Abstract
Background and Aims:
Colonoscopy is commonly performed for colorectal cancer screening in the United States. Reports are often generated in a non-standardized format and are not always integrated into electronic health records. Thus, this information is not readily available for streamlining quality management, participating in endoscopy registries, or reporting of patient- and center-specific risk factors predictive of outcomes. We aim to demonstrate the use of a new hybrid approach using natural language processing of charts that have been elucidated with optical character recognition processing (OCR/NLP hybrid) to obtain relevant clinical information from scanned colonoscopy and pathology reports, a technology co-developed by Cleveland Clinic and eHealth Technologies (West Henrietta, NY, USA).
Methods:
This was a retrospective study conducted at Cleveland Clinic, Cleveland, Ohio, and the University of Minnesota, Minneapolis, Minnesota. A randomly sampled list of outpatient screening colonoscopy procedures and pathology reports was selected. Desired variables were then collected. Two researchers first manually reviewed the reports for the desired variables. Then, the OCR/NLP algorithm was used to obtain the same variables from 3 electronic health records in use at our institution: Epic (Verona, Wisc, USA), ProVation (Minneapolis, Minn, USA) used for endoscopy reporting, and Sunquest PowerPath (Tucson, Ariz, USA) used for pathology reporting.
Results:
Compared with manual data extraction, the accuracy of the hybrid OCR/NLP approach to detect polyps was 95.8%, adenomas 98.5%, sessile serrated polyps 99.3%, advanced adenomas 98%, inadequate bowel preparation 98.4%, and failed cecal intubation 99%. Comparison of the dataset collected via NLP alone with that collected using the hybrid OCR/NLP approach showed that the accuracy for almost all variables was >99%.
Conclusions:
Our study is the first to validate the use of a unique hybrid OCR/NLP technology to extract desired variables from scanned procedure and pathology reports contained in image format with an accuracy >95%.
INTRODUCTION
Colonoscopy is a common screening modality for colorectal cancer (CRC) in the United States. The 2012 Survey of Endoscopic Capacity estimated that more than 15 million colonoscopies were performed in 2012, and an additional 10.5 million colonoscopies could be performed.1 Observation data have shown that colonoscopy screening has led to lower CRC incidence and mortality.2–4 However, increasing evidence suggests that examination quality may have an impact on its effectiveness, that is, the adenoma detection rate is inversely related to the risk of interval CRC.5 In 2015, the American Society for Gastrointestinal Endoscopy (ASGE)/American College of Gastroenterology (ACG) Task Force on Quality in Endoscopy updated the list of quality indicators for performing colonoscopy.6 Unfortunately, the information needed to assess colonoscopy examination quality is often embedded in non-standardized colonoscopy procedure reports of varying formats within electronic health records (EHRs), requiring time-consuming and costly manual data extraction for accurate reporting.
At our institution, ProVation (Minnesota, Minn, USA) software for managing procedure reporting and Sunquest PowerPath (Tucson, Ariz, USA) software for managing pathology reporting, are now integrated with our EHR Epic (Verona, Wisc, USA). However, smaller institutions and endoscopy centers continue to rely on EHRs that are not integrated, and procedure/pathology reports need to be scanned and uploaded into their EHRs to enable other physicians and ancillary health care personnel, who routinely do not have access to endoscopy reporting systems, to view these results.7 Thus, the information contained within these procedure and pathology results is not readily available for streamlining quality management, guiding clinical research initiatives, participating in endoscopy registries for meaningful use, or in reporting of patient- and center-specific risk factors predictive of poor colonoscopy outcomes, such as cecal intubation rates, quality of bowel preparation, and adenoma detection rates (ADRs). Manual extraction of important clinical variables from these procedure/pathology reports is limited by its labor-, resource-, and time-intensive nature.8,9 The lack of tools that allow error-free extraction of high-quality information has remained a major obstacle in navigating these unstructured data sources to improve the efficiency and accuracy of patient care.6
Natural language processing (NLP) is a computer-based linguistic technique that has gained prominence for its role in obtaining pertinent clinical information in an organized fashion from semi-structured and more recently, unstructured, data sources.10–12 This not only has the potential to have an impact on clinical practice but has opened pathways toward investigational and quality research that was previously unfathomable.13 The NLP-based technical approach has already shown promise in leveraging data from echocardiography reports with >90% accuracy14 and from colonoscopy reports for ADRs and advanced ADRs with >90% accuracy across multiple institutions that rely on different EHRs and use different text formats for reporting colonoscopy findings.7,15–17
Although extremely useful, the application of NLP is limited in scope because it requires machine-readable clinical text and does not work with printed or scanned documents. Optical character recognition (OCR) technology enables the conversion of scanned paper documents into editable and searchable text data. The application of NLP on documents created using OCR-based technology has not yet been studied. Our study aims to demonstrate the use of a new hybrid approach using NLP of charts that have been elucidated with OCR processing (OCR/NLP hybrid) to obtain relevant clinical information from scanned colonoscopy reports and pathology reports.
METHODS
This was a retrospective study conducted at Cleveland Clinic, Cleveland, Ohio, and the University of Minnesota. The study was approved by the institutional review board at both institutions. The NLP algorithm was used to obtain data on all patients who underwent a screening colonoscopy (N = 35,914) at Cleveland Clinic between 2010 and 2014. We had collected data for a previous study on 2530 patients after excluding those with previous colonoscopies, procedures are done for diagnostic indications, and those done on patients <40 years old. These patients were arranged in numerical order of their medical record number and every third patient was included in our study for a total of 589 procedures. This was done to use the data that was readily available to us. Data and desired variables were then collected in 3 ways. First, the colonoscopy procedure reports were manually reviewed by 2 researchers (S.L. and M.I.) to ensure that the previously collected information on the variables of interest was accurate. Any discrepancies between the datasets obtained by the 2 researchers were resolved by a third independent researcher (S.S.). Second, the NLP algorithm was used to obtain the same variables directly from the 3 EHRs being used at our institution: Epic (Verona, Wisc, USA), ProVation (Minneapolis, Minn, USA) used for endoscopy reporting, and Sunquest PowerPath (Tucson, Ariz, USA) used for pathology reporting. Third, the hybrid OCR/NLP technology, co-developed by Cleveland Clinic and eHealth Technologies (West Henrietta, NY, USA), was used to evaluate a scanned copy of each procedure and pathology report to extract our variables of interest. Because our NLP algorithm was developed using the procedure notes written only at our institution, the external applicability of our algorithm to extract data from procedure notes written at a different institution would have remained questionable due to the possibility of significant variation in procedure note templates. To overcome this, data were also obtained through manual review as well as via the OCR/NLP approach on 4 variables (indication, quality of bowel preparation, cecal intubation, and polyp detection) from 30 randomly selected colonoscopy reports of patients presenting to the gastroenterology clinic at the University of Minnesota. We only selected procedures that had been done at outside facilities and notes that were scanned into the EHR at the University of Minnesota (all of these procedure notes were written using the NextGen healthcare information systems v.5; NXGN Management, LLC, Irvine, Calif, USA).
Variables
Polyp detection rate (number of colonoscopies where single or multiple polyps were resected and successfully retrieved), location of polyps, ADR (number of colonoscopies where single or multiple adenomas were resected and successfully retrieved), advanced ADR (advanced adenoma defined as adenoma >10 mm in size, adenoma with a villous or a tubulovillous component, and/or adenoma with high-grade dysplasia), rate of inadequate bowel preparation (defined as a classification of poor, fair, and/or inadequate on the Aronchick scale), and successful cecal intubation rate (number of colonoscopies where the colonoscopy was advanced at least as far as the cecum).
Cleveland Clinic natural language processing algorithm
The NLP engine was designed and developed in Prolog, traditionally a language used for artificial intelligence, by a member of the Cleveland Clinic Digestive Disease & Surgery Institute (J.M.). Prolog is a general-purpose logic programming language, and unlike many other programming languages, it is a declarative language as opposed to a procedural language. The program logic is expressed in terms of relations, represented as facts and rules, and no distinction is made between code and data.
The engine has several components. A Structured Query Language (SQL) interface allows the NLP engine to read the colonoscopy notes from a view or table within an SQL database. The parser, developed using Prolog, tokenizes a paragraph from the procedure/pathology note into numbered sentences and sentences into numbered words, allowing them to be searched and “read” for variables or meaning. The order and position of certain words in the parsed data are evaluated to get a value or interpretation. Several algorithms then determine if a polyp was found, the size of the polyp, and the location of the polyp. Another interface then allows the NLP engine to write the parsed discreet variables back into an SQL table. The procedure data (patient ID, date, time, physical location) are written into a procedure table, and the findings are written in another table based on a relational model because the relationship between the procedure and the findings can be one to none or one to many. The polyp location is also “normalized” to standard locations within the colon. This is necessary so that the polyp findings can be associated with the pathology findings later. The pathology parser reads through the final diagnosis of the pathology report and returns the pathology findings for each polyp by location. These data (procedure details, findings, and pathology) are then “joined” to produce a report that has the procedure details, findings for each polyp with number, size, and location as well as the pathologic finding for each polyp.
Our initial iteration of the algorithm (developed in 2015) was used to parse 295,252 colonoscopy procedure notes (spanning 25 years) associated with 63,284 pathology notes. Initial manual review done by one of the authors (J.M.) showed 88% agreement with the parsed data. The parsing algorithm has evolved since its inception. The reports with missed variables were evaluated, and the algorithm was refined to start extracting the missing data and to accommodate the variability in documentation between different endoscopists. Our initial algorithm also sometimes missed the location and number of polyps, because that information was mentioned in the next sentence of the findings paragraph. The algorithm was then adapted to look at the next sentence if no location was found in the first sentence, which solved this issue.
Optical character recognition
The colonoscopy reports from our study dataset were printed from ProVation (Minneapolis, Minn, USA) along with any accompanying pathology reports from Sunquest PowerPath deidentified, randomly assigned with a computer-generated participant number for patient identification, and then scanned as image files. The image file was then reprinted and scanned again to reflect a “real-world” setting because such documents often have compromised image quality due to being faxed or photocopied multiples times. The image files were then sent to eHealth Technology (West Henrietta, NY, USA). eHealth Technology then used proprietary OCR technology to convert the scanned images into editable text files. These text files were then securely transmitted back to be analyzed by our NLP algorithm as detailed above.
Statistical analysis
Sensitivity, specificity, predictive values, and accuracy were calculated to assess the validity of NLP and the OCR/NLP hybrid for reporting the variables with manual annotation (criterion standard). Then, OCR/NLP was compared with NLP alone (proxy criterion standard). To make these comparisons, the frequency and percentage of true positives (presence noted per comparator and criterion/proxy criterion standard), false positives (presence noted per comparator, but not by criterion/proxy criterion standard), false negatives (absence noted per comparator, but present per criterion/proxy criterion standard), and true negatives (absence noted per comparator and criterion/proxy criterion standard) were calculated. Sensitivity, specificity, positive predictive value (PPV), negative predictive value, and accuracy were then calculated.
Results
A total of 589 colonoscopy procedures were included in the study and there were 262 (44.4%) corresponding pathology reports. The overall ADR was 23.1% in our study, and the rate of inadequate bowel preparation was 16.9%. The ADR was 4.1% and the failed cecal intubation rate was 1.6%. The comparison of the accuracy among the 3 different approaches are described in Figure 1. The flowchart of all patients included in our study is detailed in Figure 2.
Figure 1.
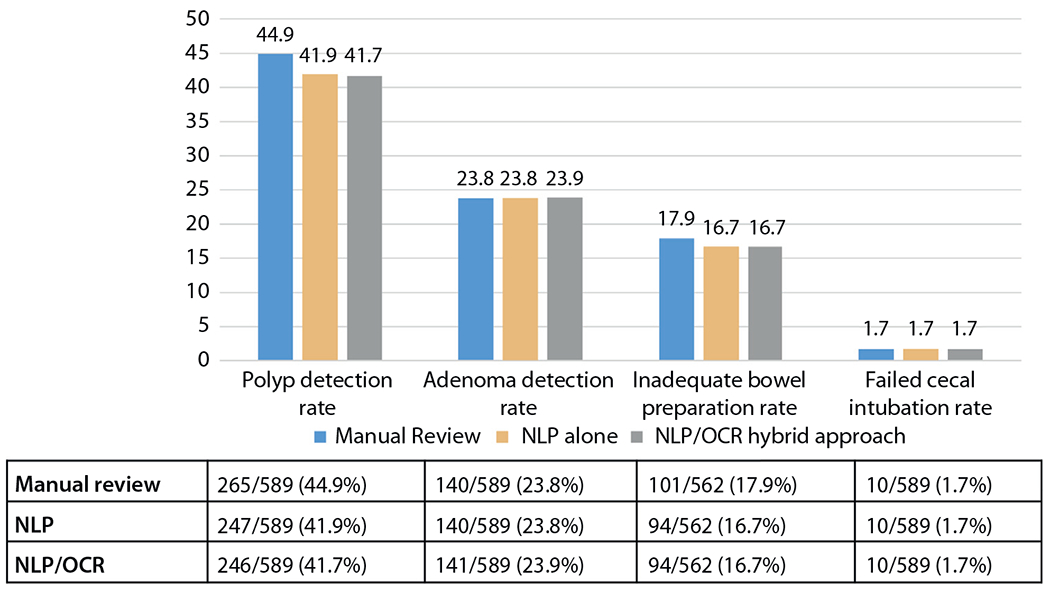
Reporting of colonoscopy quality parameters by each method: manual review, natural language processing (NLP) alone, natural language processing/optical character recognition (OCR) hybrid approach.
Figure 2.
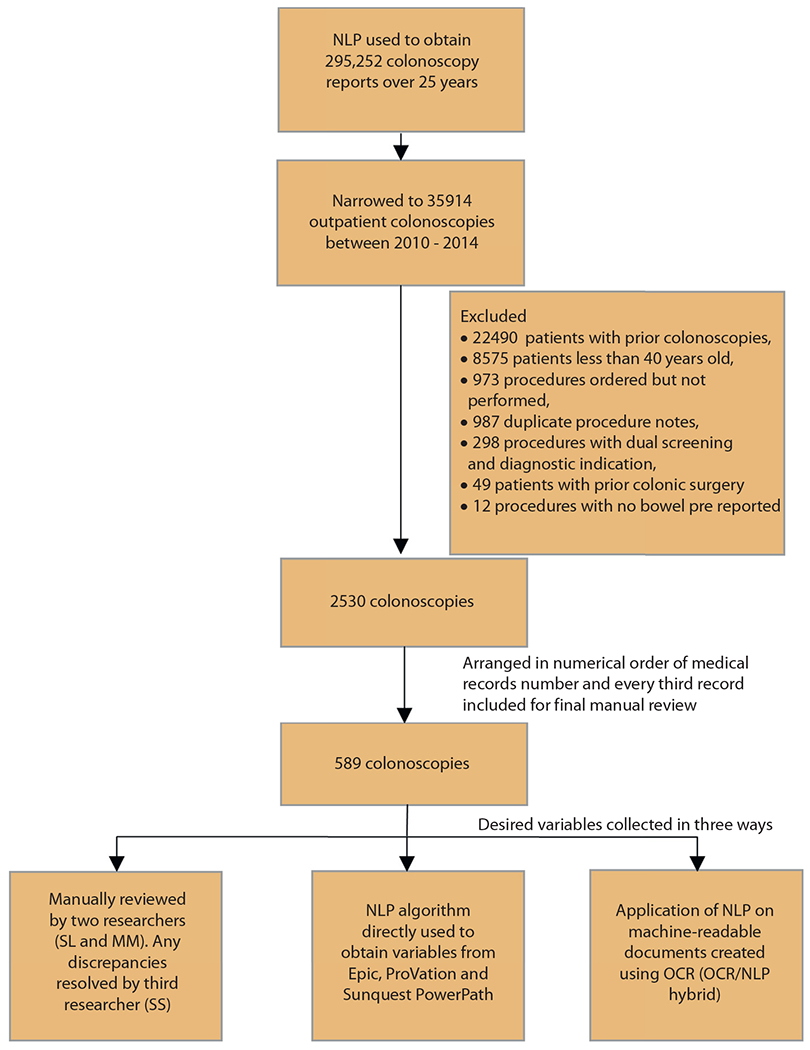
Flowchart for inclusion of patients in the study. NLP, Natural language processing; OCR, optical character recognition.
Effectiveness of NLP
Table 1 details the findings comparing our NLP findings with the manually collected dataset, which is our criterion standard. The NLP platform was able to detect polyps with an accuracy of 96%, adenomas with 98.4% accuracy, sessile serrated polyps with 99.2% accuracy, advanced adenomas with 99.3% accuracy, inadequate bowel preparation with 98.4% accuracy, and failed cecal intubation with 99% accuracy. The algorithm was able to detect high-grade dysplasia with 100% accuracy.
TABLE 1.
Comparing the validity of natural language processing with manual data collection (criterion standard)
| Variables | Sensitivity, n/N (%) | Specificity, n/N (%) | Positive predictive value, n/N (%) | Accuracy, n/N (%) |
|---|---|---|---|---|
| Polyp | 244/265 (92.1) | 321/324 (99.1) | 244/247 (98.8) | 565/589 (95.9) |
|
| ||||
| Adenoma | 136/140 (97.1) | 445/449 (99.1) | 136/140 (97.1) | 581/589 (98.6) |
|
| ||||
| Sessile serrated polyp | 24/28 (85.7) | 560/561 (99.8) | 24/25 (96) | 584/589 (99.2) |
|
| ||||
| High-grade dysplasia | 3/3 (100) | 586/586 (100) | 3/3 (100) | 589/589 (100) |
|
| ||||
| Advanced adenoma | 24/26 (92.3) | 561/563 (99.6) | 24/26 (92.3) | 585/589 (99.3) |
|
| ||||
| Inadequate preparation | 93/101 (92.1) | 460/461 (99.8) | 93/94 (98.9) | 553/562 (98.4) |
|
| ||||
| Failed cecal intubation | 7/10 (70) | 576/579 (99.5) | 7/10 (70) | 583/589 (99) |
Effectiveness of hybrid OCR/NLP technology
Table 2 details the comparison between the hybrid OCR/NLP approach for data extraction with the manually collected dataset. The accuracies in the detection of variables were similar to the use of NLP alone. The accuracy for detection of polyps was 95.6%, 98.5% for detection of adenomas, 99.3% for detection of sessile serrated polyps, 99.3% for detection of advanced adenomas, 98.4% for detection of inadequate bowel preparation, and 99% for detection of failed cecal intubation. The detection of high-grade dysplasia remained at 100%. On comparison of the dataset collected via NLP alone with the dataset collected from the hybrid OCR/NLP approach, accuracy for almost all variables of interest was >99% (Table 3 and Fig. 2).
TABLE 2.
Comparing the validity of natural language processing of charts elucidated with optical character recognition with manual data collection (criterion standard)
| Variables | Sensitivity, n/N (%) | Specificity, n/N (%) | Positive predictive value, n/N (%) | Accuracy, n/N (%) |
|---|---|---|---|---|
| Polyp | 243/265 (91.7) | 321/324 (99.1) | 243/246 (98.8) | 564/589 (95.8) |
|
| ||||
| Polyp location | ||||
|
| ||||
| Distal | 155/172 (90.1) | 416/417 (99.8) | 155/156 (99.4) | 571/589 (96.9) |
|
| ||||
| Anus | 4/4 (100) | 584/585 (99.8) | 4/5 (80) | 588/589 (99.8) |
|
| ||||
| Rectum | 41/62 (66.1) | 517/527 (98.1) | 41/51 (80.4) | 558/589 (94.7) |
|
| ||||
| Sigmoid | 89/92 (96.7) | 496/497 (99.8) | 89/90 (98.9) | 585/589 (99.3) |
|
| ||||
| Descending | 42/45 (93.3) | 543/544 (99.8) | 42/43 (97.7) | 585/589 (99.3) |
|
| ||||
| Splenic flexure | 4/4 (100) | 584/585 (99.8) | 4/5 (80) | 588/589 (99.8) |
|
| ||||
| Proximal | 152/160 (95) | 428/429 (99.8) | 152/153 (99.4) | 580/589 (98.5) |
|
| ||||
| Transverse | 67/71 (94.4) | 517/518 (99.8) | 67/68 (98.5) | 584/589 (99.2) |
|
| ||||
| Hepatic flexure | 22/24 (91.7) | 565/565 (100) | 22/22 (100) | 587/589 (99.7) |
|
| ||||
| Ascending | 66/73 (90.4) | 516/516 (100) | 66/66 (100) | 582/589 (98.8) |
|
| ||||
| Cecum | 37/39 (94.9) | 549/550 (99.8) | 37/38 (97.4) | 586/589 (99.5) |
|
| ||||
| Polyp pathology | ||||
|
| ||||
| Adenoma | 136/140 (97.1) | 444/449 (98.9) | 136/141 (96.5) | 580/589 (98.5) |
|
| ||||
| Sessile serrated polyp | 25/28 (89.3) | 560/561 (99.8) | 25/26 (96.2) | 585/589 (99.3) |
|
| ||||
| High-grade dysplasia | 3/3 (100) | 586/586 (100) | 3/3 (100) | 589/589 (100) |
|
| ||||
| Advanced adenoma | 15/26 (57.7) | 562/563 (99.8) | 15/16 (93.8) | 577/589 (98) |
|
| ||||
| Inadequate preparation | 93/101 (92.1) | 460/461 (99.8) | 93/94 (98.9) | 553/562 (98.4) |
|
| ||||
| Failed cecal intubation | 7/10 (70) | 576/579 (99.5) | 7/10 (70) | 583/589 (99) |
TABLE 3.
Comparing the validity of natural language processing of charts elucidated with optical character recognition with charts processed with natural language processing (proxy criterion standard)
| Variables | Sensitivity, n/N (%) | Specificity, n/N (%) | Positive predictive value, n/N (%) | Accuracy, n/N (%) |
|---|---|---|---|---|
| Polyp | 246/247 (99.6) | 342/342 (100) | 246/246 (100) | 588/589 (99.8) |
|
| ||||
| Adenoma | 138/140 (98.6) | 446/449 (99.3) | 138/141 (97.9) | 584/589 (99.2) |
|
| ||||
| Sessile serrated polyp | 25/25 (100) | 563/564 (99.8) | 25/26 (96.2) | 588/589 (99.8) |
|
| ||||
| High-grade dysplasia | 3/3 (100) | 586/586 (100) | 3/3 (100) | 589/589 (100) |
|
| ||||
| Advanced adenoma | 16/26 (61.5) | 563/563 (100) | 16/16 (100) | 579/589 (98.3) |
|
| ||||
| Inadequate preparation | 97/97 (100) | 464/464 (100) | 97/97 (100) | 561/561 (100) |
|
| ||||
| Failed cecal intubation | 10/10 (100) | 579/579 (100) | 10/10 (100) | 589/589 (100) |
Effectiveness of hybrid OCR/NLP technology in a different endoscopy writing software
Our hybrid OCR/NLP technology had a sensitivity, specificity, PPV, and accuracy of 100% when extracting data on the indication, quality of bowel preparation, cecal intubation, and polyp detection compared with manual annotation for 30 colonoscopy procedure notes from patients at the University of Minnesota.
DISCUSSION
Quality process and outcome metrics have been proposed by the joint ASGE-ACG taskforce around different procedure types, including colonoscopy.18 Thus, measurement and reporting of colonoscopy quality indicators have become the standard of care. Unfortunately, this has been limited by the lack of electronic tools that allow error-free extraction of these important clinical variables from procedure and pathology reports, causing us to rely on manual extraction of these data. Locally developed NLP tools have shown promise in leveraging data from colonoscopy reports with an accuracy >90% but require machine-readable clinical text. Our study is the first to demonstrate the use of a unique hybrid approach using OCR and subsequent NLP to extract desired variables from the scanned procedure and pathology reports contained in image format, with an accuracy >95%. Compared with a validated manual review of colonoscopy reports, the hybrid OCR/NLP approach yielded high levels of PPV, sensitivity, specificity, and accuracy for the location of polyps, polyp pathology, bowel preparation quality, and cecal intubation.
NLP, one of the common big data analytical tools used in health care, has allowed institutions to electronically analyze and extract information from the unstructured free text (ie, endoscopy procedure reports) as an efficient alternative to manual data extraction.19 The utility of NLP in the documentation of several quality parameters for colonoscopies has already been studied at various institutions. Mehrotra et al17 at the University of Pittsburgh Medical Center health care system used a previously validated Java-based NLP tool to extract 21 variables (eg, examination indication, examination extent, bowel preparation quality) from colonoscopy and linked pathology reports and found an average accuracy over all the variables of 89% (range, 62%-100%) compared with manual review. Imler et al,15,20 using cTAKES, a Java-based NLP system originally developed by Mayo Clinic, showed an accuracy of 98% for the highest level of pathology, with accuracy values for location, size, and number of 97%, 96%, and 84%, respectively, compared with manual review. Gawron et al12 also used a Java-based NLP and reported a PPV of 96% for screening examinations, 98% for completeness of colonoscopy, 98% for adequate bowel preparation quality, and 95% for histology of polyps compared with manual review.16 Raju and colleagues21 developed an NLP tool using the C# programming language and reported overall ADRs by NLP and manual review to be identical. Their NLP identified screening examinations with an accuracy of 91.3% (manual review, 87.8%), 99.4% for adenoma, and 100% for sessile serrated adenoma diagnosis.21,22 This is comparable with Cleveland Clinic’s NLP tool, which demonstrated >95% accuracy in identifying polyp findings and examination quality.
One of the major barriers to widespread adoption of NLP software for extraction of data is the local development and validation, which limits applicability to diverse health care settings and the predominant use of colonoscopy reports derived from highly structured template-driven software systems (eg, Pentax, EndoWorks, EndoPro, ProVation), limiting linguistic variation. Recently, Lee et al22 demonstrated the use of commercially available NLP software (Linguamatics 12E, www.linguamatics.com; United Kingdom) and its comparable accuracy in identifying examination quality and polyp findings from colonoscopy reports and unstructured progress notes across multiple medical centers with different reporting formats, addressing many of the concerns regarding locally developed NLP tools. Compared with manual review, the accuracy for screening indication was 98.2% (97.0%-99.4%), cecal intubation 99.8% (99.5%-100%), bowel preparation adequacy 100% (100%-100%), and for polyp(s) ≥10 mm, 95.2% (88.8%-100%).22 Our NLP algorithm, although developed using the procedure note formats only at our institution, was also 100% accurate in the extraction of data for 4 variables from procedure notes reported at another institution in a different state, using different endoscopy reporting software.
Another obstacle limiting the large-scale utility of NLP is the initial investment needed for the extensive and costly programming efforts for development, but we believe that the up-front one-time costs of NLP development are worth the savings in the long run. In addition, with the availability of commercial NLP software,22 the cost of using NLP for individual health care systems will reduce even further. Our initial iteration of the NLP algorithm was developed over 6 months by one of our authors (J.M.) and has since evolved with significant improvement in overall accuracy as described in our Methods section. We estimate that a total of 150 man-hours were invested in the development and subsequent changes to our algorithm. Our NLP algorithm now takes under 30 minutes to extract data on all colonoscopy procedures ever done at our institution since the introduction of EHRs. Contrasting this with manual data collection, both authors who manually extracted the data took about 6 to 8 minutes per patient, which equates to a total of 160 man-hours for annotating data from fewer than 600 patients.
An additional limiting factor that has plagued NLP is the inability to extract data from documents in image format because it requires machine-readable clinical text. We have presented a novel approach to overcome this barrier by the introduction of OCR that allows recognition/processing of the text contained in printed or scanned documents. The OCR technology performed exceptionally well in our study, but its success can be partially attributed to the use of high-quality printers and scanners to print and create scanned images of our procedure and pathology reports. This is recognizably one of the major limitations of our study. In addition, understandably, the ability of OCR to extract text from medium- and low-quality images will need to be verified before the implementation of this technology on a wider scale. However, there is great potential for this technology. Health care systems where procedure and pathology reports are scanned into the EHR, elucidation of these reports by OCR, and then subsequent NLP will assist these practices in extraction of colonoscopy (and other procedures, eg, quality measurement for ERCP using NLP) quality parameters for internal monitoring20 and allow them to report and participate in national registry programs such as GI Quality Improvement Consortium, Ltd. It will also allow practices to use a merit-based incentive payment system, which most avoid given the burdensome labor costs. Another potentially large-scale application for the OCR technology will be assistance with converting scanned records from outside the hospital into machine-readable text, allowing for easy access and subsequent data extraction for both clinical and research purposes.
In our study, the overall ADR was 23.1%, which is lower than the quality standards set by ASGE, likely a result of limited sample size and the inclusion of colonoscopies performed by endoscopists with a low number of annual colonoscopies, a known predictor for low ADR. Rates of inadequate preparation were also slightly higher than those reported in the literature at around 17%, likely a result of the small sample size used for the sole purpose of validating our NLP and hybrid OCR/NLP approaches as well as the inclusion of fair preparation on the Aronchick Scale in the inadequate bowel preparation group. Advanced adenoma detection was around 4% with 2 false positives and 2 false negatives, which are comparable with those of previous studies. On comparison of the hybrid OCR/NLP approach with NLP alone (Table 3), the accuracy in detection of almost all variables was 99% to 100%, which alludes to the fact that most of the false positives and false negatives in Table 2 (comparison of the OCR/NLP hybrid with the manually collected dataset) are likely a result of the limitation in the parsing of the data via our NLP algorithm as opposed to a limitation in the conversion of the scanned image files into readable text data using OCR technology. On further review, we found that the biggest cause for discordance between the manually annotated data and data collected with NLP alone as well as with the hybrid OCR/NLP approach was due to unstructured and/or improperly documented procedure notes. Limited discordance seen between the OCR/NLP and NLP alone approach was due to poor image quality of the scanned procedure reports.
The results of this proof-of-concept study create a new frontier in the use of large-scale data extraction from scanned reports, which was previously limited by lack of appropriate technology. The process was previously expensive and time-consuming, but can now potentially be done accurately in a time- and labor-efficient manner. Future multicenter studies elaborating the use of OCR in combination with validated commercially available NLP tools will help substantiate the use of this novel technology on a larger scale, not only for measurement of procedure quality indicators but possibly also for multiple other venues in health care.
Acknowledgments
Shashank Sarvepalli is supported by a National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Institutional National Service Award (T32) from the National Institutes of Health under Award Number T32 DK083266.
Abbreviations:
- ACG
American College of Gastroenterology
- ADR
adenoma detection rate
- ASGE
American Society for Gastrointestinal Endoscopy
- CRC
colorectal cancer
- EHR
electronic health record
- NLP
natural language processing
- OCR
optical character recognition
- PPV
positive predictive value
- SQL
Structured Query Language
- SSP
sessile serrated polyp
REFERENCES
- 1.Joseph DA, Meester RG, Zauber AG, et al. Colorectal cancer screening: estimated future colonoscopy need and current volume and capacity. Cancer 2016;122:2479–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zauber AG, Winawer SJ, O’Brien MJ, et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med 2012;366:687–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winawer SJ, Zauber AG, Ho MN, et al. Prevention of colorectal cancer by colonoscopic polypectomy. The National Polyp Study Workgroup. N Engl J Med 1993;329:1977–81. [DOI] [PubMed] [Google Scholar]
- 4.Nishihara R, Wu K, Lochhead P, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med 2013;369: 1095–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corley DA, Jensen CD, Marks AR, et al. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med 2014;370:1298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rex DK, Schoenfeld PS, Cohen J, et al. Quality indicators for colonoscopy. Am J Gastroenterol 2015;110:72–90. [DOI] [PubMed] [Google Scholar]
- 7.Raju GS, Lum PJ, Slack RS, et al. Natural language processing as an alternative to manual reporting of colonoscopy quality metrics. Gastrointest Endosc 2015;82:512–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narula J Are we up to speed?: from big data to rich insights in CV imaging for a hyperconnected world. JACC Cardiovasc Imaging 2013;6:1222–4. [DOI] [PubMed] [Google Scholar]
- 9.Murdoch TB, Detsky AS. The inevitable application of big data to health care. JAMA 2013;309:1351–2. [DOI] [PubMed] [Google Scholar]
- 10.Pakhomov SS, Hemingway H, Weston SA, et al. Epidemiology of angina pectoris: role of natural language processing of the medical record. Am Heart J 2007;153:666–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wells QS, Farber-Eger E, Crawford DC. Extraction of echocardiographic data from the electronic medical record is a rapid and efficient method for study of cardiac structure and function. J Clin Bioinform 2014;4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garvin JH, DuVall SL, South BR, et al. Automated extraction of ejection fraction for quality measurement using regular expressions in Unstructured Information Management Architecture (UIMA) for heart failure. J Am Med Inform Assoc 2012;19:859–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maddox TM, Matheny MA. Natural language processing and the promise of big data: small step forward, but many miles to go. Circ Cardiovasc Qual Outcomes 2015;8:463–5. [DOI] [PubMed] [Google Scholar]
- 14.Nath C, Albaghdadi MS, Jonnalagadda SR. A natural language processing tool for large-scale data extraction from echocardiography reports. PLoS One 2016;11:e0153749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imler TD, Morea J, Kahi C, et al. Multi-center colonoscopy quality measurement utilizing natural language processing. Am J Gastroenterol 2015;110:543–52. [DOI] [PubMed] [Google Scholar]
- 16.Gawron AJ, Thompson WK, Keswani RN, et al. Anatomic and advanced adenoma detection rates as quality metrics determined via natural language processing. Am J Gastroenterol 2014;109:1844–9. [DOI] [PubMed] [Google Scholar]
- 17.Mehrotra A, Dellon ES, Schoen RE, et al. Applying a natural language processing tool to electronic health records to assess performance on colonoscopy quality measures. Gastrointest Endosc 2012;75:1233–9.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaminski MF, Regula J, Kraszewska E, et al. Quality indicators for colonoscopy and the risk of interval cancer. N Engl J Med 2010;362: 1795–803. [DOI] [PubMed] [Google Scholar]
- 19.Kreimeyer K, Foster M, Pandey A, et al. Natural language processing systems for capturing and standardizing unstructured clinical information: a systematic review. J Biomed Inform 2017;73:14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imler TD, Sherman S, Imperiale TF, et al. Provider-specific quality measurement for ERCP using natural language processing. Gastrointest Endosc 2018;87:164–73.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raju GS, Lum PJ, Slack RS, et al. Natural language processing as an alternative to manual reporting of colonoscopy quality metrics. Gastrointest Endosc 2015;82:512–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JK, Jensen CD, Levin TR, et al. Accurate identification of colonoscopy quality and polyp findings using natural language processing. J Clin Gastroenterol 2019;53:e25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]


