Abstract
The aim of this study was to standardize a method to determine whether two strains of Cryptococcus neoformans could be considered epidemiologically linked. We hypothesized that strains isolated from the same patient were epidemiologically linked and that those isolated from different patients were unrelated. We used 17 environmental isolates and 97 clinical isolates from 31 patients diagnosed with cryptococcosis (1 to 14 isolates per patient). Using the plasmid pCnTel-1-labeled probe CENTEL, we were able to differentiate some unrelated strains that yielded the same hybridization profile with the C. neoformans middle-repetitive-element CNRE-1 probe. The genetic distances separating the strains isolated from the same patient and those separating the strains isolated from different patients were estimated, and the results obtained with the two probes were compared. Analysis of the results enabled the calculation of two Dice coefficient limits defining the zones containing the pairs of linked strains and the pairs of unrelated strains, as well as an intermediate uncertainty zone for which it was not possible to establish whether the pairs of strains were linked.
Cryptococcus neoformans is an encapsulated yeast that can cause life-threatening meningitis in immunocompromised patients (16). This encapsulated basidiomycete exists in three varieties: C. neoformans var. grubii (serotype A) (7) and C. neoformans var. neoformans (serotype D), both with worldwide distributions, and C. neoformans var. gattii (serotypes B and C), which is limited to tropical and subtropical regions (14). After the classical biotyping methods, this serotype classification was the first tool developed to study the epidemiology of cryptococcosis. The recent availability of DNA fingerprinting techniques (see reference 3 for a review) has greatly extended our knowledge of C. neoformans epidemiology over the last few years. Indeed, randomly amplified polymorphic DNA, restriction fragment length polymorphism (RFLP), karyotyping, and multilocus enzyme electrophoresis have helped answer several important questions. For example, it has been shown that patients are usually infected by a single strain, and that strain is responsible for the recurrent episodes of the infection (2, 20). Using independent molecular tools, we were recently also able to demonstrate the existence of a dormant form of crytococcosis (9). However, it was still impossible to determine with certainty whether two strains were epidemiologically linked. In our previous study, the CNRE-1 probe, which had the highest discriminatory power to date, was occasionally unable to differentiate between two strains that were obviously unrelated since they had been isolated in two different countries (9). On the other hand, using the same probe, we found that two strains isolated from the same patient sometimes generated slightly different hybridization patterns, probably due to the microevolution of strains during the infection. Thus, in some instances, two unrelated strains seemed to be genetically closer than two linked strains. This paradox was due to the poor characterization of the performances of the methods. Indeed, a common problem encountered with typing techniques used for epidemiological studies of mycoses is difficulty in distinguishing the microevolution of a strain from differences between two unrelated strains (18). We compared the abilities of CNRE-1 (19) and a new probe named CENTEL to establish whether two C. neoformans strains are linked. To assess the performances of the probes, we calculated the sensitivity and specificity for different Dice coefficient cutoff values and used a statistical method usually applied to the evaluation of diagnostic tools (receiver operating characteristic [ROC] analysis) (12). Finally, well-characterized clinical and environmental isolates were used to estimate the smallest genotypic differences separating two unrelated strains and the largest genotypic differences separating two linked strains.
MATERIALS AND METHODS
Patients and strains.
The 17 environmental isolates from different countries and the 97 clinical isolates of C. neoformans var. grubii recovered from 31 patients used in this study were described previously (9). All strains were stored frozen in 40% glycerol at −80°C and grown in YPD medium (10 g of yeast extract, 20 g of Bacto Peptone, and 20 g of glucose per liter) at 30°C.
RFLP analysis.
Genomic DNA was prepared as described elsewhere (23). The UT-4p probe has previously been shown to be a valuable molecular tool to study the epidemiology of C. neoformans (4, 10, 22). It was obtained by labeling a linear telomeric plasmid directly isolated from C. neoformans cells. In 1992, Edman described an Escherichia coli/C. neoformans shuttle plasmid containing the same telomeric repeat sequence and the same URA5 gene (5). This plasmid, named pCnTel-1 (generously provided by B. Wickes [San Antonio, Texa]), can be easily purified from E. coli transformant colonies by using a Qiagen (Hilden, Germany) plasmid kit, and we used it to label a probe that we named CENTEL. DNA from the CNRE-1 phage, a generous gift from E. Spitzer and S. Spitzer (Stony Brook, N.Y.) (19), was purified by using a Qiagen lambda kit. All isolates were typed by Southern blot analysis after labeling of the probes with digoxigenin –11-dUTP by using a DIG-High Prime kit (Boehringer Mannheim, Mannheim, Germany). Genomic DNA was digested with AccI for hybridization with CENTEL or SstI for hybridization with CNRE-1. The resulting fragments were then electrophoretically separated through a 0.8% agarose gel and transferred onto positively charged nylon membranes (Boehringer Mannheim). After overnight hybridization at 65°C (CENTEL) or 68°C (CNRE-1) and washes, bands were visualized according to the manufacturer's instructions.
Statistical analysis.
DNA fingerprint patterns were analyzed, as previously described (9), using Taxotron software (11), which compares two profiles by calculating the Dice coefficient complement (number of different bands/total number of fragments in the two profiles). As had many authors in the past (e.g., et al. [20] and Sullivan et al. [21]), we considered two strains isolated from the same patient to be genetically linked derivaties of a unique infectious strain and two strains isolated from two different patients to be unrelated. The strains recovered from different patients were isolated from unrelated individuals living in different towns in France and are unlikely to be the same, whereas reinfection by a new strain is clearly the exception (21).
To assess the capacities of the probes to determine linkages, we calculated the sensitivity and specificity for various cutoff values of the genetic distances measured by the Dice coefficient. For the sensitivity and specificity calculations, a true positive or true negative was considered to be a pair of unrelated or linked strains, respectively, within a given range of Dice coefficients. The following formulas were used: sensitivity = true positives/(true positives + false negatives), and specificity = true negatives/(true negatives + false positives). The ROC curves were then established by plotting, for various cuttoff-value ranges of Dice coefficients, the sensitivity against the value of 1 − specificity. We calculated the areas under the curves (X) to estimate the global performance of the method (12). The Dice coefficient cutoff values (Y and Z, delineating an uncertainty zone) giving X ≥ 0.99 were defined. Finally, the discriminatory power of each probe was calculated using Hunter's formula (13).
RESULTS
Study of environmental isolates.
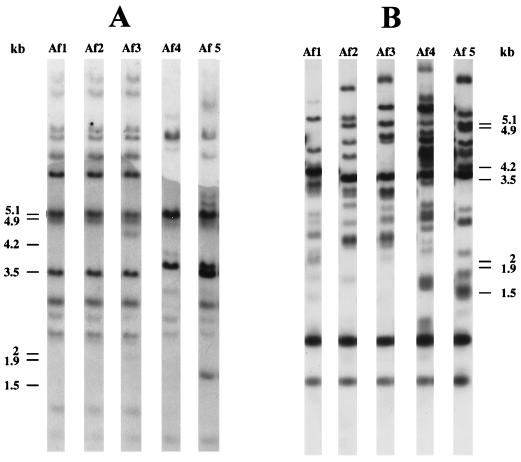
Using CENTEL to type 17 C. neoformans var. grubii strains isolated from the environment, 17 different profiles were obtained, whereas CNRE-1 generated only 15 different profiles (9). As shown in Fig. 1, strain Af2 (Togo) and strain Af1 (Morocco) had the same hybridization pattern when CNRE-1 was employed, whereas their profiles differed when CENTEL was used.
FIG. 1.
Southern blot hybridization patterns of five environmental C. neoformans isolates generated with CNRE-1 (A) or CENTEL (B). A 1-kb ladder (GibcoBRL) was used as a molecular size marker; positions of its components are indicated to the left (A) or to the right (B) of the gels.
Study of clinical isolates.
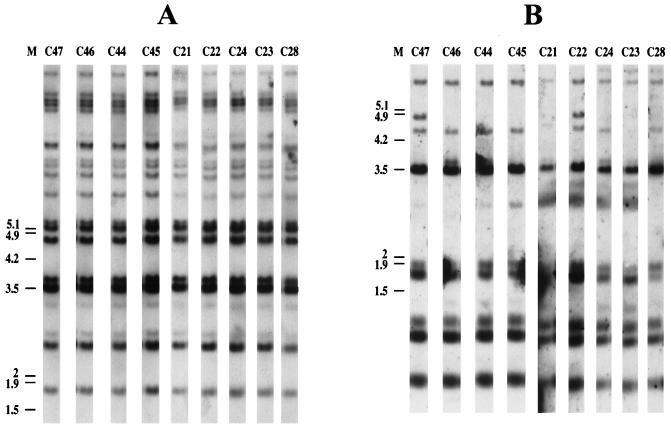
Ninety-seven clinical strains isolated from 31 different patients used in our previous study (9) were then tested with both probes; profiles specific to each patient were obtained. Sometimes hybridization patterns obtained with serial isolates from a particular patient showed some heterogeneity (Table 1). For example, all nine isolates collected from patient P6 yielded the same hybridization profile when CNRE-1 was used, but as many as seven different patterns could be distinguished with CENTEL (Fig. 2). No clear-cut relationship between the organ from which the strain had been recovered and the corresponding profiles could be established.
TABLE 1.
Numbers of profiles obtained using the CENTEL and CNRE-1 probes with isolates from 31 patients with cryptococcosis
| Patient no. | No. of isolates | No. of profiles obtained using:
|
|
|---|---|---|---|
| CENTEL | CNRE-1 | ||
| P1 | 8 | 1 | 1 |
| P2 | 6 | 1 | —a |
| P3 | 11 | 3 | 2 |
| P4 | 3 | 2 | 1 |
| P5 | 1 | 1 | 1 |
| P6 | 9 | 7 | 1 |
| P7 | 2 | 1 | 1 |
| P8 | 7 | 3 | 2 |
| P9 | 10 | 9 | 3 |
| P10 | 2 | 1 | — |
| P11 | 2 | 1 | 1 |
| P12 | 1 | 1 | 1 |
| P13 | 3 | 2 | 1 |
| P14 | 2 | 2 | 1 |
| P15 | 1 | 1 | 1 |
| P16 | 1 | 1 | 1 |
| P17 | 1 | 1 | 1 |
| P18 | 5 | 1 | 1 |
| P19 | 3 | 1 | 1 |
| P20 | 2 | 1 | 1 |
| P21 | 1 | 1 | 1 |
| P22 | 1 | 1 | 1 |
| P23 | 2 | 1 | 1 |
| P24 | 3 | 2 | 2 |
| P25 | 1 | 1 | 1 |
| P26 | 1 | 1 | 1 |
| P27 | 1 | 1 | 1 |
| P28 | 1 | 1 | 1 |
| P29 | 4 | — | 1 |
| P30 | 1 | — | 1 |
| P31 | 1 | — | 1 |
| Total | 97 | 50 | 34 |
—, results could not be interpreted for technical reasons.
FIG. 2.
Southern blot hybridization patterns of nine serial clinical C. neoformans strains isolated from patient 6, generated with CNRE-1 (A) and CENTEL (B). A 1-kb ladder (GibcoBRL) was used as a molecular size marker (M); positions of its components are indicated to the left of the gels (in kilobases).
Stability and reproducibility of the hybridization profiles.
It has been previously shown that, after subcloning of isolates, CNRE-1 was able to generate reproducible and stable hybridization patterns (19, 20). We tested the reproducibility of CENTEL profiles. Although the hybridization patterns were highly reproducible from one DNA preparation to another (data not shown), when 20 subcloned colonies from the same plate were tested simultaneously, one exhibited microevolution of the CENTEL pattern (data not shown).
Genetic distances between pairs of linked or unrelated strains.
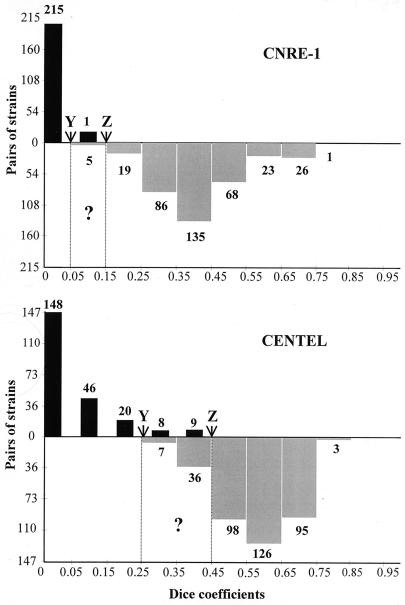
For example, the first strain isolated, from patient P1 was compared with every other strain isolated from the same patient, yielding seven pairs of related strains. The same strain was compared with the 89 strains isolated from the other patients, generating 89 pairs of unrelated strains. Thus, for CNRE-1 and CENTEL, 363 and 365 pairs of unrelated strains and 234 and 231 pairs of linked strains could be established, respectively. The small difference in strain pair numbers between the two methods was due to the exclusion from the final analysis of a few profiles because of poor-quality hybridizations. Then, using the Dice coefficient complement, we calculated the genetic distances separating the two members of every pair of epidemiologically linked strains and every pair of epidemiologically unrelated strains. The results obtained with CNRE-1 and CENTEL were compared (Fig. 3), and the discriminatory power of each probe was calculated (D = 1 and D = 0.9, respectively). Using the mean Dice coefficients to evaluate the performances of CNRE-1 and CENTEL, respectively, we found genetic distances of 0.06 and 0.11 between the linked strains and 0.46 and 0.62 for the unrelated strains. Considering the two probes, the lowest Dice coefficients separating two unrelated strains (0.349 with CENTEL and 0.049 with CNRE-1) were lower than the highest Dice coefficients separating two epidemiologically linked strains (0.449 with CENTEL and 0.149 with CNRE-1). We determined two Dice coefficient cutoff values (Y and Z) delineating the zones containing the pairs of strains that had at least a 99% chance of being genetically linked (0 to Y) or unrelated (Z to 1), as well as an intermediate uncertainty zone (Y to Z). Using CENTEL, a pair of strains with a Dice coefficient below 0.250 or above 0.449 had a 99.4% (X ≥ 0.994) chance of being genetically linked or unrelated, respectively. For CNRE-1, a pair of strains with a Dice coefficient below 0.049 or above 0.149 had a 99.9% chance (X ≥ 0.999) of being epidemiologically linked or unrelated, respectively. The uncertainty zone was wider for CENTEL (0.250 to 0.449) than for CNRE-1 (0.050 to 0.149).
FIG. 3.
Schematic representation of the genetic distances between pairs of strains recovered from the same patient (linked) (black bars), or from different patients (unrelated) (gray bars) determined with CNRE-1 or CENTEL. Three zones were established: 0 to Y, in which a pair of strains could be considered epidemiologically linked; Z to 1, in which two strains could be considered epidemiologically unrelated; and a zone of uncertainty (Y to Z). The number of pairs of strains for each of the Dice coefficient values is indicated above the corresponding column.
DISCUSSION
UT-4p is one of the previously described tools for C. neoformans epidemiological studies (4, 10, 22). With this probe, the differences seen between hybridization profiles of strains are probably due to telomeric- or subtelomeric-region length heterogeneity (15). We developed a similar tool using the telomere-containing sequence plasmid pCnTel-1 (5) as a substrate to label a probe we named CENTEL.
We showed that CENTEL profiles were reproducible (for 39 isolates typed at least twice independently) when the DNA was extracted from cultures inoculated with cell en masse, but not with a single colony, since subcloning led in some cases to microevolution of the hybridization profiles. Therefore, every strain had to be stored in 40% glycerol at −80°C and regrown without subcloning. This method of culture conservation had been already applied to Candida albicans and C. neoformans because of their natural phenotypic and genotypic variabilities (6, 17).
Our main objective was to estimate the shortest genetic distance separating two epidemiologically unrelated strains. This information had rarely been sought, since in most cases the authors tended to arbitrarily decide the minimal numbers of different bands separating two unrelated strains. Two strains yielding very similar patterns with one typing method are commonly considered to be epidemiologically linked, but the degree of similarity that should be observed between two linked strains has never been assessed or standardized.
Our working hypothesis defined two strains isolated from the same patient as being epidemiologically linked (2, 20). Two strains isolated from two different patients were considered to be epidemiologically unrelated, since transmission of cryptococcal strains between patients has never been suggested in the literature and the infectious strain seems to be acquired very early in life (1, 9).
Based on the numbers of patterns obtained with the environmental isolates using CENTEL (17 for 17 specimens) and CNRE-1 (15 for 17 specimens) and employing Hunter's formula (15), CENTEL could be classified as having the higher discriminatory power. However, when looking at the results obtained with clinical isolates, one could still wonder which tool was indeed the best since visual differences between profiles from serial (linked) isolates were sometimes larger than those between profiles obtained from isolates recovered from different patients (unrelated). Thus, to calculates the genetic distances between strains, the Dice coefficients for CNRE-1 and CENTEL were determined for each pair of clinical strains (363 and 365 pairs of unrelated and 231 and 234 pairs of linked strains, respectively). We first noticed that with both probes, the mean distance between two strains was shorter for linked than for unrelated pairs of strains, thus indicating a patient-by-patient clustering of isolates. We delineated two zones of certainty, 0 to Y and Z to 1, that differed first by cutoff values and width, depending on the true linkage or true lack of linkage, and second by the probe used (CENTEL shifted to the right). Similarly, the uncertainty zone (Y to Z) was wider for CENTEL than for CNRE-1.
Based on these findings, it could have been concluded that CENTEL would be inappropriate for the epidemiological study of cryptococcosis, but based on the calculation of the classical discriminatory power (i.e., the ability to discriminate unrelated isolates), CENTEL was more powerful than CNRE-1. This discrepancy clearly shows that one typing method alone cannot unequivocally answer all questions. In fact, none of the unrelated strains shared the same profile using CENTEL, whereas this was not true for CNRE-1, as shown with the African environmental isolates, thereby limiting the usefulness of the latter probe for the demonstration of linkage between two strains. Profile differences between linked isolates were rarely seen with CNRE-1, making it a good tool to affirm that two strains were unrelated when the profiles differed (with a Dice coefficient of >0.15 in our hands). In contrast, profile differences between linked strains were better seen with CENTEL, thereby making it a more appropriate tool for the study of what has been considered to date as microevolution during infection (8, 21).
In conclusion, every time the epidemiology of a microorganism is addressed, the epidemiological tool has to be evaluated using an adapted population of isolates for which maximum information is available. Thus, we think that the 97 clinical isolates used in the previous and present studies represent a suitable population for the assessment of a molecular typing method to study cryptococcosis epidemiology.
ACKNOWLEDGMENTS
We thank A. Casadevall (Albert Einstein College of Medicine, Bronx, N.Y.), J. M. Clauson (Western Kentucky University, Bowling Green), S. Kohno (Nagasaki University School of Medicine, Nagasaki, Japan), and D. Swinne (Institute of Tropical Medicine, Antwerp Belgium) for supplying the environmental isolates used in this study. We are grateful to E. Spitzer and S. Spitzer (State University of New York at Stony Brook) for the generous gift of the CNRE-1 probe and B. Wickes (University of Texas Health Science Center at San Antonio for providing the plasmid pCnTel-1. We also thank Olivier Ronin for serotyping the cryptococcal isolates and J. Jacobson for editorial assistance. We are grateful for the collaboration of the French Cryptococcosis Study Group, which includes clinicians and microbiologists from various hospitals in France. Those who participated in collecting strains used in this specific study are listed here (in alphabetical order by city): M. E. Bougnoux, S. Morelon, and E. Rouveix (Boulogne-Billancourt); C. Passa Gaudouen, B. Michel, G. Otterbein, and J. Roucoules (Bry-sur-Marne); J. M. Korach (Châlon-en-Champagne); B. Salles and C. Sire (Chalon/Saône); M. Gauthier and O. Salmon (Evry); F. Botterel and J. F. Delfraissy (Le Kremlin-Bicêtre); M. A. Desailly and H. Maisonneuve (La Roche/Yon); D. Bouhour, E. Dannaoui, D. Peyramond, and M. A. Piens (Lyon); O. Morin and P. Poirier (Nantes); M. Gari Toussaint and P. Dellamonica (Nice); B. Hery and J. Y. Leberre (St. Nazaire); M. F. Biava and C. Rabaud (Vandoeuvre-les-Nancy); and C. Fontier and E. Mazards (Valenciennes); and, in Paris, C. Chochillon, X. Duval, and J. L. Vildé (Hôpital Bichat); M. Kazatchkine, V. Lavarde, and C. Piketty (Hôpital Broussais); B. Dupont (Institut Pasteur); L. Baril, F. Bricaire, J. Carrière, A. Datry, S. Herson, and C. Trivalle (Hôpital Pitié-Salpêtrière); G. Delzant and G. Kac (Hôpital Tenon); J. Gilquin (Hôpital St.-Joseph); and D. Toubas (Reims).
Financial support for this work was provided by SIDACTION and the Pasteur Institute (Contrat de Recherche Clinique).
REFERENCES
- 1.Abadi J, Pirofski L. Antibodies reactive with the cryptococcal polysaccharide glucuronoxylomannan are present in sera from children with and without human immunodeficiency virus infection. J Infect Dis. 1999;180:915–919. doi: 10.1086/314953. [DOI] [PubMed] [Google Scholar]
- 2.Brandt M E, Hutwagner L C, Klug L A, Baughman W S, Rimland D, Graviss E A, Hamill R J, Thomas C, Pappas P G, Reingold A L, Pinner R W the Cryptococcal Disease Active Surveillance Group. Molecular subtype distribution of Cryptococcus neoformans in four areas of the United States. J Clin Microbiol. 1996;34:912–917. doi: 10.1128/jcm.34.4.912-917.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadevall A, Perfect J R. Cryptococcus neoformans. Washington, D.C.: ASM Press; 1998. [Google Scholar]
- 4.Dromer F, Varma A, Ronin O, Mathoulin S, Dupont B. Molecular typing of Cryptococcus neoformans serotype D clinical isolates. J Clin Microbiol. 1994;32:2364–2371. doi: 10.1128/jcm.32.10.2364-2371.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edman J C. Isolation of telomerelike sequences from Cryptococcus neoformans and their use in high-efficiency transformation. Mol Cell Biol. 1992;12:2777–2783. doi: 10.1128/mcb.12.6.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franzot S P, Mukherjee J, Cherniak R, Chen L-C, Hamdan J S, Casadevall A. Microevolution of a standard strain of Cryptococcus neoformans resulting in differences in virulence and other phenotypes. Infect Immun. 1998;66:89–97. doi: 10.1128/iai.66.1.89-97.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franzot S P, Salkin I F, Casadevall A. Cryptococcus neoformans var. grubii: separate varietal status for Cryptococcus neoformans serotype A isolates. J Clin Microbiol. 1999;37:838–840. doi: 10.1128/jcm.37.3.838-840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fries B, Casadevall A. Serial isolates of Cryptococcus neoformans from patients with AIDS differ in virulence for mice. J Infect Dis. 1998;178:1761–1766. doi: 10.1086/314521. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Hermoso D, Janbon G, Dromer F. Epidemiological evidence for dormant Cryptococcus neoformans infection. J Clin Microbiol. 1999;37:3204–3209. doi: 10.1128/jcm.37.10.3204-3209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Hermoso D, Mathoulin-Pélissier S, Couprie B, Ronin O, Dupont B, Dromer F. DNA typing suggests pigeon droppings as a source of pathogenic Cryptococcus neoformans serotype D. J Clin Microbiol. 1997;35:2683–2685. doi: 10.1128/jcm.35.10.2683-2685.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimont P A D. Taxotron's user's manual. Paris, France: Institut Pasteur; 1998. [Google Scholar]
- 12.Hanley J A, McNeil B J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 13.Hunter P R. Reproducibility and indices of discriminatory power of microbial typing methods. J Clin Microbiol. 1990;28:1903–1905. doi: 10.1128/jcm.28.9.1903-1905.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon-Chung K J, Bennett J E. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am J Epidemiol. 1984;120:123–130. doi: 10.1093/oxfordjournals.aje.a113861. [DOI] [PubMed] [Google Scholar]
- 15.Louis E J. The chromosome ends of Saccharomyces cerevisiae. Yeast. 1995;11:1553–1573. doi: 10.1002/yea.320111604. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell T G, Perfect J R. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rustchenko-Bulgac E P. Variations of Candida albicans electrophoretic karyotypes. J Bacteriol. 1991;173:6586–6596. doi: 10.1128/jb.173.20.6586-6596.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soll D R. The ins and outs of DNA fingerprinting the infectious fungi. Clin Microbiol Rev. 2000;13:332–370. doi: 10.1128/cmr.13.2.332-370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spitzer E D, Spitzer S G. Use of a dispersed repetitive DNA element to distinguish clinical isolates of Cryptococcus neoformans. J Clin Microbiol. 1992;30:1094–1097. doi: 10.1128/jcm.30.5.1094-1097.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spitzer E D, Spitzer S G, Freundlich L F, Casadevall A. Persistance of initial infection in recurrent Cryptococcus neoformans meningitis. Lancet. 1993;341:595–596. doi: 10.1016/0140-6736(93)90354-j. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan D, Haynes K, Moran G, Shanley D, Coleman D. Persistance, replacement, and microevolution of Cryptococcus neoformans strains in recurrent meningitis in AIDS patients. J Clin Microbiol. 1996;34:1739–1744. doi: 10.1128/jcm.34.7.1739-1744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varma A, Kwon-Chung K J. DNA probe for strain typing of Cryptococcus neoformans. J Clin Microbiol. 1992;30:2960–2967. doi: 10.1128/jcm.30.11.2960-2967.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varma A, Kwon-Chung K J. Rapid method to extract DNA from Cryptococcus neoformans. J Clin Microbiol. 1991;29:810–812. doi: 10.1128/jcm.29.4.810-812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]