ABSTRACT
Exosomes mediate the interaction between cancer cells and their microenvironment, and play a key role in tumor development. Although exosomes can package lncRNAs to mediate extracellular communication, the role of exosomal lncRNA AY927529 in prostate cancer (PCa) remains unclear. Exosomes were extracted from normal human prostatic epithelial cell lines (BPH-1 and RWPE-1) and PCa cell lines (VCaP and LNCaP, DU145, PC3) by ultrahigh speed centrifugation. Results of Western blot indicated that Alix, HSC70 and TSGl01 protein levels were upregulated in exosomes derived from PCa cells. LncAY927529 level was upregulated in PCa cells and exosomes derived from PCa patient serum and human PCa cells. CCK-8, Transwell and Flow cytometry assays demonstrated that bone marrow stromal cell line (ST2) conditioned medium (ST2-CM), treated with exosomes derived from PCa cells with high lncAY927529 level, promoted proliferation and invasion of PC3 and DU145 cells, and inhibited cell apoptosis. RT-qPCR assay indicated that lncAY927529 level was downregulated in PC3 and DU145 cells, exosomes derived from PCa cells (PCa-Exo) and ST2-CM treated with PCa-Exo with low expression of lncAY927529, and overexpression of lncAY927529 had the opposite results. In addition, Western blot assay showed that the autophagy related protein LC3II level was increased in ST2 cells treated with exosomes derived from DU145 cells with high expression of lncAY927529, and LC3I protein level was decreased. CXCL14 acted as a RNA-binding protein of lncAY927529, and exosome-mediated lncAY927529 positively regulated CXCL14 levels in ST2 cells. In general, exosome-mediated lncAY927529 could promote PCa cell proliferation and invasion by regulating bone microenvironment, suggesting that exosomal lncAY927529 may be a potential molecular diagnostic marker of PCa.
KEYWORDS: Prostate cancer, lncAY927529, exosomes, apoptosis, autophagy
Introduction
Prostate cancer (PCa) is the most common malignant tumor of male reproductive system in European and American countries. According to a report, there were approximately 174,650 new male cases in the United States in 2019, and the estimated death toll was 31,620 [1]. At present, prostate specific antigen (PSA) is the only clinical screening marker for PCa, but its low specificity, high false-positive and lack of tumor type specificity can easily lead to overtreatment of PCa [2,3]. Tumor biopsy is a common diagnostic method, but it can cause local damage to patients. Although several new agents, including abiraterone, sipuleucel-T, radium-223, enzalutamide and cabazitaxel were used to treat PCa, there were disadvantages of drug resistance [4]. Therefore, more accurate biomarkers of PCa are needed clinically. Previous studies have shown that the development of tumors is closely related to the exosomes secreted by cancer cells [5,6]. Therefore, the study of exosomes may contribute to the early diagnosis of tumors and the analysis of tumor prognosis.
Exosomes are small membranous vesicles that can be secreted by most cells. They have a lipid bilayer structure with a diameter of about 30 ~ 200 nm [7]. Exosomes are secreted into the extracellular environment through the fusion of intracellular multivesicular bodies (MVB) and cell membranes, and they can pass through the extracellular matrix and blood vessel walls, and are widely present in body fluids. Exosomes in body fluids can fuse with the cell membranes of surrounding cells to realize the circulation of biofilms and the transmission of information between cells [8]. Recent studies have shown that exosomes play an important role in many physiological and pathological aspects, such as tumor growth and metastasis, antigen presentation and repair of tissue damage [9–12]. Exosomes secreted by different cells have different components and functions, so they can be used as biomarkers for disease diagnosis. Therefore, exosomes, as a means of liquid biopsy, have important clinical significance in the diagnosis of breast cancer, PCa, lung cancer and other malignant tumors [13].
Long non-coding RNAs (lncRNAs) are non-coding RNAs with a length of more than 200 nucleotides and have no protein coding ability. Importantly, in many cases, lncRNAs have been proved to be the main regulators of gene expression, and they play a key role in various biological functions and disease processes [14]. LncRNAs have a significant tissue specificity, and can regulate the function of protein coding genes by recruiting chromatin modified complexes and interacting with miRNAs, mRNAs or proteins. LncAY927529, as a new lncRNA, located on chromosome X: 76,173,062–76,178,314, has not largely been reported in previous studies. A previous sequencing report reported that lncAY927529 was enriched in the exosomes of all four prostate cancer cell lines (VCaP, LNCaP, DU145 and PC3) [15]. Exosomes promote cancer development by mediating the interaction between cancer cells and their microenvironment. The aim of this study was to investigate the biological function of lncAY927529 in prostate cancer (PCa)-derived exosomes and its role in the development of PCa.
Materials and methods
Clinical samples
Serum samples of PCa (n = 10) and male healthy volunteers (n = 10) were collected from the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China) during March 2017 to September 2019. Ten Patients (49 ± 7.4 years old) were diagnosed with PCa in the Hospital without medical treatment or surgical treatment. In addition, 10 male volunteers who had normal physical examinations in the hospital during the same period were selected to participate in the study. The inclusion criteria included patients age 49 ± 7.4 years old, which met the diagnostic criteria for PCa; tumor staging was in Tumor 1-Tumor 3. The exclusion criteria included patients with severe cardiac insufficiency or other malignant tumors or incomplete clinical data. In short, 5 mL of venous blood from each patient and participant was collected by cubital venous blood collection. Within 2 h after collection, the serum was separated by centrifugation at 2, 000 × g for 10 min at room temperature, and the supernatant of each serum was transferred to store in a centrifuge tube without RNase and stored at −80°C for later use. All participants received informed consent, and the experiment was approved by the institutional review committee and ethics committee of the First Affiliated Hospital of Zhengzhou University.
Cell culture and cell treatment
In this study, human benign prostatic hyperplasia cells BPH1 (# CL0350) and immortalized prostate epithelial cell RWPE1 (#CL0343) were purchased from Fenghui biological Co., Ltd. (Changsha, Hunan, China); human PCa cells VCap (# bncc341358), LNCaP (# bncc261381), DU145 (# bncc338240) and PC3 (# BNCC337715) were purchased from American Type Culture Collection (ATCC, Manassas, VA). Cells were all cultured in Roswell Park Memorial Institute-1640 medium (RPMI-1640) (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA), 100 units/mL penicillin, and 100 μg/mL streptomycin (Thermo Fisher Scientific, Waltham, MA, USA) at 37°C with 5% CO2 atmosphere. The medium was changed every 2–3 days. When the fusion rate reached 70%, RPMI-1640 and non heterologous serum (Termo Fisher scientifc, Waltham, MA, USA) were used to replace the medium for 2 days. The supernatant of cell culture was collected to separate the extracellular bodies.
The mouse bone marrow stromal cell line ST-2 was derived from the cell line preserved in our laboratory. ST-2 cells were cultured in MEM alpha medium (Termo Fisher scientifc, Waltham, MA, USA) containing 10% FBS serum (Termo Fisher scientifc, Waltham, MA, USA) and 1% antibiotics. 293 T cells (#BNCC281963, Fenghui biological Co., Ltd, Changsha, Hunan, China) were cultured in DMEM medium containing 10% FBS serum and 1% antibiotics in 5% CO2 incubator at 37 oC.
ST2 cells were seeded in a 6-well plate with 2 × 104 cells per well. The old medium was discarded the next day. After washing twice with PBS, the medium without FBS was added, and exosomes secreted by different PCa cells were added for treatment (2 x 1010 particles/well), respectively. The cell culture supernatant was collected and used as ST2-CM-Exo; the supernatant without exosomes was used as control conditioned medium (ST2-CM).
AY927529 siRNA, pcDNA-AY927529 and corresponding negative controls (Scramble and pcDNA3.1) were purchased from GenePharma Co., Ltd. (Shanghai, China). The cells were inoculated into the culture plate. When the cell grew to about 70% confluence, cells were transfected with plasmid according to the instructions of Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA).
Exosome isolation
Exosomes were separated by ultracentrifugation[16]. When the fusion degree of prostate cells reached 70%, the original medium was discarded and washed twice with sterile DPBS (Termo Fisher Scientific, Waltham, Ma, USA), and the corresponding 0.5% Exo-FBSTMExosome-depleted FBS medium was added. The supernatant was collected after two days of culture at 37°C, and the dead cells and debris were removed. Then, the supernatant using centricon plus-70 (milipore, American) was centrifuged at 4°C at 3, 500 x g for 30 min and 1, 000 x g for 2 min to concentrate the cell supernatant. Then 100,000 x g ultra-high speed centrifugation was performed twice for 70 min. The precipitates were collected and suspended in 0.5 mL DPBS and stored at – 80°C.
After thawing, the serum samples were placed in a 1.5 mL of centrifuge tube, centrifuged at 300 x g for 10 min, and the supernatant was centrifuged at 2, 000 x g for 10 min at 4°C. The supernatant was filtered by 0.22-μm PVDF filter, and then centrifuged twice at 4°C100, 000 x g for 70 min. The exosomes were collected and resuspended in 0.5 mL of DPBS and stored at −80°C.
Transmission electron microscopy (TEM)
The exosome pellets were resuspended in 50 μL PBS, and a drop of the suspension was placed on a sheet of parafilm. A carbon-coated copper grid was floated on the drop for 5 min at room temperature. Then, the grid was removed, and excess liquid was drained by touching the grid edge against a piece of clean filter paper. The grid was then placed onto a drop of 2% phosphotungstic acid with pH 7.0 for approximately 5 s, and excess liquid was drained off. The grid was allowed to dry for several minutes and then examined using a JEM-1200 EX microscope (JEOL, Akishima, Japan) at 80 kV.
Real‑time reverse transcription polymerase chain reaction (RT-qPCR)
Trizol (Invitrogen, Carlsbad, CA, USA) kits were used to extract total RNA from cells and tissues, and primescript ™ (Takara Biotechnology, Dalian, China) kits were used for reverse transcription to synthesize cDNA. The SYBR® Premix Ex Taq™ (Takara biotechnology, Dalian, China) kits were used for RT-qPCR detection. The reaction conditions were as follows: 95°C for 1 min; 94°C for 15 s, 54°C for 30 s, 72°C for 20 s, a total of 35 cycles. U6 and GAPDH were used as endogenous controls, and the results were calculated with 2−ΔΔCt method. The primer sequences were as follows: lncRNA AY927529 forward, 5ʹ-GCC TAC GCA ACA GTA CTC CA-3ʹ, and reverse, 5ʹ-ATT GGA AAG CTG AGC CGG AT-3ʹ; CXCL14 forward, 5ʹ-CTG CGA GGA GAA GAT GGT TA-3ʹ, and reverse, 5ʹ-CTT TGC ACA AGT CTC CCA AC-3ʹ; GAPDH forward, 5ʹ-CTG CGA GGA GAA GAT GGT TA-3ʹ, and reverse, 5ʹ-CTT TGC ACA AGT CTC CCA AC-3ʹ.
Cell proliferation and invasion
After 48 h of transfection, the cell density was adjusted to 2 × 105 cells/mL and inoculated into 96 well plate. 15 μL of CCK-8 solution was added at 24, 48 and 72 h after culture. The culture medium was discarded after 4 h of incubation. 150 μL of dimethyl sulfoxide (DMSO) was added to each well for 15 min. The absorbance at 450 nm wavelength was determined with a microplate reader (Molecular Devices, Shanghai, China).
Matrigel was diluted 1:5 with PBS, and the concentration was 3.9 μg/μL. Matrigel was coated on Transwell plate and incubated for 2–3 h. Then, 200 μL of 2 × 105 cells/mL cell suspension prepared by serum-free medium and pre-treated with 1 μg/mL mitomycin C (Sigma, St. Louis, MO, USA) were added into the upper chamber to inhibit cell proliferation, and 800 μL of medium containing 10% FBS was added to the lower chamber. The cells were cultured in 37°C incubator for 24 h. After the cells were fixed with formaldehyde solution, the cells were stained with crystal violet and observed and counted under the microscope.
Cell apoptosis
Cell apoptosis assay was carried out according to the instructions of Annexin V-FITC/PI cell apoptosis detection kit (Sigma, St. Louis, MO, USA). Briefly, cells (5 × 105 cells/mL) were resuspended in 195 μL of binding buffer. Then 10 μL of Annexin V-FITC and 10 μL of PI were added, and cells were incubated in the dark at 4°C for 15 min. Cell apoptosis was measured with a flow cytometer (BD Bioscience, San Jose, CA, USA) according to the manufacturer’s instruction. Images were taken using a fluorescence microscopy (Nikon Ti-S, Tokyo, Japan).
Western blot assay
After 48 h of transfection, the total protein of tissues or cells was extracted by RIPA lysate buffer (Beyotime, Shanghai, China), and the protein concentration of each sample was determined by BCA Protein Assay Kit (Beyotime, Shanghai, China). The equivalent protein was passed through 10% polyacrylamide gel electrophoresis (SDS-PAGE), constant pressure 80 V, and running gel for about 30 min. When the protein entered the separation gel, the voltage was adjusted to 120 V. After separation, the protein bands was transferred to the PVDF membranes (Millipore, Bedford, MA, USA) at 300 mA for 2 h. Then the membranes were sealed with 5% bovine serum albumin (BSA) at room temperature for 1 h, then incubated overnight at 4°C with the following primary antibodies: rabbit polyclonal to ALIX (ab88388, 1:300), rabbit monoclonal to TSG101 (ab125011, 1:400), Hsc70 (ab112549, 1:400), CXCL14 (ab36622, 1:300) (Abcam, Cambridge, UK), p-ERK (ab229912, 1:500), LC3I (ab232940, 1:300), and LC3II (ab232940, 1:300) antibody. After washing 3 times with TBST, the membranes were incubated with goat anti-rabbit horseradish peroxidase (HRP) (ab205718, 1:1000) IgG antibody at room temperature for 1 h. All the antibodies were from Abcam (Cambridge, UK). After TBST rinsing, ECL developer (Advansta, CA, USA) was exposed for development. Image J software (Wayne Rasband, MD, USA) was used for protein quantitative analysis. GAPDH was used as an internal reference, and the ratio of gray value of electrophoresis band to GAPDH was used as the relative expression amount of target protein. The experiment was repeated three times and the average value was taken.
Pull-down assay
In order to confirm the relationship between lncAY927529 and CXCL14, biotin-labeled AY927529 was synthesized and transfected into HEK293T cells. Bio- AY927529 probe was transcribed and purified by GenePharma Company (Shanghai, China) through the AmpliScribe™ T7-Flash™ Biotin-RNA Transcription Kit (Epicenter, Madison, Wisconsin, USA). After 48 h, the cells were washed and lysed, and then the extract was incubated with anti-rabies magnetism at 4°C for 3 h. Then, the beads were washed twice with ice-cold buffer, three times with low salt buffer, and once with high salt buffer. Finally, the RNA-RNA complex was eluted and the RNA pull-down product was detected by specific RT-qPCR.
ELISA
The protein lysate was added to each group of cell samples to homogenize cells. The supernatant was collected after centrifugation at 8,000 × g at 4°C. The contents of IL-6, IL-8 and IL-10 in ST2 cells were measured with ELISA kits (R&D Systems, Minneapolis, MN, USA), and the absorbance values were measured at 450 nm (ELx800, BioTek, Vermont, USA).
Statistical analysis
Statistics were carried out using SPSS 22.0 (IBM, NY, USA). Results were presented as means ± SEM. Data comparison was carried out through Student’s t test. More than two groups were statistically compared through one-way ANOVA, and multiple comparisons were performed with Tukey-Kramer correction. P < 0.05 was considered as statistically significant difference.
Results
Exosomes derived from PCa cells and serum were isolated and detected
Electron microscopic images showed that the purifed exosomes were mainly at 50 ~ 90 nm with a clear membrane structure (Figure 1(a)). Alix, TSG101 and HSC70 were the marker proteins in exosomes [17]. Exosomes were isolated from prostate cells and serum by ultracentrifugation. In order to identify these exosomes, we measured the protein levels of specific marker proteins (Alix, TSG101 and HSC70) in exosomes derived from two normal prostate cell lines (BPH1-derived exosomes and RWPE1-derived exosomes) and exosomes derived from four PCa cell lines (VCaP-derived exosomes, LNCaP-derived exosomes, DU145-derived exosomes and PC3-derived exosomes). As shown in Figure 1(b-e), Alix, TSG101 and HSC70 protein levels were upregulated in exosomes derived from four PCa cell lines compared with exosomes derived from two normal prostate cell lines (P < 0.05). In addition, we found that Alix, TSG101 and HSC70 protein levels were also upregulated in exosomes derived from PCa patient serum compared with exosomes derived from normal volunteer serum (Figure 1(f,g)) (P < 0.05). These results suggested that we did isolate exosomes from the samples.
Figure 1.
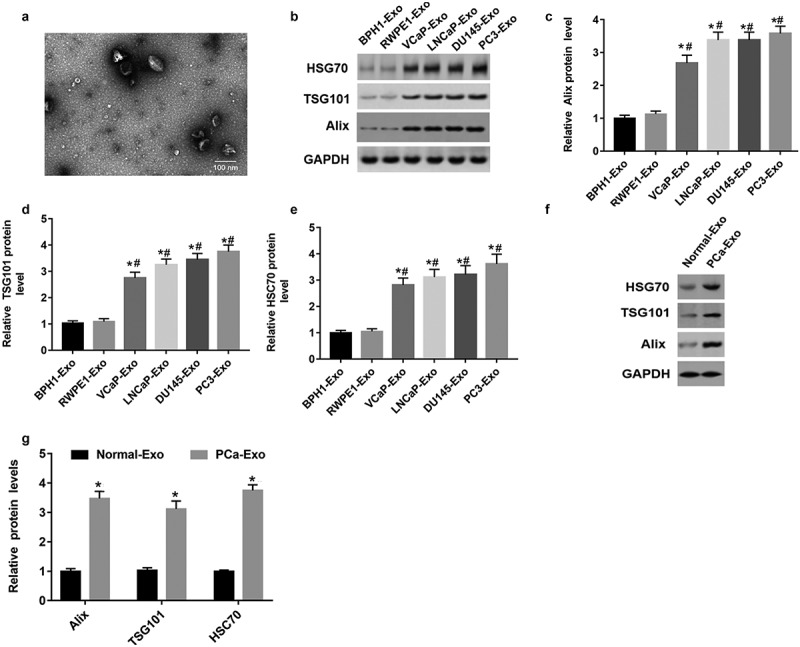
Alix, TSG101 and HSC70 protein levels were upregulated in exosomes derived from PCa cells and PCa patient serum. (a) Electron microscopic images of exosomes isolated from PCa cells. (b-e) The protein levels of Alix, TSG101 and HSC70 in the exosomes of normal prostatic epithelial cells (BPH1 and RWPE1) and PCa cells (VCap and LNCap, DU145, PC3) were determined by Western blot assay. *P < 0.05 compared with BPH1-Exo; #P < 0.05 compared with RWPE1-Exo group. (f and g) The protein levels of Alix, TSG101 and HSC70 were measured in the exosomes derived from serum of healthy volunteers (n = 10) and PCa patients (n = 10) with Western blot assay. *P < 0.05 compared with normal-Exo group. N = 10, data were expressed as mean ± SEM; Student’s t test or one-way ANOVA was used for analyzing data
LncAY927529 level was upregulated in PCa cells and exosomes derived from PCa cell lines and patient serum
Inspired by the sequencing results of others [15], we selected lncAY927529, which was highly expressed in exosomes secreted by PCa cells. In Figure 2(a,b), the results of RT-qPCR showed that lncAY927529 was upregulated in exosomes derived from PCa cell lines (VCaP-Exo, LNCaP-Exo, DU145-Exo and PC3-Exo) and it also upregulated in PCa cell lines (VCaP, LNCaP, DU145 and PC3) (P < 0.05). To determine the potential of lncAY927529 as a molecular diagnostic marker of PCa, we extracted exosomes from PCa patients (N = 10) and normal serum samples (N = 10), and detected the expression level of lncAY927529 in exosomes. And we found that lncAY927529 was also upregulated in the serum of prostate patients (n = 10) compared with male healthy volunteers (n = 10) (Figure 2(c)). These results showed that AY927529 may be a predictor of PCa.
Figure 2.
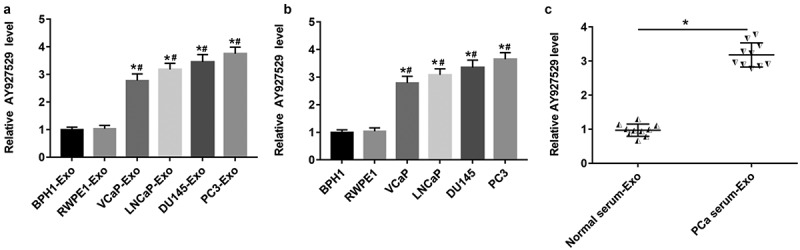
Expression of lncAY927529 in exosomes secreted by different prostate cells, prostate cells and clinical serum samples. (a) RT-qPCR was used to detected the lncAY927529 level in the exosomes of normal prostatic epithelial cells (BPH1 and RWPE1) and PCa cells (VCap, LNCap, DU145 and PC3). *P < 0.05 compared with BPH1-Exo; #P < 0.05 compared with RWPE1-Exo group. (b) RT-qPCR was used to detected the lncAY927529 level in normal prostatic epithelial cells (BPH1 and RWPE1) and PCa cells (VCap and LNCap, DU145, PC3). *P < 0.05 compared with BPH1; #P < 0.05 compared with RWPE1 group. (c) The lncAY927529 level in the exosomes of serum of healthy volunteers (n = 10) and PCa patients (n = 10). *P < 0.05 compared with normal serum-Exo group. N = 4, data were expressed as mean ± SEM; Student’s t test or one-way ANOVA was used for analyzing data
ST2-CM treated with exosomes secreted by PC3 and DU145 cell lines promoted proliferation and invasion of PC3 cells, and inhibited PC3 cell apoptosis
In order to reveal the role of exosomes in bone microenvironment, we treated mouse bone marrow stromal cells (BMSCs) (ST2 cells) with exosomes derived from different PCa cell lines (VCaP, LNCaP, DU145 and PC3), collected the conditioned medium of ST2 pretreated with PC3-Exo and DU145-Exo, and then ST2-CM was used to treat PC3 cells. As shown in Figure 3(a), lncAY927529 was upregulated in ST2 cells treated with exosomes derived from PCa cell lines (VCaP, LNCaP, DU145 and PC3) compared with exosomes derived from normal prostate cell lines (BPH1 and RWPE1) and control groups (P < 0.05). In Figure 3(b), lncAY927529 was upregulated in PC3 cells treated with ST2-CM pretreated with PC3-Exo and DU145-Exo (P < 0.05). In addition, the results of CCK-8 and Transwell assays revealed that ST2-CM treated with exosomes secreted by PC3 and DU145 cell lines promoted proliferation and invasion of PC3 cells, and inhibited PC3 cell apoptosis (Figure 3(c-e)).
Figure 3.
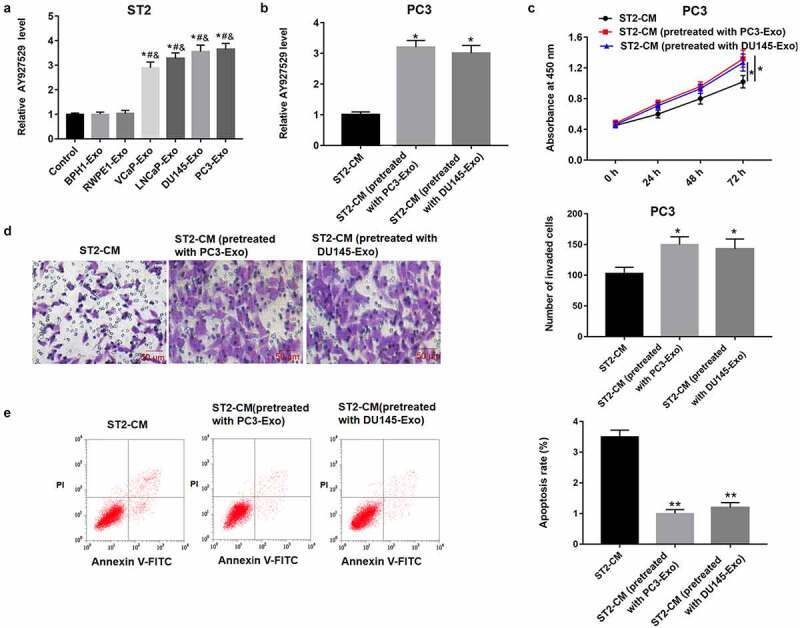
LncAY927529 level was detected in ST2 cells treated with exosomes derived from different PCa cell lines, and PC3 cell proliferation, invasion and apoptosis were measured treated with ST2-CM pretreated with PC3-Exo and DU145-Exo. (a) ST2 cells were treated with exosomes derived from normal prostatic epithelial cells (BPH1 and RWPE1) and PCa cell lines (VCap, LNCap, DU145 and PC3), and RT-qPCR assay was used to detect lncAY927529 level in ST2 cells. *P < 0.05 compared with control group; #P < 0.05 compared with BPH1-Exo group; &P < 0.05 compared with RWPE1-Exo group. (b) LncAY927529 level in PC3 cells was measured treated with ST2-CM pretreated with PC3-Exo and DU145-Exo, which was detected with RT-qPCR assay. *P < 0.05 compared with control conditioned medium of ST2 cells (ST2-CM) group. CCK-8 and Transwell assays were used to detect proliferation (c), invasion (d) and apoptosis (e) of PC3 cells treated with ST2-CM pretreated with PC3-Exo and DU145-Exo. *P < 0.05 or **P < 0.01 compared with ST2-CM group. N = 4, data were expressed as mean ± SEM; Student’s t test or one-way ANOVA was used for analyzing data
Construction of PCa cell lines with low or high expression of lncAY927529
LncAY927529 siRNA was transfected into PC3 and DU145 cells to knock down of lncAY927529, and pcDNA-lncAY927529 was transfected into PC3 and DU145 cells to over expression of lncAY927529. As shown in Figure 4(a,b), lncAY927529 level was downregulated in PC3 and DU145 cells transfected with lncAY927529 siRNA. And lncAY927529 level was also downregulated in exosomes derived from PC3 and DU145 cells with low expression of lncAY927529 (Figure 4(c,d)). LncAY927529 level was upregulated in PC3 and DU145 cells transfected with pcDNA-lncAY927529, and it also upregulated in exosomes derived from PC3 and DU145 cells with high expression of lncAY927529 (Figure 4(e-h)).
Figure 4.
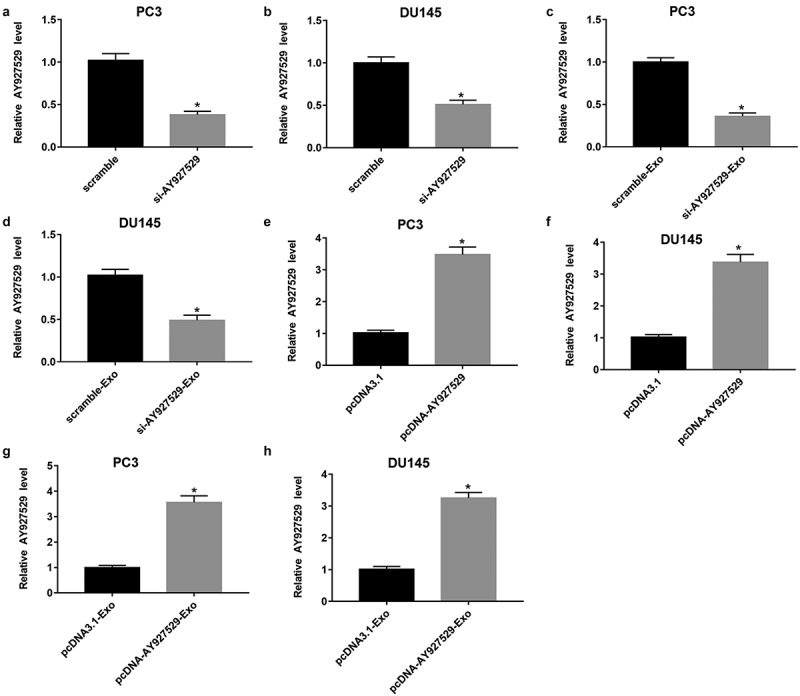
Effect of overexpression or knockdown of lncAY927529 on the level of lncAY927529 in PCa cells and exosomes derived from PCa cells. LncAY927529 siRNA, scramble (a negative control of lncAY927529 siRNA), pcDNA-lncAY927529 and pcDNA3.1 (a negative control of pcDNA-lncAY927529) were transfected into PC3 and DU145 cells: (a and b) LncAY927529 level was detected in PC3 and DU145 cells with RT-qPCR transfected with lncAY927529 siRNA or scramble. *P < 0.05 compared with scramble group. (c and d) LncAY927529 level was detected in PC3-Exo and DU145-Exo with RT-qPCR when PC3 and DU145 cells were transfected with lncAY927529 siRNA or scramble. *P < 0.05 compared with scramble-Exo group. (e and f) LncAY927529 level was detected in PC3 and DU145 cells with RT-qPCR transfected with pcDNA-lncAY927529 or pcDNA3.1. *P < 0.05 compared with pcDNA3.1 group. (g and h) LncAY927529 level was detected in PC3-Exo or DU145-Exo with RT-qPCR when PC3 and DU145 cells were transfected with pcDNA-lncAY927529 or pcDNA3.1. *P < 0.05 compared with pcDNA3.1-Exo group. N = 4, data were expressed as mean ± SEM; Student’s t test or one-way ANOVA was used for analyzing data
LncAY927529 in exosomes mediated the effect of bone stromal cells on cytokines in the supernatant of ST2 cells
As shown in Figure 5(a,c), the contents of IL-6, IL-8 and IL-10 were increased in ST2 cell supernatant when ST2 cells were treated with exosomes derived from DU145 cells with high expression of lncAY927529, and they were decreased in ST2 cell supernatant when ST2 cells were treated with exosomes derived from DU145 cells with low expression of lncAY927529.
Figure 5.
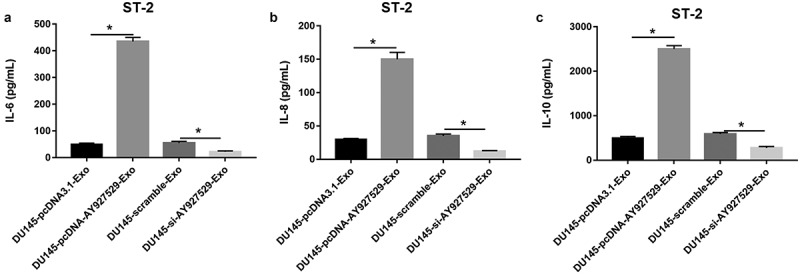
The contents of inflammatory factors were detected in ST2 cells. DU145 cells were transfected with lncAY927529 siRNA, scramble, pcDNA-lncAY927529 and pcDNA3.1: (a-c) ELISA assay was used to detect the contents of IL-6, IL-8 and IL-10 in ST2 cells treated with exosomes derived from DU145 cells with low and high expression of AY927529.*P < 0.05 compared with DU145-pcDNA3.1-Exo group or DU145-scramble-Exo. N = 4, data were expressed as mean ± SEM; Student’s t test or one-way ANOVA was used for analyzing data
LncAY927529 level was upregulated in ST2 cells treated with PC3 and DU145 cells with high expression of lncAY927529, and it was downregulated in ST2 cells treated with PC3 and DU145 cells with low expression of lncAY927529
To determine whether lncAY927529 was transferred into bone marrow stromal cells by exosomes secreted by cancer cells, PC3 and DU145 cells were transfected with pcDNA-lncAY927529 or lncAY927529 siRNA, and then ST2 cells were treated with exosomes derived from DU145 and PC3 cells. As shown in Figure 6(a,b), lncAY927529 level was upregulated in ST2 cells treated with exosomes derived from DU145 and PC3 cells with high expression of lncAY927529, and it was downregulated in ST2 cells treated with exosomes derived from DU145 and PC3 cells with low expression of lncAY927529. These results suggested that lncAY927529 was transferred into bone marrow stromal cells through exosomes derived from PCa cells.
Figure 6.

LncAY927529 level was upregulated in ST2 cells transfected with PC3-pcDNA-AY927529-Exo, and lncAY927529 level was downregulated in ST2 cells transfected with PC3-AY927529 siRNA-Exo. (a and b) RT-qPCR assay was used to detect lncAY927529 level when ST2 cells were treated with PC3-pcDNA-AY927529-Exo or PC3-scramble-Exo. *P < 0.05 compared with PC3-pcDNA3.1-Exo group, PC3-scramble-Exo group, DU145-pcDNA3.1-Exo group or DU145-scramble-Exo group. N = 4, data were expressed as mean ± SEM; Student’s t test or one-way ANOVA was used for analyzing data
ST-CM treated with exosomes derived from PC3 and DU145 cells with high lncAY927529 promoted cell proliferation and invasion and inhibited cell apoptosis
The results of CCK-8 and Transwell assays showed that ST2-CM treated with PC3-Exo and DU145-Exo with high lncAY927529 expression significantly promoted PC3 and DU145 cell proliferation (Figure 7(a,b)) and invasion (Figure 7(c,d)) (P < 0.05), and ST2-CM treated with PC3-Exo or DU145-Exo with low lncAY927529 level significantly suppressed PC3 or DU145 cell proliferation (Figure 7(a,b)) and invasion (Figure 7(c,d)) (P < 0.05). As shown in Figure 7(e,f), the results of Flow cytometry assay presented that ST2-CM treated with PC3-Exo or DU145-Exo with high lncAY927529 expression significantly inhibited PC3 cell apoptosis, and ST2-CM treated with PC3-Exo or DU145-Exo with low lncAY927529 expression significantly enhanced PC3 cell apoptosis (P < 0.01).
Figure 7.
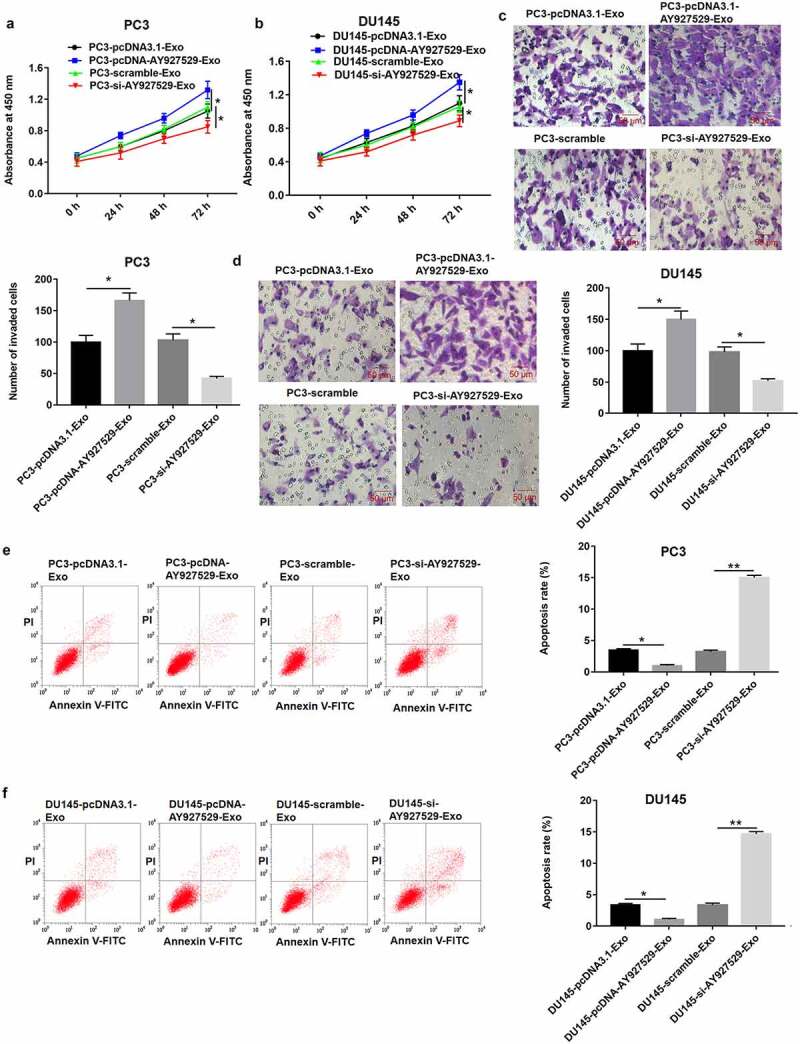
ST2-CM enhanced proliferation and invasion of PC3 and DU145 cells. ST2 cells were treated with exosomes derived from PC3 and DU145 with knockdown of AY927529 or overexpression of AY927529 for 48 h, ST2-CM was collected, and then PC3 and DU145 cells were treated with ST2-CM. CCK-8, transwell assay and flow cytometry assays were uesd to detect proliferation (a and b), invasion (c and d) and apoptosis (e and f) of PC3 and DU145 cells. *P < 0.05 compared with PC3-pcDNA3.1-Exo group, PC3-scramble-Exo group, DU145-pcDNA3.1-Exo group or DU145-scramble-Exo group; **P < 0.01 compared with PC3-scramble-Exo or DU145-scramble-Exo group. N = 4, data were expressed as mean ± SEM; Student’s t test or one-way ANOVA was used for analyzing data
CXCL14 acted as a RBP of lncAY927529
RPIseq (http://ctsb.is.wfubmc.edu/projects/rpi-pred/rpipred1.php) and Pull down assay were used to predict and verify the relation of lncAY927529 and CXCL14 (Figure 8(a,b)). As shown in Figure 8(c,d), CXCL14 protein level or ERK phosphorylated protein were upregulated in ST2 cells treated with DU145-Exo with high lncAY927529 expression and downregulated in ST2 cells treated with DU145-Exo with low lncAY927529 expression. CXCL14 mRNA level was upregulated in ST2-CM treated with exosomes derived from PC3 and DU145 cells with high expression of AY927529, it was downregulated treated with exosomes derived from PC3 and DU145 cells with low expression of AY927529 (Supplementary Figure 1).
Figure 8.
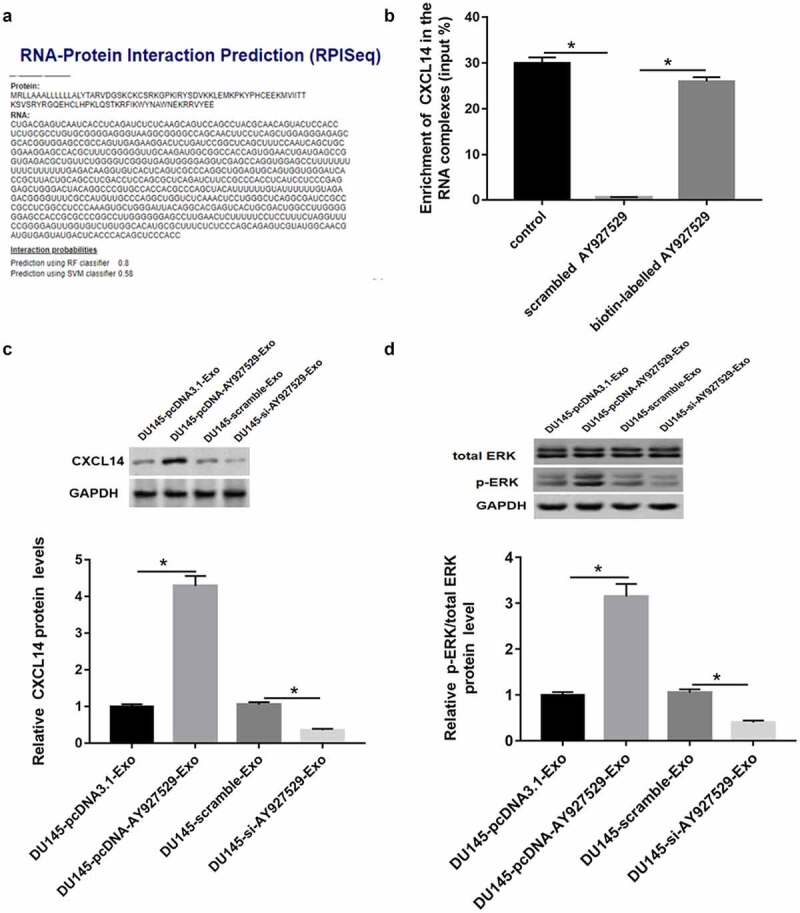
Prediction and verification the relationship between CXCL14 and lncAY927529. RPIseq (http://ctsb.is.wfubmc.edu/projects/rpi-pred/rpipred1.php) (a) and Pull down assay (b) were used to predict and verify the relation of lncAY927529 and CXCL14. *P < 0.05 compared with control or scramble AY927529 group. (c and d) Western blot assay was used to detected CXCL14 and ERK phosphorylated protein levels in ST2 cells treated with exosomes derived from DU145 cells with knockdown of AY927529 or overexpression of AY927529 for 48 h, and the conditioned medium (ST2-CM) was collected. *P < 0.05 compared with DU145-pcDNA3.1-Exo group or DU145-scramble-Exo group. N = 4, data were expressed as mean ± SEM; Student’s t test or one-way ANOVA was used for analyzing data
LncAY927529 in exosomes regulated bone microenvironment by mediating autophagy of bone stromal cells
As shown in Figure 9(a), LC3I protein level was downregulated in ST2 cells treated with DU145-Exo with high lncAY927529 level and was upregulated in ST2 cells treated with DU145-Exo with low lncAY927529 level. And LC3II protein level was upregulated in ST2 cells treated with DU145-Exo with high lncAY 927529 level and downregulated in ST2 cells treated with DU145-Exo with low lncAY927529 level. The autophagic vacuoles was increased in ST2 cells treated with DU145-Exo with high lncAY927529 level and decreased in ST2 cells treated with DU145-Exo with low lncAY927529 level (Figure 9(b)) (P < 0.05).
Figure 9.
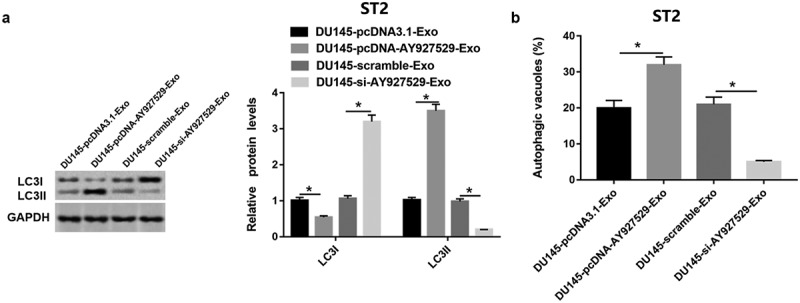
LncAY927529 in exosomes regulated bone microenvironment by mediating autophagy of bone stromal cells. (a) Western blot assay was uesd to detect LC3I and LC3II protein levels in ST2 cells treated with exosomes derived from DU145 with knockdown of AY927529 or overexpression of AY927529 for 48 h, and the conditioned medium (ST2-CM) was collected. (b) Autophagic vacuoles (%) was measured by immunofluorescence staining assay. *P < 0.05 compared with DU145-pcDNA3.1-Exo group or DU145-scramble-Exo group. N = 4, data were expressed as mean ± SEM; Student’s t test or one-way ANOVA was used for analyzing data
Autophagy mediated recycling maintained growth and proliferation of DU145 cells
As shown in Figure 10(a), we found that CXCL14 level was upregulated in DU145 cells treated with high expression of AY927529, and it was downregulated in cells treated with low expression of AY927529. Next, we detected the proliferation and invasion of DU145 cells. CCK-8 and Transwell assays showed that overexpression of CXCL14 enhanced cell proliferation and invasion, and knockdown of CXCL14 suppressed cell proliferation and invasion (Figure 10(b,c)).
Figure 10.
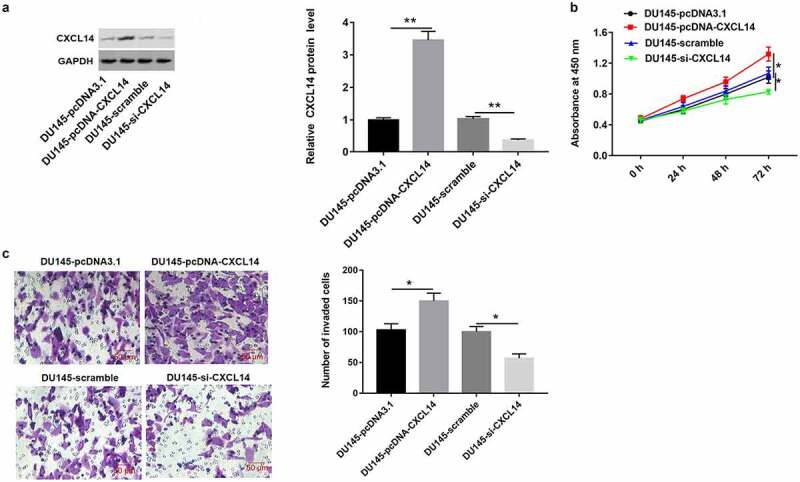
Overexpression of CXCL1 enhanced the proliferation and invasion of DU145 cells. DU145 cells were transfected with pcDNA-CXCL14, pcDNA3.1 (a negative control of pcDNA-CXCL14), CXCL14 siRNA or scramble (a negative control of CXCL14 siRNA): (a) Western blot assay was used to measure CXCL14 protein level in DU145 cells. (b and c) The proliferation and invasion of DU145 cells were detected with CCK-8 and transwell assays. *P < 0.05 compared with DU145-pcDNA3.1 group or DU145-scramble group. N = 4, data were expressed as mean ± SEM; Student’s t test or one-way ANOVA was used for analyzing data
Discussion
PCa cells PC3-derived exosomes and DU145-derived exosomes have been described previously [18,19], while whether or how these vesicles take part in osteogenesis remain to be elucidated. In this research, it was reported that PC3-derived exosomes or DU145-derived exosomes entered into BMSCs and enhanced cell proliferation and invasion, suggesting that PC3-derived exosomes or DU145-derived exosomes had the same function as their parent cells.
As a new lncRNA, lncAY927529 has not been largely reported in PCa cells and exosomes secreted by PCa cells. Therefore, our experiment focused on the role of lncAY92752 in the bone microenvironment of exosomes secreted by cancer cells. In our previous work, we collected the conditioned culture supernatant of mice treated with exosomes secreted by PCa cells. When ST2 cells were recruited into bone microenvironment, under the interaction and regulation of conditioned culture supernatant and peripheral cells, the growth of cancer cells was finally promoted.
Chemokines are a kind of small molecule secreted proteins with directional migration of chemotactic cells, which can cause the directional migration of lymphocytes and play an important role in many physiological and pathological processes such as embryonic development, inflammation, angiogenesis, tumor, AIDS and so on [20–22]. A study has shown that chemokines and their receptors were involved in the occurrence and metastasis of tumors [23], and the complex chemokine system in tumor tissues affected the growth of tumor cells. CXCL14 is a member of CXC chemokine family, and CXCL14 is a chemokine (C-X-C Motif) ligand 14 gene (also known as BRAK, BMAC or Mip-2γ), located on human chromosome 5q31. CXCL14 was initially identified from breast and kidney cells, and was also widely expressed in epithelial cells. Previous sequencing results showed that CXCL14 was upregulated in PCa tissues, which was consistent with our results [24,25]. However, CXCL14 plays a unique role in different tumors. It has been reported that it inhibits the growth of lung cancer and head and neck cancer cells in vivo, while CXCL14 promotes the invasiveness of breast and PCa cells. CXCL14 inhibits human papillomavirus (HPV) positive head and neck cancer by restoring the expression of MHC-I in tumor cells and promoting antigen-specific CD8+ T cell response [26]. CXCL14 is an important candidate gene downstream of Notch signaling pathway in inducing metastasis in breast cancer [27]. In this study, we also found that AY927529 was overexpressed in PCa cells, and the upregulation of CXCL14 protein levels in the bone microenvironment was due to tumor exosomes-mediated AY927529 regulating cell permeability to promote prostate cancer metastasis. Subsequently, it was found that upregulation of CXCL14 protein level activated the ERK signaling pathway in bone microenvironment and stimulated autophagy of bone marrow stromal cells, thus regulating bone microenvironment.
Conditioned media from PC3 cells or DU145 cells have previously been described to promote osteogenesis, which may be the result of osteoblasts secreting enzymes or growth factors [28,29]. Our results showed that lncAY927529 level was upregulated when exosomes derived from DU145 with overexpression of AY927529 for 48 h, and the conditioned medium (ST2-CM) was collected. In addition, we found that overexpression of AY927529 enhanced CXCL14 protein level in ST2 cells, and it activated ERK signal pathway through promoting phosphorylation of ERK protein level. Next, we found that CXCL14 protein level was also upregulated in PC3 cells when lncAY927529 promoted autophagy in ST2 cells through CXCL14 dependent pathway. Autophagy mediated recycling maintains cell growth and proliferation, and eventually led to the occurrence and development of PCa. Thus, there seems to be an exosome-mediated communication between stromal stem cells and PC3 cells in the local bone marrow microenvironment.
Autophagy is the basic catabolic process of cells in physiological and biological activities [30]. Under certain survival threatening conditions, such as when extracellular nutrients are limited, cells can digest their contents through lysosome mediated intracellular degradation pathway, a process called autophagy. Autophagy plays a variety of important roles, such as maintaining amino acid pool during starvation, renewing damaged proteins and organelles, preventing neurodegeneration, tumor inhibition, cell differentiation, clearance of intracellular microorganisms and regulation of innate and adaptive immunity [31–34]. Autophagy is also considered to be a dynamic process that can be regulated both positively and negatively in multiple steps [30]. Previously, some researchers reported that exosomes from cancer cells can induce autophagy to create a favorable microenvironment for tumor growth [35,36]. However, the relationship between exosomes and bone marrow stromal cells has not largely been reported in PCa. Our results showed that the level of light chain LC3I protein decreased, while LC3II protein level increased after ST2 cells were treated with exosomes secreted by DU145 cells overexpressing AY927529, which suggested that the autophagy process was activated. The light chain LC3I forms a coupled light chain LC3II when conjugated with phosphatidylethanolamine (PE), which is then recruited into the autophagy membrane. These results suggested that AY927529 in the exosomes secreted by PCa cells could activate autophagy of bone marrow mesenchymal stem cells, thus regulating bone microenvironment.
In conclusion, AY927529 in exosome can promote the occurrence and development of PCa by regulating bone microenvironment, suggesting that lncAY927529 in exosomes has the potential to become a molecular diagnostic marker of PCa.
Supplementary Material
Acknowledgments
We would like to acknowledge the reviewers for their helpful comments on this paper, and the project was supported by Key scientific research projects of Institutions in Henan province, China [No.: 18A320014].
Funding Statement
This work was supported by Key Scientific Research Projects of the Higher Education Institutions in Henan Province, China [No.: 18A320014].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Authors’ contributions
guarantor of integrity of the entire study:Qi Li
study concepts: Qi Li
study design: Qi Li, Jinhao Hu
experimental studies: Yibo Shi, Mulun Xiao, Tianxiang Bi,
statistical analysis: Yibo Shi, Chaoliang Wang, Liang Yan
manuscript editing: Qi Li, Xiaoyan Li
Supplementary material
Supplemental data for this article can be accessed here.
Ethical statements
Ethical Approval
This study was approved by the Ethics Committee at the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China).
Statement of Human and Animal Rights
All procedures in this study were conducted in accordance with the First Affiliated Hospital of Zhengzhou University of ethics committee’s or institutional review board’s (APPROVAL NUMBER: 0022) approved protocols.
References
- [1].Siegel RL, Miller KD, Jemal A.. Cancer statistics. CA Cancer J Clin. 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- [2].Nordström T, Akre O, Aly M, et al. Prostate-specific antigen (PSA) density in the diagnostic algorithm of prostate cancer. Prostate Cancer Prostatic Dis. 2018;21(1):57–63. [DOI] [PubMed] [Google Scholar]
- [3].Li HF, Xie Q, Nie QW, et al. Prostate specific antigen as a biomarker for breast cancer: a meta-analysis study. Eur Rev Med Pharmacol Sci. 2018;22(13):4188–4195. [DOI] [PubMed] [Google Scholar]
- [4].Komura K, Sweeney CJ, Inamoto T, et al. Current treatment strategies for advanced prostate cancer. Int J Urol. 2018;25(3):220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kang M, Ren M, Li Y, et al. Exosome-mediated transfer of lncRNA PART1 induces gefitinib resistance in esophageal squamous cell carcinoma via functioning as a competing endogenous RNA. J Exp Clin Cancer Res. 2018;37(1):171–187. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [6].Ai Y, Wei H, Wu S, et al. Exosomal lncRNA LBX1-AS1 derived from RBPJ overexpressed-macrophages inhibits oral squamous cell carcinoma progress via miR-182-5p/FOXO3. Front Oncol. 2021;11:605884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88(1):487–514. [DOI] [PubMed] [Google Scholar]
- [8].Meldolesi J. Exosomes and ectosomes in intercellular communication. Curr Biol. 2018;28(8):R435–r444. [DOI] [PubMed] [Google Scholar]
- [9].Whiteside TL. Tumor-derived exosomes and their role in cancer progression. Adv Clin Chem. 2016;74:103–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Li K, Chen Y, Li A, et al. Exosomes play roles in sequential processes of tumor metastasis. Int J Cancer. 2019;144(7):1486–1495. [DOI] [PubMed] [Google Scholar]
- [11].Lindenbergh MFS, Stoorvogel W. Antigen Presentation by extracellular vesicles from professional antigen-presenting cells. Annu Rev Immunol. 2018;36(1):435–459. [DOI] [PubMed] [Google Scholar]
- [12].Zhao T, Sun F, Liu J, et al. Emerging role of mesenchymal stem cell-derived exosomes in regenerative medicine. Curr Stem Cell Res Ther. 2019;14(6):482–494. [DOI] [PubMed] [Google Scholar]
- [13].Lin J, Li J, Huang B, et al. Exosomes: novel biomarkers for clinical diagnosis. The Scientific World Journal. 2015;2015:657086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ahadi A, Brennan S, Kennedy PJ, et al. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Sci Rep. 2016;6(1):24922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Greening DW, Xu R, Ji H, et al. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol Biol. 2015;1295:179–209. [DOI] [PubMed] [Google Scholar]
- [17].Bobrie A, Colombo M, Krumeich S, et al. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J Extracell Vesicles. 2012;1(1):18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hasegawa T, Glavich GJ, Pahuski M, et al. Characterization and evidence of the miR-888 cluster as a novel cancer network in prostate. Mol Cancer Res. 2018;16(4):669–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Muñoz-Moreno L, Carmena MJ, Schally AV, et al. Stimulation of neuroendocrine differentiation in prostate cancer cells by GHRH and its blockade by GHRH antagonists. Invest New Drugs. 2020;38(3):746–754. [DOI] [PubMed] [Google Scholar]
- [20].Vilgelm AE, Richmond A. Chemokines modulate immune surveillance in tumorigenesis, metastasis, and response to immunotherapy. Front Immunol. 2019;10:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dimberg A. Chemokines in Angiogenesis . Berlin Heidelberg: Springer; 2010. ;341:59-80. [DOI] [PubMed] [Google Scholar]
- [22].Animesh C, Anurag R, Sanjukta V, et al. Chemokines and chemokine receptors in susceptibility to HIV-1 infection and progression to AIDS. Dis Markers. 2012;32(3):143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pellegrino A, Vacca A, Scavelli C, et al. Chemokines and tumors. Recenti Prog Med. 2002;93(11):642. [PubMed] [Google Scholar]
- [24].Gerashchenko GV, Grygoruk OV, Kononenko OA, et al. Expression pattern of genes associated with tumor microenvironment in prostate cancer. Exp Oncol. 2018;40(4):315–322. [PubMed] [Google Scholar]
- [25].Williams KA, Lee M, Hu Y, et al. A systems genetics approach identifies CXCL14, ITGAX, and LPCAT2 as novel aggressive prostate cancer susceptibility genes. PLoS Genet. 2014;10(11):e1004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Westrich JA, Vermeer DW, Silva A, et al. CXCL14 suppresses human papillomavirus-associated head and neck cancer through antigen-specific CD8+ T-cell responses by upregulating MHC-I expression. Oncogene. 2019;38(2):7166–7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].KÜÇÜKKÖSE C, YALÇIN ÖZUYSAL Ö. Effects of notch signalling on the expression of SEMA3C, HMGA2, CXCL14, CXCR7, and CCL20 in breast cancer. Turk J Biol. 2018;43(1):70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Duan Y, Tan Z, Yang M, et al. PC-3-derived exosomes inhibit osteoclast differentiation by downregulating miR-214 and blocking NF-kappaB signaling pathway. Biomed Res Int. 2019;2019:8650846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Borel M, Lollo G, Magne D, et al. Prostate cancer-derived exosomes promote osteoblast differentiation and activity through phospholipase D2. Biochim Biophys Acta Mol Basis Dis. 2020;1866(12):165919. [DOI] [PubMed] [Google Scholar]
- [30].Ravanan P, Srikumar IF, Talwar P. Autophagy: the spotlight for cellular stress responses. Life Sci. 2017;188:53–67. [DOI] [PubMed] [Google Scholar]
- [31].Menzies FM, Fleming A, Caricasole A, et al. Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron. 2017;93(5):1015–1034. [DOI] [PubMed] [Google Scholar]
- [32].Kuo CJ, Hansen M, Troemel E. Autophagy and innate immunity: insights from invertebrate model organisms. Autophagy. 2018;14(2):233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Deretic V, Levine B. Autophagy balances inflammation in innate immunity. Autophagy. 2018;14(2):243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Luo Z, Xu X, Sho T, et al. ROS-induced autophagy regulates porcine trophectoderm cell apoptosis, proliferation, and differentiation. Am J Physiol Cell Physiol. 2019;316(2):C198–c209. [DOI] [PubMed] [Google Scholar]
- [35].Milane L, Singh A, Mattheolabakis G, et al. Exosome mediated communication within the tumor microenvironment. J Control Release. 2015;219:278–294. [DOI] [PubMed] [Google Scholar]
- [36].Zhang X, Shi H, Yuan X, et al. Tumor-derived exosomes induce N2 polarization of neutrophils to promote gastric cancer cell migration. Mol Cancer. 2018;17(1):146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


