Significance
Cells coordinate the rates of major biological processes in response to environmental conditions such as nutrient availability and temperature. Coordination mechanisms in which effects on one process are compensated for by effects on another process maintain an appropriate cellular balance. Here, we uncover a compensatory link between cleavage/polyadenylation, which generates poly(A) tails at mRNA 3′ ends, and mRNA decay. This compensatory link suggests that during cleavage/polyadenylation in the nucleus, mRNA isoforms are marked in a manner that persists upon translocation to the cytoplasm. This mark affects the activity of mRNA degradation machinery, thus influencing mRNA stability and allowing cells to maintain relatively similar levels of mRNA isoforms even when cleavage/polyadenylation rates change in response to environmental conditions.
Keywords: mRNA isoforms, polyadenylation, mRNA decay, mRNA stability
Abstract
Cells have compensatory mechanisms to coordinate the rates of major biological processes, thereby permitting growth in a wide variety of conditions. Here, we uncover a compensatory link between cleavage/polyadenylation in the nucleus and messenger RNA (mRNA) turnover in the cytoplasm. On a global basis, same-gene 3′ mRNA isoforms with twofold or greater differences in half-lives have steady-state mRNA levels that differ by significantly less than a factor of 2. In addition, increased efficiency of cleavage/polyadenylation at a specific site is associated with reduced stability of the corresponding 3′ mRNA isoform. This inverse relationship between cleavage/polyadenylation and mRNA isoform half-life reduces the variability in the steady-state levels of mRNA isoforms, and it occurs in all four growth conditions tested. These observations suggest that during cleavage/polyadenylation in the nucleus, mRNA isoforms are marked in a manner that persists upon translocation to the cytoplasm and affects the activity of mRNA degradation machinery, thus influencing mRNA stability.
Cells coordinate the rates of major biological processes in accord with their growth rate, which can vary greatly depending on environmental conditions such as nutrients and temperature. As part of this coordination, cells have mechanisms in which effects on one process are compensated for by effects on another process to maintain an appropriate cellular balance. For example, yeast cells have a mechanism in which synthesis and degradation of messenger RNA (mRNA) are linked in a circular manner (1–3). This mechanism coordinates mRNA synthesis and degradation to maintain overall steady-state mRNA levels at the desired amounts. For example, mutant strains with reduced global levels of mRNA synthesis typically have steady-state mRNA levels that are comparable to those in wild-type cells (4–6), presumably due to reduced mRNA degradation.
In the yeast Saccharomyces cerevisiae, the average gene yields ∼50 3′ mRNA isoforms arising from different cleavage/polyadenylation sites in its 3′ untranslated region (3′UTR) (7–9). We and others previously measured the half-lives of >20,000 3′ mRNA isoforms originating from thousands of genes in yeast cells grown in YPD (yeast extract–peptone–dextrose) medium (10, 11). This was accomplished by measuring the relative levels of 3′ isoforms at multiple time points after blocking RNA polymerase (Pol) II transcription. The Pol II shutoff was achieved either by rapidly depleting Rpb1, the catalytic subunit of Pol II, from the nucleus (10) by the anchor-away method (12) or by thermally inactivating a temperature-sensitive Rpb1 derivative (11). The 3′ mRNA isoforms, even when they end just one or a few nucleotides apart, can differ dramatically with respect to mRNA stability (10, 11), structure throughout the 3′UTR (10, 13), and association with Pab1, the poly(A)-binding protein (10, 13).
The steady-state level of an mRNA isoform is determined by its rate of production and its rate of decay (3, 14, 15). In absolute terms, the rate of production depends on transcription, splicing, and cleavage/polyadenylation. In yeast, same-gene 3′ mRNA isoforms are transcribed from a common promoter through the coding region, and splicing is limited to a small subset of genes. As such, the relative rates of production of same-gene 3′ mRNA isoforms in yeast are determined by the relative rates of cleavage/polyadenylation at the sites that correspond to the specific isoform endpoints. Here, we use steady-state levels and half-lives of individual 3′ mRNA isoforms in yeast cells to calculate their relative rates of production. Unexpectedly, we find that, in four different growth conditions, the efficiency of cleavage/polyadenylation at a specific site is inversely related to stability of the corresponding 3′ mRNA isoform. This compensatory link between cleavage/polyadenylation and mRNA decay acts to dampen the variability of steady-state mRNA levels.
Results
Calculating Production, and Hence, Cleavage/Polyadenylation, Rates for Individual 3′ mRNA Isoforms.
The steady-state level of an mRNA isoform is determined by its rates of production and decay (3, 14, 15). For same-gene 3′ mRNA isoforms in yeast, the relative rates of production reflect the relative rates of cleavage/polyadenylation at sites corresponding to the isoform endpoints. For each isoform i, the relationship between production (βi), decay (αi), and steady-state level (Mi) is given by the equation Mi = βi/αi (see Materials and Methods). Because isoform decay αi is proportional to the computed half-life λi (αi = ln2/ λi; see Materials and Methods), production (and hence cleavage/polyadenylation) rates for individual 3′ isoforms can be calculated from measurements of their steady-state levels and half-lives. Using similar methodology, relative mRNA half-lives have been calculated based on steady-state levels and production rates obtained by metabolic labeling or precision run-on sequencing (15–17).
Efficiency of Cleavage/Polyadenylation Is Inversely Related to Stability of 3′ mRNA Isoforms.
To address the relationship between cleavage/polyadenylation and mRNA stability, we compared the relative half-lives, steady-state transcript levels (defined at the initial time point), and production rates (see above) of the most and least stable isoforms for thousands of yeast genes. For each 3′UTR, we identified the two isoforms with the most widely diverging half-lives and then determined the following three ratios (all downstream/upstream) for the isoforms: that of their steady-state levels, that of their half-lives, and that of their production rates. We then considered three categories of 3′UTRs: “downstream less stable,” in which the downstream isoform is greater than twofold less stable (i.e., has a greater than twofold shorter half-life) than the upstream isoform; “downstream more stable,” in which the downstream isoform is greater than twofold more stable than the upstream isoform; and a control group in which the isoforms are similarly spaced and have comparable half-lives. According to the above equations, if two mRNA isoforms are generated at the same rate and differ in their half-lives by a factor of 2, the steady-state levels of these isoforms should also differ by a factor of 2. In addition, the production and half-life ratios should be independent of each other.
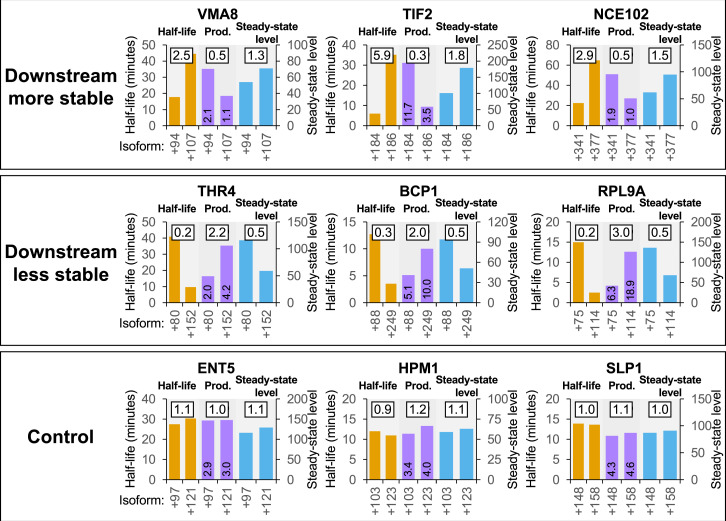
We tested this idea on our transcriptome-scale dataset of 3′ mRNA isoform half-lives and steady-state levels in YPD medium (10). Unexpectedly, 3′ mRNA isoform pairs with at least a twofold difference in half-life exhibit a smaller difference in steady-state level, indicating that the production levels of these paired isoforms are not equivalent (examples shown in Fig. 1 and SI Appendix, Fig. S1). The production ratios are not independent of, but rather are inversely related to, the half-life ratios, indicating a relationship between cleavage/polyadenylation and mRNA stability. These observations do not depend upon whether the more stable isoform is upstream or downstream of the less-stable isoform. In contrast, isoform pairs with similar half-lives have similar ratios for steady-state levels and production rates.
Fig. 1.
Examples from a published dataset (10) showing compensation between cleavage/polyadenylation and mRNA stability in YPD medium. For each gene, the most- and least-stable isoforms are compared with respect to isoform stability (orange bars, values on left y axis), relative production rate (purple bars, values printed on the bars), and steady-state level (blue bars, values on right y axis). Three examples are shown for each of the “downstream less stable,” “downstream more stable,” and “control” 3′UTR categories. The boxed number above each pair of bars gives the ratio of values for the two isoforms (always expressed as downstream/upstream).
On the transcriptome level, if cleavage/polyadenylation and mRNA stability are independent of each other, the simple expectation is that the median half-life ratio should be the same as the median steady-state ratio, and the median production ratio should be 1 (log2 = 0). However, isoform pairs with at least a twofold difference in half-life show significantly less than a twofold median difference in steady-state levels, and they have a median production ratio that significantly differs from 1 (Fig. 2; median values shown as horizontal lines within box plots). Interestingly, for these classes of isoform pairs, the magnitude of the isoform half-life ratios is larger than that of the production ratios, which reduces the steady-state ratios to below 1.
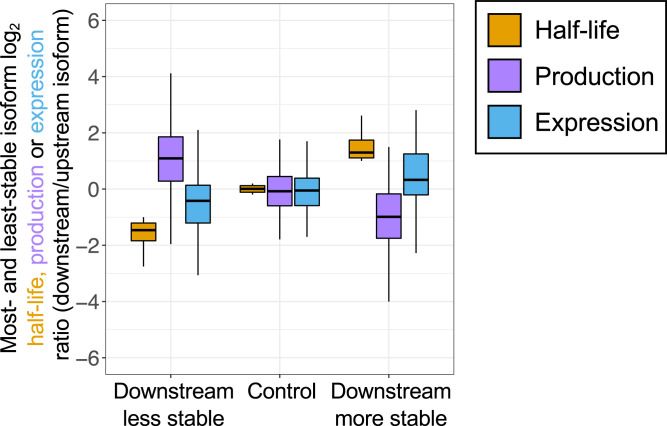
Fig. 2.
Median production and stability ratios for most- and least-stable same-gene isoforms are inversely related in YPD medium. Three classes of most- and least-stable isoform pairs are considered, based on whether the downstream isoform is more stable, less stable, or comparably stable to the upstream isoform. For each group, distributions of relative production rate ratios (purple boxes) and relative steady state ratios (at t = 0; blue boxes) are shown alongside the half-life ratio of the same isoforms (gold boxes). Ratios are plotted as log2 downstream/upstream isoform values. Data (10) are tabulated in SI Appendix.
These observations hold, to comparable extents, for isoform pairs in either the “downstream less stable” category (log2 median half-life ratio = −1.46, log2 median steady-state ratio = −0.44, and median log2 production ratio = 1.07) or the “downstream more stable” group (log2 median half-life ratio = 1.30, log2 median steady-state ratio = 0.33, and median log2 production ratio = −0.99). The relative levels of the downstream isoforms are higher than expected when they are less stable and lower than expected when they are more stable, relative to the upstream isoform (P < 2 × 10−16 for both groups, Mann–Whitney U test). In accord with the difference in median ratios for half-lives and steady-state levels, the distributions of the steady-state ratios (blue boxes representing values between the 25th and 75th percentile) are also shifted. In contrast, isoform pairs having similar half-lives have median steady-state levels that are essentially the same (log2 median half-life ratio = 0.01 vs. log2 median steady-state ratio = −0.08; P > 0.05, Mann–Whitney U test).
We validated these results by analyzing an independent dataset comprising half-lives and steady-state levels of 3′ mRNA isoforms in yeast cells (11). This dataset involved a different method of Pol II inactivation (shifting a temperature-sensitive Pol II strain to the restrictive temperature), a different method of converting poly(A) RNA into a library for sequencing, and genotypic differences between strains. Nevertheless, the results obtained with this independently generated dataset (SI Appendix, Fig. S2 A and B) are remarkably similar to the results shown in Figs. 1 and 2. Thus, these results suggest that yeast cells have a compensatory link in which increased frequency of cleavage/polyadenylation is linked to decreased mRNA half-life, thereby reducing the variation in steady-state 3′ mRNA isoform levels.
The Compensatory Link between Cleavage/Polyadenylation and mRNA Stability Is Observed in Multiple Growth Conditions.
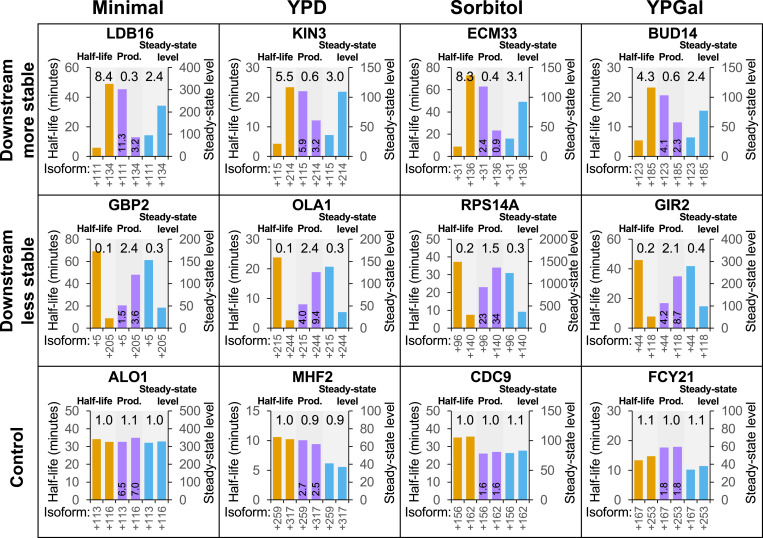
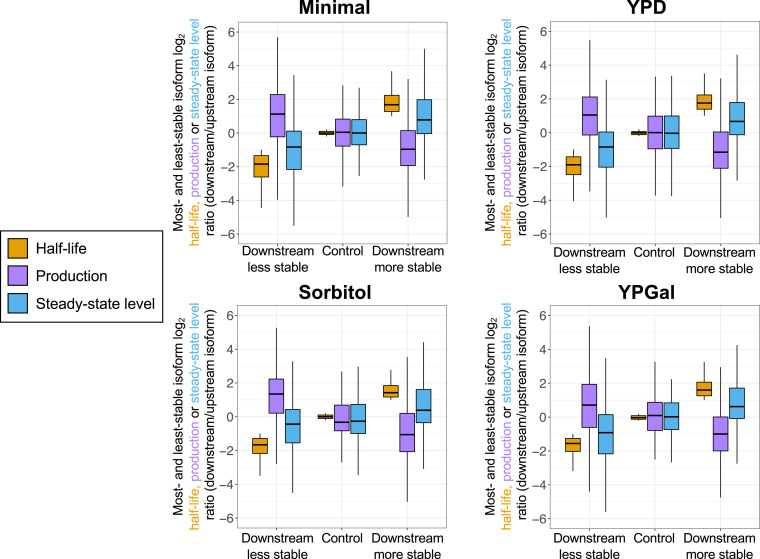
To address the generality of this compensatory link, we performed similar analyses on yeast cells grown in the following conditions: glucose-containing rich medium (YPD), galactose-containing rich medium (YPGal), nutrient-poor minimal medium (Minimal), and YPD containing 1 M sorbitol (Sorbitol), an inducer of osmotic stress. It should be noted that the method for RNA analysis in these experiments (3′ READS) differs from that used in the experiments shown in Figs. 1 and 2 (direct RNA sequencing). Similar results are obtained in all four growth conditions (see Fig. 3 and SI Appendix, Figs. S3–S6 for specific examples and Fig. 4 for transcriptome-scale analysis), indicating that this compensatory link is a general phenomenon.
Fig. 3.
Examples showing compensation between cleavage/polyadenylation and mRNA stability in Minimal, YPD, Sorbitol, and YPGal media. For each gene, the most- and least-stable isoforms are compared with respect to stability (orange bars, values on left y axis), relative production rate (purple bars, values printed on bars), and steady-state expression level (blue bars, values on right y axis). Examples are shown for each of the “downstream less stable,” “downstream more stable,” and “control” categories for every growth condition. Ratios (numbers above the bars for each isoform pair) are expressed as downstream/upstream isoform values.
Fig. 4.
Median production and stability ratios for most- and least- stable same-gene isoforms are inversely related in Minimal, YPD, Sorbitol, and YPGal media. Three classes of most- and least-stable same-gene isoform pairs are considered, based on whether the downstream isoform is more stable, less stable, or comparably stable to the upstream isoform. For each group, distributions of relative production rate ratios (purple boxes) and relative steady-state ratios (at t = 0; blue boxes) are shown alongside the half-life distributions of the same isoforms (gold boxes). Ratios are plotted as log2 downstream/upstream isoform values. Data (10, 30) (GEO accession no. GSE191091) are tabulated in SI Appendix.
Discussion
We demonstrate an inverse relationship between mRNA stability and the efficiency of cleavage/polyadenylation in S. cerevisiae. For cells grown in YPD medium, very similar results are observed from analysis of three independent datasets that involve two different laboratories, three different methods of RNA analysis, two different methods for inactivating Pol II and generating 3′ isoform half-lives, and two different yeast strains. In addition, very similar results are observed when yeast cells are grown under different environmental conditions. The fact that the same phenomenon is observed, both qualitatively and quantitatively, in the six very different datasets makes it exceedingly unlikely that these results arise from an ad hoc technical or analytical artifact. This inverse relationship indicates that yeast cells have a compensatory link between cleavage/polyadenylation and mRNA stability that reduces the variation in steady-state 3′ mRNA isoform levels.
The compensation between cleavage/polyadenylation and mRNA stability is not complete, because median half-life differences are greater than median production differences. In addition, the magnitude of this inverse relationship, and hence the degree of compensation, varies among 3′UTRs. The high reproducibility among replicates indicates that the variable level of compensation is not due to statistical variation. These observations indicate that cleavage/polyadenylation and mRNA stability are not strictly linked and that the relationship between these parameters can differ among same-gene isoforms. We performed Gene Ontology analysis on genes showing high levels of compensation but did not find any biological categories that are significantly enriched.
This compensatory link is surprising, because cleavage/polyadenylation is a rapid event that occurs in the nucleus upon passage of Pol II through the 3′UTR, whereas mRNA turnover takes place in the cytoplasm and is relatively slow. This suggests that during the cleavage/polyadenylation process, the mRNA is imbued with a “mark” that persists upon translocation to the cytoplasm, where it affects the efficiency of mRNA degradation and hence mRNA stability. This “marking” process could involve the length of the poly(A) tail, a chemical modification of the RNA itself, or a protein bound to the mRNA, presumably at or near the site of cleavage/polyadenylation.
The concept of nuclear-generated marking of mRNA leading to increased mRNA degradation has been established for nonsense-mediated decay, the surveillance pathway whose main function is to target mRNAs containing premature translational termination codons that encode truncated proteins for rapid degradation (18, 19). The prevailing model is that UPF proteins associate with nascent mRNA in the nucleus. In mammalian cells, UPF proteins are also part of the exon–exon junction complex bound to mRNA after splicing, but this is not the case in yeast, where few genes are subject to mRNA splicing. Upon transport to the cytoplasm, translation of the UPF-marked mRNA causes dissociation of the UPF proteins. Untranslated regions downstream of the nonsense codon retain UPF proteins, which act as signals for rapid and specific degradation of mRNAs containing premature stop codons.
Whatever its molecular nature, placement of the mark that influences mRNA decay must be linked to the efficiency of cleavage/polyadenylation. For example, as cleavage/polyadenylation is carried out by the same complex, more efficient cleavage might result in longer poly(A) tails, a phenomenon observed in vitro (20). mRNAs with longer poly(A) tails are likely to be more efficient at binding Pab1, the poly(A)-binding protein that stimulates the Ccr4-Not deadenylase and the degradation process (10, 21–27). Alternatively, variation in cleavage/polyadenylation among sites within mRNA might result in different levels of RNA modifications (28) and/or an associated protein that could influence (either positively or negatively) the rate of mRNA degradation. In this regard, Pab1 (10) and other proteins (11, 29) bind differentially to 3′ mRNA isoforms. Biologically, this compensation mechanism allows cells to maintain relatively similar levels of mRNAs even when the cleavage/polyadenylation rate, whether of individual isoforms or on a global level, changes in response to environmental conditions.
Materials and Methods
Strains, Growth Conditions, and Library Construction.
The analyses in this paper are based on data obtained previously (10, 30) (Gene Expression Omnibus [GEO] accession nos. GSE52286, GSE151196, and GSE191091) using the following methods. Most of the data (10, 25) were generated using the yeast strain JGY2000 (MATa, his3Δ0, leu2Δ0, met15Δ0, ura3Δ0, rpb1::RPB1–FRB, rpl13::RPL13–FK512), which contains an anchor-away allele (12) of the Pol II catalytic subunit Rpb1 that permits rapidly sequestration of Pol II in the cytoplasm upon rapamycin treatment (10). Cells were grown in YP + 2% dextrose (YPD), YP medium + 2% galactose (YPGal), YPD + 1 M sorbitol (Sorbitol), or nutrient-poor minimal medium (MM; 2% dextrose, yeast nitrogen base with ammonium sulfate and without amino acids, supplemented with uracil and essential amino acids) to optical density at 600 nm = 0.3 to 0.4 at 30 °C. Cells were treated with rapamycin (1 μg/mL final concentration) and samples taken prior to (t = 0) and at various times after treatment. For all samples, 2 × 106 Schizosaccharomyces pombe cells were added to 50 mL of cells to serve as a spike-in control.
To obtain 3′ mRNA isoform profiles on a transcriptome scale, total RNA was isolated using the hot acid phenol method followed by Qiagen RNeasy (13), and 3′ READS was performed with 17 cycles of amplification (31). Barcoded libraries were quantified on an Agilent Bioanalyzer 2100, pooled, and sequenced on the Illumina NextSEq 500 platform.
The MIST-Seq dataset obtained by thermal inactivation of a strain harboring a temperature-sensitive allele of Rpb1 (rpb1-1) has been described previously (11).
Read Processing and Normalization.
Sequence analysis was performed as previously described (10, 30). In summary, the first four nucleotides, representing random sequences added during library construction, were removed from the 5′ end of each sequence read. Next, consecutive T residues (representing potential poly(A) tail residues) at the 5′ end of each read were counted, their number was recorded, and they were removed from the read. Sequences not having at least one initial T were discarded. The trimmed reads were mapped using Bowtie (32) to a merged reference genome consisting of both S. cerevisiae (Sac Cer 3 genome build) and S. pombe (EF2 build), discarding any reads not mapping uniquely. The reference genome adjacent to each mapped read was examined, and any reads for which the previously recorded number of initial Ts did not exceed the number of As in the neighboring reference genome sequence were discarded. The remaining reads were deemed to be true poly(A) reads.
Within each biological replicate, isoform levels were corrected to account for a time-dependent global decrease in mRNA levels upon Pol II shutoff. For individual (non-steady-state) time points, this was performed by first multiplying the noncorrected isoform read values by the fraction of total S. pombe spike-in reads at t = 0 and then dividing by the fraction of all S. pombe spike-in reads at the given (non-steady-state) time point. Previously unpublished datasets have been deposited to GEO (accession no. GSE191091).
Computation of Isoform Half-Lives.
Isoform half-lives were calculated as described previously (10). To ensure reliable measurements, isoforms were required to have ≥30 reads at the initial time point. After identifying the first time point at which the number of reads was >10-fold lower than at t = 0, half-lives were calculated half-lives using the Excel LOGEST function using the data within that time interval. For very stable isoforms in which the number of reads never dropped below the 10-fold threshold, half-lives were calculated using all the data up to t = 120 min. Isoforms with unreliable half-life measurements (R2 < 0.7), those with maximal reads at non-steady-state time points, and those that mapped to genes containing fewer than two isoforms with reliable half-lives were eliminated from the analysis. For the dataset involving the Pol II temperature-sensitive derivative (11), reads from individual bedgraph files were consolidated, normalized to S. pombe inputs and mapped to the genome (reads outside +1 to +400 regions of 3′UTRs were not analyzed).
Calculation of Isoform Production Rates.
If production rate and turnover are both constant, then an isoform i’s steady-state levels (Μi) are linked to its production (βi) and decay (αi) rates by the following equation:
| [1] |
The decay rate αi for each isoform is related to its calculated half-life (λi) as follows:
| [2] |
To obtain each isoform’s production rate, we multiplied each isoform’s steady-state expression level by its decay rate:
| [3] |
Classification of Genes into Downstream More Stable, Downstream Less Stable, and Control Groups.
In each dataset, for every gene with at least two isoforms that fit the criteria above, the most- and least-stable isoforms were first identified. Half-life ratios were computed by dividing the half-life of the distal isoform by that of the proximal isoform. If the log2 half-life ratio of the distal/proximal isoform for a given gene was ≥1, the 3′UTR is classified as “downstream more stable.” Conversely, if the log2 half-life ratio of the distal/proximal isoform was ≤−1, the 3′UTR is categorized as “downstream less stable.” A 3′UTR was assigned to the “control” group if its log2 half-life (distal/proximal isoform) was >−0.2 and <0.2. For these same isoform pairs, steady-state expression ratios and production rates were computed by dividing either the steady-state level or production rate of the distal isoform by the steady-state level or the production rate of the proximal isoform. Log2 production and steady-state expression ratios for “downstream less stable,” “downstream more stable,” and similarly spaced “control” categories were then plotted in Figs. 2 and 4 and SI Appendix, Fig. S2B.
Supplementary Material
Acknowledgments
We thank Steve Buratowski for suggesting the relationship to nonsense-mediated decay and Jim Manley for pointing us to his 1983 observation that cleavage efficiency is associated with increased poly(A) length in vitro. This work was supported by grants to K.S. from the NIH (GM 30186 and GM181801).
Footnotes
Reviewers: M.C., Technion; and B.T., Rutgers University.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2121488119/-/DCSupplemental.
Data Availability
RNA-sequencing data have been deposited in the Gene Expression Omnibus (accession no. GSE191091).
References
- 1.Haimovich G., et al. , Gene expression is circular: Factors for mRNA degradation also foster mRNA synthesis. Cell 153, 1000–1011 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Fischer J., et al. , The yeast exoribonuclease Xrn1 and associated factors modulate RNA polymerase II processivity in 5′ and 3′ gene regions. J. Biol. Chem. 295, 11435–11454 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun M., et al. , Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol. Cell 52, 52–62 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Schwabish M. A., Struhl K., Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell 22, 415–422 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Schwabish M. A., Struhl K., The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol. Cell. Biol. 27, 6987–6995 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esberg A., et al. , Iwr1 protein is important for preinitiation complex formation by all three nuclear RNA polymerases in Saccharomyces cerevisiae. PLoS One 6, e20829 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozsolak F., et al. , Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143, 1018–1029 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pelechano V., Wei W., Steinmetz L. M., Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 497, 127–131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moqtaderi Z., Geisberg J. V., Jin Y., Fan X., Struhl K., Species-specific factors mediate extensive heterogeneity of mRNA 3′ ends in yeasts. Proc. Natl. Acad. Sci. U.S.A. 110, 11073–11078 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geisberg J. V., Moqtaderi Z., Fan X., Ozsolak F., Struhl K., Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell 156, 812–824 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta I., et al. , Alternative polyadenylation diversifies post-transcriptional regulation by selective RNA-protein interactions. Mol. Syst. Biol. 10, 719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haruki H., Nishikawa J., Laemmli U. K., The anchor-away technique: Rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell 31, 925–932 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Moqtaderi Z., Geisberg J. V., Struhl K., Extensive structural differences of cloesly related 3′ mRNA isoforms: Links to Pab1 binding and mRNA stability. Mol. Cell 72, 849–861.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwanhäusser B., et al. , Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Blumberg A., et al. , Characterizing RNA stability genome-wide through combined analysis of PRO-seq and RNA-seq data. BMC Biol. 19, 30 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller C., et al. , Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol. Syst. Biol. 7, 458 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun M., et al. , Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Res. 22, 1350–1359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He F., Jacobson A., Nonsense-mediated mRNA decay: Degradation of defective transcripts is only part of the story. Annu. Rev. Genet. 49, 339–366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurosaki T., Popp M. W., Maquat L. E., Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 20, 406–420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manley J. L., Accurate and specific polyadenylation of mRNA precursors in a soluble whole-cell lysate. Cell 33, 595–605 (1983). [DOI] [PubMed] [Google Scholar]
- 21.Simón E., Séraphin B., A specific role for the C-terminal region of the poly(A)-binding protein in mRNA decay. Nucleic Acids Res. 35, 6017–6028 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker R., RNA degradation in Saccharomyces cerevisae. Genetics 191, 671–702 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang C., et al. , The RRM1 domain of the poly(A)-binding protein from Saccharomyces cerevisiae is critical to control of mRNA deadenylation. Mol. Genet. Genomics 288, 401–412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webster M. W., et al. , mRNA deadenylation is coupled to translation rates by the differential activities of Ccr4-NOT nucleases. Mol. Cell 70, 1089–1100.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi H., et al. , PABP cooperates with the CCR4-NOT complex to promote mRNA deadenylation and block precocious decay. Mol. Cell 70, 1081–1088.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Brambilla M., Martani F., Bertacchi S., Vitangeli I., Branduardi P., The Saccharomyces cerevisiae poly (A) binding protein (Pab1): Master regulator of mRNA metabolism and cell physiology. Yeast 36, 23–34 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Turtola M., et al. , Three-layered control of mRNA poly(A) tail synthesis in Saccharomyces cerevisiae. Genes Dev. 35, 1290–1303 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorenz D. A., Sathe S., Einstein J. M., Yeo G. W., Direct RNA sequencing enables m6A detection in endogenous transcript isoforms at base-specific resolution. RNA 26, 19–28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciolli Mattioli C., et al. , Alternative 3′ UTRs direct localization of functionally diverse protein isoforms in neuronal compartments. Nucleic Acids Res. 47, 2560–2573 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geisberg J. V., Moqtaderi Z., Struhl K., The transcriptional elongation rate regulates alternative polyadenylation in yeast. eLife 9, e59810 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin Y., et al. , Mapping 3′ mRNA isoforms on a genomic scale. Curr. Protoc. Mol. Biol. 110, 4.23.21-24.23.17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data have been deposited in the Gene Expression Omnibus (accession no. GSE191091).