Significance
Bacteria have evolved appendages called flagella that are spun by an ingenious rotary motor that harnesses electrochemical energy to power rotation. To uncover and understand nature's blueprint of this nanoscale engine, an integrative structural biology approach is required. We used a combination of mutagenesis, cryogenic electron tomography, and crystallography to reveal the architecture of a circle of rings scaffold that likely serves to organize and stabilize individual power-generating units of the flagellar motor in their active form. The knowledge about the structure–function relationships within the bacterial flagella motor is a source of inspiration for nanotechnology and can be one of the first steps toward making artificial motors on the same scale or controlling motility for medical applications.
Keywords: bacterial motility, flagellar motor, structure and function
Abstract
The flagellar motor stator is an ion channel nanomachine that assembles as a ring of the MotA5MotB2 units at the flagellar base. The role of accessory proteins required for stator assembly and activation remains largely enigmatic. Here, we show that one such assembly factor, the conserved protein FliL, forms an integral part of the Helicobacter pylori flagellar motor in a position that colocalizes with the stator. Cryogenic electron tomography reconstructions of the intact motor in whole wild-type cells and cells lacking FliL revealed that the periplasmic domain of FliL (FliL-C) forms 18 circumferentially positioned rings integrated with the 18 MotAB units. FliL-C formed partial rings in the crystal, and the crystal structure–based full ring model was consistent with the shape of the rings observed in situ. Our data suggest that each FliL ring is coaxially sandwiched between the MotA ring and the dimeric periplasmic MotB moiety of the stator unit and that the central hole of the FliL ring has density that is consistent with the plug/linker region of MotB in its extended, active conformation. Significant structural similarities were found between FliL-C and stomatin/prohibitin/flotillin/HflK/C domains of scaffolding proteins, suggesting that FliL acts as a scaffold. The binding energy released upon association of FliL with the stator units could be used to power the release of the plug helices. The finding that isolated FliL-C forms stable partial rings provides an insight into the putative mechanism by which the FliL rings assemble around the stator units.
Bacteria use rotating flagella to swim in liquids or swarm over wet surfaces. Motility by flagella is essential for the survival and virulence of many pathogenic bacteria associated with human, animal, and plant diseases (1, 2). The macromolecular structure of the flagellum consists of the basal body, hook, and filament. The cell wall–embedded basal body acts as an ion-motive force-driven rotary motor; it generates a turning force that is applied through the hook to the filament, making it spin (3–5). The flagellar motor is made up of the stator, the bushing, and the rotor, which is comprised of the rod, the MS-ring, and the cytoplasmic C-ring (also known as the switch complex).
The stator is a key component of the flagellar motor which converts the energy of an electrochemical ion gradient across the inner membrane (IM) into mechanical rotation force on the rotor. It is composed of ∼10 to 20 identical units that form a ring around the rotor. Each stator unit is an asymmetric complex of five MotA or PomA subunits surrounding two MotB or PomB subunits (6, 7), each containing a periplasmic peptidoglycan-binding domain that helps to anchor the stator to the cell wall (6–11).
Although it is well understood that the MotAB complexes function as ion channels that remain closed and inactive until they incorporate into the motor (12), little is known about how they are recruited to the motor and how they switch into an open, ion-conducting state. In situ imaging of intact flagellar motors by cryogenic electron tomography (cryo-ET) revealed that many of them contain various additional proteinaceous structures (disks/rings) next to the MotAB stator, without which the motor does not function (reviewed in ref. 13). The determination of the molecular composition of these structures and their role in the stator assembly and function is critical to our understanding of the motor function. The observation that in some bacteria, such as spirochetes Treponema phagedenis and Borrelia burgdorferi, the motA and motB genes are next to the gene encoding a conserved flagellar protein FliL (14, 15) raised a question about the role of this protein in relation to the stator function.
FliL is a single-span IM protein with the majority of the polypeptide chain in the periplasm. FliL has been shown to be required for flagellar function: mutants in multiple microbes are nonmotile under some or all conditions (15–21). There appears to be a trend for fliL mutants to have greater motility defects under swarming conditions (15–18) in which the propulsive force and the force of viscous drag on the moving cell body are significantly higher compared to swimming in liquid (22). Additionally, there is evidence that FliL promotes structural integrity of the flagellar motor: mutants in some microbes had an abnormal orientation of the flagella (15) or flagella that broke off within the rod and detached from the cell bodies (17). Cryo-ET of B. burgdorferi showed that FliL may be part of the motor (15). Suppressor studies of fliL mutants of Rhodobacter sphaeroides (20) indicated that FliL and the stator may function together as parts of a complex—a notion supported by a crystallographic study of the periplasmic domain of Vibrio alginolyticus FliL that revealed a fold similar to that of the stomatin/prohibitin/flotillin/HflK/C (SPFH) domain of mouse stomatin, a protein that functions together with acid-sensing ion channels (16). Cumulatively, the results of the previous studies indicate that FliL plays a stabilizing role in the function of the flagellar motor, possibly by forming interactions with the MotAB stator that are particularly critical under high viscous drag conditions.
In this work, we have examined the structural rationale for the stabilizing role of FliL in the motor. As a model organism, we chose the gastric carcinogenic bacterium Helicobacter pylori that naturally resides in a highly viscous liquid environment, the gel-like mucous layer of the stomach (23). It represents an ideal study system because, compared to other motile bacteria, it demonstrates unusually high motility in viscous media (24) under conditions in which the function of FliL appears to be particularly important. A comparison of cryo-ET reconstructions of the intact H. pylori motor in wild-type and fliL mutant cells enabled localization of FliL in the motor in situ. The determination of the crystal structures of the C-terminal periplasmic domain of H. pylori FliL (FliL-C) in three different crystal forms and their docking into the corresponding density in the motor confirmed the identity of the structures observed in situ. Our findings shed significant light on the flagellar motor architecture and on the process of the stator activation and support the notion that FliL acts as a scaffold protein.
Results and Discussion
fliL Is Essential for Motility but Not Flagellation in H. pylori.
To investigate the role of FliL in the H. pylori flagellar motor, we inactivated the fliL gene by replacing part of it with the chloramphenicol resistance cassette (SI Appendix, Fig. S1A). To minimize the possibility of polar effects, the beginning and the end of the fliL gene were kept intact. The mutation was confirmed by Sanger sequencing and the resultant strain used for cryo-ET.
We next determined the motility and flagellation phenotype of the parent wild-type (WT) strain and the fliL mutant. Transmission electron microscopy analysis of the mutant strain showed that it retained flagella. Soft agar motility assays (SI Appendix, Fig. S1B) and phase-contrast microscopy analysis showed that, in contrast to the motile WT parent, the fliL mutant cells were nonmotile. These results suggested that FliL is required for flagellar motor function but not assembly of the H. pylori flagella, similar to what has been reported in other microbes.
Localization of FliL in the Intact H. pylori Motor.
To determine whether FliL forms part of the flagellar motor in H. pylori and to pinpoint its putative location, we visualized the flagellar motor in intact WT and mutant cells using cryo-ET. We previously published a tomography reconstruction of the H. pylori motor in situ at low (4-nm) resolution (25). In this work, we have significantly improved the resolution of the reconstructions (to ∼2.5 nm, SI Appendix, Fig. S2) by collecting more data at higher magnification.
An analysis of the electron density maps obtained by cryo-ET reconstruction and subtomogram averaging allowed unambiguous identification of the core components of the motor, including the C-ring, export apparatus, L-, P- and MS-rings, basal disk, and the putative 18-membered MotAB stator ring (Fig. 1 A and B). Each individual stator unit was easily recognizable by its IM-embedded moiety of a truncated cone shape of a size close to that of the purified Campylobacter jejuni MotAB complex [50 × 90 × 75 Å (7)]. In its shape, size, position, and its association with the rotor C-ring via its base, it was highly similar to the bell-like stator units previously observed in situ in Borrelia burgdorferi (26). Furthermore, the shape and dimensions of this moiety were similar to those of the MotAB complexes isolated from Clostridium sporogenes, Bacillus subtilis, and Vibrio mimicus (6). External to the stator, an additional proteinaceous structure termed the cage (25) was also clearly visible.
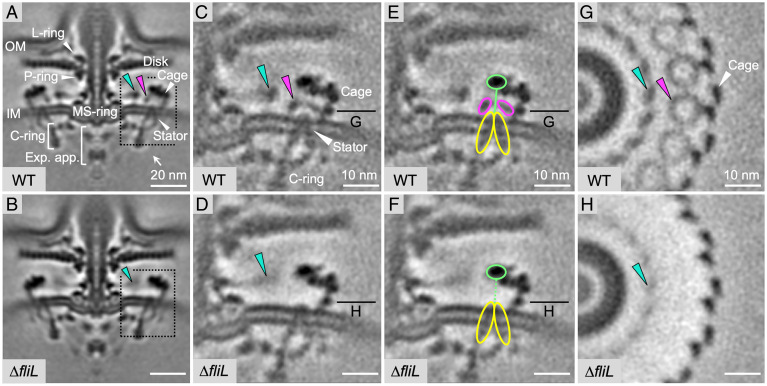
Fig. 1.
Structures of intact H. pylori flagellar motor in WT and fliL mutant cells. (A and B) Medial slices through in situ structures of WT and mutant motors determined by subtomogram averaging over several hundred particles. FliL and collar-like rings are indicated with magenta and cyan arrows, respectively. (C-H) Region containing rings (dashed frame in A and B) after focused refinement showing close-up view of medial slice (C–F) and perpendicular cross-sections (G and H), with densities for putative FliL ring, MotA ring and MotB dimer indicated in magenta, yellow, and light green, respectively.
An examination of the periplasmic space between the stator/cage and the rotor MS-ring in WT cells revealed two concentric rings in the periplasm (cyan and magenta arrows in Fig. 1), the 18-fold symmetry of which matched that of the stator. Their relative location was close to that of FliL in the B. burgdorferi motor (15), prompting a hypothesis that either or both rings were formed by FliL. A comparison of the reconstructions for the motor in WT cells (Fig. 1A) and the fliL mutant (Fig. 1B) suggested these rings are less dense or lost in the mutant, thus supporting our hypothesis. To resolve the architecture of the two rings, we performed focused refinement of this region (dash framed in Fig. 1 A and B). A comparison of the refined reconstructions for the motor in WT cells and the fliL mutant revealed that the inner ring forms a periplasmic collar-like structure around the MS-ring (Fig. 1 C–H). The collar is still present in the fliL mutant, albeit in a less-ordered conformation or at a partial occupancy as evidenced by the significantly weaker density (Fig. 1 D and H). The larger radius circle of density is composed of 18 circumferentially positioned smaller periplasmic rings that appear to be integrated with the 18 stator units (Fig. 1 C, E and G). The absence of the rings in the mutant (Fig. 1 D and H) strongly suggests they are formed by FliL. Each ring is coaxially sandwiched between the IM-embedded truncated cone moiety of the respective stator unit and a separate density in the periplasm, corresponding to the putative dimeric peptidoglycan (PG)-binding domain of MotB (Fig. 1E). The assignment of the density as the PG-binding domain of MotB is supported by both its position and a good match with the dimensions of its crystal structure (8, 9). The IM-embedded stator moiety and the PG-binding domain of MotB are connected by the MotB linker, which likely accounts for a separate small-volume density seen in the central hole of each FliL ring (Fig. 1G).
Determination of the Crystal Structure of the Periplasmic Domain of H. pylori FliL-C.
To test the hypothesis that FliL self-assembles into rings that fully account for the density seen in the cryo-ET reconstruction of the intact motor (Fig. 1) and to obtain the structural rationale for the reported stabilizing role played by FliL in the flagellar motor, we undertook a crystallographic study of the FliL ordered part—the periplasmic domain FliL-C (residues 81 to 183). The crystallization of FliL-C yielded three different crystal forms. Forms A, B, and C crystals contained 12, six, and one subunit per asymmetric unit, respectively. The form C crystal structure was determined to 2.1-Å resolution by ab initio phasing (27). The form A and form B crystal structures were determined to 2.8- and 2.7-Å resolution by molecular replacement.
Overall Fold of H. pylori FliL-C and Comparison to Other Proteins.
The polypeptide chain of FliL-C folds into a single α/β globular domain with a three-stranded, curved antiparallel β-sheet on one side and four consecutive helices on the other side (Fig. 2A). FliL-C has an ellipsoid shape with dimensions of 53 × 24 × 24 Å. The strands and helices are arranged in the topological order β1β2α1α2η1α3β3 (Fig. 2B). The N-terminal β-strand 1 is subdivided into β1a and β1b by a short loop 90 to 92. Helix α2 is formed by residues 127 to 139 and runs approximately antiparallel to helix α3, which is the longest helix of the structure (18 residues, 148 to 165). Short helices α1 (residues 118 to 125) and η1 (310 helix, residues 143 to 146) lie at the two opposite ends of the β-sheet, nearly orthogonal to helices α2 and α3.
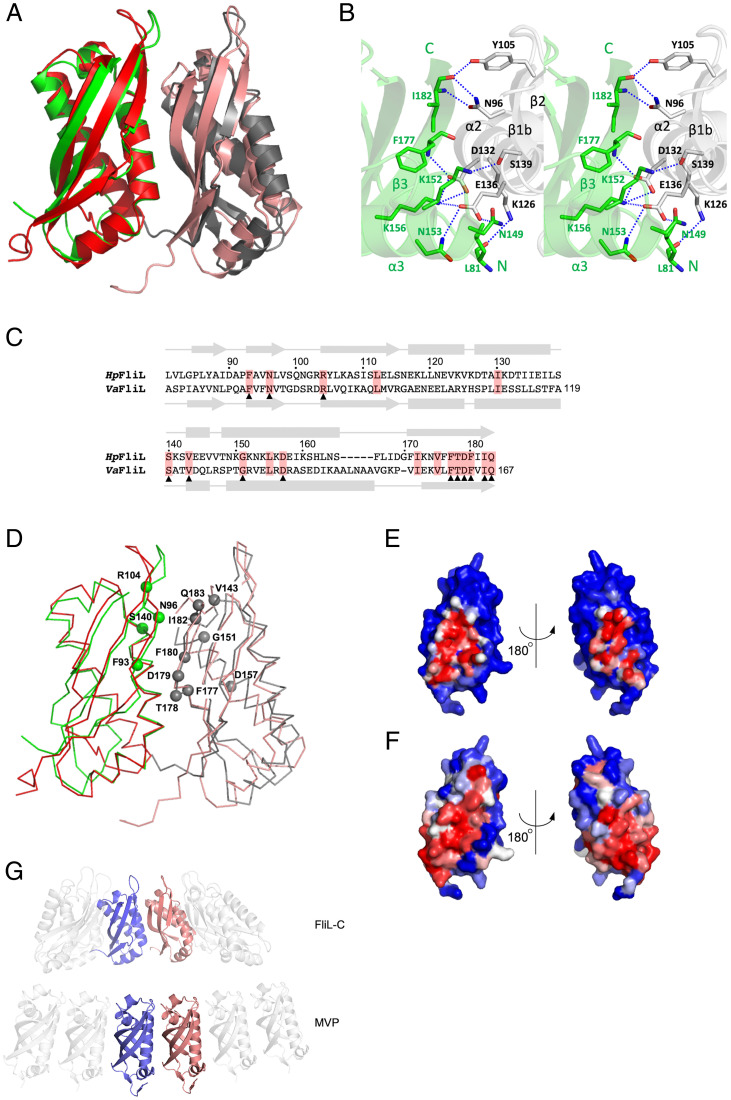
Fig. 2.
The overall fold of H. pylori FliL-C and comparison with the structures of SPFH domains. (A) Stereo representation of FliL-C. (B) Topology of secondary structure elements of FliL-C. The α-helices are represented by rods, 310-helix by a shaded rod, and β-strands by arrows. (C–E) Comparisons of the structures of other SPFH domains. (C) Stereo view of the superposition of the crystal structures of H. pylori FliL-C (blue, residues 82 to 183) and the C-terminal domain of V. alginolyticus FliL (red, residues 63 to 167, PDB identification [ID] 6ahp). (D) Stereo view of the superposition with the mouse stomatin domain (orange, PDB ID 4fvj, residues 95 to 199) and stomatin-like domain of P. horikoshii protein PH0470 (PDB ID 2rpb, residues 68 to 171). (E) Side-by-side comparison of the structures of FliL-C from H. pylori (residues 82 to 183), the mouse stomatin domain (PDB ID 4fvj, residues 95 to 199), stomatin-like domains of P. horikoshii proteins PH1511 (PDB ID 3bk6, residues 65 to 170) and PH0470 (PDB ID 2rpb, residues 68 to 171), the shoulder domain of the rat liver MVP (PDB ID 4hl8, residues 515 to 646), and the band-7 domain of mouse flotilin-2 (PDB ID 1win, residues 12 to 125).
The superposition of the crystal structure of H. pylori FliL-C with the recently published crystal structure of the 40 to 167 fragment of V. alginolyticus FliL (16) reveals that, although the two domains share only 17% amino acid sequence identity, 85 Cα atoms can be overlapped with an rmsd of 2.3 Å (Fig. 2C), indicating the significant overall similarity of the C-terminal domains of the two proteins. Structural similarity extends over the entire fold and includes all the secondary structure elements.
We also compared the structure of H. pylori FliL-C against all structures deposited in the Protein Data Bank (PDB) using the DALI server (28) and found that it closely resembles those of the members of the SPFH domain family. Similarities were found to the mouse stomatin domain [mmStomatin (29)], stomatin-like domains of proteins PH0470 and PH1511 from the archaeon Pyrococcus horikoshii [phStomatin0470 and phStomatin1511 (30, 31)], the shoulder domain of the rat liver major vault protein [MVP (32, 33)], and the band-7 domain of mouse flotilin-2 (PDB identification 1win). The FliL-C structure can be superimposed over the SPFH domains of mmStomatin, phStomatin0470 (Fig. 2D), phStomatin1511, MVP, and flotilin-2 with an rmsd of 2.4, 2.2, 2.4, 2.6, and 2.6 Å for 92, 93, 94, 87, and 89 aligned Cα atoms, respectively. Fig. 2E illustrates that FliL-C and SPFH domains adopt a very similar fold, despite the significant differences in their amino acid sequences (<13% identity with FliL-C). Domains with the SPFH fold are very diverse in their amino acid sequence and often function as membrane scaffolds that provide stable platforms for the assembly of protein complexes (34). The observed structural similarity between FliL-C and SPFH domains suggests that FliL also functions as a membrane scaffolding protein.
H. pylori FliL-C Forms Ring Segments in the Crystal.
An analysis of the crystal packing in the form C crystals (1 subunit per asymmetric unit) using the PDBePISA server (https://www.ebi.ac.uk/pdbe/pisa/) revealed no quaternary structure, indicating that, under those conditions, FliL-C crystallizes as a monomer. In contrast, the asymmetric unit of the form A crystals contains two 6-subunit ring segments (Fig. 3A) that are essentially identical (607 Cα atoms can be superimposed with an rmsd of 0.4 Å). The 12 subunits in the asymmetric unit adopt highly similar conformations, with the 102 aligned Cα atoms superimposing with rmsd values in the range 0.3 to 0.6 Å. The form C (monomeric) crystal structure can be superimposed with any subunit in the form A crystal structure with an rmsd of <0.7 Å for 102 aligned Cα atoms, indicating that the overall protein structure does not undergo any significant conformational changes upon formation of the super-molecular complex. The six subunits in the asymmetric unit of the form B crystals are also essentially identical. An analysis of the molecular packing revealed that in the form B crystal lattice, the FliL-C subunits assemble into five-membered ring segments (Fig. 3B) plus one associated subunit that is not part of the ring.
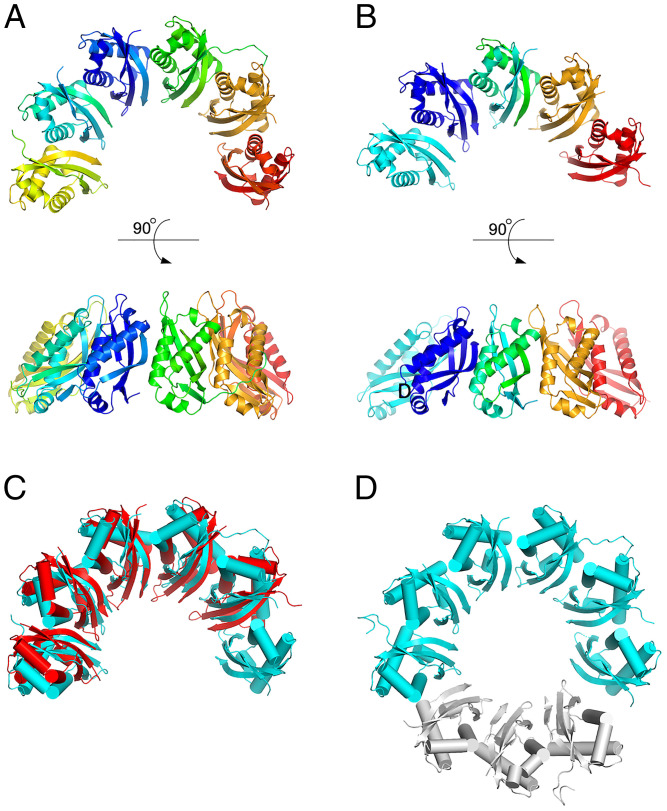
Fig. 3.
The structure of the ring segments formed by H. pylori FliL-C in the crystal. (A and B) 6-subunit and 5-subunit ring segments observed in the form A and form B crystals, respectively. (C) Superposition of the five-membered and six-membered segments. (D) Model of the full ring revealing a ninefold rotational pseudosymmetry within the ring segment. The three modeled subunits are shown in gray.
The five-membered partial ring (form B) can be superimposed onto the six-membered partial ring (form A) with an rmsd of 1.4 Å for 493 aligned Cα atoms, showing that the two arc-shaped structures are very similar (Fig. 3C). The arc has an overall diameter of 96 Å and height of 45 Å. The α-helices of all subunits are positioned on the outside, while the β-sheets are positioned on the inside, forming an incomplete β-barrel. The modeling of the full ring by overlapping subunits 1 to 3 over subunits 4 to 6 reveals that the FliL-C molecules within the arc structure are related to each other by a ninefold rotational pseudosymmetry axis (Fig. 3D). Of note, the V. alginolyticus homolog of H. pylori FliL-C formed full 10-membered rings or pseudohelices with 12 members per turn in crystals (16). Our analysis shows that H. pylori FliL-C subunits associate to form ring segments in a fashion that is remarkably similar to that seen in the V. alginolyticus decameric rings (Fig. 4A)—an unexpected finding given that the sequence identity between the two proteins (17%) is close to that of unrelated sequences.
Fig. 4.
Conserved residues cluster at the interface between the FliL-C subunits of the arch. (A) Superposition of the two adjacent H. pylori FliL-C subunits from the six-membered ring segment (green and gray) with the two adjacent V. alginolyticus FliL-C subunits from the 10-membered ring (red and salmon), illustrating the conserved mode of oligomerization. (B) The hydrogen bonding network at the subunit interface in H. pylori FliL-C. (C) Structure-guided sequence alignment of FliL-C from H. pylori and V. alginolyticus, shown in A and in Fig. 1C. The two sequences share only 17% identity. Conserved residues are highlighted in red. Conserved surface-exposed residues are indicated by triangles. (D) A ribbon representation of the superposition of the two adjacent subunits of FliL-C from H. pylori and V. alginolyticus (same view and color scheme as in A) showing location of the conserved surface-exposed residues in the H. pylori protein. (E) Molecular surface of H. pylori FliL-C, colored according to the accessible surface area buried at the intersubunit interface. The color gradient runs from blue (fully exposed) to red (fully buried). (F) Molecular surface of H. pylori FliL-C, colored according to the evolutionary conservation of each residue among sequences with ≥26% identity to H. pylori FliL-C. The color gradient runs from blue (not conserved) to red (fully conserved). (G) Comparison of the mode of oligomerization into ring structures in H. pylori FliL-C and the shoulder domain of the rat liver MVP.
Conserved FliL-C Residues Cluster at the Interface between the Subunits of the Arch.
To obtain a structural rationale for the remarkably similar mode of association between the FliL-C subunits in H. pylori and V. alginolyticus, we performed a comparative analysis of the respective intermolecular contacts. Except for the two flanking molecules, each FliL-C subunit within the partial ring in H. pylori is involved in extensive interactions with its two neighboring subunits, with 24% (∼1,500 Å2) of the subunit accessible surface area buried by the two interfaces. Every β-strand and two out of the three α-helices contribute to this interface (Fig. 4B). The subunits associate via multiple van der Waals contacts and hydrogen bonds. The latter form a network with many residues on one molecule bound to more than one residue on the other. The side chain of E136 of helix α2 forms hydrogen bonds to the side chains of N149’, N153’, and K156’ of the adjacent subunit. K156’ makes an additional hydrogen bond contact with the side chain of D132 (helix α2), which, in turn, also forms a hydrogen bond with the backbone amide of F177’. The side chains of N96 of strand β1b and Y105 of strand β2 form hydrogen bonds with the backbone amide and carbonyl of I182’. The subunit interface is further stabilized by hydrogen bonds between the side chains of S139 from helix α2 and K152’ and between the side chain of K126 and the backbone carbonyl of the N-terminal residue L81’ (Fig. 4B). Most of these interactions are observed between every two subunits of the partial ring.
We performed structure-guided sequence alignment of H. pylori and V. alginolyticus FliL-C (Fig. 4B) and mapped the common surface-exposed residues on the structure of the two neighboring FliL-C subunits (Fig. 4D). This analysis revealed that most (11 out of 13) surface-exposed side chains that are conserved between FliL-C from H. pylori and V. alginolyticus are located at the subunit interface. We have then extended this analysis to include a total of 250 amino acid sequences homologous to H. pylori FliL-C, found using a PSI-BLAST search, with sequence identity to the H. pylori protein of ≥ 26%. The sequences were aligned by COBALT (35) and used to generate a conservation profile with Consurf (36). The molecular surface of H. pylori FliL-C was then color-coded to highlight residues buried at the intersubunit interface (Fig. 4E) or to show residue conservation scores (Fig. 3F). This analysis clearly showed that the conserved FliL-C residues cluster at the interface between the subunits of the arch. Cumulatively, our findings suggest that FliL from different species share a conserved mode of oligomerization into partial or full rings and that the rings seen in the crystals therefore represent the functional, physiologically relevant form of FliL.
The shoulder domain of the rat liver MVP (Fig. 2E) is an example of an SPFH domain that also forms a ring within the vault organelle, albeit a much larger one (39 subunits) (32, 33). Although it shares no significant sequence identity with FliL, its subunits associate in a surprisingly similar manner, with α-helices positioned on the outside and β-strands lining the surface on the inside (Fig. 4G). This similarity suggests that the SPFH fold has evolved to retain the ability to assemble into rings in these proteins, which further strengthens the notion that FliL self-assembly into rings is important for its function.
Implications for FliL Function as Structural Scaffold Supporting Active Conformation of Stator.
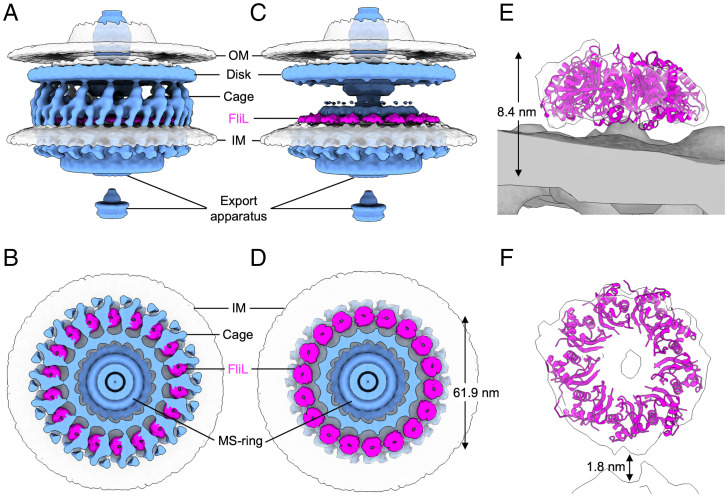
The overall shape of the crystal structure–based model of the full nine-membered FliL-C ring (Fig. 3D) resembles a truncated hollow cone. The N termini of all FliL-C subunits localize to the wider base of the cone, indicating that in the physiological oligomer, the wider opening, tethered to the transmembrane moiety, faces the IM. When docked into the cryo-ET map in this orientation, the atomic model was in good agreement with the density for the 18 rings that are lost in the mutant, supporting their identification as complete FliL rings (Fig. 5). However, the precise stoichiometry of the FliL rings in situ remains to be established, as fitting of the nine-membered ring of H. pylori FliL-C and the 10-membered ring of V. alginolyticus FliL-C resulted in very similar correlation coefficient values (SI Appendix, Fig. S3), and the cryo-ET map did not have sufficient resolution to allow a rotational symmetry analysis of the relatively small FliL rings.
Fig. 5.
Isosurface renderings showing periplasmic circle of 18 rings formed by FliL-C. (A and B) Side view and perpendicular cross-section of WT motor with FliL colored in magenta and all other protein components in light blue. (C and D) Same as A and B but with cage and MotB densities removed to show FliL rings clearly. The diameter of the FliL ring (61.9 nm) has been measured as the distance between the centers of the opposing rings. (E and F) Close-up side and top views showing crystal structure–based model of full nine-membered FliL-C ring docked into cryo-ET density.
We did not observe any significant differences between the densities for the MotAB stator in H. pylori WT and fliL mutant cells, suggesting that the MotAB complexes still incorporated into the motor in the absence of FliL. Our results are thus consistent with the idea that MotAB instead recruit FliL in agreement with previous studies in V. alginolyticus that showed FliL was not recruited when PomAB were missing or had an altered conformation (18). Our finding that isolated FliL-C forms stable partial rings provides an insight into the putative mechanism by which the FliL rings assemble around the stator units: partial five- to six-membered rings likely form first and latch onto the MotB linker/plug region via their central cavity, following which additional FliL subunits are added, completing the ring seen in the intact motors.
It was previously shown that because of the linker flexibility, H. pylori MotB can exist in a variety of conformations, with the distance spanned by its periplasmic region (comprising the plug, linker, and C-terminal domain) ranging between 50 (compact) to 115 Å (fully extended) (11). The height of the periplasmic part of MotB in the intact H. pylori motor, measured as the average distance between the outer leaflet of the inner membrane and the top of the globular MotB domain in our cryo-ET maps (∼129 Å), is close to the top of this range, indicating that upon incorporation into the motor, the MotB linker adopts an extended conformation. This observation is in agreement with the previous crystallographic, biochemical, and mutagenesis studies that showed that activation of the MotAB stator units upon insertion into the motor involves linker extension (9, 37).
Our results show that H. pylori is nonmotile without FliL. One possibility is that the MotAB stator is not functional in the absence of FliL. The MotB linker appears to be in the extended, activated conformation in the fliL mutant, suggesting there must be another deficit. One possibility is that the MotAB channel is not open because the MotB plug helices have not been released from the IM (12), a step that may depend on FliL. Our observation that each FliL-C ring is sandwiched between a MotA ring and a globular MotB domain, with the MotB plug/linker segment passing through its central hole, therefore suggests that the circle of FliL rings serves as a structural scaffold for the stator units, with the binding energy released upon association of the stator units with the scaffold used to power the release of the plug helices. This conclusion is supported by the previous studies that showed that motility defects in mutants lacking FliL were alleviated in strains with unplugged mutant stators (20, 38). It is also strongly supported by the detected significant structural similarities between FliL-C and SPFH domains of proteins, some of which (e.g., prohibitins) are known to form membrane-anchored rings which serve as scaffolds that control the organization of other proteins (39, 40). Our results also suggest that the interactions between FliL and the MotAB complex stabilize the MotB linker in its extended conformation, which, in turn, has been shown to be important for holding the periplasmic domain of MotB in a conformation that is able to bind PG (11). The additional role of the FliL scaffold therefore appears to be to strengthen the attachment of MotB to PG, which is consistent with the previous observations that, in many bacteria, FliL becomes indispensable for the motor function only under high viscous drag conditions when a higher motor torque, and hence a more secure attachment of the stator to the cell wall, are required.
Materials and Methods
Construction of Isogenic ΔfliL Mutant in H. pylori Strain SS1 and Analysis of Swimming Behavior.
The deletion of fliL (gene identification HPYLSS1_00526 in SS1, HPG27_765 in G27) was achieved by transforming strain SS1 (41) to chloramphenicol resistance with genomic DNA from strain G27 that contained a ΔfliL::cat mutation. The original mutation was created by splicing by overlapping extension to insert a cat gene. The insertion replaced 228 internal base pairs in the middle of the 552-bp fliL, leaving 151 bases at the 5′ end and 173 bases at the 3′ end (42). The SS1 mutation was verified by PCR and Sanger sequencing.
Soft agar motility assays were carried out as described (43) using 4-d incubation on plates composed of Brucella broth and 0.35% agar under microaerobic conditions (5% O2, 10% CO2, and 85% N2). Motility in liquid media was assessed by growing the bacteria in Brucella broth with 10% heat-inactivated serum (BB10) to an OD600 of ∼0.3 and visualizing by microscopy.
Cryo-ET Sample Preparation and Imaging.
H. pylori strain SS1 and its fliL deletion mutant (ΔfliL) were cultured on tryptic soy agar plates supplemented with 5% sheep blood under microaerobic conditions. To prepare frozen hydrated H. pylori samples, the cells were washed off and suspended in phosphate buffer saline mixed with 10 nm fiducial gold particles, deposited on cryo–electron microscopy grids, and plunge frozen using liquid ethane. Tilt series were collected using the Titan Krios G2 electron microscope (Thermo Fisher Scientific) equipped with a field emission gun and a direct detection camera (Gatan) and operated at 300 kV and a magnification corresponding to a pixel size of 2.2 Å at the specimen level. SerialEM software (44) was used to record a single-axis tilt series with a phase plate at -0.5-µm defocus and a cumulative dose of ∼50 e/Å2 distributed over 35 stacks covering an angle range of −51 to 51° in 3° increments.
Three-Dimensional Image Processing and Subtomogram Averaging.
Drift correction of the image stacks and alignment of the tilt series were performed using Motioncorr2 (45) and IMOD (46), respectively. Tomograms were reconstructed using Tomo3D (47). A total of 143 and 89 tomograms of the flagellated cell poles were reconstructed for WT H. pylori and the ΔfliL mutant, respectively. The i3 suite (48, 49) was used to manually pick 855 and 388 flagellar motor particles in the WT and mutant tomograms, respectively, for subtomogram extraction and averaging. Binned 4 × 4 × 4 subtomograms were used for the refinement of the intact flagella motor structures. The right side of motor regions in 2 × 2 × 2 binned subtomograms was used for additional focused refinement.
Crystallization of FliL-C and X-ray Diffraction Data Collection.
The form A crystals of Helicobacter pylori SS1 FliL-C were grown as previously described (50). The crystals belong to space group P1, with unit cell parameters a = 62.5, b = 82.6, c = 97.8 Å, α = 67.7, β = 83.4, and γ = 72.8° (SI Appendix, Table S1), and contain 12 subunits in the asymmetric unit. The form B crystals of FliL-C were obtained using the hanging-drop vapor diffusion method, with 0.35 M ammonium phosphate monobasic and 0.2 M trisodium citrate dihydrate, pH 5.0, as a reservoir solution. These crystals belong to space group C2 with unit cell parameters a = 161.4, b = 42.5, c = 140.6 Å, and β = 93.3° and contain six subunits in the asymmetric unit. The form C crystals were obtained by using an Additive Screen HR2-138 (Hampton Research) in combination with reservoir conditions similar to those of the form A and form B crystals. The form C crystals grew from 0.3 M ammonium phosphate monobasic, 0.1 M trisodium citrate dihydrate, pH 5.6, and 0.2 M sodium thiocyanate (an additive). The form C crystals belong to space group P212121 with cell parameters a = 41.5, b = 60.0, and c = 61.0 Å and with a monomer in the asymmetric unit. X-ray diffraction data for the cryo-cooled forms A, B, and C crystals were collected to 2.8-, 2.7-, and 2.1-Å resolution, respectively, using the Macromolecular Crystallography beamlines MX1 and MX2 at the Australian Synchrotron. All the data were processed and scaled using XDS (51) and AIMLESS from the Collaborative Computational Project, Number 4 suite (52). Data collection and processing statistics are summarized in SI Appendix, Table S1.
Determination of FliL-C Crystal Structure.
Secondary structure prediction from sequence analysis was performed using the Jpred4 server (http://www.compbio.dundee.ac.uk/jpred4/index.html) (53). The phase problem for the orthorhombic (form C) crystal structure was solved by ab initio phasing with ARCIMBOLDO (27) using an initial search model comprising one 20-residue and three 10-residue polyalanine α-helices. A complete model of FliL-C except for the N-terminal residue 81 was built from the model produced by ARCIMBOLDO using the programs ARP/wARP (54) and COOT (55). Refinement and automated placement of ordered water molecules was performed using PHENIX (56) and REFMAC (57). The form A and form B crystal structures were solved by molecular replacement using PHASER (58) with the refined form C structure as a search model. The individual subunits in the asymmetric units of form A and form B crystals were refined independently. Because of the disorder of several N-terminal residues in each subunit, the final refined form A crystal structure contains 1,232 of the expected 1,308 amino acid residues, and the form B crystal structure contains 605 of the expected 654 amino acid residues. Refinement statistics and stereochemistry are given in SI Appendix, Table S2. Representations of crystal structures were prepared using PYMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC).
Three-Dimensional Visualization of the Motor and Docking Crystal Structures into Cryo-ET Map.
UCSF ChimeraX (59) was used to generate isosurface representations of the averaged structure of the intact flagella motor from WT H. pylori. Computational docking of the crystal structure–based model of the full nine-membered FliL-C ring (Fig. 3D) into the electron density for the intact motor obtained by cryo-ET was performed using the function “fit in map” in UCSF ChimeraX.
Supplementary Material
Acknowledgments
This research was undertaken in part using the MX2 beamline at the Australian Synchrotron. We thank Nina Salama for providing the genomic DNA isolated from the H. pylori G27 fliL mutant. This work was supported by the Australian Research Council Grant DP210103056 (to A.R.) and the National Institute of Allergy and Infectious Diseases Grants R01AI087946 (to J.L.) and RO1AI116946 (to K.M.O). Cryo-ET data were collected using the Yale Cryo-EM resources funded in part by the NIH grant 1S10OD023603-01A1.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2118401119/-/DCSupplemental.
Data Availability
The coordinates and structure factors for H. pylori FliL-C (forms A, B, and C) have been deposited in the PDB (http://www.rcsb.org) with accession codes 7sgn, 7sgo, and 7sgp, respectively. The cryo-ET maps of the motor in WT and fliL mutant cells have been deposited in the Electron Microscopy Data Bank under accession codes EMD-25123 and EMD-25124.
References
- 1.Josenhans C., Suerbaum S., The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291, 605–614 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Ottemann K. M., Miller J. F., Roles for motility in bacterial-host interactions. Mol. Microbiol. 24, 1109–1117 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Manson M. D., Tedesco P., Berg H. C., Harold F. M., Van der Drift C., A protonmotive force drives bacterial flagella. Proc. Natl. Acad. Sci. U.S.A. 74, 3060–3064 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao X., Norris S. J., Liu J., Molecular architecture of the bacterial flagellar motor in cells. Biochemistry 53, 4323–4333 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takekawa N., Imada K., Homma M., Structure and energy-conversion mechanism of the bacterial Na+-driven flagellar motor. Trends Microbiol. 28, 719–731 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Deme J. C., et al. , Structures of the stator complex that drives rotation of the bacterial flagellum. Nat. Microbiol. 5, 1553–1564 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santiveri M., et al. , Structure and function of stator units of the bacterial flagellar motor. Cell 183, 244–257.e16 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Roujeinikova A., Crystal structure of the cell wall anchor domain of MotB, a stator component of the bacterial flagellar motor: Implications for peptidoglycan recognition. Proc. Natl. Acad. Sci. U.S.A. 105, 10348–10353 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Neill J., Xie M., Hijnen M., Roujeinikova A., Role of the MotB linker in the assembly and activation of the bacterial flagellar motor. Acta Crystallogr. D Biol. Crystallogr. 67, 1009–1016 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Reboul C. F., Andrews D. A., Nahar M. F., Buckle A. M., Roujeinikova A., Crystallographic and molecular dynamics analysis of loop motions unmasking the peptidoglycan-binding site in stator protein MotB of flagellar motor. PLoS One 6, e18981 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrews D. A., Nesmelov Y. E., Wilce M. C., Roujeinikova A., Structural analysis of variant of Helicobacter pylori MotB in its activated form, engineered as chimera of MotB and leucine zipper. Sci. Rep. 7, 13435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosking E. R., Vogt C., Bakker E. P., Manson M. D., The Escherichia coli MotAB proton channel unplugged. J. Mol. Biol. 364, 921–937 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Zhou X., Roujeinikova A., The structure, composition, and role of periplasmic stator scaffolds in polar bacterial flagellar motors. Front. Microbiol. 12, 639490 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Limberger R. J., Slivienski L. L., El-Afandi M. C., Dantuono L. A., Organization, transcription, and expression of the 5′ region of the fla operon of Treponema phagedenis and Treponema pallidum. J. Bacteriol. 178, 4628–4634 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Motaleb M. A., Pitzer J. E., Sultan S. Z., Liu J., A novel gene inactivation system reveals altered periplasmic flagellar orientation in a Borrelia burgdorferi fliL mutant. J. Bacteriol. 193, 3324–3331 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takekawa N., et al. , Structure of Vibrio FliL, a new stomatin-like protein that assists the bacterial flagellar motor function. MBio 10, e00292–e00219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Attmannspacher U., Scharf B. E., Harshey R. M., FliL is essential for swarming: Motor rotation in absence of FliL fractures the flagellar rod in swarmer cells of Salmonella enterica. Mol. Microbiol. 68, 328–341 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Zhu S., Kumar A., Kojima S., Homma M., FliL associates with the stator to support torque generation of the sodium-driven polar flagellar motor of Vibrio. Mol. Microbiol. 98, 101–110 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Jenal U., White J., Shapiro L., Caulobacter flagellar function, but not assembly, requires FliL, a non-polarly localized membrane protein present in all cell types. J. Mol. Biol. 243, 227–244 (1994). [DOI] [PubMed] [Google Scholar]
- 20.Suaste-Olmos F., et al. , The flagellar protein FliL is essential for swimming in Rhodobacter sphaeroides. J. Bacteriol. 192, 6230–6239 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mengucci F., et al. , Characterization of FliL proteins in Bradyrhizobium diazoefficiens: Lateral FliL supports swimming motility, and subpolar FliL modulates the lateral flagellar system. J. Bacteriol. 202, e00708–e00719 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramia M., Tullock D. L., Phan-Thien N., The role of hydrodynamic interaction in the locomotion of microorganisms. Biophys. J. 65, 755–778 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Celli J. P., et al. , Rheology of gastric mucin exhibits a pH-dependent sol-gel transition. Biomacromolecules 8, 1580–1586 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Hazell S. L., Lee A., Brady L., Hennessy W., Campylobacter pyloridis and gastritis: Association with intercellular spaces and adaptation to an environment of mucus as important factors in colonization of the gastric epithelium. J. Infect. Dis. 153, 658–663 (1986). [DOI] [PubMed] [Google Scholar]
- 25.Qin Z., Lin W. T., Zhu S., Franco A. T., Liu J., Imaging the motility and chemotaxis machineries in Helicobacter pylori by cryo-electron tomography. J. Bacteriol. 199, e00695–e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang Y., et al. , Molecular mechanism for rotational switching of the bacterial flagellar motor. Nat. Struct. Mol. Biol. 27, 1041–1047 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodríguez D. D., et al. , Crystallographic ab initio protein structure solution below atomic resolution. Nat. Methods 6, 651–653 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Holm L., Laakso L. M., Dali server update. Nucleic Acids Res. 44, W351–W355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brand J., et al. , A stomatin dimer modulates the activity of acid-sensing ion channels. EMBO J. 31, 3635–3646 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuwahara Y., et al. , Unusual thermal disassembly of the SPFH domain oligomer from Pyrococcus horikoshii. Biophys. J. 97, 2034–2043 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yokoyama H., Fujii S., Matsui I., Crystal structure of a core domain of stomatin from Pyrococcus horikoshii illustrates a novel trimeric and coiled-coil fold. J. Mol. Biol. 376, 868–878 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Tanaka H., et al. , The structure of rat liver vault at 3.5 angstrom resolution. Science 323, 384–388 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Casañas A., et al. , New features of vault architecture and dynamics revealed by novel refinement using the deformable elastic network approach. Acta Crystallogr. D Biol. Crystallogr. 69, 1054–1061 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Langhorst M. F., Reuter A., Stuermer C. A. O., Scaffolding microdomains and beyond: The function of reggie/flotillin proteins. Cell. Mol. Life Sci. 62, 2228–2240 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papadopoulos J. S., Agarwala R., COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N., ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kojima S., et al. , Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol. Microbiol. 73, 710–718 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Partridge J. D., Nieto V., Harshey R. M., A new player at the flagellar motor: FliL controls both motor output and bias. MBio 6, e02367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tatsuta T., Model K., Langer T., Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol. Biol. Cell 16, 248–259 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osman C., Merkwirth C., Langer T., Prohibitins and the functional compartmentalization of mitochondrial membranes. J. Cell Sci. 122, 3823–3830 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Lee A., et al. , A standardized mouse model of Helicobacter pylori infection: Introducing the Sydney strain. Gastroenterology 112, 1386–1397 (1997). [DOI] [PubMed] [Google Scholar]
- 42.Yang D. C., et al. , A genome-wide Helicobacter pylori morphology screen uncovers a membrane-spanning helical cell shape complex. J. Bacteriol. 201, e00724–e00718 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowenthal A. C., et al. , A fixed-time diffusion analysis method determines that the three cheV genes of Helicobacter pylori differentially affect motility. Microbiology (Reading) 155, 1181–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kremer J. R., Mastronarde D. N., McIntosh J. R., Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 47.Agulleiro J.-I., Fernandez J.-J., Tomo3D 2.0--exploitation of advanced vector extensions (AVX) for 3D reconstruction. J. Struct. Biol. 189, 147–152 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Winkler H., 3D reconstruction and processing of volumetric data in cryo-electron tomography. J. Struct. Biol. 157, 126–137 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Winkler H., et al. , Tomographic subvolume alignment and subvolume classification applied to myosin V and SIV envelope spikes. J. Struct. Biol. 165, 64–77 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan K. L., et al. , Purification, crystallization and preliminary X-ray crystallographic studies on the C-terminal domain of the flagellar protein FliL from Helicobacter pylori. Biosci. Trends 12, 630–635 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drozdetskiy A., Cole C., Procter J., Barton G. J., JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 43, W389–W394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langer G., Cohen S. X., Lamzin V. S., Perrakis A., Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murshudov G. N., et al. , REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goddard T. D., et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors for H. pylori FliL-C (forms A, B, and C) have been deposited in the PDB (http://www.rcsb.org) with accession codes 7sgn, 7sgo, and 7sgp, respectively. The cryo-ET maps of the motor in WT and fliL mutant cells have been deposited in the Electron Microscopy Data Bank under accession codes EMD-25123 and EMD-25124.