Abstract
Phospholipase C plays a key role in the pathogenesis of several bacterial infections, for example, those caused by Clostridium perfringens and Listeria monocytogenes. Previous studies have reported multiple copies of plc genes homologous to Pseudomonas aeruginosa plcH and plcN genes encoding the hemolytic and nonhemolytic phospholipase C enzymes in the genomes of Mycobacterium tuberculosis, M. marinum, M. bovis, and M. ulcerans. In this study we analyzed the possible relationship between phospholipase C and hemolytic activity in 21 strains of nontuberculous mycobacteria representing nine different species. Detection of phospholipase C enzymatic activity was carried out using thin-layer chromatography to detect diglycerides in the hydrolysates of radiolabeled phosphatidylcholine. DNA sequences of M. kansasii and M. marinum homologous to the genes encoding phospholipase C from M. tuberculosis and M. ulcerans were identified by DNA-DNA hybridization and sequencing. Finally, we developed a direct and simple assay to detect mycobacterial hemolytic activity. This assay is based on a modified blood agar medium that allows the growth and expression of hemolysis of slow-growing mycobacteria. Hemolytic activity was detected in M. avium, M. intracellulare, M. ulcerans, M. marinum, M. tuberculosis, and M. kansasii mycobacteria with phospholipase C activity, but not in M. fortuitum. No hemolytic activity was detected in M. smegmatis, M. gordonae, and M. vaccae. Whether or not phospholipase C enzyme plays a role in the pathogenesis of nontuberculous mycobacterial diseases needs further investigation.
Phospholipases have a wide spectrum of in vivo and in vitro effects, ranging from minor alterations in cell membrane composition and function to lethality. The most important classes of phospholipases that have been shown to play a significant role in bacterial pathogenesis are phospholipase C and phospholipase D (31). Phospholipase C has been demonstrated to be an important virulence factor in an increasing number of bacteria, including Clostridium perfringens (22, 23), Bacillus cereus (25), the intracellular pathogen Listeria monocytogenes (32, 37), the extracellular pathogen Pseudomonas aeruginosa (24, 34), and other bacteria (20, 35).
Phospholipase genes homologous to the hemolytic plcH and nonhemolytic plcN genes from P. aeruginosa have been described in Mycobacterium tuberculosis, M. bovis, M. marinum, and recently in M. ulcerans (3, 11, 16). Fusion of mpcA or mpcB plc genes of M. tuberculosis encoding glutathione S-transferase produced beta-hemolytic activity when expressed in Escherichia coli (19), and phospholipase C sphingomyelinase activity was detected in cell extracts from M. smegmatis harboring recombinant mpcA or mpcB genes (16).
Nontuberculous mycobacteria are free-living saprophytes that have been detected and isolated from a variety of environmental sources, including water, soil, and dust (6), and some of these mycobacteria are opportunistic pathogens.
Nontuberculous mycobacteria, including M. kansasii and M. avium-M. intracellulare complex, cause pulmonary disease (1, 4, 41). M. avium causes cervical lymphadenitis in children (14, 15), while M. marinum and M. ulcerans have been found to cause skin disease (5, 17, 43). The incidence of nontuberculous mycobacteria has increased since the first reports of nontuberculous mycobacterial disease in immunocompromised hosts in 1982 (40, 42). The impact of nontuberculous mycobacterial infections in AIDS patients poses a major challenge to physicians and mycobacteriologists. These challenges include the development of antimicrobial agents with efficacy in prophylaxis and therapy for nontuberculous mycobacterial infections and the identification of the virulence mechanisms of these mycobacteria. In some infections caused by opportunistic mycobacteria, anemia has been found to be an important negative predictor for survival (2, 10, 29).
In the present study we have investigated the possible relationship between phospholipase C and hemolytic activity in nontuberculous mycobacteria by attempting to detect genes encoding phospholipase C as well as phospholipase C and phospholipase D activities in vitro in nine isolates of these mycobacteria. Finally, we have devised a blood agar test as a simple procedure to screen for the presence of hemolytic activity either produced by phospholipase C or produced by other equivalent enzymes related to pathogenicity.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The mycobacterial strains used in this study are listed in Table 1. Identification at the species level was made by conventional tests (38). M. avium and M. intracellulare were identified according to the 16S rRNA signature sequence as previously described (9). For M. intracellulare, M. marinum, M. kansasii, M. fortuitum, and M. vaccae, only single isolates were employed. Homogeneous suspensions of each strain of mycobacteria, including 1 mg (wet weight) per ml of phosphate-buffered saline (PBS), were prepared from organisms subcultured on Lowenstein-Jensen medium. Then, 10 ml of this bacterial suspension was inoculated into 90 ml of Dubos broth (Difco, Detroit, Mich.). Incubation was performed at 37°C, except for M. ulcerans and M. marinum, which were incubated at 33°C. The bacterial growth period was 8 weeks for M. ulcerans, 6 weeks for M. tuberculosis, M. bovis, M. kansasii, M. gordonae, M. avium, and M. intracellulare, and 1 week for M. marinum, M. smegmatis, M. fortuitum, and M. vaccae.
TABLE 1.
List of mycobacterial strains studieda
| ITM strain no. | Species | Origin | Resultb with:
|
|||
|---|---|---|---|---|---|---|
| PLC | PLD | SB | HA | |||
| 1104 | M. avium | United States | + | + | N | + |
| 98-920 | M. avium | Poland | + | + | N | + |
| 98-922 | M. avium | Poland | + | + | N | + |
| 98-924 | M. avium | Poland | + | + | N | + |
| 98-925 | M. avium | Poland | + | + | N | + |
| 98-926 | M. avium | Poland | + | + | N | + |
| 98-927 | M. avium | Poland | + | + | N | + |
| 4199 | M. intracellulare | Australia | + | + | N | + |
| 5142 | M. ulcerans | ATCC 19423 | + | + | + | + |
| 5146 | M. ulcerans | DRC | + | + | + | + |
| 5147 | M. ulcerans | Australia | + | + | + | + |
| 94-511 | M. ulcerans | Ivory Coast | + | + | + | + |
| 94-1317 | M. ulcerans | Australia | + | + | + | + |
| 94-1326 | M. ulcerans | Australia | + | + | + | + |
| 7732 | M. marinum | ATCC 927 | + | + | + | + |
| 8251 | M. tuberculosis | Rwanda | + | + | + | + |
| 4995 | M. smegmatis | ATCC 607 | N | + | N | N |
| MC2/155 | M. smegmatis | United States | N | + | N | N |
| 96-1073 | M. kansasii | Italy | + | + | + | + |
| 97-461 | M. fortuitum | ATCC 12790 | N | + | N | + |
| 5077 | M. gordonae | Australia | N | + | N | N |
| 4978 | M. vaccae | Belgium | N | + | N | N |
ITM, Institute of Tropical Medicine; ATCC, American Type Culture Collection; DRC, Democratic Republic of Congo. PLC, phospholipase C; PLD, phospholipase D; SB, Southern blot; HA, hemolytic activity.
+, positive; N, negative.
Preparation of whole-cell extracts.
Cultures were centrifuged for 15 min at 3,000 × g. Supernatants and pellets were processed separately. To obtain whole-cell extracts, 1 g (wet weight) of pellet was resuspended in 1 ml of buffer A (50 mM Tris, pH 7.4; 50 mM NaCl; 5% glycerol) and ultrasonicated for 1 min using a Branson Sonifier 250 (Branson Sonic Power Company, Danbury, Conn.). Extracts were centrifuged and stored at −70°C until phospholipase assays were performed.
Detection of phospholipase C and phospholipase D activity.
Phospholipase C enzymes cleave phosphatidylcholine to yield phosphorylcholine and diacylglycerol (DAG), while phosphatidylcholine hydrolysis by phospholipase D leads to the production of choline and phosphatidic acid (PA) (8, 16). Phospholipase C and D activities were determined as previously described by Johansen et al. (16). Briefly, 100-μl portions of whole-cell extracts or culture filtrates were mixed with 100 μl of buffer (250 mM Tris, pH 7.4; 0.25% deoxycholate) containing 13.5 μg of phosphatidylcholine (Sigma-Aldrich SA, Bornem, Belgium) and 0.045 μCi of [14C]phosphatidylcholine (NEN, Boston, Mass.) with the fatty acid side chain moieties labeled. The mixture was vortexed and sonicated for 1 min in a Branson bath sonicator 1200 before overnight incubation at 37°C. Then, 40 μl of each sample was loaded onto a silica gel glass plate (Merck, KGaA, Darmstadt, Germany) and allowed to ascend using petroleum ether-ethyl ether-acetic acid (50:50:1, vol/vol) until the solvent front was within 2 cm of the top of the plate. The plates were air dried and subjected to autoradiography for 1 week.
Search for putative plc genes by PCR.
The design of degenerate primers was based on comparison between mpcA and mpcB gene sequences from M. tuberculosis (GenBank accession no. U49511), (16) and genes encoding nonhemolytic plcN and hemolytic plcH from P. aeruginosa (GenBank accession no. M59304 and M13047) (24, 26). Primers PLC1 (5′-CGGACATTTGACNGAYAT-3′; amino acids 135 to 152 in the sequence of mpcA), and PLC2 (5′-TGCTCGGGGTCGTTNACRCA-3′; amino acids 355 to 374 in the sequence of mpcA were used, where “N” in the sequence stands for A, G, C, or T, “Y” stands for C or T, and “R” stands for A or G. A total of 1 μl of the mycobacterial crude lysates was subjected to PCR. Reactions were taken to a final volume of 50 μl containing 1× buffer (10 mM Tris-HCl, pH 8.4; 50 mM KCl; 1.65 mM MgCl2; 0.1% Triton X-100), 1 U of Taq polymerase, a 0.2 mM concentration of each triphosphate nucleotide, and 20 pmol of each primer. PCRs were performed in a Thermojet thermocycler (Eurogentec, Seraing, Belgium) programmed for 35 amplification cycles as follows: denaturation at 94°C for 1 min, annealing at 56°C for 45 s, and extension at 72°C for 1 min. The products were analyzed by agarose gel electrophoresis and visualized by UV illumination after ethidium bromide staining.
PCR products of 239 bp amplified with primers PLC1 and PLC2 from M. kansasii 4987 and M. marinum 7732 (Table 1) were ligated into PCR II TOPO vector according to the manufacturer's instructions (Invitrogen, Leek, The Netherlands). Recombinant E. coli Top 10 F′ cells were produced by calcium chloride transformation and plasmid preparations using Mini and Maxi Prep systems from Qiagen (Qiagen, Hilden, Germany).
Sequencing of the inserts was performed by the dideoxy-chain termination method of Sanger et al. as previously described (11), with an ABI PRISM 377 DNA sequencer (PE Applied Biosystems, Foster City, Calif.). Multiple-sequence alignments were done with the PILEUP program from the Genetics Computer Group (Madison, Wis.), version 6.2, available at the Belgian Node of the EMBnet (http://ben.vub.ac.be).
Southern blot analysis.
The ulcerans plc probe was prepared as previously described (11). Genomic DNA extraction was performed as previously described by Hermans et al. (13). Extracted DNA was digested with PvuII (Boehringer, Mannheim, Germany), separated by 0.8% agarose gel electrophoresis, and transferred onto a Hybond-N membrane (Amersham-Pharmacia Biotech. Roosendael). The blot was prehybridized and then exposed to a peroxidase-labeled ulcerans plc probe prepared with the enhanced chemiluminescence (ECL) system of Amersham-Pharmacia Biotech Roosendael. The hybridization, stringent washing, and development stages of filter processing were performed according to the manufacturer's instructions provided with the ECL direct nucleic acid labeling and detection kit (Amersham-Pharmacia Biotech Roosendael).
Hemolytic assays on solid media.
A total of 90 ml of citrate buffer (pH 6.6) was mixed with 80 ml of sodium hydroxide (0.1 N). Then, 3.8 g of Middlebrook 7H10 dehydrated broth (Difco Laboratories, Detroit, Mich.) was added to the mixture and shaken vigorously. Next, 1 ml of glycerol was added to the solution, and the mixture was heated at 80°C and stirred for 30 min. The medium was supplemented with 20 ml of 5% bovine serum albumin; 10 ml of defibrinated horse or sheep blood was added and mixed by gentle shaking. Then, 5 ml of the medium was dispensed into each petri dish quadrant and left to dry. The plates were stored at 4°C until inoculation. We prepared mycobacterial suspensions in PBS (pH 7.3) with an inoculum from the Lowenstein-Jensen medium. Aliquots of 100 ml corresponding to McFarland standards 1, 5, and 10 (10) were spread on the blood agar plates. All mycobacteria were incubated at 37°C, except for M. ulcerans and M. marinum, for which a temperature of 33°C was used. All bacteria were grown under aerobic, microaerophilic (CampyPAK Plus, microaerophilic system envelope with a palladium catalyst system; Becton Dickinson, Cockeysville, Md.), and anaerobic conditions (Gas Pak Plus anaerobic system, envelopes with a palladium catalyst; Becton Dickinson). Incubation periods were 1 week for fast-growing mycobacteria and 3 to 4 weeks for slow-growing mycobacteria. Hemolytic activity was considered to be positive when a clear zone was observed around isolated colonies. The hemolytic assay was repeated three times for each mycobacterial strain.
RESULTS
Detection of phospholipase C and D activities.
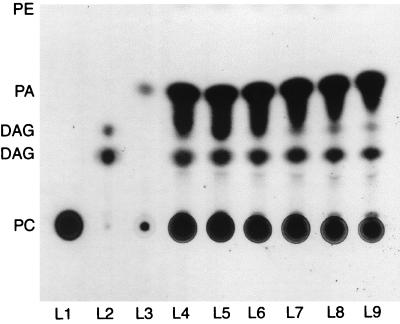
Thin-layer chromatography was performed in order to detect lipid hydrolysis products resulting from incubation of whole-cell extracts from each mycobacterial isolates with phosphatidylcholine. The whole-cell extracts from M. marinum, M. kansasii, M. tuberculosis, M. intracellulare, and M. avium (Table 1) produced DAG upon incubation with phosphatidylcholine with 14C labeling of the lipid moiety. M. avium seems to be very efficient at converting phospholipase C to DAG and PA compared to the positive phospholipase C control from C. perfringens and type I phospholipase D control from cabbage (Fig. 1).
FIG. 1.
Autoradiography of thin-layer chromatography results showing phospholipase C and phospholipase D activity of whole-cell extracts on the radiolabeled phosphatidylcholine l-α-1-palmitoyl-2-linoleoyl-(linoleyl-1-14C). The solvent system included petroleum ether, ethyl ether, and acetic acid (50:50:1). Lanes: L1, H2O (negative control); L2, phospholipase C control (phospholipase C from C. perfringens [Sigma, Bornem, Belgium]); L3, phospholipase D control (phospholipase D type I from cabbage [Sigma]); L4 to L9, M. avium isolates 98-920, 98-922, 98-924, 98-925, 98-926, and 98-927, respectively.
Whole-cell extracts from M. smegmatis, M. fortuitum, M. gordonae, and M. vaccae did not show any evidence of phospholipase C enzymatic activity since no DAG was detected (Table 1). Phospholipase D enzymatic activity was detected in all mycobacterial strains tested (Table 1); Fig. 1 shows the results of a “run” in which the presence of PA in M. avium is demonstrated. These results show that all of the slow-growing mycobacteria, except M. gordonae, possess both phospholipase C and phospholipase D activities, while fast-growing bacilli have only phospholipase D.
Hybridization analysis.
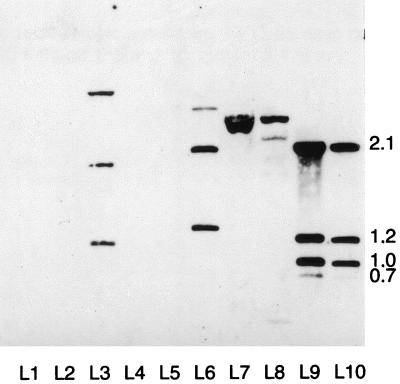
DNA hybridization analysis of Southern blots containing mycobacterial DNA was performed under high-stringency washing conditions. The 239-bp M. ulcerans plc probe hybridized with M. kansasii, M. marinum, M. tuberculosis, M. bovis, and M. ulcerans (Fig. 2). A pattern of three bands was observed in M. kansasii (1.2, 2.0, and 4.5 kb; Fig. 2, lane 3) and M. marinum ATCC 927 (1.5, 2.1, and 3.7 kb; lane 6), while M. bovis (lane 7) and M. tuberculosis (lane 8) showed similar patterns of two bands each (2.5 and 3.5 kb). The M. ulcerans strains (lanes 9 and 10) showed hybridization patterns with four bands (0.75, 1.0, 1.2, and 2.1 kbp).
FIG. 2.
Southern hybridization using the plc probe. Genomic DNA from each mycobacterial strain was digested with PvuII and probed with the plc probe. There is evidence of hybridization with M. kansasii 96-1073 (lane 3) M. marinum 7732 (lane 6), M. bovis 96-319 (lane 7), M. tuberculosis 8251 (lane 8), and M. ulcerans strains 5147 and 94-1326 (lanes 9 and 10). No hybridization is seen with M. smegmatis 4995 (lane 2), M. avium 1104 (lane 4), and M. intracellulare 4199 (lane 5). λ HindIII DNA marker is in lane 1. Sizes are given in kilobase pairs.
No hybridization patterns were observed with M. smegmatis (lane 2), M. avium (lane 4), M. intracellulare (lane 5) and with M. fortuitum, M. gordonae, and M. vaccae (data not shown) with the M. ulcerans plc probe.
Analysis of putative partial phospholipase C sequences.
DNA sequence analysis of the PCR products amplified from M. marinum and M. kansasii confirmed the presence of putative plc genes highly homologous (71.6 and 84% identity in 201 overlap, respectively) to M. ulcerans probe sequence (EMBL accession no. AJ272336).
Hemolytic assays in solid media.
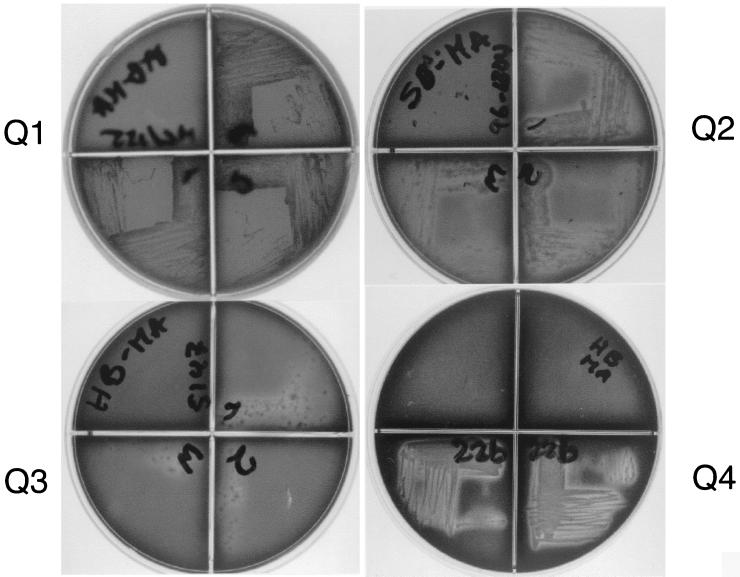
Optimal conditions for bacterial growth and screening of hemolytic activity were determined by preliminary assays using aerobic, microaerophilic, and anaerobic conditions. When evaluated under aerobic conditions, the hemolytic activity was weak even after extended growth. A microaerophilic atmosphere was best for screening hemolytic activity in all mycobacteria. There was no bacterial growth under anaerobic conditions. Hemolytic activity was observed in 8 of 11 mycobacterial isolates screened (Table 1). Among these, only M. fortuitum has hemolytic activity without also having phospholipase C activity.
For M. smegmatis (Fig. 3, Q1), M. gordonae, and M. vaccae isolates, hemolytic activity was not detected despite evidence of extensive bacterial growth. As shown in Fig. 3 (Q2, Q3, and Q4). M. tuberculosis, M. ulcerans, and M. avium produced the widest clear zones around colonies, with M. ulcerans and M. avium apparently having the strongest hemolytic activity. Hemolytic activity was observed in both sheep and horse blood media (Fig. 3).
FIG. 3.
Hemolytic activity in horse and sheep blood agar medium. The hemolytic activity of M. tuberculosis 8251 (Q2, sheep blood), M. ulcerans 5147 (Q3, horse blood), and M. avium 98-922 (Q4, horse blood), with clear zones of hemolysis surrounding the growth of the bacteria, is shown. Although extensive growth is observed in M. smegmatis MC2/155 (Q1, horse blood), there are no clear zones and, hence, there is no hemolytic activity.
DISCUSSION
In the present study, we report for the first time phospholipase C activity in whole-cell extracts of slow-growing M. avium, M. intracellulare, and M. kansasii isolates. Phospholipase C activity in M. tuberculosis, M. ulcerans, and M. marinum has also been confirmed (Table 1) (11, 16, 19). We were not able to detect phospholipase C activity in whole-cell extracts of another slow grower, M. gordonae, nor in the fast growers M. smegmatis, M. vaccae, and M. fortuitum, even upon prolonged exposure of the autoradiograph (Table 1). Phospholipases produced by pathogenic bacteria have been shown to play a variety of roles in the pathogenesis of disease (30, 33, 35).
The plc genes identified in M. tuberculosis have significant homology with the P. aeruginosa plcH and plcN genes encoding the hemolytic and nonhemolytic phospholipases C (16, 19).
In this study a DNA-DNA hybridization assays using a phospholipase C probe from M. ulcerans showed putative plc genes in three mycobacterial isolates with phospholipase C enzymatic activity; however, we were not able to detect hybridization patterns for M. avium and M. intracellulare (Fig. 2). A preliminary hybridization assay using a phospholipase C probe from M. tuberculosis with 76% identity to the phospholipase C probe from M. ulcerans was negative as well (data not shown). These findings may be related to the fact that the M. avium genome is phylogenetically more distant from M. tuberculosis than the other slow growers tested here, when compared by 16S rRNA sequences.
When screened for hemolysis, all phospholipase C-positive mycobacterial isolates were found to be hemolytic (Table 1); however, among the phospholipase C-negative isolates only M. fortuitum was hemolytic. Interestingly, M. fortuitum was shown by Udou (36) to possess an extracellular hemolytic activity that was stable after heating and proteinase treatment, suggesting the presence of a nonprotein hemolysin. The design of our phospholipase C probe has been based on the comparison between mpcA and mpcB gene sequences from M. tuberculosis (GenBank accession no. U49511) (19) and genes coding nonhemolytic plcN and hemolytic plcH from P. aeruginosa (GenBank accession no. M59304 and M13047) (24, 26). We speculate that the phospholipase C produced by mycobacterial isolates with plc genes homologous to P. aeruginosa may have a similar function. Furthermore, several studies suggest that the ability of some bacterial phospholipases to cause hemolysis is due to their ability to hydrolyze both phosphatidylcholine and sphingomyelin (24, 35). The fact that mycobacterial phospholipase C is able to hydrolyze membrane phospholipids, yielding DAG (Fig. 1), suggests a role for these enzymes in the pathogenesis of some mycobacterial infections (7, 21). Furthermore, ceramides are key mediators in coordinating cellular responses and are generated via the sphingomyelinase pathway which, in turn, is activated by the phospholipase C product DAG (32). The hemolytic activity assay that we have developed for slow-growing mycobacteria is a qualitative test, which we believe can detect weak hemolytic substances that may require several days for erythrocyte lysis. However, the hemolysis detected does not depend exclusively on the time of incubation since strong hemolysis was detected in the fast-growing organism M. fortuitum after 4 days and no hemolytic activity was detected with the slow-growing organism M. gordonae after 6 weeks of growth on blood agar.
Among several comparative reports on hemolytic activity between mycobacterial isolates (18, 36), only one showed a possible link between hemolytic activity and pathogenicity (28). Rudnicka et al. observed that weak hemolytic M. avium strains induced more intensive production of nitric oxide and were more susceptible to bactericidal effects caused by infected murine macrophages. In contrast, strongly hemolytic isolates induced significantly stronger tumor necrosis factor alpha production by infected macrophages than weakly hemolytic bacilli. In our study, phospholipase D activity was detected in all mycobacterial isolates analyzed (Table 1); however, whether or not phospholipase D enzymatic activity in mycobacteria plays a role in pathogenesis is, as yet, not established.
In conclusion, we analyzed the possible relationship between phospholipase C and hemolytic activity in nontuberculous mycobacteria. We demonstrated for the first time the production of hemolysis of various mycobacterial isolates in horse and sheep blood agar solid medium. This simple visual assay for hemolytic activity could provide an efficient tool for screening mycobacterium genome libraries for hemolytic and cytolytic factors. Mycobacterial plc gene expression could be related to hemolysis and pathogenesis, as has been demonstrated for similar genes in P. aeruginosa. We found a positive correlation between the presence of phospholipase C enzymatic activity and the hemolytic activity in the mycobacterium isolates we evaluated; however, since the hemolysis induced by mycobacteria appears to result from multiple factors, further investigations on this topic are required.
ACKNOWLEDGMENTS
We gratefully acknowledge Carolina Garcia, Juan Carlos Palomino, and David Davis for their helpful suggestions and Pim de Rijk for his technical assistance.
This work was supported by Colombian government grant COL/453 from the Instituto Colombiano de la Ciencia y la Tecnología (COLCIENCIAS).
REFERENCES
- 1.Ahn C H, Lowell J R, Onstad G D, Shuford E H, Hurst G A. A demographic study of disease due to Mycobacterium kansasii or M. intracellulare-avium in Texas. Chest. 1979;75:120–125. doi: 10.1378/chest.75.2.120. [DOI] [PubMed] [Google Scholar]
- 2.Bogner J R, Gathof B, Heinrich B, Matuschke A, Backer U, Goebel F D. Erythrocyte antibodies in AIDS are associated with mycobacteriosis and hypergammaglobulinemia. Klin Wochenschr. 1990;68:1050–1053. doi: 10.1007/BF01649303. [DOI] [PubMed] [Google Scholar]
- 3.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 4.Corbett E L, Churchyard G J, Hay M, Herselman P, Clayton T, Williams B, Hayes R, Mulder D, De Cock K M. The impact of HIV infection on Mycobacterium kansasii disease in South African gold miners. Am J Respir Crit Care Med. 1999;160:10–14. doi: 10.1164/ajrccm.160.1.9808052. [DOI] [PubMed] [Google Scholar]
- 5.Dobos K M, Frederick D Q, Ashford D A, Horsburgh R C, King C H. Emergence of a unique group of necrotizing mycobacterial diseases. Emerg Infect Dis. 1999;5:367–378. doi: 10.3201/eid0503.990307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falkinham J O., III Epidemiology of infection by nontuberculous mycobacteria. Clin Microbiol Rev. 1996;9:177–215. doi: 10.1128/cmr.9.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Firth J D, Putnins E E, Larjava H, Uitto V J. Bacterial phospholipases C upregulates matrix metalloproteinase expression by cultured epithelial cells. Infect Immun. 1998;65:4931–4936. doi: 10.1128/iai.65.12.4931-4936.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flieger A, Gong S, Faigle M, Neumeister B. Critical evaluation of p-nitrophenylphosphorylcholine (p-NPPC) as artificial substrate for the detection of phospholipase C. Enzyme Microb Technol. 2000;26:451–458. doi: 10.1016/s0141-0229(99)00190-8. [DOI] [PubMed] [Google Scholar]
- 9.Fonteyne P A, Kunze Z M, De Beenhouwer H, Dumonceau J M, Dawson D J, McFadden J J, Portaels F. Characterization of Mycobacterium avium complex related mycobacteria isolated from an African environment and patients with AIDS. Trop Med Int Health. 1997;2:200–207. doi: 10.1046/j.1365-3156.1997.d01-244.x. [DOI] [PubMed] [Google Scholar]
- 10.Gascon P, Sathe S S, Rameshwar P. Impaired erythropoiesis in the acquired immunodeficiency syndrome with disseminated Mycobacterium avium complex. Am J Med. 1993;94:41–48. doi: 10.1016/0002-9343(93)90118-9. [DOI] [PubMed] [Google Scholar]
- 11.Gomez A, Mve-Obiang A, Vray B, Remacle J, Chemlal K, Meyers W M, Portaels F, Fonteyne P A. Biochemical and genetic evidence for phospholipase C activity in Mycobacterium ulcerans. Infect Immun. 2000;68:2995–2997. doi: 10.1128/iai.68.5.2995-2997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendrickson D A, Krenz M M. Reagents and stains. In: Ballows A, Hausler W J, Herrmann K L, Isenberg H D, Shadomy H J, editors. Manual of clinical microbiology. 5th ed. Washington, D.C.: American Society for Microbiology; 1991. p. 1296. [Google Scholar]
- 13.Hermans P W M, van Soolingen D, Dale J W, Schuitema A R J, McAdam R A, Catty D, van Embden J D A. Insertion element IS986 from Mycobacterium tuberculosis: a useful tool for diagnosis and epidemiology of tuberculosis. J Clin Microbiol. 1990;28:2051–2058. doi: 10.1128/jcm.28.9.2051-2058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horsburgh C R., Jr Epidemiology of disease caused by nontuberculous mycobacteria. Semin Respir Infect. 1996;11:244–251. [PubMed] [Google Scholar]
- 15.Hsueh P R, Hsiue T R, Jarn J J, Ho S W, Hsieh W C. Disseminated infection due to Mycobacterium scrofulaceum in an immunocompetent host. Clin Infect Dis. 1996;22:159–161. doi: 10.1093/clinids/22.1.159. [DOI] [PubMed] [Google Scholar]
- 16.Johansen K A, Ronald E G, Vasil L M. Biochemical and molecular analysis of phospholipase C and phospholipase D activity in mycobacteria. Infect Immun. 1996;64:3259–3266. doi: 10.1128/iai.64.8.3259-3266.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson P D R, Stinear T P, Hayman H A. Mycobacterium ulcerans a minireview. J Med Microbiol. 1999;48:511–513. doi: 10.1099/00222615-48-6-511. [DOI] [PubMed] [Google Scholar]
- 18.King C H, Mundayoor S, Crawford J T, Shinnick T M. Expression of contact-dependent cytolytic activity by Mycobacterium tuberculosis and isolation of the genomic locus that encodes the activity. Infect Immun. 1993;61:2708–2712. doi: 10.1128/iai.61.6.2708-2712.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leâo S C, Rocha C L, Murillo L A, Parra C A, Patarroyo M E. A species-specific nucleotide sequence of Mycobacterium tuberculosis encodes a protein that exhibits hemolytic activity when expressed in Escherichia coli. J Bacteriol. 1995;63:4301–4306. doi: 10.1128/iai.63.11.4301-4306.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merino S, Aguilar A, Nogueras M M, Regue M, Swift S, Tomás J M. Cloning, sequencing, and role in virulence of two phospholipases (A1 and C) from mesophilic Aeromonas sp. serogroup O:34. Infect Immun. 1999;67:4008–4013. doi: 10.1128/iai.67.8.4008-4013.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monick M M, Carter A B, Gudmundsson G, Mallampalli R, Powers L S, Hunninghake G W. A phosphatidylcholine-specific phospholipase C regulates activation of p42/44 mitogen-activated protein kinase in lipopolysaccharide-stimulated human alveolar macrophages. J Immunol. 1999;162:3005–3012. [PubMed] [Google Scholar]
- 22.Naylor C E, Jepson M, Crane D T, Titball R W, Miller J, Basak A K, Bolgiano B. Characterization of the calcium-binding C-terminal domain of Clostridium perfringens alpha-toxin. J Mol Biol. 1999;294:757–770. doi: 10.1006/jmbi.1999.3279. [DOI] [PubMed] [Google Scholar]
- 23.Ninomiya M, Matsushita O, Minami J, Sakamoto H, Nakano M, Okabe A. Role of alpha-toxin in Clostridium perfringens infection determined by using recombinants of C. perfringens and Bacillus subtilis Infect. Immun. 1994;62:5032–5039. doi: 10.1128/iai.62.11.5032-5039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ostroff R M, Vasil A I, Vasil M L. Molecular comparison of a non-hemolytic and a hemolytic phospholipase C from Pseudomonas aeruginosa. J Bacteriol. 1990;172:5915–5923. doi: 10.1128/jb.172.10.5915-5923.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otnaess A B, Little C, Sletten K, Wallin R, Johansen S, Flensgrud R, Prydz H. Some characteristic of phospholipase C from Bacillus cereus. Eur J Biochem. 1977;79:459–468. doi: 10.1111/j.1432-1033.1977.tb11828.x. [DOI] [PubMed] [Google Scholar]
- 26.Pritchard A E, Vasil M L. Nucleotide sequence and expression of a phosphate-regulated gene encoding a secreted hemolysin of Pseudomonas aeruginosa. J Bacteriol. 1986;167:291–298. doi: 10.1128/jb.167.1.291-298.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rahme L G, Stevens E J, Wolfort S F, Shao J, Tompkins R G, Ausubel F M. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 28.Rudnicka W, Brzychcy M, Kink M, Lopez A G, Fonteyne P A, Rusch-Gerdes S, Rozalska B. The production of nitric oxide and tumor necrosis factor by murine macrophages infected with mycobacterial strains differing by hemolytic activity. Microbiol Immunol. 1999;43:637–644. doi: 10.1111/j.1348-0421.1999.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 29.Sathe S S, Gascon P, Lo W, Pinto R, Reichman L B. Severe anemia is an important negative predictor for survival with disseminated Mycobacterium avium-intracellulare in acquired immunodeficiency syndrome. Am Rev Respir Dis. 1990;142:1306–1312. doi: 10.1164/ajrccm/142.6_Pt_1.1306. [DOI] [PubMed] [Google Scholar]
- 30.Schluter D, Domann E, Buck C, Hain T, Hof H, Chakraborty T, Deckert-Schluter M. Phosphatidylcholine-specific phospholipase C from Listeria monocytogenes is an important virulence factor in murine cerebral listeriosis. Infect Immun. 1998;66:5930–5938. doi: 10.1128/iai.66.12.5930-5938.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmiel D H, Miller V L. Bacterial phospholipases and pathogenesis. Microbes Infect. 1999;1:1103–1112. doi: 10.1016/s1286-4579(99)00205-1. [DOI] [PubMed] [Google Scholar]
- 32.Schwarzer N, Nöst R, Seybold J, Parida S K, Fuhrmann O, Krull M, Schmidt R, Newton R, Hippenstiel S, Domann E, Chakraborty T, Suttorp N. Two distinct phospholipases of Listeria monocytogenes induce ceramide generation, nuclear factor-κB activation and E-selectin expression in human endothelial cells. J Immunol. 1998;161:3010–3018. [PubMed] [Google Scholar]
- 33.Songer J G. Bacterial phospholipases and their role in virulence. Trends Microbiol. 1997;5:156–161. doi: 10.1016/S0966-842X(97)01005-6. [DOI] [PubMed] [Google Scholar]
- 34.Terada L S, Johansen K A, Nowbar S, Vasil A I, Vasil M L. Pseudomonas aeruginosa hemolytic phospholipase C suppresses neutrophil respiratory burst activity. Infect Immun. 1999;67:2371–2376. doi: 10.1128/iai.67.5.2371-2376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Titball R W. Bacterial phospholipases C. Microbiol Rev. 1993;57:347–366. doi: 10.1128/mr.57.2.347-366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Udou T. Extracellular hemolytic activity in rapidly growing mycobacteria. Can J Microbiol. 1994;40:318–321. doi: 10.1139/m94-052. [DOI] [PubMed] [Google Scholar]
- 37.Vazquez-Boland J A, Kocks C, Dramsi S, Ohayon H, Geoffroy C, Mengaud J, Cossart P. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role lecithinase in cell-cell spread. Infect Immun. 1992;60:219–230. doi: 10.1128/iai.60.1.219-230.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vincent L-F, Portaels F. Proposed minimal standards for the genus mycobacteria and for description of a new slowly growing Mycobacterium species. Int J Syst Bacteriol. 1992;42:315–323. doi: 10.1099/00207713-42-2-315. [DOI] [PubMed] [Google Scholar]
- 39.Wadsworth S J, Goldfine H. Listeria monocytogenes phospholipase C-dependent calcium signaling modulates bacterial entry into J774 macrophage-like cells. Infect Immun. 1999;67:1770–1778. doi: 10.1128/iai.67.4.1770-1778.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolinsky E J. Nontuberculous mycobacteria associated disease. Am Rev Respir Dis. 1979;119:107–159. doi: 10.1164/arrd.1979.119.1.107. [DOI] [PubMed] [Google Scholar]
- 41.Yamazaki Y, Kubo K, Takamizawa A, Yamamoto H, Honda T, Sone S. Markers indicating deterioration of pulmonary Mycobacterium avium-intracellulare infection. Am J Respir Crit Care Med. 1999;160:1851–1855. doi: 10.1164/ajrccm.160.6.9902019. [DOI] [PubMed] [Google Scholar]
- 42.Zakowski P, Fligiel S, Berlin G W, Johnson L., Jr Disseminated Mycobacterium avium-intracellulare infection in homosexual men dying of acquired immunodeficiency. JAMA. 1982;248:2980–2982. doi: 10.1001/jama.1982.03330220024029. [DOI] [PubMed] [Google Scholar]
- 43.Zeligman I. Mycobacterium marinum granuloma: a disease acquired in the tributaries of the Chesapeake Bay. Arch Dermatol. 1972;106:26–31. doi: 10.1001/archderm.106.1.26. [DOI] [PubMed] [Google Scholar]