Abstract
Background:
To identify additional at-risk groups for lung cancer screening, which targets persons with long history of smoking and thereby misses younger or non-smoking cases, we evaluated germline pathogenic variants (PVs) in lung adenocarcinoma patients for association with accelerated onset.
Methods:
We assembled a retrospective cohort (1999–2018) of oncogenetic clinic patients with lung adenocarcinoma. Eligibility required family history of cancer, data on smoking, and germline biospecimen to screen via multi-gene panel. Germline PVs (TP53/EGFR; BRCA2; other Fanconi anemia (FA) pathway genes; non-FA DNA repair genes) were interrogated for association with age at diagnosis using an accelerated failure-time model.
Results:
Subjects (n=187, age 28–89 years, female 72.7%, Hispanic 11.8%) included “smokers” (minimum 5 pack-years, n=65) and “non-smokers” (lighter ever-smokers n=18, never-smokers n=104). Overall, 26.7% of subjects carried 1–2 germline PVs: TP53 (n=5), EGFR (n=2), BRCA2 (n=6), other FA gene (n=11), other DNA repair gene (n=28). Adjusted for smoking, sex, and ethnicity, lung adenocarcinoma diagnosis was accelerated 12.2 (95% confidence limits 2.5, 20.6) years by BRCA2 PVs, 9.0 (0.5, 16.5) years by TP53/EGFR PVs, and 6.1 (−1.0, 12.6) years by PVs in other FA genes. PVs in other DNA repair genes showed no association. Germline associations did not vary by smoking.
Conclusions:
Among lung adenocarcinoma cases, germline PVs (TP53, EGFR, BRCA2, possibly other FA genes) may be associated with earlier onset. With further study, criteria for lung cancer screening may need to include carriers of high-risk PVs, and findings could influence precision therapy and reduce lung cancer mortality by earlier stage diagnosis.
Keywords: Adenocarcinoma, BRCA2, EGFR, TP53, Germline pathogenic variants, hereditary lung cancer
Precis:
Among lung adenocarcinoma cases, we found germline pathogenic variants (TP53, EGFR, BRCA2, other FA genes) in 26.7% of our cohort. These pathogenic variants were associated with earlier onset of lung cancer with or without tobacco exposure.
Introduction
Lung cancer is the leading cause of cancer mortality in men and women, accounting for more deaths than breast, colon, and prostate cancers combined.1 Because early-stage lung cancer rarely causes symptoms, most lung cancers are not diagnosed until the disease is advanced. As a result, the 5-year survival remains below 20%.2
Most lung cancers are attributable to carcinogens in tobacco smoke. However, according to a recent study in the United States,3 an increasing proportion (up to 15%) of non-small cell lung cancers (NSCLC) is diagnosed in patients who have never smoked. Yet current guidelines do not suggest lung cancer screening for never-smokers, greatly reducing their opportunity for early detection.
Individuals with an affected relative have been shown to have a higher risk of lung cancer.4 Screening for inherited predisposition may prove useful for identifying individuals who are at elevated risk of lung cancer5 and thus may be appropriate candidates for lung cancer screening. Recent studies showed that a minority of patients with lung adenocarcinoma carry germline PVs in cancer-associated genes, especially those in the Fanconi Anemia (FA) or DNA-repair pathways.6 For example, EGFR, harbors the most common somatic mutation observed in NSCLC,7 and a rare variant of EGFR, p.T790M,8,9 is associated with a hereditary lung cancer syndrome that affects predominantly female never-smokers.10 TP53 is one of a dozen genome-maintenance genes whose transcript levels in normal bronchial epithelial cells are the basis for a biomarker-based risk score for lung cancer.5 Inherited in germline, PVs in TP53 cause a severe autosomal dominant susceptibility to multiple primary cancers (Li Fraumeni syndrome); after the core cancers (sarcoma, brain tumors, breast cancer, and adrenal cortical cancer), lung cancer is among the most frequent observed in TP53 carriers.11–13 PVs in genes within the FA and DNA-repair pathways are suspected of contributing to lung cancer but remain to be definitively linked to the disease. A known polymorphic nonsense variant in BRCA2 (rs11571833; p.K3326*) has been associated with increased risk for lung cancer, especially squamous cell NSCLC, in genome wide association study (GWAS) data analyses14,15 despite being non-pathogenic for breast or ovarian cancer.16 To date, highly-penetrant BRCA2 PVs that are associated with breast or ovarian cancer have not been associated with excess risk of lung cancer in family studies.17,18
Typically, germline PVs that increase the risk of cancer also cause earlier age at diagnosis (acceleration) of disease.19 Accordingly, the current study evaluated the hypothesis that the disease occurs significantly earlier in carriers versus non-carriers of germline PVs in genes that are either commonly mutated in lung tumors (EGFR, TP53) or belong to pathways involved in DNA repair. Because effects of PVs may be specific to histologic subtypes of lung cancer, the current study focused exclusively on the most common histologic subtype, adenocarcinoma. The statistical analysis considered potential confounding factors and the possibility that associations with germline PVs might vary by history of smoking.
Materials and Methods
Eligibility and Recruitment
We assembled a retrospective cohort of patients with primary lung adenocarcinoma from an ongoing, IRB-approved registry of patients from oncogenetic clinics in the United States (ClinicalTrials.gov Identifier: NCT04185935; Clinical Cancer Genomics Community Research Network, CCGCRN20,21). The registry-derived cohort was supplemented with lung cancer patients from the City of Hope medical oncology clinic. Regardless of recruitment site, eligibility for the current study required a personal diagnosis of primary lung adenocarcinoma and a family history of cancer, defined as having a first- or second-degree relative ever diagnosed with cancer (other than non-melanoma skin cancer or intraepithelial carcinoma of the uterine cervix). Such family history was documented in a multi-generational pedigree. Further eligibility criteria included enrollment in the registry during 1999–2018, confirmation of lung adenocarcinoma by pathology report, known age at diagnosis, data on history of smoking, and availability of blood sample for comprehensive multi-gene panel testing. Selection into the cohort is summarized in the Supplemental Figure.
Genomic Sequencing
Genomic DNA was extracted from the blood samples and sequenced for coding variants using a custom-designed Agilent SureSelect targeted gene capture with 789 genes, that included EGFR and all those known to be associated with cancer (commonly on clinical panels), Fanconi anemia, as well as candidate genes involved in DNA repair and damage response, cell cycle regulation, apoptosis, and the mTOR, JAK–STAT, and RAS–MAPK pathways, as well as frequently mutated tumor suppressor and oncogenes from the Catalog of Somatic Mutations in Cancer (COSMIC) database,20,21 The bait design included full exon, 5’ and 3’ untranslated regions, and splicing region coverage. Reaction products were evaluated using paired-end sequencing at an average of 100x coverage depth, as previously described.20,21 Variants with an allele fraction ≥ 35% and read depth 5 were annotated using Ingenuity Variant Analysis using American College of Medical Genetics and Genomics Guidelines.22 Pathogenic and likely pathogenic variants were analyzed together as pathogenic variants (PVs).
Statistical Analysis
Subjects with a history of smoking at least 5 pack-years were termed “smokers”, whether current or former. Included in that group were ever-smokers whose pack-year data were unavailable, because they had similar age at onset of lung cancer. The remaining subjects, termed “non-smokers”, included never-smokers as well as ever-smokers with fewer than 5 pack-years, because they had similar age of onset tothe never-smokers.
An accelerated failure-time model of age at diagnosis, specifying a Weibull distribution, was used to explore primary associations with 4 categories of germline PVs. Three categories of PVs were mutually exclusive: those in genes commonly mutated in lung cancer (TP53, EGFR); in BRCA2; and in FA pathway genes23 other than BRCA2 (of which current subjects carried PVs in BLM, BRCA1, BRIP1, FAN1, MLH1, PMS2). The fourth category of PVs, those in DNA repair genes24 (of which current subjects carried PVs in APTX, ATM, CHEK2, ERCC2, FH, MRE11A, MSH2, MUTYH, OGG1, PARP4, PRKDC, RAD50, RAD54B, RAD54L, RECQL, RECQL4, RRM2B, WRN, XPA) that are not included in the 3 preceding categories. The DNA repair category was not mutually exclusive with the preceding categories, because some subjects carried PVs in one of the latter DNA repair genes as well as in one of the 3 other categories. Because some subjects carried PVs in more than one DNA repair gene, this variable was coded as the number (0,1,2) of mutated genes in this category.
Potential covariates in the model were status as smoker versus non-smoker, sex, smoker-by-sex interaction, gene-by-smoker interaction, race (Asian vs non-Asian), and ethnicity (Hispanic vs non-Hispanic). Any covariate that did not improve the model’s fit to the observed data, according to Aikake’s Information Criteria, was omitted from the final analysis. Because of the exploratory nature of the analysis, statistical significance was not adjusted for multiple hypothesis testing.
Results
The demographic characteristics of subjects (n=187) were age 28–89 years, female 72.7%, Asian 26.7%, and Hispanic 11.8% (Table 1). Present in numbers too small to analyze separately were subjects who identified as African American (n=5), Native American (n=1), or Pacific Islander (n=3). Subjects included smokers (5-<20 pack-years, n=22; 20–50 pack-years, n=21; unrecorded pack-years, n=22) and non-smokers (never-smoker, n=104; light ever-smoker, <5 pack-years, n=18).
Table 1.
Subjects with Lung Adenocarcinoma
| Characteristic | All Patients N=187 (Column %) | Patients with PVs N=50 (Column %) |
|---|---|---|
|
| ||
| History of Smoking | ||
| Smoker | ||
| Ever smoked at least 5 Pack-years | 65 (34.8) | 24 (48.0) |
| Nonsmoker | ||
| Never smoked | 104 (55.6) | 22 (44.0) |
| Ever smoked less than 5 Pack-years | 18 (9.6) | 4 (8.0) |
| Sex | ||
| Female | 136 (72.7) | 35 (70.0) |
| Male | 51 (27.3) | 15 (30.0) |
| Primary Ancestry (Self-identified) | ||
| European | 128 (68.4) | 38 (76.0) |
| Asian | 50 (26.7) | 10 (20.0) |
| African | 5 (2.7) | 1 (2.0) |
| Pacific Islander | 3 (1.6) | 1 (2.0) |
| Native American | 1 (0.5) | 0 |
| Ethnicity (Self-identified) | ||
| Hispanic | 22 (11.8) | 7 (14.0) |
| Non-Hispanic | 165 (87.2) | 43 (86.0) |
Overall, 26.7% (binomial 95% confidence interval: 20.4%−33.1%) carried at least 1 of the germline PVs under study. The number of PV carriers (listed by category in Table 2) is exceeded by the number of germline PVs (listed in the Supplemental Table), because 6 subjects carried more than one of these PVs. Specifically, one subject with BRCA2 PV also carried a PV in DNA repair gene ERCC2; one subject with PV in another FA gene (BRIP1) also carried a PV in DNA repair gene POLG; and 4 subjects whose PVs were exclusively in other DNA repair genes each carried PVs in 2 such genes (either ATM and MUTYH, ATM and RAD50, MUTYH and RAD54B, or MSH2 and RAD54L).
Table 2.
Lung Adenocarcinoma Cohort (N=187): Multivariable Model-estimated Age at Diagnosis and Acceleration in Onset
| N Subjects | Age at Diagnosis (95% Confidence Interval, CI) | p* | Acceleration in Age at Diagnosis (95% CI), Relative to Referent Group | |
|---|---|---|---|---|
|
| ||||
| Referent Group: Non-Hispanic Male Smoker Without Germline Pathogenic Variant Below | 9 | 73.9 (68.5, 79.8) | ----- | ----- |
| Pathogenic Variants (Mutually Exclusive) | ||||
| In BRCA2 | 6 | 61.7 (53.3, 71.4) | 0.015 | −12.2 (−2.5, −20.6) |
| In TP53 or EGFR | 7 | 64.9 (57.4, 73.4) | 0.039 | −9.0 (−0.5, −16.5) |
| In FA Gene other than BRCA2 | 11 | 67.8 (61.4, 74.9) | 0.090 | −6.1 (+1.0, −12.6) |
| None of the above | 163 | referent | ||
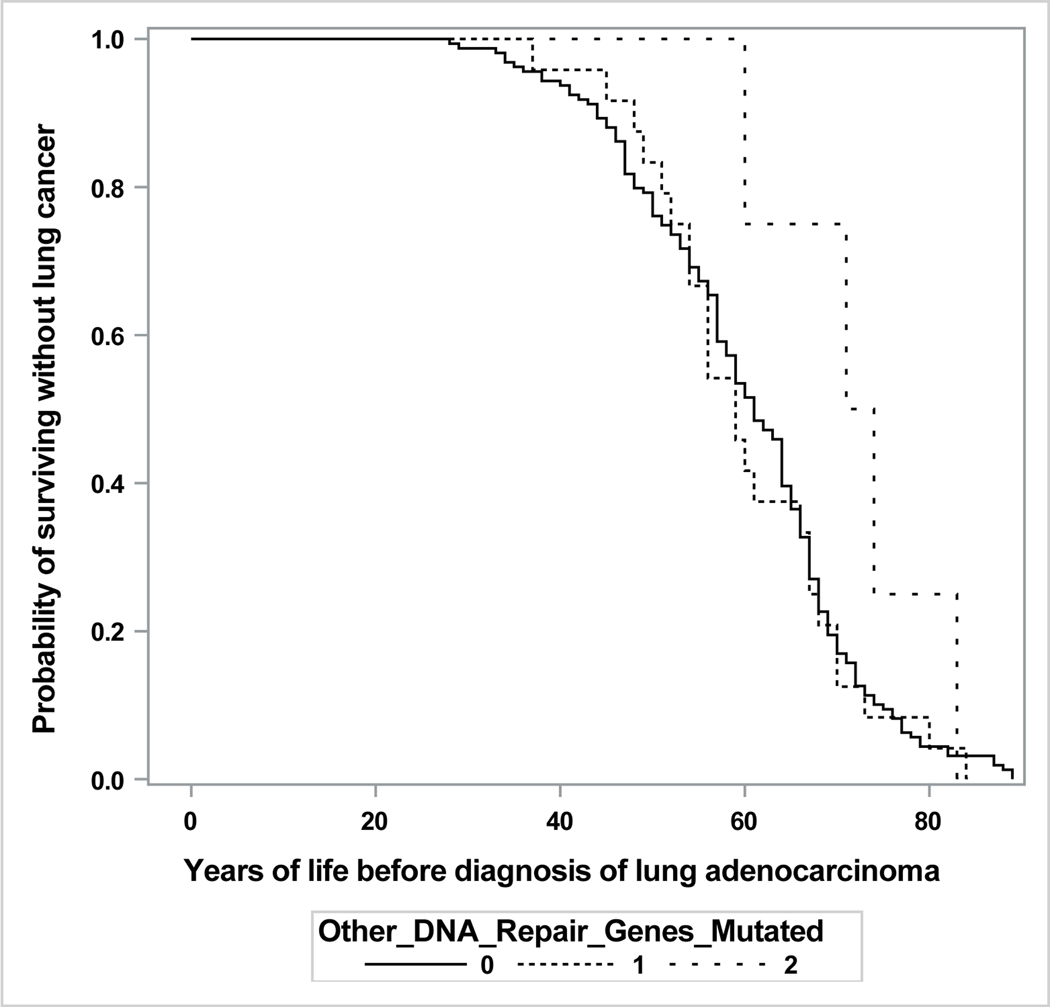
| Pathogenic Variants in Other DNA Repair Genes | ||||
| No | 159 | referent | ||
| Yes, 1 Mutated Gene | 24 | 72.2 (67.2, 77.6) | 0.525 | −1.7 (+3.7, −6.7) |
| Yes, 2 Mutated Genes | 4 | 85.7 (72.7, 101.0) | 0.078 | +11.8 (+27.1, −1.2) |
| 5+ Pack-Year Smoker, by Sex | ||||
| Smoker, Male | 19 | referent | ||
| Non-Smoker, Male | 32 | 61.4 (55.8, 67.5) | ----- | −12.6 (−6.5, −18.1) |
| Smoker, Female | 46 | 67.0 (61.3, 73.2) | ----- | −7.0 (−0.8, −12.6) |
| Non-Smoker, Female | 90 | 66.9 (59.9, 74.8) | ----- | −7.0 (+0.9, −14.1) |
| Ethnicity | ||||
| Hispanic | 22 | 65.8 (60.8–71.2) | ----- | −8.1 (−2.7, −13.1) |
| Non-Hispanic | 165 | referent | ||
P values are calculated for hypothesized risk factors but not for covariates.
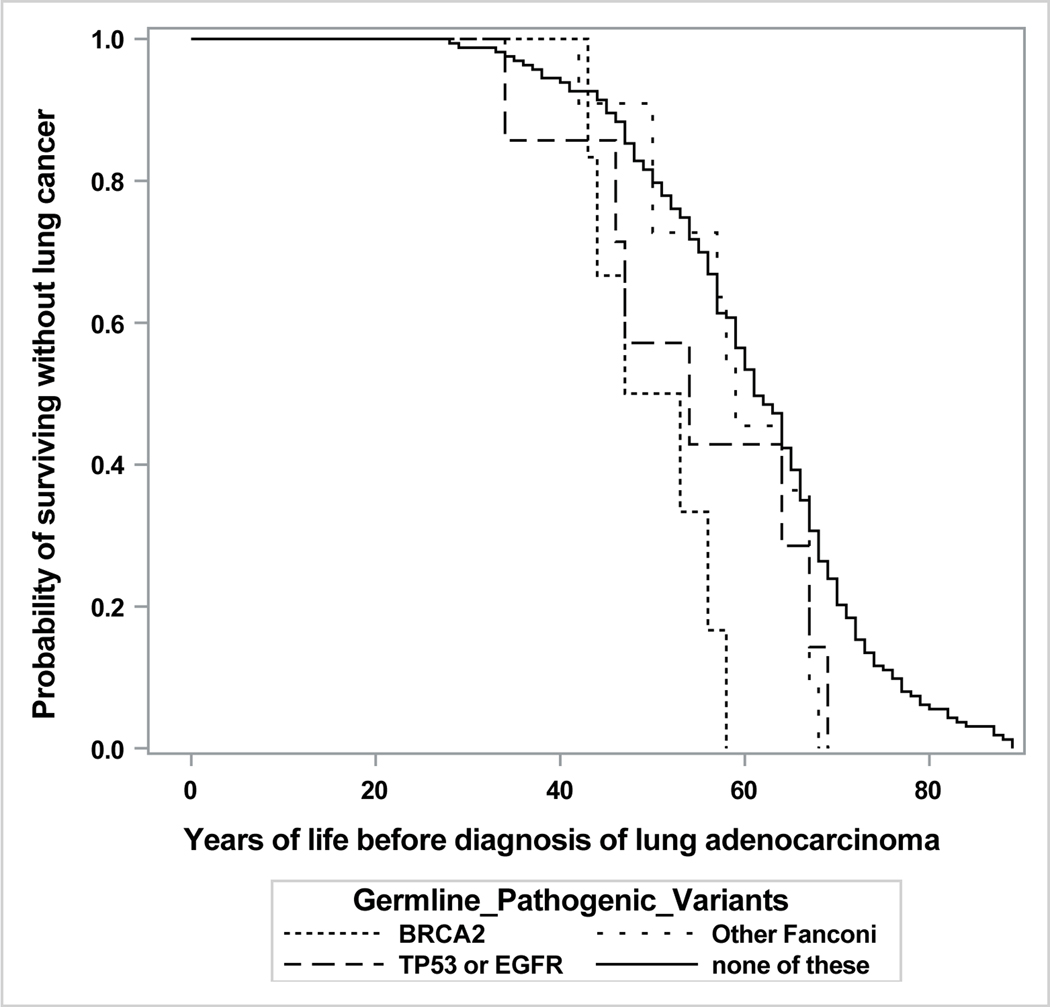
A plot of years lived until diagnosis of lung adenocarcinoma, by PV category, is shown in Figure 1. This plot is unadjusted for potential confounding factors, which are considered in the following multivariable model.
Figure 1.
Lung Adenocarcinoma Cohort (N=187): Years of Life until Diagnosis, by Germline Pathogenic Variant Status
The model was adjusted for ethnicity, smoking history, sex, and their interaction. As shown in Table 2, the age at diagnosis of lung adenocarcinoma was accelerated by PVs in BRCA2, TP53/EGFR, and FA genes other than BRCA2 but was unaffected by PVs in other DNA-repair genes. The associations with germline PVs did not vary by smoking history or by sex (as indicated by non-significant interaction terms, not shown). The model’s fit to the observed data was not improved by separating light ever-smokers from never-smokers. Asian ancestry was unassociated with age at diagnosis and thus was omitted from the final model.
Discussion
Using comprehensive multi-gene panel screening of patients with lung adenocarcinoma, the current study explores the contribution of germline PVs in genes having an established or proposed association with lung cancer. As we report, heritable mutations in these genes appear to be fairly common among lung adenocarcinoma patients who have a family history of cancer. When present, certain of these PVs can accelerate the onset of lung adenocarcinoma, according to our exploratory model.
In that model, the gene whose pathogenic mutation shows the strongest association with age at onset is BRCA2. This novel finding suggests that carriers of PVs in BRCA2 need to be monitored from early adulthood not only for breast and ovarian cancer but possibly also for lung cancer. As additional support for a BRCA2 association with lung cancer, we cite our previous identification of BRCA2 as the most common germline finding among lung cancer patients undergoing commercial genomic analyses of cell-free DNA.25
Another current association, linking PVs in TP53 and EGFR with accelerated onset of lung adenocarcinoma, is consistent with the established roles of somatic mutations in these genes in the pathophysiology of lung cancer. Of note, the EGFR PVs detected in our subjects (p.Thr790Met and p.Arg776His) have been rarely reported to date.8,9,19 In particular, to our knowledge, the germline PV p.Arg776His has been reported only once before.26
Of similar magnitude to the current associations with PVs in BRCA2 and TP53/EGFR, but of lesser statistical significance, is the current association with PVs in other genes within the Fanconi anemia pathway.
Incidental findings from the current model include the observation that, regardless of smoking history, the onset of lung adenocarcinoma is accelerated among Hispanic individuals. However, there were relatively few Hispanic subjects in our cohort, and with one exception, all were female. Hence, further study will be required to confirm this observation, to verify that it extends to males also, and to identify environmental, occupational, lifestyle, genetic or epigenetic factors that may explain the association.
Additional incidental observations from the current model are that non-smokers have earlier onset of lung cancer than smokers, but only among males, and that females have earlier onset of lung cancer than males, but only among smokers. These observations, while they might be surprising, are consistent with the intensifying effect of women’s narrower airways on the established risk of lung cancer from tobacco smoking. Though smoking is a well-established risk factor for lung cancer, it does not necessarily follow that smokers develop lung cancer earlier than non-smokers do; in fact, our data indicate that the opposite is true.
This exploratory study has several limitations. The study is hypothesis generating and warrants validation from a separate cohort study. Restricted by design to cases of lung adenocarcinoma, the current study cohort cannot be used to estimate the relative risk of this disease among carriers of germline PVs. Instead, we have explored the extent to which germline PVs accelerate onset of this disease, a surrogate for predisposition.
The sample size, determined by the contents of the source registry, is not powered to include substantial numbers of PV carriers for each gene of interest, a goal that would have required studying thousands of subjects. Nor is the study powered to support the adjustment of statistical significance necessary to control the risk of error inherent in testing multiple hypotheses (here, the 4 groups of germline PVs). The small numbers of PVs among our subjects result in wide confidence intervals around the acceleration estimates we report and limit our ability to investigate whether PV-associated acceleration of lung cancer varies by history of smoking. Our multi-gene panel, although it included hundreds of cancer-associated genes, did not include all of the genes within the FA and DNA repair pathways; as a result, some subjects may have been misclassified as non-carriers of PVs in such genes. Finally, tobacco exposure was incompletely ascertained for many subjects: the number of pack-years could not be calculated for one third of the subjects classified as smokers, and data on non-smokers’ exposure to environmental tobacco smoke (i.e., within the home) were not collected. As a result, a few subjects may have been misclassified on their status as smokers or non-smokers because of missing years of smoking data.
In conclusion, current findings propose a set of germline PVs, including BRCA2, for further investigation as heritable risk factors for lung adenocarcinoma. The disease acceleration we report for several PVs, when confirmed by additional studies, may warrant the modification of two sets of clinical guidelines. Specifically, eligibility criteria for lung cancer screening may need to include genetically predisposed individuals regardless of smoking history, and germline screening may be warranted for patients with lung cancer to screen for BRCA2 and potentially other genes within the FA pathway. Ultimately, identification of an EGFR or BRCA2 PV could influence precision therapy of lung cancer, and identification of at-risk persons through germline PVs could reduce lung cancer mortality by diagnosing the disease at an earlier, more curative stage.
Supplementary Material
Supporting Figure 1: Consort Diagram - Study Recruitment
Funding Acknowledgements:
The research reported in this publication was supported by the National Cancer Institute (NCI) of 7the National Institutes of Health (NIH) under award numbers P30CA33572 (Integrative Genomics, Bioinformatics, and Biostatistical cores), City of Hope Comprehensive Cancer Center Cancer Control and Population Science Program Pilot Award (PI: K. Reckamp), RC4CA153828 (PI: J. Weitzel), R01CA242218 (PI: J. Weitzel), K08CA234394 (PI: T. Slavin), as well as the Jacqueline Rudman Research Fund (PI: K. Reckamp), the Breast Cancer Research Foundation (PI: J. Weitzel). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Conflict of Interest Statement: None of the authors declared a conflict of interest directly relevant or directly related to the work in the manuscript.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: a cancer journal for clinicians. 2018;68(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2.Campling BG, Hwang WT, Zhang J, et al. A population-based study of lung carcinoma in Pennsylvania: comparison of Veterans Administration and civilian populations. Cancer. 2005;104(4):833–840. [DOI] [PubMed] [Google Scholar]
- 3.Pelosof L, Ahn C, Gao A, et al. Proportion of Never-Smoker Non-Small Cell Lung Cancer Patients at Three Diverse Institutions. J Natl Cancer Inst. 2017;109(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matakidou A, Eisen T, Houlston RS. Systematic review of the relationship between family history and lung cancer risk. Br J Cancer. 2005;93(7):825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeo J, Crawford EL, Zhang X, et al. A lung cancer risk classifier comprising genome maintenance genes measured in normal bronchial epithelial cells. BMC cancer. 2017;17(1):301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parry EM, Gable DL, Stanley SE, et al. Germline Mutations in DNA Repair Genes in Lung Adenocarcinoma. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2017;12(11):1673–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suh JH, Johnson A, Albacker L, et al. Comprehensive Genomic Profiling Facilitates Implementation of the National Comprehensive Cancer Network Guidelines for Lung Cancer Biomarker Testing and Identifies Patients Who May Benefit From Enrollment in Mechanism-Driven Clinical Trials. Oncologist. 2016;21(6):684–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37(12):1315–1316. [DOI] [PubMed] [Google Scholar]
- 9.Yu HA, Arcila ME, Harlan Fleischut M, et al. Germline EGFR T790M mutation found in multiple members of a familial cohort. J Thorac Oncol. 2014;9(4):554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gazdar A, Robinson L, Oliver D, et al. Hereditary lung cancer syndrome targets never smokers with germline EGFR gene T790M mutations. J Thorac Oncol. 2014;9(4):456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malkin D, Li FP, Strong LC, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1237. [DOI] [PubMed] [Google Scholar]
- 12.Hwang SJ, Cheng LS, Lozano G, Amos CI, Gu X, Strong LC. Lung cancer risk in germline p53 mutation carriers: association between an inherited cancer predisposition, cigarette smoking, and cancer risk. Hum Genet. 2003;113(3):238–243. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez KD, Noltner KA, Buzin CH, et al. Beyond Li Fraumeni Syndrome: clinical characteristics of families with p53 germline mutations. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27(8):1250–1256. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, McKay JD, Rafnar T, et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet. 2014;46(7):736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Lusk CM, Cho MH, et al. Rare Variants in Known Susceptibility Loci and Their Contribution to Risk of Lung Cancer. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2018;13(10):1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazoyer S, Dunning AM, Serova O, et al. A polymorphic stop codon in BRCA2. Nature genetics. 1996;14(3):253–254. [DOI] [PubMed] [Google Scholar]
- 17.Breast Cancer Linkage C. Cancer risks in BRCA2 mutation carriers. Journal of the National Cancer Institute. 1999;91(15):1310–1316. [DOI] [PubMed] [Google Scholar]
- 18.van Asperen CJ, Brohet RM, Meijers-Heijboer EJ, et al. Cancer risks in BRCA2 families: estimates for sites other than breast and ovary. Journal of medical genetics. 2005;42(9):711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weitzel JN, Blazer KR, MacDonald DJ, Culver JO, Offit K. Genetics, genomics and cancer risk assessment: state of the art and future directions in the era of personalized medicine. CA Cancer J Clin. 2011;61(5):327–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slavin T, Neuhausen SL, Rybak C, et al. Genetic Gastric Cancer Susceptibility in the International Clinical Cancer Genomics Community Research Network. Cancer Genet. 2017;216–217:111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slavin TP, Neuhausen SL, Nehoray B, et al. The spectrum of genetic variants in hereditary pancreatic cancer includes Fanconi anemia genes. Fam Cancer. 2018;17(2):235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine : official journal of the American College of Medical Genetics. 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M. Fanconi anemia pathway, KEGG (Kyoto Encyclopedia of Genes and Genomes) PATHWAY database. https://www.genome.jp/kegg-bin/show_pathway?ko03460 Web site. Accessed October 28, 2020.
- 24.Broad Institute I, Massachusetts Institute of Technology, and Regents of the University of California,. Molecular Signatures Database v7.2, Gene Set: GO_DNA_REPAIR. https://www.gsea-msigdb.org/gsea/msigdb/cards/GO_DNA_REPAIR.html. Published 2020. Accessed October 28, 2020.
- 25.Slavin TP, Banks KC, Chudova D, et al. Identification of Incidental Germline Mutations in Patients With Advanced Solid Tumors Who Underwent Cell-Free Circulating Tumor DNA Sequencing. J Clin Oncol. 2018;36(35):3459-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Noesel J, van der Ven WH, van Os TA, et al. Activating germline R776H mutation in the epidermal growth factor receptor associated with lung cancer with squamous differentiation. J Clin Oncol. 2013;31(10):e161–164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figure 1: Consort Diagram - Study Recruitment