
- 1.Abele-Horn M, Busch U, Nitschko H, Jacobs E, Bax R, Pfaff F, Schaffer B, Heesemann J. Molecular approaches to diagnosis of pulmonary diseases due to Mycoplasma pneumoniae. J Clin Microbiol. 1998;36:548–551. doi: 10.1128/jcm.36.2.548-551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernet C, Garret M, De Barbeyrac B, Bebear C, Bonnet J. Detection of Mycoplasma pneumoniae by using the polymerase chain reaction. J Clin Microbiol. 1989;27:2492–2496. doi: 10.1128/jcm.27.11.2492-2496.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck G E, O'Hara L C, Summersgill J T. Rapid, sensitive detection of Mycoplasma pneumoniae in simulated clinical specimens by DNA amplifications. J Clin Microbiol. 1992;30:3280–3283. doi: 10.1128/jcm.30.12.3280-3283.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Barbeyrac B, Bernet-Poggi C, Fébrer F, Renaudin H, Dupont M, Bébéar C. Detection of Mycoplasma pneumoniae and Mycoplasma genitalium in clinical samples by polymerase chain reaction. Clin Infect Dis. 1993;17(Suppl. 1):S83–S89. doi: 10.1093/clinids/17.supplement_1.s83. [DOI] [PubMed] [Google Scholar]
- 5.Dorigo-Zetsma J W, Zaat S A J, Wertheim-Van Dillen P M E, Spanjaard L, Rijntjes J, Van Wavern G, Jensen J S, Angulo A F, Dankert J. Comparison of PCR, culture, and serological tests for diagnosis of Mycoplasma pneumoniae respiratory tract infection in children. J Clin Microbiol. 1999;37:14–17. doi: 10.1128/jcm.37.1.14-17.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehrlich G D, Sirko D A. PCR and its role in clinical diagnostics. In: Ehrlich G D, Greenberg S J, editors. PCR-based diagnostics in infectious disease. Boston. Mass: Blackwell Scientific Publications; 1994. pp. 3–18. [Google Scholar]
- 7.Falguera M, Nogues A, Ruiz-Gonzalez A, Garcia M, Puig T. Detection of Mycoplasma pneumoniae by polymerase chain reaction in lung aspirates from patients with community-acquired pneumonia. Chest. 1996;110:972–976. doi: 10.1378/chest.110.4.972. [DOI] [PubMed] [Google Scholar]
- 8.Foy H M. Infections caused by Mycoplasma pneumoniae and possible carrier state in different populations of patients. Clin Infect Dis. 1993;17(Suppl. 1):S37–S46. doi: 10.1093/clinids/17.supplement_1.s37. [DOI] [PubMed] [Google Scholar]
- 9.Gray G C, McPhate D C, Leinonen M, Cassell G H, Deperalta E P, Putnam S D, Karcher J A, Sawyer M H, Laurila A, Connor J D. Weekly oral azithromycin as prophylaxis for agents causing acute respiratory disease. Clin Infect Dis. 1998;26:103–110. doi: 10.1086/516275. [DOI] [PubMed] [Google Scholar]
- 10.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B-L, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honda J, Yano T, Kusaba M, Yonemitsu J, Kitajima H, Masuoka M. Clinical use of capillary PCR to diagnose Mycoplasma pneumonia. J Clin Microbiol. 2000;38:1382–1384. doi: 10.1128/jcm.38.4.1382-1384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ieven M, Ursi D, Van Bever H, Quint W, Niesters H G M, Goossens H. Detection of Mycoplasma pneumoniae by two polymerase chain reactions and role of M. pneumoniae in acute respiratory tract infections in pediatric patients. J Infect Dis. 1996;173:1445–1452. doi: 10.1093/infdis/173.6.1445. [DOI] [PubMed] [Google Scholar]
- 13.Klausner J D, Passaro D, Rosenberg J, Thacker W L, Talkington D F, Werner S B, Vugia D J. Enhanced control of an outbreak of Mycoplasma pneumoniae pneumonia with azithromycin prophylaxis. J Infect Dis. 1998;177:161–166. doi: 10.1086/513818. [DOI] [PubMed] [Google Scholar]
- 14.Kleemola S R M, Karjalainen J E, Räty R K H. Rapid diagnosis of Mycoplasma pneumoniae infection: clinical evaluation of a commercial probe test. J Infect Dis. 1990;162:70–75. doi: 10.1093/infdis/162.1.70. [DOI] [PubMed] [Google Scholar]
- 15.Persing D H. In vitro nucleic acid amplification techniques. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C.: American Society for Microbiology; 1993. pp. 51–87. [Google Scholar]
- 16.Ruland K, Wenzel R, Herrmann R. Analysis of three different repeated DNA elements present in the P1 operon of Mycoplasma pneumoniae: size, number and distribution on the genome. Nucleic Acids Res. 1990;18:6311–6317. doi: 10.1093/nar/18.21.6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma S, Brousseau R, Kasatiya S. Detection and confirmation of Mycoplasma pneumoniae in urogenital specimens by PCR. J Clin Microbiol. 1998;36:277–280. doi: 10.1128/jcm.36.1.277-280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skakni L, Sardet A, Just J, Landman-Parker J, Costil J, Moniot-Ville N, Bricout F, Garbarg-Chenon A. Detection of Mycoplasma pneumoniae in clinical samples from pediatric patients by polymerase chain reaction. J Clin Microbiol. 1992;30:2638–2643. doi: 10.1128/jcm.30.10.2638-2643.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su C J, Tryon V V, Baseman J B. Cloning and sequence analysis of cytadhesin P1 gene from Mycoplasma pneumoniae. Infect Immun. 1987;55:3023–3029. doi: 10.1128/iai.55.12.3023-3029.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tully J G, Rose D L, Whitcomb R F, Wenzel R P. Enhanced isolation of Mycoplasma pneumoniae from throat washing with a newly modified culture medium. J Infect Dis. 1979;139:478–482. doi: 10.1093/infdis/139.4.478. [DOI] [PubMed] [Google Scholar]
- 21.Ursi D, Ursi J-P, Ieven M, Docx M, Van Reempts P, Pattyn S R. Congenital pneumonia due to Mycoplasma pneumoniae. Arch Dis Child. 1995;72:F118–F120. doi: 10.1136/fn.72.2.f118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waites K B, Taylor-Robinson D. Mycoplasma and ureaplasma. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: ASM Press; 1999. pp. 782–794. [Google Scholar]
- 23.Wenzel R P, Craven R B, Davies J A, Hendley J O, Hamory B H, Gwaltney J M., Jr Protective efficacy of an inactivated Mycoplasma pneumoniae vaccine. J Infect Dis. 1977;136(Suppl.):S204–S207. doi: 10.1093/infdis/136.supplement.s204. [DOI] [PubMed] [Google Scholar]
- 24.Williamson J, Marmion B P, Worswick D A, Kok T W, Tannock G, Herd R, Harris J. Laboratory diagnosis of Mycoplasma pneumoniae infection. Antigen capture and PCR-gene amplification for detection of the Mycoplasma: problems of clinical correlation. Epidemiol Infect. 1992;109:519–537. doi: 10.1017/s0950268800050512. [DOI] [PMC free article] [PubMed] [Google Scholar]