Abstract
Accumulating evidence has shown that long intergenic non-protein-coding RNA 346 (LINC00346) functions as an oncogene in the tumorigenesis of several cancers. The expression level of LINC00346 has been shown to be obviously correlated with prognosis, lymphoma metastasis, histological grade, TNM stage, tumor size and pathologic stage. LINC00346 has been found to regulate specific cellular functions by interacting with several molecules and signaling pathways. In this review, we summarize recent evidence concerning the role of LINC00346 in the occurrence and development of diseases. We also discuss the potential clinical utility of LINC00346, thereby providing new insight into the diagnosis and treatment of diseases. In addition, we further discuss the potential clinical utility of LINC00346 in the diagnosis, prognostication, and treatment of diseases.
Keywords: lncRNA, LINC00346, biological function, mechanism, clinical utility
Introduction
Human genome sequencing data have revealed that less than 2% of the human genome contains protein-coding genes, and the vast majority of genes give rise to noncoding RNAs (ncRNAs) (Li et al., 2018; He et al., 2019; Wei et al., 2019). ncRNAs, originally considered transcriptional noise (Yang X. et al., 2020; Wu et al., 2020), are considered essential regulators of gene expression; for example, they regulate transcription, mRNA stability, and mRNA translation (Fabian et al., 2010; Chen and Huang, 2018; Panni et al., 2020). There are multiple types of ncRNAs, such as microRNAs (miRNAs), small interfering RNAs (siRNAs), long noncoding RNAs (lncRNAs), and circular RNAs (Luo et al., 2017; Liu X. et al., 2019).
LncRNAs, typically longer than 200 nucleotides in length (Xu H. et al., 2020), lack an open reading frame of significant length (Liu X. et al., 2018; Li Y. et al., 2019; Parolia et al., 2019). Emerging evidence suggests that lncRNAs are key regulators of gene expression levels, posttranscriptional modifications, and binding to transcription factors or miRNAs (Zhao et al., 2019; Guo et al., 2021). LncRNAs are abnormally expressed in a plethora of diseases (Liu S. et al., 2019; Huang L. et al., 2019; Chen et al., 2019; Shen et al., 2019; Xing et al., 2019; Radhakrishnan and Kowluru, 2021). LncRNAs are correlated with a variety of clinical characteristics and are considered indispensable regulators of many cell activities, including cell proliferation, migration, invasion, and apoptosis (Yang et al., 2019; Li B. et al., 2021; Lei et al., 2021). LncRNAs play important roles in the occurrence and development of human diseases. These ideas provide new perspectives on the diagnosis and treatment of human diseases (Li L. et al., 2021; Ma et al., 2021; Morgan et al., 2021). Long intergenic non-protein-coding RNA 346 (LINC00346), a novel lncRNA, is encoded on chromosome 13q34. Several studies have revealed that LINC00346 is abnormally expressed in a variety of diseases and that aberrant LINC00346 expression is associated with many clinical features. LINC00346 has been found to regulate specific cellular functions by interacting with several molecular and signaling pathways. LINC00346 has also been identified as a potential biomarker in the diagnosis, prognostication, and treatment of diseases. In this review, we summarize current evidence concerning the expression, clinical characteristics, functions, and related mechanisms of LINC00346 in the occurrence and development of diseases. We also discuss the potential clinical utility of LINC00346, thereby providing new insight into the diagnosis and treatment of diseases.
Role of LINC00346 in Disease
Expression of LINC00346 in Disease
Increasing evidence has revealed that the expression level of LINC00346 is significantly upregulated in schizophrenia (Ghafouri-Fard et al., 2021), nasopharyngeal carcinoma (Cui et al., 2020), lung cancer (Wang et al., 2017), hepatocellular carcinoma (HCC) (Jin et al., 2020; Yin et al., 2020; Zhang and Chen, 2020), glioma (Yang C. et al., 2020; Chen X. et al., 2020), colorectal cancer (CRC) (Tong et al., 2020), cutaneous squamous cell carcinoma (Piipponen et al., 2020), breast cancer (Li et al., 2020d), gastric cancer, and pancreatic cancer (Table 1). LINC00346 may have a pathogenic role in disease progression. Interestingly, Gheliji et al. (2020) found that LINC00346 expression was decreased in lung cancer tissues compared with adjacent normal tissues. The expression level of LINC00346 needs to be further explored in lung cancer.
TABLE 1.
The expression and clinical features of LINC00346 in disease.
| Type | Expression | Feature | Refs |
|---|---|---|---|
| Epilepsy | — | Vitamin D level | Mazdeh et al. (2019) |
| Schizophrenia | Upregulated | Sex difference | Ghafouri-Fard et al. (2021) |
| Gastric cancer | Upregulated | Tumor size, pathologic stage, and disease-free survival | Xu et al. (2019) |
| Colorectal cancer | — | Lymphoma metastasis, histological grade, and TNM stage | Li et al. (2020b) |
| Hepatocellular carcinoma | Upregulated | — | Jin et al. (2020) |
| Hepatocellular carcinoma | Upregulated | — | Zhang and Chen, (2020) |
| Hepatocellular carcinoma | Upregulated | — | Yin et al. (2020) |
| Pancreatic cancer | Upregulated | Disease-free survival and overall survival | Peng et al. (2019) |
| Pancreatic cancer | Upregulated | — | Shi et al. (2019) |
| Pancreatic cancer | Upregulated | Overall survival | Zhang et al. (2018) |
| Glioma | — | Overall survival | Geng et al. (2020) |
| Glioma | Upregulated | Disease-free survival and overall survival | Chen et al. (2020b) |
| Glioma | Upregulated | — | Yang et al. (2020a) |
| Lung cancer | Upregulated | Sex difference | Gheliji et al. (2020) |
| Lung cancer | Upregulated | — | Wang et al. (2017) |
| Lung cancer | — | Overall survival | Wang et al. (2021c) |
| Breast cancer | Upregulated | — | Li et al. (2020d) |
| Breast cancer | — | Overall survival | Liu et al. (2016) |
| Nasopharyngeal carcinoma | Upregulated | Overall survival and recurrence-free survival | Cui et al. (2020) |
| Cutaneous squamous cell carcinoma | Upregulated | — | Piipponen et al. (2020) |
LINC00346 and Clinical Characteristics
Some research groups have reported a potential relationship between LINC00346 expression and clinicopathological features (Table 1). The expression of LINC00346 has the potential to indicate the prognosis of numerous diseases, such as nasopharyngeal carcinoma (Cui et al., 2020), lung cancer (Wang et al., 2017), glioma (Yang C. et al., 2020; Chen X. et al., 2020), breast cancer (Li et al., 2020d), and pancreatic cancer (Zhang et al., 2018; Peng et al., 2019; Shi et al., 2019). In addition, LINC00346 expression was found to be strongly correlated with metastasis, histological grade, and TNM stage in (Li T. et al., 2020) CRC. An increased LINC00346 level predicted larger tumor size and poorer pathologic stage in gastric cancer (Xu et al., 2019). The level of LINC00346 was relatively correlated with sex in lung cancer and schizophrenia patients (Gheliji et al., 2020; Ghafouri-Fard et al., 2021). The LINC00346 expression level was negatively associated with the vitamin D level in epileptic patients (Mazdeh et al., 2019).
Functional Roles of LINC00346 in Disease
A growing amount of evidence has shown that lncRNAs play an important role in human disease (Zheng et al., 2019; Yu et al., 2020; Luo et al., 2021). LINC00346 exerts a vital role in the development of the disease by regulating various cellular functions. The regulatory functions of LINC00346 are related to cell proliferation, migration, invasion, and apoptosis. In this section, we summarize the current findings on the functions of LINC00346 (Table 2).
TABLE 2.
The functions and mechanisms of LINC00346 in disease.
| Type | Role | Function | Related genes | Refs |
|---|---|---|---|---|
| Schizophrenia | Oncogene | — | — | Ghafouri-Fard et al. (2021) |
| Atherosclerosis | — | Inflammatory factors and functional injury | miR-148a-3p, HUVECs, and KLF5 | Wang et al. (2021a) |
| Gastric cancer | Oncogene | Cell proliferation, cell migration, cell invasion, and cell cycle | KLF5, MYC and miR-34a-5p | Xu et al. (2019) |
| Colorectal cancer | — | Cell proliferation, apoptosis, cell migration, and cell invasion | miR-148b | Li et al. (2020b) |
| Colorectal cancer | — | Cancer stemness properties | miR-509-5p and WBSCR22 | Zhao et al. (2020) |
| Hepatocellular carcinoma | Oncogene | Apoptosis, cell migration, and cell cycle | miR-199a-3p, CDK1, CCNB1, and p53 | Jin et al. (2020) |
| Hepatocellular carcinoma | Oncogene | Cell proliferation, cell migration, cell invasion | miR-542-3p, WDR18, Wnt/β-catenin pathway, and MYC | Zhang and Chen, (2020) |
| Hepatocellular carcinoma | Oncogene | Cell proliferation, apoptosis, cell migration, and cell invasion | JAK and STAT3 | Yin et al. (2020) |
| Pancreatic cancer | Oncogene | Cell proliferation, cell migration, cell invasion | C-Myc and CTCF | Peng et al. (2019) |
| Pancreatic cancer | Oncogene | Cell proliferation, cell cycle, chemoresistance | miR-188-3p and BRD4 | Shi et al. (2019) |
| Pancreatic cancer | Oncogene | Cell proliferation | — | Zhang et al. (2018) |
| Glioma | — | — | miR-128-3p and SZRD1 | Geng et al. (2020) |
| Glioma | oncogene | Cell proliferation, apoptosis, cell migration, and cell invasion | miR-340-5p and ROCK1 | Chen et al. (2020b) |
| Glioma | Oncogene | Angiogenesis | ANKHD1, LINC00346, and ZNF655 | Yang et al. (2020a) |
| Lung cancer | Tumor suppressor | — | — | Gheliji et al. (2020) |
| Lung cancer | Oncogene | Cell proliferation, apoptosis, and cell cycle | JAK and STAT3 | Wang et al. (2017) |
| Lung cancer | — | Cell proliferation, apoptosis, cell migration, cell invasion, and cell cycle | miR-30c-2-3 and MYBL2 | Xu et al. (2021) |
| Lung cancer | — | Chemoresistance | — | Wang et al. (2021c) |
| Breast cancer | Oncogene | Cell proliferation, apoptosis, and glycolysis | miR-148a/b and GLUT1 | Li et al. (2020d) |
| Breast cancer | — | — | — | Liu et al. (2016) |
| Bladder cancer | — | Cell proliferation, apoptosis, cell migration, and cell cycle | — | Ye et al. (2017) |
| Nasopharyngeal carcinoma | Oncogene | Chemoresistance | miR-342-5p | Cui et al. (2020) |
| Cutaneous squamous cell carcinoma | Oncogene | Cell invasion | STAT3 and MMP | Piipponen et al. (2020) |
The Role of LINC00346 in Cellular Growth
Malignant diseases are often caused by unregulated cell growth (Huang et al., 2016; Emmanuel et al., 2018; Tan et al., 2019). Controlling cell growth is critical for the treatment of some diseases. The upregulation of LINC00346 has been found to promote cell proliferation and inhibit cell apoptosis in many diseases. Silencing LINC00346 has been found to obviously inhibit cell proliferation and promote cell apoptosis in bladder cancer (Ye et al., 2017), lung cancer (Wang et al., 2017; Xu et al., 2021), (Jin et al., 2020; Yin et al., 2020) HCC, glioma (Chen X. et al., 2020), (Li T. et al., 2020) CRC, and breast cancer (Li et al., 2020d). Additionally, several studies have revealed that elevated LINC00346 expression enhances cell proliferation in gastric cancer (Xu et al., 2019) and pancreatic cancer (Zhang et al., 2018; Peng et al., 2019; Shi et al., 2019). Cell cycle arrest induced by LINC00346 has been observed in bladder cancer (Ye et al., 2017), lung cancer (Wang et al., 2017; Xu et al., 2021), (Jin et al., 2020) HCC, gastric cancer (Xu et al., 2019), and pancreatic cancer (Shi et al., 2019). Knockdown of LINC00346 facilitates G1/G0 cell cycle arrest in bladder cancer (Ye et al., 2017), non-small-cell lung cancer (Wang et al., 2017; Xu et al., 2021), and (Jin et al., 2020)HCC. In gastric cancer, increased LINC00346 levels suppress cell cycle arrest at the G1–S phase (Xu et al., 2019) transition. The cell cycle is blocked in the G2/M phase with upregulated LINC00346 expression in pancreatic cancer (Shi et al., 2019) and lung adenocarcinoma (Xu et al., 2021).
The Role of LINC00346 in Cell Motility
Metastasis to adjacent and distant sites, indicating poor prognosis, is a sign of malignant disease (Chakraborty et al., 2008; Okugawa et al., 2014; Lin et al., 2020). Cell motility is a highly regulated mechanical process important in wound healing, metastasis, and embryogenesis (Selmeczi et al., 2005; Yip et al., 2007; Mejean et al., 2009). Increased expression of LINC00346 has been found to promote tumor cell metastasis in bladder cancer (Ye et al., 2017), lung cancer (Xu et al., 2021), (Jin et al., 2020; Yin et al., 2020; Zhang and Chen, 2020) HCC, glioma (Chen X. et al., 2020), (Li T. et al., 2020; Zhao et al., 2020) CRC, cutaneous squamous cell carcinoma (Piipponen et al., 2020), gastric cancer (Xu et al., 2019), and pancreatic cancer (Peng et al., 2019). LINC00346 significantly enhances cell migration and invasion in lung adenocarcinoma (Xu et al., 2021), (Jin et al., 2020; Yin et al., 2020; Zhang and Chen, 2020) HCC, glioma (Chen X. et al., 2020), (Li T. et al., 2020; Zhao et al., 2020)CRC, gastric cancer (Xu et al., 2019), and pancreatic cancer (Peng et al., 2019). LINC00346 also promotes cell migration in bladder cancer (Ye et al., 2017) and cell invasion in cutaneous squamous cell carcinoma (Piipponen et al., 2020).
The Role of LINC00346 in Drug Resistance
The increasing frequency of DR has prompted substantial research interest (Shehata, 2005; Shi and Gao, 2016; Carné Trécesson et al., 2017). DR remains a significant obstacle in the treatment of cancer. It is important to investigate the underlying mechanisms of chemoresistance for cancer treatment. The expression of LINC00346 was found to be markedly correlated with DR in nasopharyngeal carcinoma (Cui et al., 2020) and pancreatic cancer (Shi et al., 2019). Knockdown of LINC00346 enhanced the sensitivity to cisplatin in nasopharyngeal carcinoma cell lines (Cui et al., 2020). Silencing LINC00346 attenuated the gemcitabine tolerance in pancreatic cancer cell lines (Shi et al., 2019). Monitoring and modulating LINC00346 expression may be potential therapeutic strategies for guiding clinical treatment.
The Role of LINC00346 in Other Functions
Inflammation is considered a key factor in the pathophysiology of atherosclerosis (Sivapalaratnam et al., 2011; Paramel et al., 2020; Good et al., 2021). The upregulation of LINC00346 promoted inflammatory factor expression and functional injury in human umbilical vein endothelial cells (HUVECs) stimulated by OX-LDL (Wang F. et al., 2021). LINC00346 facilitated angiogenesis of glioma-associated endothelial cells (GECs), and in vitro LINC00346 knockdown experiments further verified this result (Yang C. et al., 2020). Glycolysis is a vital feature of tumor cells (Wang et al., 2013; Deng et al., 2020; Zhang et al., 2021). Inhibition of glycolysis is a promising therapeutic strategy for inhibiting tumors (Lee et al., 2017; Du et al., 2020). Increased LINC00346 levels were found to significantly promote glycolysis ability in breast cancer cell lines (Li et al., 2020d). In CRC, LINC00346 was found to regulate cancer stemness properties in vitro (Zhao et al., 2020).
LINC00346 Regulatory Mechanisms in Disease
Mechanisms of LINC00346 in Tumors
Mechanisms of LINC00346 in Digestive System Tumors
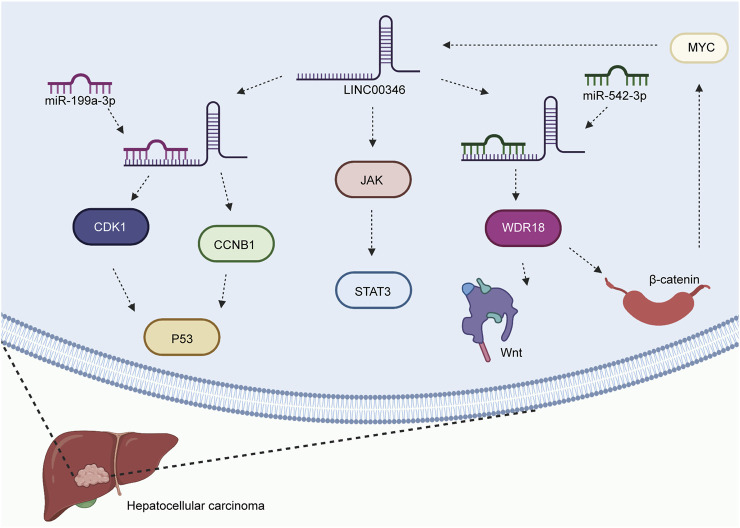
HCC is the most common type of liver cancer and has increasing mortality worldwide (Gomha et al., 2015; Shen et al., 2018; Yapasert et al., 2020). The underlying signaling mechanisms of HCC progression are poorly defined. A growing amount of evidence has shown that LINC00346 is significantly associated with the progression of HCC. Upregulation of LINC00346 was found to promote cancer progression by regulating various biological functions in HCC. LINC00346 promoted the expression levels of CDK1 and CCNB1 by acting as a sponge of miR-199a-3p in HCC (Jin et al., 2020) (Figure 1). In addition, LINC00346 inhibited cell invasion and apoptosis and controlled the cell cycle by regulating p53 and the miR-199a-3p/CDK1/CCNB1 axis. LINC00346 facilitated WDR18 expression and the Wnt/β-catenin pathway by sponging miR-542-3p in HCC (Zhang and Chen, 2020). Researchers also observed that β-catenin and LINC00346 form a positive feedback loop by interacting with MYC. LINC00346 was also found to affect cell proliferation and survival by regulating the JAK-STAT3 signaling pathway (Yin et al., 2020).
FIGURE 1.
Mechanisms of LINC00346 in HCC (HCC). LINC00346 upregulates the expression levels of CDK1 and CCNB1 by sponging miR-199a-3p in HCC. It attenuates cell invasion and apoptosis and to regulates the cell cycle by regulating p53 and the miR-199a-3p/CDK1/CCNB1 axis. LINC00346 facilitates WDR18 expression and activates the Wnt/β-catenin pathway by acting as a sponge of miR-542-3p in HCC. β-catenin and LINC00346 form a positive feedback loop by interacting with MYC. Finally, LINC00346 affects cell proliferation and survival by activating the JAK-STAT3 signaling pathway.
Pancreatic carcinoma is one of the most malignant tumors and has an extremely poor prognosis (Xu D. et al., 2020; Wang et al., 2020; Chen et al., 2021). Chemotherapy is an important method of adjuvant therapy in the comprehensive treatment of pancreatic cancer (Rahman et al., 2017; Zhu et al., 2019). Therefore, the chemoresistance and pathogenesis of pancreatic cancer urgently need to be explored. LINC00346 was found to facilitate the transcription and expression of c-Myc by interacting with CTCF in pancreatic cancer (Peng et al., 2019). LINC00346, acts as a sponge of miR-188-3p and downregulated the level of BRD4 to increase gemcitabine resistance in pancreatic cancer (Shi et al., 2019). In CRC, LINC00346 promotes cell migration and invasion by reducing miR-148b levels (Li T. et al., 2020). LINC00346 also regulates the biological functions of CRC stem cells by activating the Linc00346/miR-509-5p/wbscr22 pathway (Zhao et al., 2020). LINC00346 suppresses miR-34a-5p expression to affect CD44, Notch1, and AXL expression in gastric cancer (Xu et al., 2019). In addition, the expression of LINC00346 was found to be markedly upregulated by KLF5 and MYC in gastric cancer.
Mechanisms of LINC00346 in Central Nervous System Tumors
Gliomas are the most common type of tumor of the central nervous system (Ceresa et al., 2019; Liu Y. et al., 2019; Stępniak et al., 2021).
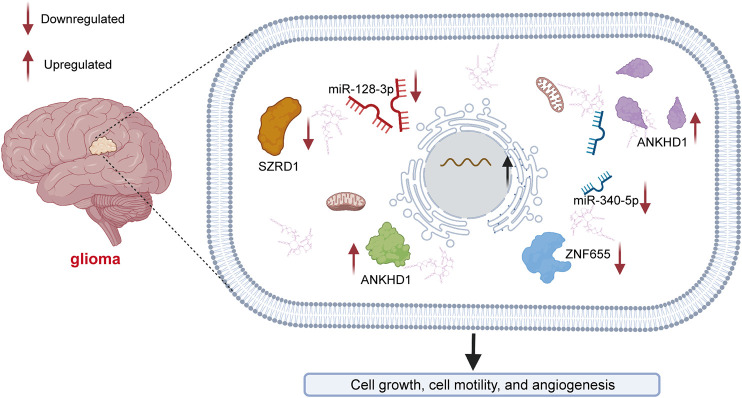
Gliomas are the most aggressive type of brain tumor and have an extremely poor prognosis (Seliger and Hau, 2018; Chen W. et al., 2020). Glioma can be divided into astrocytoma, glioblastoma multiforme (GBM), oligodendroglioma and mixed tumors (Peng et al., 2018). Geng et al. (2020) found that LINC00346 inhibited the expression of miR-128-3p to upregulate SZRD1 levels in glioma (Figure 2). LINC00346 was found to affect cell proliferation and apoptosis through the regulation of miR-128-3p/SZRD1. LINC00346 was also found to act as a ceRNA (competing endogenous RNA) of miR-340-5p to suppress the expression of ROCK1 in glioma progression (Chen X. et al., 2020). Exploring the potential angiogenesis mechanisms is essential for the development of novel strategies for glioma treatment. LncRNAs play an essential role in tumor angiogenesis (Ruan et al., 2019; Cheng et al., 2020; Wang X. et al., 2021). In glioma, the expression level of LINC00346 was found to be positively regulated by ankyrin repeat and KH domain-containing protein 1 (Yang C. et al., 2020) (ANKHD1). The activation of ANKHD1/LINC00346/znf655 was found to facilitate angiogenesis in association with glioma-associated endothelial cells (GECs).
FIGURE 2.
Mechanisms of LINC00346 in glioma. LINC00346 upregulates SZRD1 levels by sponging miR-128-3p in glioma. LINC00346 modulates cell proliferation and apoptosis by regulating miR-128-3p/SZRD1. LINC00346 also functions as a ceRNA of miR-340-5p to reduce the expression level of ROCK1 in glioma. The expression level of LINC00346 is positively regulated by ankyrin repeat and KH domain-containing protein 1 (ANKHD1). Angiogenesis associated with glioma-associated endothelial cells (GECs) is facilitated by the ANKHD1/LINC00346/znf655 pathway.
Mechanisms of LINC00346 in Tumors of Other Systems
Lung cancer is the leading cause of cancer-related deaths worldwide (Liu J. et al., 2018; Harikrishnan et al., 2020; To et al., 2021). It can be divided into small-cell lung cancer and non-small-cell lung cancer (NSCLC) (Du et al., 2019; Li H. et al., 2020). LINC00346 was found to facilitate NSCLC progression via regulation of the JAK-STAT3 signaling pathway (Wang et al., 2017). LINC00346 promotes the expression of MYBL2 to regulate the cell cycle by acting as a sponge of miR-30c-2-3p in lung adenocarcinoma (Xu et al., 2021). Nasopharyngeal carcinoma (NPC) is a head and neck malignancy with a high incidence (Sun and Xu, 2015; Yao et al., 2020). Chemoresistance remains an obstacle in the treatment of NPC. LINC00346 attenuates cisplatin sensitivity by sponging miR-342-5p in NPC (Cui et al., 2020). In breast cancer, the expression of LINC00346 upregulates glucose transporter 1 levels by targeting miR-148a/b (Li et al., 2020d). Cutaneous squamous cell carcinoma is the second most frequent malignant skin cancer, and the incidence is increasing. LINC00346 increases the expression of matrix metalloproteinase by activating STAT3 signaling in cutaneous squamous cell carcinoma (Piipponen et al., 2020).
Mechanisms of LINC00346 in Nontumor Disease
Atherosclerosis is a major cause of multiple diseases, such as coronary artery disease (CHD) (Martinus and Goldsbury, 2018; Xu et al., 2018), peripheral artery disease (PAD) (Biscetti et al., 2019), and atherosclerotic cerebrovascular disease (Sascău et al., 2021). However, the pathogenesis of atherosclerosis remains unclear. There is an urgent need to explore the mechanism of atherosclerosis. The level of LINC00346 was found to be negatively correlated with Krüppel-like factor 5 (KLF5) expression in atherosclerosis (Wang F. et al., 2021). Knockdown of LINC00346 inhibited inflammatory reactions and functional injury in the progression of atherosclerosis. LINC00346 was found to affect the initiation and development of atherosclerosis by regulating the KLF5/LINC00346/miR-148a-3p pathway (Wang F. et al., 2021).
Clinical Utility of LINC00346 in Disease
Despite improved technology and advances in modern medicine, malignant disease, especially cancer, remains one of the leading causes of death. Abnormal cell growth, metastasis, and drug assistance result in poor disease prognoses. Early diagnosis and targeted treatment are important for improving the prognosis of disease. LncRNAs may be potential diagnostic biomarkers and therapeutic targets. In this section, we will further discuss the potential clinical utility of LINC00346 in the diagnosis, prognostication, and treatment of diseases.
LINC00346 as a Diagnostic Biomarker
The detection and diagnosis of disease are essential for disease management (Li et al., 2020c; Álvarez et al., 2020; Adhikari et al., 2021). Increasing evidence has revealed that lncRNAs are potential diagnostic biomarkers for several diseases (Wang et al., 2016; Jin et al., 2017; Huang G.-z. et al., 2019; Cheng et al., 2019). LINC00346 has found to be obviously upregulated in many tumors and nontumor diseases. Gheliji et al. (2020) observed that LINC00346 expression was downregulated in lung cancer tissues. The level of LINC00346 in lung cancer needs to be further explored. The sensitivity was 83.3%, and the specificity was 52.4% in an analysis of the ability to distinguish between lung cancer tissues and adjacent tissues (Gheliji et al., 2020). Importantly, the expression of LINC00346 in venous blood was found to be markedly upregulated in patients with schizophrenia or pancreatic cancer (Zhang et al., 2018; Ghafouri-Fard et al., 2021). LINC00346 has great value in the diagnosis of schizophrenia (Ghafouri-Fard et al., 2021) and pancreatic cancer (Zhang et al., 2018). Substances that are stably expressed in body fluids are more likely to be used as biomarkers in disease diagnosis.
LINC00346 as a Prognostic Biomarker
Individualized therapy requires the identification of biomarkers to predict patient prognosis (Li Z. et al., 2019; Melling et al., 2019). The expression of LINC00346 was found to be significantly correlated with the poor prognosis of nasopharyngeal carcinoma (Cui et al., 2020), lung cancer (Wang et al., 2017), glioma (Yang C. et al., 2020; Chen X. et al., 2020), breast cancer (Li et al., 2020d), and pancreatic cancer (Zhang et al., 2018; Peng et al., 2019; Shi et al., 2019). The level of LINC00346 was found to be negatively associated with overall survival in nasopharyngeal carcinoma (Cui et al., 2020), lung adenocarcinoma (Wang Z. et al., 2021), glioma (Chen X. et al., 2020; Geng et al., 2020), breast cancer (Liu et al., 2016), and pancreatic cancer (Zhang et al., 2018; Peng et al., 2019). LINC00346 was also found to affect disease-free survival in patients with gastric cancer (Xu et al., 2019) and pancreatic cancer (Peng et al., 2019). The upregulation of LINC00346 was found to be correlated with shorter recurrence-free survival in nasopharyngeal carcinoma (Cui et al., 2020).
LINC00346 as a Biomarker of Targeted Therapy
Molecular targeted therapy shows advantages for many diseases, especially malignancies (Baba et al., 2019; Becker et al., 2020; Xu P. et al., 2020). LncRNAs contribute to disease progression through the regulation of cellular pathways (Li Z.-W. et al., 2020; Sun and Wu, 2020). They serve as an important therapeutic targets in the treatment of diseases. In Section 2.4, we introduced the mechanisms of LINC00346 in tumor and nontumor diseases. LINC00346 is a meaningful therapeutic biomarker in disease treatment. It is also considered an oncogene that contributes to tumorigenesis. Knockdown or silencing of LINC00346 can inhibit cell biological functions to suppress cancer progression in several cancers, such as bladder cancer (Ye et al., 2017), lung cancer (Wang et al., 2017), and glioma (Chen X. et al., 2020; Geng et al., 2020). The association of LINC00346 and chemoresistance has implications for treatment.
Conclusions and Future Perspectives
LINC00346, a novel lncRNA, is encoded on chromosome 13q34. It is significantly upregulated in many diseases. However, some researchers have found that the level of LINC00346 is reduced in lung cancer. Therefore, the expression of LINC00346 in lung cancer needs to be further explored. Importantly, the expression of LINC00346 in venous blood was found to be elevated in patients with schizophrenia or pancreatic cancer. This finding is crucial for the successful clinical application of LINC00346. Substances stably expressed in bodily fluids have strong potential in the diagnosis of disease. The expression level of LINC00346 was found to be obviously correlated with prognosis, lymphoma metastasis, histological grade, TNM stage, tumor size and pathologic stage. LINC00346 expression has important guiding significance in the management of patients. Different strategies can be used for different patients if the prognosis can be accurately predicted. LINC00346 exerts a vital role by regulating cellular growth, cell motility, chemoresistance, and other functions in diseases. LINC00346 affects these biological functions by interacting with several pathways. Knockdown or silencing of LINC00346 inhibits the progression of several cancers. In conclusion, LINC00346 is a potential biomarker in the diagnosis, prognostication, and treatment of diseases. In terms of clinical applications, further basic experiments and multicenter research data are needed.
Author Contributions
LL and JL designed and guided the study. JL and ZX wrote and edited the manuscript. MX helped with reference collection. All authors read and approved the final manuscript.
Funding
This work was funded by the National Nature Science Foundation of China (U20A20343) and the Independent Project Fund of the State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, the National Key Research and Development Program of China (2016YFC1101404/3).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Adhikari M., Devkota H. R., Cesuroglu T. (2021). Barriers to and Facilitators of Diabetes Self-Management Practices in Rupandehi, Nepal- Multiple Stakeholders' Perspective. BMC Public Health 21, 1269. 10.1186/s12889-021-11308-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez D., Cerezo-Hernández A., Crespo A., Gutiérrez-Tobal G. C., Vaquerizo-Villar F., Barroso-García V., et al. (2020). A Machine Learning-Based Test for Adult Sleep Apnoea Screening at home Using Oximetry and Airflow. Sci. Rep. 10, 5332. 10.1038/s41598-020-62223-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba K., Fujita T., Tasaka S., Fujii H. (2019). Simultaneous Detection of the T790M and L858R Mutations in the EGFR Gene by Oligoribonucleotide Interference-PCR. Ijms 20, 4020. 10.3390/ijms20164020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker L., Verdurmen W. P. R., Plückthun A. (2020). Reengineering Anthrax Toxin Protective Antigen for Improved Receptor-specific Protein Delivery. BMC Biol. 18, 100. 10.1186/s12915-020-00827-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biscetti F., Nardella E., Bonadia N., Angelini F., Pitocco D., Santoliquido A., et al. (2019). Association between Plasma Omentin-1 Levels in Type 2 Diabetic Patients and Peripheral Artery Disease. Cardiovasc. Diabetol. 18, 74. 10.1186/s12933-019-0880-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carné Trécesson S. d., Souazé F., Basseville A., Bernard A.-C., Pécot J., Lopez J., et al. (2017). BCL-XL Directly Modulates RAS Signalling to Favour Cancer Cell Stemness. Nat. Commun. 8, 1123. 10.1038/s41467-017-01079-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceresa D., Alessandrini F., Bosio L., Marubbi D., Reverberi D., Malatesta P., et al. (2019). Cdh4 Down-Regulation Impairs In Vivo Infiltration and Malignancy in Patients Derived Glioblastoma Cells. Ijms 20, 4028. 10.3390/ijms20164028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty G., Jain S., Patil T. V., Kundu G. C. (2008). Down-regulation of Osteopontin Attenuates Breast Tumour Progressionin Vivo. J. Cel. Mol. Med. 12, 2305–2318. 10.1111/j.1582-4934.2008.00263.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Huang S. (2018). Circular RNA: an Emerging Non-coding RNA as a Regulator and Biomarker in Cancer. Cancer Lett. 418, 41–50. 10.1016/j.canlet.2018.01.011 [DOI] [PubMed] [Google Scholar]
- Chen J.-X., Cheng C.-S., Gao H.-F., Chen Z.-J., Lv L.-L., Xu J.-Y., et al. (2021). Overexpression of Interferon-Inducible Protein 16 Promotes Progression of Human Pancreatic Adenocarcinoma through Interleukin-1β-Induced Tumor-Associated Macrophage Infiltration in the Tumor Microenvironment. Front. Cel Dev. Biol. 9, 640786. 10.3389/fcell.2021.640786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Hong L., Hou C., Wang Y., Wang F., Zhang J. (2020a). MicroRNA-585 Inhibits Human Glioma Cell Proliferation by Directly Targeting MDM2. Cancer Cel Int 20, 469. 10.1186/s12935-020-01528-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Li D., Chen L., Hao B., Gao Y., Li L., et al. (2020b). Long Noncoding RNA LINC00346 Promotes Glioma Cell Migration, Invasion and Proliferation by Up‐regulating ROCK1. J. Cel. Mol. Med. 24, 13010–13019. 10.1111/jcmm.15899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xiong D., Yang H., Ye L., Mei S., Wu J., et al. (2019). Long Noncoding RNA OPA‐interacting Protein 5 Antisense Transcript 1 Upregulated SMAD3 Expression to Contribute to Metastasis of Cervical Cancer by Sponging miR‐143‐3p. J. Cel. Physiol. 234, 5264–5275. 10.1002/jcp.27336 [DOI] [PubMed] [Google Scholar]
- Cheng C., Zhang Z., Cheng F., Shao Z. (2020). Exosomal lncRNA RAMP2-AS1 Derived from Chondrosarcoma Cells Promotes Angiogenesis through miR-2355-5p/VEGFR2 axis. Ott Vol. 13, 3291–3301. 10.2147/ott.S244652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J.-T., Wang L., Wang H., Tang F.-R., Cai W.-Q., Sethi G., et al. (2019). Insights into Biological Role of LncRNAs in Epithelial-Mesenchymal Transition. Cells 8, 1178. 10.3390/cells8101178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Pu T., Zhang Y., Wang J., Zhao Y. (2020). Long Non-coding RNA LINC00346 Contributes to Cisplatin Resistance in Nasopharyngeal Carcinoma by Repressing miR-342-5p. Open Biol. 10, 190286. 10.1098/rsob.190286 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Deng X., Deng J., Yi X., Zou Y., Liu H., Li C., et al. (2020). Ubiquitin-like Protein FAT10 Promotes Osteosarcoma Glycolysis and Growth by Upregulating PFKFB3 via Stabilization of EGFR. Am. J. Cancer Res. 10, 2066–2082. [PMC free article] [PubMed] [Google Scholar]
- Du T., Li H., Fan Y., Yuan L., Guo X., Zhu Q., et al. (2019). The Deubiquitylase OTUD3 Stabilizes GRP78 and Promotes Lung Tumorigenesis. Nat. Commun. 10, 2914. 10.1038/s41467-019-10824-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Zhang M. j., Li L. l., Xu X. L., Chen H., Feng Y. b., et al. (2020). ATPR Triggers Acute Myeloid Leukaemia Cells Differentiation and Cycle Arrest via the RARα/LDHB/ERK‐glycolysis Signalling axis. J. Cel. Mol. Med. 24, 6952–6965. 10.1111/jcmm.15353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanuel N., Lofgren K. A., Peterson E. A., Meier D. R., Jung E. H., Kenny P. A. (2018). Mutant GATA3 Actively Promotes the Growth of normal and Malignant Mammary Cells. Anticancer Res. 38, 4435–4441. 10.21873/anticanres.12745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian M. R., Sonenberg N., Filipowicz W. (2010). Regulation of mRNA Translation and Stability by microRNAs. Annu. Rev. Biochem. 79, 351–379. 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- Geng Y. B., Pan C. C., Xu C., Zuo P. C., Wang Y., Li X. O., et al. (2020). Long Non-coding RNA LINC00346 Regulates Proliferation and Apoptosis by Targeting miR-128-3p/SZRD1 axis in Glioma. Eur. Rev. Med. Pharmacol. Sci. 24, 9581–9590. 10.26355/eurrev_202009_23046 [DOI] [PubMed] [Google Scholar]
- Ghafouri-Fard S., Eghtedarian R., Seyedi M., Pouresmaeili F., Arsang-Jang S., Taheri M. (2021). Upregulation of VDR-Associated lncRNAs in Schizophrenia. J. Mol. Neurosci. 10.1007/s12031-021-01901-y [DOI] [PubMed] [Google Scholar]
- Gheliji T., Oskooei V. K., Ashrafi Hafez A., Taheri M., Ghafouri-Fard S. (2020). Evaluation of Expression of Vitamin D Receptor Related lncRNAs in Lung Cancer. Non-coding RNA Res. 5, 83–87. 10.1016/j.ncrna.2020.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomha S., Salaheldin T., Hassaneen H., Abdel-Aziz H., Khedr M. (2015). Synthesis, Characterization and Molecular Docking of Novel Bioactive Thiazolyl-Thiazole Derivatives as Promising Cytotoxic Antitumor Drug. Molecules 21, 3. 10.3390/molecules21010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good E., Ochoa-Figueroa M., Ziegler M., Ressner M., Warntjes M., Dyverfeldt P., et al. (2021). 18Fluorodeoxyglucose Uptake in Relation to Fat Fraction and R2* in Atherosclerotic Plaques, Using PET/MRI: a Pilot Study. Sci. Rep. 11, 14217. 10.1038/s41598-021-93605-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Lv B., Liu R., Dai Z., Zhang F., Liang Y., et al. (2021). Role of LncRNAs in Regulating Cancer Amino Acid Metabolism. Cancer Cel Int 21, 209. 10.1186/s12935-021-01926-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikrishnan K., Joshi O., Madangirikar S., Balasubramanian N. (2020). Cell Derived Matrix Fibulin-1 Associates with Epidermal Growth Factor Receptor to Inhibit its Activation, Localization and Function in Lung Cancer Calu-1 Cells. Front. Cel Dev. Biol. 8, 522. 10.3389/fcell.2020.00522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A., He S., Li X., Zhou L. (2019). ZFAS1: a Novel Vital Oncogenic lncRNA in Multiple Human Cancers. Cell Prolif 52, e12513. 10.1111/cpr.12513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G.-z., Wu Q.-q., Zheng Z.-n., Shao T.-r., Lv X.-Z. (2019a). Identification of Candidate Biomarkers and Analysis of Prognostic Values in Oral Squamous Cell Carcinoma. Front. Oncol. 9, 1054. 10.3389/fonc.2019.01054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Lin H., Kang L., Huang P., Huang J., Cai J., et al. (2019b). Aberrant Expression of Long Noncoding RNA SNHG15 Correlates with Liver Metastasis and Poor Survival in Colorectal Cancer. J. Cel. Physiol. 234, 7032–7039. 10.1002/jcp.27456 [DOI] [PubMed] [Google Scholar]
- Huang N., Cheng S., Mi X., Tian Q., Huang Q., Wang F., et al. (2016). Downregulation of Nitrogen Permease Regulator Like-2 Activates PDK1-AKT1 and Contributes to the Malignant Growth of Glioma Cells. Mol. Carcinog. 55, 1613–1626. 10.1002/mc.22413 [DOI] [PubMed] [Google Scholar]
- Jin J., Xu H., Li W., Xu X., Liu H., Wei F. (2020). LINC00346 Acts as a Competing Endogenous RNA Regulating Development of Hepatocellular Carcinoma via Modulating CDK1/CCNB1 axis. Front. Bioeng. Biotechnol. 8, 54. 10.3389/fbioe.2020.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Chen X., Hu Y., Ying F., Zou R., Lin F., et al. (2017). LncRNA-TCONS_00026907 Is Involved in the Progression and Prognosis of Cervical Cancer through Inhibiting miR-143-5p. Cancer Med. 6, 1409–1423. 10.1002/cam4.1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. Y., Jeong E. K., Ju M. K., Jeon H. M., Kim M. Y., Kim C. H., et al. (2017). Induction of Metastasis, Cancer Stem Cell Phenotype, and Oncogenic Metabolism in Cancer Cells by Ionizing Radiation. Mol. Cancer 16, 10. 10.1186/s12943-016-0577-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei G.-L., Niu Y., Cheng S.-J., Li Y.-Y., Bai Z.-F., Yu L.-X., et al. (2021). Upregulation of Long Noncoding RNA W42 Promotes Tumor Development by Binding with DBN1 in Hepatocellular Carcinoma. Wjg 27, 2586–2602. 10.3748/wjg.v27.i20.2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Zhou Y., Chen J., Wang T., Li Z., Fu Y., et al. (2021a). Long Noncoding RNA H19 Acts as a miR‐29b Sponge to Promote Wound Healing in Diabetic Foot Ulcer. FASEB j. 35, e20526. 10.1096/fj.201900076RRRRR [DOI] [PubMed] [Google Scholar]
- Li H., Liu F., Qin W. (2020a). Circ_0072083 Interference Enhances Growth-Inhibiting Effects of Cisplatin in Non-small-cell Lung Cancer Cells via miR-545-3p/CBLL1 axis. Cancer Cel Int 20, 78. 10.1186/s12935-020-1162-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li Z., Leng K., Xu Y., Ji D., Huang L., et al. (2018). ZEB 1‐ AS 1: A Crucial Cancer‐related Long Non‐coding RNA. Cel Prolif 51, e12423. 10.1111/cpr.12423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wei H., Zhang Y. W., Zhao S., Che G., Wang Y., et al. (2021b). Differential Expression of Long Non-coding RNAs as Diagnostic Markers for Lung Cancer and Other Malignant Tumors. Aging 13, 23842–23867. 10.18632/aging.203523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Wang B., Zhang L., Cui M., Sun B. (2020b). Silencing of Long Noncoding RNA LINC00346 Inhibits the Tumorigenesis of Colorectal Cancer through Targeting microRNA-148b. Ott Vol. 13, 3247–3257. 10.2147/ott.24271510.2147/ott.s242715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Han X., Feng H., Han J. (2019a). Long Noncoding RNA OIP5-AS1 in Cancer. Clinica Chim. Acta 499, 75–80. 10.1016/j.cca.2019.08.031 [DOI] [PubMed] [Google Scholar]
- Li Y., Li D., Zhao P., Nandakumar K., Wang L., Song Y. (2020c). Microfluidics-based Systems in Diagnosis of Alzheimer's Disease and Biomimetic Modeling. Micromachines 11, 787. 10.3390/mi11090787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li H., Wang W., Yu X., Xu Q. (2020d). LINC00346 Regulates Glycolysis by Modulation of Glucose Transporter 1 in Breast Cancer Cells. Mol. Cell Probes 54, 101667. 10.1016/j.mcp.2020.101667 [DOI] [PubMed] [Google Scholar]
- Li Z.-W., Zhang T.-Y., Yue G.-J., Tian X., Wu J.-Z., Feng G.-Y., et al. (2020e). Small Nucleolar RNA Host Gene 22 (SNHG22) Promotes the Progression of Esophageal Squamous Cell Carcinoma by miR-429/SESN3 axis. Ann. Transl. Med. 8, 1007. 10.21037/atm-20-5332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Han C., Wang L., Zhu J., Yin Y., Li B. (2019b). Prognostic Value of Texture Analysis Based on Pretreatment DWI-Weighted MRI for Esophageal Squamous Cell Carcinoma Patients Treated with Concurrent Chemo-Radiotherapy. Front. Oncol. 9, 1057. 10.3389/fonc.2019.01057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Zhang Y., Zeng X., Xue C., Lin X. (2020). CircRNA circRIMS Acts as a microRNA Sponge to Promote Gastric Cancer Metastasis. ACS Omega 5, 23237–23246. 10.1021/acsomega.0c02991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Li J., Koirala P., Ding X., Chen B., Wang Y., et al. (2016). Long Non-coding RNAs as Prognostic Markers in Human Breast Cancer. Oncotarget 7, 20584–20596. 10.18632/oncotarget.7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Bian T., Feng J., Qian L., Zhang J., Jiang D., et al. (2018a). miR-335 Inhibited Cell Proliferation of Lung Cancer Cells by Target Tra2β. Cancer Sci. 109, 289–296. 10.1111/cas.13452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Liu L. H., Hu W. W., Wang M. (2019a). Long Noncoding RNA TUG1 Regulates the Development of Oral Squamous Cell Carcinoma through Sponging miR‐524‐5p to Mediate DLX1 Expression as a Competitive Endogenous RNA. J. Cel. Physiol. 234, 20206–20216. 10.1002/jcp.28620 [DOI] [PubMed] [Google Scholar]
- Liu X., Chen R., Liu L. (2019b). SP1-DLEU1-miR-4429 Feedback Loop Promotes Cell Proliferative and Anti-apoptotic Abilities in Human Glioblastoma. Biosci. Rep. 39, BSR20190994. 10.1042/bsr20190994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., She Y., Wu H., Zhong D., Zhang J. (2018b). Long Non-coding RNA Gas5 Regulates Proliferation and Apoptosis in HCS-2/8 Cells and Growth Plate Chondrocytes by Controlling FGF1 Expression via miR-21 Regulation. J. Biomed. Sci. 25, 18. 10.1186/s12929-018-0424-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Tang Z. G., Yang J. Q., Zhou Y., Lin Y., Lv W., et al. (2019c). Effect of Silencing S-phase Kinase-Associated Protein 2 on Chemosensitivity to Temozolomide of Human Glioma Cells U251. Am. J. Transl. Res. 11, 2470–2476. [PMC free article] [PubMed] [Google Scholar]
- Luo Q., Xu C., Li X., Zeng L., Ye J., Guo Y., et al. (2017). Comprehensive Analysis of Long Noncoding RNA and mRNA Expression Profiles in Rheumatoid Arthritis. Exp. Ther. Med. 14, 5965–5973. 10.3892/etm.2017.5284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Zheng S., Wu Q., Wu J., Zhou R., Wang C., et al. (2021). Long Noncoding RNA (lncRNA) EIF3J-DT Induces Chemoresistance of Gastric Cancer via Autophagy Activation. Autophagy 17, 4083–4101. 10.1080/15548627.2021.1901204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Yan W., Sun X., Chen P. (2021). Long Noncoding RNA VPS9D1-AS1 Promotes Esophageal Squamous Cell Carcinoma Progression via the Wnt/β-Catenin Signaling Pathway. J. Cancer 12, 6894–6904. 10.7150/jca.54556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinus R. D., Goldsbury J. (2018). Endothelial TNF-α Induction by Hsp60 Secreted from THP-1 Monocytes Exposed to Hyperglycaemic Conditions. Cell Stress and Chaperones 23, 519–525. 10.1007/s12192-017-0858-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazdeh M., Zamani M., Eftekharian M. M., Komaki A., Arsang-Jang S., Taheri M., et al. (2019). Expression Analysis of Vitamin D Receptor-Associated lncRNAs in Epileptic Patients. Metab. Brain Dis. 34, 1457–1465. 10.1007/s11011-019-00446-9 [DOI] [PubMed] [Google Scholar]
- Mejean C. O., Schaefer A. W., Millman E. A., Forscher P., Dufresne E. R. (2009). Multiplexed Force Measurements on Live Cells with Holographic Optical Tweezers. Opt. Express 17, 6209–6217. 10.1364/oe.17.006209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melling N., Norrenbrock S., Kluth M., Simon R., HubeMagg C., Steurer S., et al. (2019). p53 Overexpression Is a Prognosticator of Poor Outcome in Esophageal Cancer. Oncol. Lett. 17, 3826–3834. 10.3892/ol.2019.10020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan R., da Silveira W. A., Kelly R. C., Overton I., Allott E. H., Hardiman G. (2021). Long Non-coding RNAs and Their Potential Impact on Diagnosis, Prognosis, and Therapy in Prostate Cancer: Racial, Ethnic, and Geographical Considerations. Expert Rev. Mol. Diagn. 21, 1257–1271. 10.1080/14737159.2021.1996227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okugawa Y., Toiyama Y., Hur K., Toden S., Saigusa S., Tanaka K., et al. (2014). Metastasis-associated Long Non-coding RNA Drives Gastric Cancer Development and Promotes Peritoneal Metastasis. Carcinogenesis 35, 2731–2739. 10.1093/carcin/bgu200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panni S., Lovering R. C., Porras P., Orchard S. (2020). Non-coding RNA Regulatory Networks. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1863, 194417. 10.1016/j.bbagrm.2019.194417 [DOI] [PubMed] [Google Scholar]
- Paramel G. V., Karadimou G., Eremo A. G., Ljungberg L. U., Hedin U., Olofsson P. S., et al. (2020). Expression of CARD8 in Human Atherosclerosis and its Regulation of Inflammatory Proteins in Human Endothelial Cells. Sci. Rep. 10, 19108. 10.1038/s41598-020-73600-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolia A., Venalainen E., Xue H., Mather R., Lin D., Wu R., et al. (2019). The Long Noncoding RNA HORAS 5 Mediates Castration‐resistant Prostate Cancer Survival by Activating the Androgen Receptor Transcriptional Program. Mol. Oncol. 13, 1121–1136. 10.1002/1878-0261.12471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W.-X., He R.-Z., Zhang Z., Yang L., Mo Y.-Y. (2019). LINC00346 Promotes Pancreatic Cancer Progression through the CTCF-Mediated Myc Transcription. Oncogene 38, 6770–6780. 10.1038/s41388-019-0918-z [DOI] [PubMed] [Google Scholar]
- Peng Z., Liu C., Wu M. (2018). New Insights into Long Noncoding RNAs and Their Roles in Glioma. Mol. Cancer 17, 61. 10.1186/s12943-018-0812-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piipponen M., Nissinen L., Riihilä P., Farshchian M., Kallajoki M., Peltonen J., et al. (2020). p53-regulated Long Noncoding RNA PRECSIT Promotes Progression of Cutaneous Squamous Cell Carcinoma via STAT3 Signaling. Am. J. Pathol. 190, 503–517. 10.1016/j.ajpath.2019.10.019 [DOI] [PubMed] [Google Scholar]
- Radhakrishnan R., Kowluru R. A. (2021). Long Noncoding RNA MALAT1 and Regulation of the Antioxidant Defense System in Diabetic Retinopathy. Diabetes 70, 227–239. 10.2337/db20-0375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman F. A. u., Ali S., Saif M. W. (2017). Update on the Role of Nanoliposomal Irinotecan in the Treatment of Metastatic Pancreatic Cancer. Therap. Adv. Gastroenterol. 10, 563–572. 10.1177/1756283x17705328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Z., Xu Z., Li Z., Lv Y. (2019). Integral Analyses of Survivalrelated Long Noncoding RNA MIR210HG and its Prognostic Role in colon Cancer. Oncol. Lett. 18, 1107–1116. 10.3892/ol.2019.10435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sascău R., Clement A., Radu R., Prisacariu C., Stătescu C. (2021). Triglyceride-rich Lipoproteins and Their Remnants as Silent Promoters of Atherosclerotic Cardiovascular Disease and Other Metabolic Disorders: a Review. Nutrients 13, 1774. 10.3390/nu13061774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seliger C., Hau P. (2018). Drug Repurposing of Metabolic Agents in Malignant Glioma. Ijms 19, 2768. 10.3390/ijms19092768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmeczi D., Mosler S., Hagedorn P. H., Larsen N. B., Flyvbjerg H. (2005). Cell Motility as Persistent Random Motion: Theories from Experiments. Biophysical J. 89, 912–931. 10.1529/biophysj.105.061150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata M. F. (2005). Rel/Nuclear Factor-Kappa B Apoptosis Pathways in Human Cervical Cancer Cells. Cancer Cel Int 5, 10. 10.1186/1475-2867-5-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M., Cao J., Shi H. (20182018). Effects of Estrogen and Estrogen Receptors on Transcriptomes of HepG2 Cells: a Preliminary Study Using RNA Sequencing. Int. J. Endocrinol. 2018, 1–16. 10.1155/2018/5789127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S. N., Li K., Liu Y., Yang C. L., He C. Y., Wang H. R. (2019). Down‐regulation of Long noncodingRNA PVT1 Inhibits Esophageal Carcinoma Cell Migration and Invasion and Promotes Cell Apoptosis via microRNA‐145‐mediated Inhibition ofFSCN1. Mol. Oncol. 13, 2554–2573. 10.1002/1878-0261.12555 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Shi W.-J., Gao J.-B. (2016). Molecular Mechanisms of Chemoresistance in Gastric Cancer. Wjgo 8, 673–681. 10.4251/wjgo.v8.i9.673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Zhang C., Ning Z., Hua Y., Li Y., Chen L., et al. (2019). Long Non-coding RNA LINC00346 Promotes Pancreatic Cancer Growth and Gemcitabine Resistance by Sponging miR-188-3p to Derepress BRD4 Expression. J. Exp. Clin. Cancer Res. 38, 60. 10.1186/s13046-019-1055-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivapalaratnam S., Farrugia R., Nieuwdorp M., Langford C. F., van Beem R. T., Maiwald S., et al. (2011). Identification of Candidate Genes Linking Systemic Inflammation to Atherosclerosis; Results of a Human in vivoLPS Infusion Study. BMC Med. Genomics 4, 64. 10.1186/1755-8794-4-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stępniak K., Machnicka M. A., Mieczkowski J., Macioszek A., Wojtaś B., Gielniewski B., et al. (2021). Mapping Chromatin Accessibility and Active Regulatory Elements Reveals Pathological Mechanisms in Human Gliomas. Nat. Commun. 12, 3621. 10.1038/s41467-021-23922-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Xu M. (2015). Matrine Inhibits the Migratory and Invasive Properties of Nasopharyngeal Carcinoma Cells. Mol. Med. Rep. 11, 4158–4164. 10.3892/mmr.2015.3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G., Wu C. (2020). ZFPM2-AS1 Facilitates Cell Growth in Esophageal Squamous Cell Carcinoma via Up-Regulating TRAF4. Biosci. Rep. 40, BSR20194352. 10.1042/bsr20194352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q., Li J., Peng J., Liu Z., Liu J., Zhang H., et al. (2019). E4F1 Silencing Inhibits the Cell Growth through Cell‐cycle Arrest in Malignant Transformed Cells Induced by Hydroquinone. J. Biochem. Mol. Toxicol. 33, e22269. 10.1002/jbt.22269 [DOI] [PubMed] [Google Scholar]
- To K. K. W., Fong W., Cho W. C. S. (2021). Immunotherapy in Treating EGFR-Mutant Lung Cancer: Current Challenges and New Strategies. Front. Oncol. 11, 635007. 10.3389/fonc.2021.635007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W. H., Mu J. F., Zhang S. P. (2020). LINC00346 Accelerates the Malignant Progression of Colorectal Cancer via Competitively Binding to miRNA-101-5p/MMP9. Eur. Rev. Med. Pharmacol. Sci. 24, 6639–6646. 10.26355/eurrev_202006_21650 [DOI] [PubMed] [Google Scholar]
- Wang F., Chen J. G., Wang L. L., Yan Z. Z., Chen S. P., Wang X. G. (2017). Up-regulation of LINC00346 Inhibits Proliferation of Non-small Cell Lung Cancer Cells through Mediating JAK-STAT3 Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 21, 5135–5142. 10.26355/eurrev_201711_13830 [DOI] [PubMed] [Google Scholar]
- Wang F., Ge J., Huang S., Zhou C., Sun Z., Song Y., et al. (2021a). KLF5/LINC00346/miR148a3p axis Regulates Inflammation and Endothelial Cell Injury in Atherosclerosis. Int. J. Mol. Med. 48, 152. 10.3892/ijmm.2021.4985 [DOI] [PubMed] [Google Scholar]
- Wang M., Xu Y., Yang M., Jiang D., Chen Y., Jiang J., et al. (2020). Conversion Therapy for Advanced Pancreatic Cancer: the Case Series and Literature Review. Front. Pharmacol. 11, 579239. 10.3389/fphar.2020.579239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Guo Y., Wang C., Wang Q., Yan G. (2021b). Long Noncoding RNA ZEB1-AS1 Downregulates miR-23a, Promotes Tumor Progression, and Predicts the Survival of Oral Squamous Cell Carcinoma Patients. Ott 14, 2699–2710. 10.2147/ott.s297209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xue D., Li Y., Pan X., Zhang X., Kuang B., et al. (2016). The Long Noncoding RNA MALAT-1 Is a Novel Biomarker in Various Cancers: a Meta-Analysis Based on the GEO Database and Literature. J. Cancer 7, 991–1001. 10.7150/jca.14663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Ren Z., Li R., Ge J., Zhang G., Xin Y., et al. (2021c). Multi-omics Integrative Bioinformatics Analyses Reveal Long Non-coding RNA Modulates Genomic Integrity via Competing Endogenous RNA Mechanism and Serves as Novel Biomarkers for Overall Survival in Lung Adenocarcinoma. Front. Cel Dev. Biol. 9, 691540. 10.3389/fcell.2021.691540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wang D., Han S., Wang N., Mo F., Loo T. Y., et al. (2013). Bioactivity-guided Identification and Cell Signaling Technology to Delineate the Lactate Dehydrogenase A Inhibition Effects of Spatholobus Suberectus on Breast Cancer. PLoS One 8, e56631. 10.1371/journal.pone.0056631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Wang X., Lv L., Zheng Y., Zhang N., Yang M. (2019). The Emerging Role of Noncoding RNAs in Colorectal Cancer Chemoresistance. Cell Oncol. 42, 757–768. 10.1007/s13402-019-00466-8 [DOI] [PubMed] [Google Scholar]
- Wu L., Liu S., Qi H., Cai H., Xu M. (2020). Research Progress on Plant Long Non-coding RNA. Plants 9, 408. 10.3390/plants9040408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Zheng X., Fu Y., Qi J., Li M., Ma M., et al. (2019). Long Noncoding RNA-Maternally Expressed Gene 3 Contributes to Hypoxic Pulmonary Hypertension. Mol. Ther. 27, 2166–2181. 10.1016/j.ymthe.2019.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Wang Y., Zhou K., Wu J., Zhang Z., Zhang J., et al. (2020a). Identification of an Extracellular Vesicle-Related Gene Signature in the Prediction of Pancreatic Cancer Clinical Prognosis. Biosci. Rep. 40, BSR20201087. 10.1042/bsr20201087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Brown A. N., Waddell N. J., Liu X., Kaplan G. J., Chitaman J. M., et al. (2020b). Role of Long Noncoding RNA Gas5 in Cocaine Action. Biol. Psychiatry 88, 758–766. 10.1016/j.biopsych.2020.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Wu Q., Lu D., Yu J., Rao Y., Kou Z., et al. (2020c). A Systematic Study of Critical miRNAs on Cells Proliferation and Apoptosis by the Shortest Path. BMC Bioinformatics 21, 396. 10.1186/s12859-020-03732-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Xu Z., Zhu K., Lin J., Ye B. (2021). LINC00346 Sponges miR-30c-2-3p to Promote the Development of Lung Adenocarcinoma by Targeting MYBL2 and Regulating CELL CYCLE Signaling Pathway. Front. Oncol. 11, 687208. 10.3389/fonc.2021.687208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T.-p., Ma P., Wang W.-y., Shuai Y., Wang Y.-f., Yu T., et al. (2019). KLF5 and MYC Modulated LINC00346 Contributes to Gastric Cancer Progression through Acting as a Competing Endogeous RNA and Indicates Poor Outcome. Cell Death Differ 26, 2179–2193. 10.1038/s41418-018-0236-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W.-M., Yang K., Jiang L.-J., Hu J.-Q., Zhou X.-Z. (2018). Integrated Modules Analysis to Explore the Molecular Mechanisms of Phlegm-Stasis Cementation Syndrome with Ischemic Heart Disease. Front. Physiol. 9, 7. 10.3389/fphys.2018.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Zheng J., Liu X., Xue Y., He Q., Dong Y., et al. (2020a). Role of ANKHD1/LINC00346/ZNF655 Feedback Loop in Regulating the Glioma Angiogenesis via Staufen1-Mediated mRNA Decay. Mol. Ther. - Nucleic Acids 20, 866–878. 10.1016/j.omtn.2020.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Liu M., Li M., Zhang S., Hiju H., Sun J., et al. (2020b). Epigenetic Modulations of Noncoding RNA: a Novel Dimension of Cancer Biology. Mol. Cancer 19, 64. 10.1186/s12943-020-01159-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Liu D., Wu D., Liu F., Liu C. (2019). RETRACTED ARTICLE: The Biological Function of Long Noncoding RNA FAL1 in Oesophageal Carcinoma Cells. Artif. Cell Nanomedicine, Biotechnol. 47, 896–903. 10.1080/21691401.2019.1573738 [DOI] [PubMed] [Google Scholar]
- Yao H., Yang L., Tian L., Guo Y., Li Y. (2020). LncRNA MSC-AS1 Aggravates Nasopharyngeal Carcinoma Progression by Targeting miR-524-5p/nuclear Receptor Subfamily 4 Group A Member 2 (NR4A2). Cancer Cel Int 20, 138. 10.1186/s12935-020-01202-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yapasert R., Lertprasertsuk N., Subhawa S., Poofery J., Sripanidkulchai B., Banjerdpongchai R. (2020). Antitumor Efficacy of the Herbal Recipe Benja Amarit against Highly Invasive Cholangiocarcinoma by Inducing Apoptosis Both In Vitro and In Vivo . Ijms 21, 5669. 10.3390/ijms21165669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye T., Ding W., Wang N., Huang H., Pan Y., Wei A. (2017). Long Noncoding RNA Linc00346 Promotes the Malignant Phenotypes of Bladder Cancer. Biochem. Biophysical Res. Commun. 491, 79–84. 10.1016/j.bbrc.2017.07.045 [DOI] [PubMed] [Google Scholar]
- Yin Y. Z., Zheng W. H., Zhang X., Chen Y. H., Tuo Y. H. (2020). LINC00346 Promotes Hepatocellular Carcinoma Progression via Activating the JAK‐STAT3 Signaling Pathway. J. Cel. Biochem. 121, 735–742. 10.1002/jcb.29319 [DOI] [PubMed] [Google Scholar]
- Yip S.-C., El-Sibai M., Coniglio S. J., Mouneimne G., Eddy R. J., Drees B. E., et al. (2007). The Distinct Roles of Ras and Rac in PI 3-kinase-dependent Protrusion during EGF-Stimulated Cell Migration. J. Cel Sci. 120, 3138–3146. 10.1242/jcs.005298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Yang Y., Xu Z., Lan C., Chen C., Li C., et al. (2020). Long Noncoding RNA Ahit Protects against Cardiac Hypertrophy through SUZ12 (Suppressor of Zeste 12 Protein Homolog)-Mediated Downregulation of MEF2A (Myocyte Enhancer Factor 2A). Circ. Heart Fail. 13, e006525. 10.1161/circheartfailure.119.006525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Li C., Sun Z. (2018). Long Non-coding RNA LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 Are Novel Prognostic Markers for Pancreatic Cancer. Am. J. Transl. Res. 10, 2648–2658. [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Chen X. (2020). A Positive Feedback Loop Involving the LINC00346/β-Catenin/MYC axis Promotes Hepatocellular Carcinoma Development. Cel Oncol. 43, 137–153. 10.1007/s13402-019-00478-4 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Liu Q., Zheng S., Liu T., Yang L., Han X., et al. (2021). Shikonin Inhibits Tumor Growth of ESCC by Suppressing PKM2 Mediated Aerobic Glycolysis and STAT3 Phosphorylation. J. Cancer 12, 4830–4840. 10.7150/jca.58494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Su W., Sun Y., Wu Z. (2020). WBSCR22 Competes with Long Non-coding RNA Linc00346 for miR-509-5p Binding Site to Regulate Cancer Stem Cell Phenotypes of Colorectal Cancer. Biochem. Genet. 58, 384–398. 10.1007/s10528-020-09949-y [DOI] [PubMed] [Google Scholar]
- Zhao X., Gan L., Yan C., Li C., Sun Q., Wang J., et al. (2019). Genome-wide Identification and Characterization of Long Non-coding RNAs in Peanut. Genes 10, 536. 10.3390/genes10070536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Hong W., Gao M., Yi E., Zhang J., Hao B., et al. (2019). Long Noncoding RNA COPDA1 Promotes Airway Smooth Muscle Cell Proliferation in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Cel Mol. Biol. 61, 584–596. 10.1165/rcmb.2018-0269OC [DOI] [PubMed] [Google Scholar]
- Zhu J., Li B., Ji Y., Zhu L., Zhu Y., Zhao H. (2019). Βelemene Inhibits the Generation of Peritoneum Effusion in Pancreatic Cancer via Suppression of the HIF1AVEGFA Pathway Based on Network Pharmacology. Oncol. Rep. 42, 2561–2571. 10.3892/or.2019.7360 [DOI] [PMC free article] [PubMed] [Google Scholar]