Abstract
Clostridium difficile isolates recovered from patients with C. difficile-associated diarrhea (CDAD) at three hospitals located in diverse areas of Japan were analyzed by three typing systems, PCR ribotyping, pulsed-field gel electrophoresis (PFGE), and Western immunoblotting. At the three hospitals examined, a single PCR ribotype strain (type smz) was predominant and accounted for 22 (65%) of 34, 18 (64%) of 28, and 11 (44%) of 25 isolates, respectively. All of the 51 isolates that represented PCR ribotype smz were nontypeable by PFGE because of DNA degradation. Since the type smz strain did not react with any of the antisera against 10 different serogroups (A, B, C, D, F, G, H, I, K, and X), we prepared a new antiserum against a type smz isolate. All 51 type smz isolates presented identical banding patterns, reacting with the newly prepared antiserum (designated subserogroup JP-0 of serogroup JP). These results were compared with those of a strain from a hospital outbreak that occurred in New York, which has been identified as type J9 by restriction enzyme analysis and type 01/A by arbitrarily primed PCR but was nontypeable by PFGE because of DNA degradation. This strain was reported to be epidemic at multiple hospitals in the United States. The J9 strain represented a PCR ribotype pattern different from that of a type smz strain and was typed as subserogroup G-1 of serogroup G by immunoblot analysis. A single outbreak type causing nosocomial CDAD in Japan was found to be different from the strain causing multiple outbreaks in the United States, even though the outbreak strains from the two countries were nontypeable by PFGE because of DNA degradation.
Clostridium difficile is the most frequently identified cause of nosocomial diarrhea. Numerous systems for the typing of C. difficile strains have been evaluated and employed for epidemiological studies of outbreaks of C. difficile-associated diarrhea (CDAD) (1, 4, 7, 12, 17). Recently obtained data suggest that strain differences play some role in the pathogenicity of this organism. Multicenter studies in the United States (16) and the United Kingdom (3) indicate that a single type may be responsible for outbreaks at geographically widely separated hospitals. In Belgium, strains belonging to serogroup C were reported to be the most frequently implicated in outbreaks (20). These studies indicate that particular types of C. difficile are associated with active disease and nosocomial outbreaks. Little is known about the significance of C. difficile types in the epidemiology and etiology of CDAD in Japan.
In the present study, we analyzed C. difficile strains isolated from three geographically widely separated hospitals in Japan by three typing systems, PCR amplification of rRNA intergenic spacer regions (PCR ribotyping), pulsed-field gel electrophoresis (PFGE), and Western immunoblotting, and identified a single type that was predominant at these three different hospitals.
MATERIALS AND METHODS
Bacterial strains.
Eighty-seven isolates recovered at three hospitals geographically separated in Japan and 33 isolates from an outbreak at a hospital in the United States were used in this study. Thirty-four isolates from Nagoyashi-Koseiin Geriatric Hospital, Nagoya, Japan (hospital A), represented 34 episodes from 28 CDAD patients (2 patients had two episodes, and 2 patients had three episodes) during the study period, February 1996 through November 1999; 28 isolates came from 28 CDAD patients hospitalized at Ogaki Municipal Hospital, Ogaki, Japan (hospital B), between April 1997 and March 1998; and 25 isolates were recovered from 25 CDAD patients at Yamaguchi Prefectural Hospital, Bofu, Japan (hospital C), from March 1996 through July 1997. An outbreak which occurred at a hospital in New York (hospital D), between October 1989 and May 1990 has already been described and analyzed by immunoblotting and arbitrarily primed PCR (AP-PCR) (7, 12). Thirty-three strains from the outbreak at hospital D in the United States were examined in the present study.
C. difficile isolates from the hospitals in Japan were identified at the Institute of Anaerobic Bacteriology, Gifu University School of Medicine, Gifu, Japan, and the U.S. isolates were identified at the Centers for Disease Control and Prevention, Atlanta, Ga. Isolation and identification of C. difficile were performed as previously described (8). Toxigenicity of isolates was determined by amplification of the nonrepeating and repeating sequences of the toxin A gene (8, 11) and the nonrepeating sequences of the toxin B gene (8).
Genotypic and phenotypic typing.
PCR ribotying was performed by the method described by Stubbs et al. (18), with the reaction volume for PCR scaled down to 50 μl. Resultant PCR products were concentrated by heating at 75°C for 30 min and separated in 3% agarose (Nacalai Tesque, Inc., Kyoto, Japan) at a constant voltage of 120 V for 4 h. PFGE analysis was performed as previously described (10). SmaI (New England Biolabs Inc., Beverly, Mass.) was used for digestion of DNA in the inserts, and resulting macrorestriction fragments were resolved by PFGE at a constant voltage of 6 V/cm with 25-s pulses for 3 h, followed by 50-s pulses for 20 h. Major PFGE types were defined by more than three fragment differences, and these major types were subtyped by three and fewer than three fragment differences in accordance with the criteria described by Tenover et al. (19). Typing by Western immunoblotting using antisera against the reference strains of 10 different serogroups (serogroups A, B, C, D, F, G, H, I, K, and X) established by slide agglutination test (5) was performed as previously described (7). Isolates were typed into serogroups (5) and subserogroups according to band variations (7). One strain (GAI 97660), which was isolated from a patient with pseudomembranous colitis (PMC) at hospital A, was used to prepare new polyclonal antiserum by immunizing rabbits as previously described (5, 15).
Testing of susceptibility to clindamycin and PCR assay for detection of the erythromycin ribosomal methylase B gene (ermB).
Testing of susceptibility to clindamycin was performed by Etest (AB BIODISK, Solna, Sweden) as directed by the manufacturer using a Brucella HK agar plate supplemented with 5% laked sheep blood. The MIC was measured after anaerobic incubation at 37°C for 48 h. A PCR assay for detection of the ermB gene was performed by using the primer set 2980–2981 as described previously (6).
RESULTS
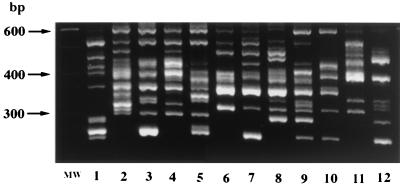
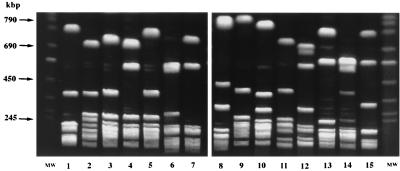
Toxigenicity and typing results of C. difficile isolates recovered from four hospitals are presented in Table 1. All of 120 isolates from four hospitals were typeable and differentiated into 15 types by PCR ribotyping. Only 33 of the 120 isolates were typeable by PFGE and resolved into 16 major types and 18 subtypes (a and b). The remaining 87 isolates were nontypeable by PFGE because of DNA degradation during sample processing. Figures 1 and 2 represent 12 PCR ribotype patterns and 15 PFGE patterns (15 subtypes and 13 major types) of isolates recovered from CDAD patients at the three hospitals in Japan.
TABLE 1.
Typing results of C. difficile isolates from CDAD patients in three hospitals in Japan and a hospital in the United States
| Toxigenicity | Type
|
No. of strains
|
|||||
|---|---|---|---|---|---|---|---|
| PCR ribotype | PFGE type | IBa type | Hospital A (Japan) | Hospital B (Japan) | Hospital C (Japan) | Hospital D (United States) | |
| Toxin A+, toxin B+ | ud | NK1003 | C-1 | 3 | |||
| smz | NTb | JP-0 | 22 | 18 | 11 | 3 | |
| gr | NT | G-1 | 3 | 4 | 26 | ||
| okz | NK09 | G-2 | 1 | ||||
| ny279 | NY279 | G-2 | 1 | ||||
| hr | Hr-ac | H-0 | 1 | 2 | 1 | ||
| hr | Hr-b | H-0 | 2 | ||||
| hr | Y11 | H-0 | 1 | ||||
| yok | OG02-a | NT | 7 | 1 | |||
| yok | OG02-b | NT | 1 | ||||
| og39 | OG39 | NT | 1 | ||||
| og45 | OG45 | NT | 1 | ||||
| y02 | Y02 | NT | 1 | ||||
| y05 | Y05 | NT | 1 | ||||
| y32 | Y32 | NT | 1 | ||||
| ny232 | NY232 | NT | 1 | ||||
| ny342 | A30 | NT | 1 | ||||
| Toxin A−, toxin B+ | fr | fr | F-0 | 1 | 2 | ||
| fr | OG57 | F-0 | 1 | ||||
| fr | Y30 | F-0 | 1 | ||||
| Total | 34 | 28 | 25 | 33 | |||
IB typing, immunoblot typing.
NT, nontypeable.
The isolates were assigned to major types when they had more than three fragment differences and to subtypes (a and b) when they had three or fewer than three fragment differences (19).
FIG. 1.
PCR ribotype patterns of 12 isolates recovered from CDAD patients in Japan (lanes 1 to 12). Lane MW, standard 100-bp DNA ladder used as a molecular weight standard.
FIG. 2.
PFGE patterns of SmaI-digested genomic DNA of 15 isolates recovered from CDAD patients in Japan (lanes 1 to 15). The isolates assigned to PCR ribotypes smz and gr could not be analyzed by PFGE because of DNA degradation. Lanes MW, chromosomal DNA of Saccharomyces cerevisiae used as molecular weight markers.
Twenty-two (65%) of 34 isolates from hospital A had the same PCR ribotype pattern, designated type smz (Fig. 3A), and the next common PCR ribotype (designated type yok) was found in seven episodes. PCR ribotype smz was the most common type found at hospitals B and C, as well as at hospital A, and accounted for 64 and 44% of the isolates at hospitals B and C, respectively (Table 1 and Fig. 3A). Ribotype smz was also isolated in three episodes (9%) at hospital D. When analyzed by PFGE, all 54 PCR ribotype smz isolates, including three isolates from hospital D, exhibited DNA degradation. In contrast, the strain that caused the outbreak at hospital D in the United States was ribotype gr, a pattern identical to that of the reference strain of serogroup G (Fig. 3A) and accounted for 26 (79%) of the 33 isolates. PCR ribotype gr isolates were nontypeable by PFGE, as were type smz isolates. Three and four isolates from hospitals B and C, respectively, represented PCR ribotype gr.
FIG. 3.
PCR ribotype patterns (A) and immunoblot patterns with serogroup JP antiserum (B-1) and serogroup G antiserum (B-2) of epidemic strains from four hospitals. Lanes: MW, 100-bp DNA ladder used as a molecular weight standard; 1, epidemic strain from hospital A (strain GAI 97660, the reference strain of serogroup JP); 2, epidemic strain from hospital B; 3, epidemic strain from hospital C; 4, epidemic strain from hospital D; G, reference strain of serogroup G (used as a control for immunoblotting).
All 120 C. difficile isolates were examined by immunoblot analysis. Of these, 70 isolates, including PCR ribotype smz isolates, did not react with any of the antisera against 10 serogroups. One type smz isolate (GAI 97660), recovered from a patient with PMC at hospital A, was used to prepare a new antiserum. All 54 PCR ribotype smz isolates showed a blotting profile identical to that of strain GAI 97660 when the newly prepared antiserum was used. This immunoblot type was designated subserogroup JP-0 of serogroup JP (Fig. 3B-1). No PCR ribotypes other than type smz reacted with serogroup JP antiserum. The epidemic strain at hospital D reacted with serogroup G antiserum, presented a banding pattern different from that of the reference strain of serogroup G in the region beyond 60 kDa, and was typed in subserogroup G-1 (Fig. 3B-2) (7). PCR ribotype gr strains recovered from hospitals B and C showed an immunoblot pattern identical to that of the PCR ribotype gr strain that caused the epidemic at hospital D.
Eight of 12 isolates from 12 patients diagnosed as having PMC at hospitals A and B were typed as PCR ribotype smz. Analysis of the four patients with recurrences of CDAD at hospital A showed that three patients acquired new strains. In two patients with two episodes, PCR ribotype yok from the first episode and type smz from the second episode were identified. One patient with three episodes had type smz from the first and second episodes and acquired a yok strain in the third episode. In the patient with three episodes, PCR ribotype smz was isolated from all of her three episodes.
Susceptibility to clindamycin was examined in all 120 isolates. Among isolates assigned to type smz, high-level resistance to clindamycin (MIC, >256 μg/ml) was found in 22 (100%) of 22, 15 (83%) of 18, and 3 (27%) of 11 isolates recovered at hospitals A, B, and C, respectively. Eleven (92%) of 12, 6 (60%) of 10, and 5 (36%) of 14 non-smz isolates from hospitals A, B, and C, respectively, were highly resistant to clindamycin. All 26 isolates typed as PCR ribotype gr, PFGE-nontypeable subserogroup G-1 of serogroup G from hospital D were highly resistant to clindamycin, while the MICs for 7 nonepidemic isolates ranged from 8 to 32 μg/ml. Six of seven PCR ribotype gr isolates from hospitals B and C were highly resistant to clindamycin, but the MIC for the remaining isolate of PCR ribotype gr was 6 μg/ml. A PCR assay for detection of the ermB gene was carried out on type smz isolates. Of 54 isolates tested, all 40 that were highly resistant to clindamycin were PCR positive and the remaining 14 isolates, for which the MICs ranged from 8 to 32 μg/ml, were PCR negative. Data on antibiotics implicated in CDAD were available in 28 episodes from hospital A and 16 episodes from hospital B. The antibiotic agents most commonly used at both hospitals were a variety of cephalosporins. No specific antibiotic use was associated with CDAD caused by the type smz strain (data not shown).
Toxin A−, toxin B+ isolates were recovered from five patients with intestinal symptoms at three hospitals in Japan (Table 1). All of these five isolates represented a single type by both PCR ribotyping (PCR ribotype fr, with a pattern identical to that of the reference strain of serogroup F) and immunoblot typing (subserogroup F-0 of serogroup F). The five isolates were resolved into three different major types by PFGE analysis.
DISCUSSION
In the present study, a single type predominant at three hospitals located in diverse areas of Japan was identified. All of the C. difficile isolates belonging to type smz according to PCR ribotyping appeared to be serologically identical, reacting only with the antiserum newly prepared in this study. The type smz strain was nontypeable by PFGE because of DNA degradation. Interestingly, the epidemic strain from hospital D in United States had the same DNA degradation problem, although it was type gr according to PCR ribotyping and serogroup G according to immunoblot analysis. It has already been noted that isolates belonging to some subgroups of serogroup G exhibit DNA degradation (2, 9, 10). Samore et al. (16) investigated six outbreaks in the United States and reported that a single type was responsible for outbreaks at five of the six hospitals (one of these five hospitals corresponds to hospital D in the present study) and that the epidemic type was identified as type J9 by restriction enzyme analysis and type 01/A by AP-PCR (16). This type corresponds to PCR ribotype gr, PFGE-nontypeable subserogroup G-1 of serogroup G in this study. Preliminary results of an international typing study indicated that the epidemic strain from the Boston outbreak identified as restriction enzyme analysis type J9 and AP-PCR type 01/A (16) corresponds to O'Neill's PCR ribotype 1, which caused multiple outbreaks in England and Wales (3, 18). The data show that the isolates typed as PCR ribotype smz, PFGE-nontypeable subserogroup JP-0 of serogroup JP that caused multiple outbreaks in Japan are different from the outbreak type at multiple hospitals in the United States and possibly in the United Kingdom. Type smz was found in three isolates recovered from hospital D, suggesting the existence of the type smz strain not only in Japan but also in the United States. On the other hand, three and four isolates from hospitals B and C, respectively, showed the same typing pattern as the outbreak type in the United States, but this type did not appear to be epidemic at the Japanese hospitals.
Particular virulence factors associated with this smz type strain have not been elucidated. Johnson et al. documented that all of the epidemic strain isolates from four hospitals in the United States were highly resistant to clindamycin and possessed the ermB gene, while only 15% of nonepidemic strains were resistant to clindamycin (6). They concluded that use of clindamycin was one of the specific risk factors for nosocomial outbreaks in the United States (6). This finding is not consistent with observations on epidemics caused by type smz in Japanese hospitals. High-level resistance to clindamycin did not appear in all isolates of type smz. The isolation rate of high-level clindamycin-resistant strains among type smz strains was similar to that among non-smz isolates at hospitals A and C. These findings suggest that clindamycin resistance does not affect the epidemic potential of type smz. Moreover, PCR analysis showed that the ermB gene was found exclusively in type smz isolates which were highly resistant to clindamycin. These results suggest that during passage through the guts of hospitalized patients, the smz strain may acquire or lose the ermB gene, which has been reported to be transferable in the absence of detectable plasmids (14). More studies are needed to clarify the roles of antibiotic resistance in nosocomial epidemics of CDAD. It has been documented that an epidemic type in a hospital is frequently represented among isolates associated with heavy environmental contamination and carriage by hospital personnel (17). Pathogenicity factors promoting transmission and colonization of epidemic strains compared to nonepidemic strains warrant further study. The relationship between pseudomembrane formation and types was not defined because sigmoidoscopic examination was not routinely carried out at any of the three hospitals.
Toxin A−, toxin B+ strains recovered from symptomatic patients were included in this study. Although the human intestinal pathogenicity of toxin A−, toxin B+ strains has not been fully defined, current reports (1, 13) support the clinical significance of toxin A−, toxin B+ strains. All five toxin A−, toxin B+ isolates from three different hospitals represented identical PCR ribotyping and immunoblotting patterns, suggesting a high degree of similarity in toxin A−, toxin B+ strains (3, 8).
PFGE has proved to be highly discriminatory and to provide a highly reproducible profile for typing of C. difficile; however, its significant problem is DNA degradation, which may be caused by a particular endonuclease activity in some isolates (2, 10, 16). It should be noted that epidemic strains found both in the United States (16) and in Japan exhibited DNA degradation. The relationship between DNA degradation and virulence factors affecting epidemic potential remains unknown.
In conclusion, we have identified a strain with a genotypic and phenotypic character which is likely to be epidemic in Japan. Detection and identification of particular types may be of help in the control and prevention of nosocomial infections.
ACKNOWLEDGMENT
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture, Japan.
REFERENCES
- 1.Alfa M J, Kabani A, Lyerly D, Moncrief S, Neville L M, Al-Barrak A, Harding G K, Dyck B, Olekson K, Embil J M. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile responsible for a nosocomial outbreak of Clostridium difficile-associated diarrhea. J Clin Microbiol. 2000;38:2706–2714. doi: 10.1128/jcm.38.7.2706-2714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bidet P, Lalande V, Salauze B, Burghoffer B, Avesani V, Delmee M, Rossier A, Barbut F, Petit J C. Comparison of PCR-ribotyping, arbitrarily primed PCR, and pulsed-field gel electrophoresis for typing Clostridium difficile. J Clin Microbiol. 2000;38:2484–2487. doi: 10.1128/jcm.38.7.2484-2487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brazier J S. The epidemiology and typing of Clostridium difficile. J Antimicrob Chemother. 1998;41(Suppl. C):47–57. doi: 10.1093/jac/41.suppl_3.47. [DOI] [PubMed] [Google Scholar]
- 4.Cartmill T D, Panigrahi H, Worsley M A, McCann D C, Nice C N, Keith E. Management and control of a large outbreak of diarrhoea due to Clostridium difficile. J Hosp Infect. 1994;27:1–15. doi: 10.1016/0195-6701(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 5.Delmée M, Laroche Y, Avesani V, Cornelis G. Comparison of serogrouping and polyacrylamide gel electrophoresis for typing Clostridium difficile. J Clin Microbiol. 1986;24:991–994. doi: 10.1128/jcm.24.6.991-994.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson S, Samore M H, Farrow K A, Killgore G E, Tenover F C, Lyras D, Rood J I, DeGirolami P, Baltch A L, Rafferty M E, Pear S M, Gerding D N. Epidemics of diarrhea caused by a clindamycin-resistant strain of Clostridium difficile in four hospitals. N Engl J Med. 1999;341:1645–1651. doi: 10.1056/NEJM199911253412203. [DOI] [PubMed] [Google Scholar]
- 7.Kato H, Cavallaro J J, Kato N, Bartley S L, Killgore G E, Watanabe K, Ueno K. Typing of Clostridium difficile by Western immunoblotting with 10 different antisera. J Clin Microbiol. 1993;31:413–415. doi: 10.1128/jcm.31.2.413-415.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato H, Kato N, Watanabe K, Iwai N, Nakamura H, Yamamoto T, Suzuki K, Kim S-M, Chong Y, Wasito E B. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR J. Clin Microbiol. 1998;36:2178–2182. doi: 10.1128/jcm.36.8.2178-2182.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato H, Kato N, Watanabe K, Ueno K, Sakata Y, Fujita K. Relapses or reinfections: analysis of a case of Clostridium difficile-associated colitis by two typing systems. Curr Microbiol. 1996;33:220–223. doi: 10.1007/s002849900103. [DOI] [PubMed] [Google Scholar]
- 10.Kato H, Kato N, Watanabe K, Ueno K, Ushijima H, Hashira S, Abe T. Application of typing by pulsed-field gel electrophoresis to the study of Clostridium difficile in a neonatal intensive care unit. J Clin Microbiol. 1994;32:2067–2070. doi: 10.1128/jcm.32.9.2067-2070.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato N, Ou C Y, Kato H, Bartley S L, Luo C C, Killgore G E, Ueno K. Detection of toxigenic Clostridium difficile in stool specimens by the polymerase chain reaction. J Infect Dis. 1993;167:455–458. doi: 10.1093/infdis/167.2.455. [DOI] [PubMed] [Google Scholar]
- 12.Killgore G E, Kato H. Use of arbitrary primer PCR to type Clostridium difficile and comparison of results with those by immunoblot typing. J Clin Microbiol. 1994;32:1591–1593. doi: 10.1128/jcm.32.6.1591-1593.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Limaye A P, Turgeon D K, Cookson B T, Fritsche T R. Pseudomembranous colitis caused by a toxin A− B+ strain of Clostridium difficile. J Clin Microbiol. 2000;38:1696–1697. doi: 10.1128/jcm.38.4.1696-1697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mullany P, Wilks M, Tabaqchali S. Transfer of macrolide-lincosamide-streptogramin B (MLS) resistance in Clostridium difficile is linked to a gene homologous with toxin A and is mediated by a conjugative transposon, Tn5398. J Antimicrob Chemother. 1995;35:305–315. doi: 10.1093/jac/35.2.305. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura S, Serikawa T, Mikawa M, Nakashio S, Yamakawa K, Nishida S. Agglutination, toxigenicity and sorbitol fermentation of Clostridium difficile. Microbiol Immunol. 1981;25:863–870. doi: 10.1111/j.1348-0421.1981.tb00091.x. [DOI] [PubMed] [Google Scholar]
- 16.Samore M, Killgore G, Johnson S, Goodman R, Shim J, Venkataraman L, Sambol S, DeGirolami P, Tenover F, Arbeit R, Gerding D. Multicenter typing comparison of sporadic and outbreak Clostridium difficile isolates from geographically diverse hospitals. J Infect Dis. 1997;176:1233–1238. doi: 10.1086/514117. [DOI] [PubMed] [Google Scholar]
- 17.Samore M H, Venkataraman L, DeGirolami P C, Arbeit R D, Karchmer A W. Clinical and molecular epidemiology of sporadic and clustered cases of nosocomial Clostridium difficile diarrhea. Am J Med. 1996;100:32–40. doi: 10.1016/s0002-9343(96)90008-x. [DOI] [PubMed] [Google Scholar]
- 18.Stubbs S L J, Brazier J S, O'Neill G L, Duerden B I. PCR targeted to the 16S–23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J Clin Microbiol. 1999;37:461–463. doi: 10.1128/jcm.37.2.461-463.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Dijck P, Avesani V, Delmée M. Genotyping of outbreak-related and sporadic isolates of Clostridium difficile belonging to serogroup C. J Clin Microbiol. 1996;34:3049–3055. doi: 10.1128/jcm.34.12.3049-3055.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]