Significance
Current high-throughput cell screening tools do not select cells based on their behavior in production or commercial environments, making it difficult to translate selected cells from the laboratory to commercialization. With PicoShells, we are able to perform high-throughput sorting of cells based on their phenotypic behavior in production-relevant environments, like stirred flasks, and in the presence of background cells, potentially speeding up the development of new biotechnology products by several months to years. In particular, PicoShells enable the selection of clonal colonies based on their overall accumulated growth or production of, for example, chlorophyll over a set period of time, potentially creating a cell selection tool that will improve yields of desired bioproducts.
Keywords: microfluidics, high-throughput screening, biofuel, selection, biomaterials
Abstract
Production of high-energy lipids by microalgae may provide a sustainable energy source that can help tackle climate change. However, microalgae engineered to produce more lipids usually grow slowly, leading to reduced overall yields. Unfortunately, culture vessels used to select cells based on growth while maintaining high biomass production, such as well plates, water-in-oil droplet emulsions, and nanowell arrays, do not provide production-relevant environments that cells experience in scaled-up cultures (e.g., bioreactors or outdoor cultivation farms). As a result, strains that are developed in the laboratory may not exhibit the same beneficial phenotypic behavior when transferred to industrial production. Here, we introduce PicoShells, picoliter-scale porous hydrogel compartments, that enable >100,000 individual cells to be compartmentalized, cultured in production-relevant environments, and selected based on growth and bioproduct accumulation traits using standard flow cytometers. PicoShells consist of a hollow inner cavity where cells are encapsulated and a porous outer shell that allows for continuous solution exchange with the external environment. PicoShells allow for cell growth directly in culture environments, such as shaking flasks and bioreactors. We experimentally demonstrate that Chlorella sp., Saccharomyces cerevisiae, and Chinese hamster ovary cells, used for bioproduction, grow to significantly larger colony sizes in PicoShells than in water-in-oil droplet emulsions (P < 0.05). We also demonstrate that PicoShells containing faster dividing and growing Chlorella clonal colonies can be selected using a fluorescence-activated cell sorter and regrown. Using the PicoShell process, we select a Chlorella population that accumulates chlorophyll 8% faster than does an unselected population after a single selection cycle.
With the heightened interest in cell-derived bioproducts (e.g., high-energy lipids, recombinant proteins, antibody therapies, and nutraceuticals) and cell therapies (chimeric antigen receptor T cell and stem cell therapies), the selection of desired cells based on their phenotypic properties has become increasingly important. In particular, the selection of microalgae strains for use as factories that convert light into biofuels has a long history because of their potential to be used as a carbon-neutral energy source. Specifically, high-energy lipids such as triacylglycerols extracted from microalgae strains can be processed into biodiesel that can serve as an alternative energy source to power transportation (1, 2). CO2 emissions from the burning of biodiesel can later be fixed by microalgae and used to produce more high-energy lipids, creating a carbon-neutral mechanism to power today’s economy (3). Microalgae are preferred over terrestrial plants because they have much faster biomass accumulation rates, and particular strains do not compete for resources that are important for agriculture (4). Specifically, algae will occupy less land, and certain strains can survive within recycled waste or seawater, eliminating potential competition for fresh water. In order to scale the microalgae industry to a point where microalgal-derived biofuels can be used ubiquitously, it is important to identify novel algae populations with enhanced biomass and lipid accumulation rates (5). However, algal populations that are selected to overaccumulate high-energy lipids often have reduced cell division rates (6), making it necessary to develop technologies that can select algae populations based on their coupled division rate and lipid production.
Unfortunately, high-throughput screening tools for selection based on growth and bioproduct accumulation have not been readily available for scientists engineering cell strains. Fluorescence-activated cell sorting (FACS) methods to select microalgal strains are only capable of selecting based on lipid content and not growth rate. This is because FACS traditionally has measured single cells at a single time point rather than assaying colonies that are growing over time. Growth-based selection has been limited to low-throughput techniques such as using microtiter plates (7) or small-scale bioreactors.
Microwell (8), microcapillary (9), droplet (10, 11), and gel microdrop technologies (12, 13) are capable of compartmentalizing single cells into nanoliter-sized compartments and allowing cells to grow into small clonal colonies for selection, but they do have some key limitations. The microfluidic approaches can have automated high-throughput selection mechanisms that make it possible to screen populations greater than 100,000 colonies or single cells per screen. Unfortunately, these microfluidic compartments have physical or chemical barriers that inhibit transport between the compartment and the external environment. In consequence, enclosed cells may deplete nutrients within the compartment, can accumulate secreted cytotoxic elements, can change the pH of the environment, and may have reduced ability to communicate with other cells via secreted factors. So, these high-throughput screening technologies may not be suitable for long-term (e.g., >24-h) growth assays, yielding selection pressures that are not aligned with final growth environments. This is because over these timescales, the compartments do not provide an environment that is physiologically similar to what is expected in scaled production cultures. As a result, selected cells may not behave the same way when scaled up for real-world applications as they did when cultured within these nanoliter-sized compartments. This behavior has been recently observed for yeast cells, where the size of the droplet affects overall cell morphology following culture (14). Scientists often need to perform further experiments and do additional genetic manipulation of selected strains to adapt them to scaled-up industrial cultures, a process that can take several months or years without guaranteed success. Nanopen technology (15) does have nanoliter-sized compartments that can have their solution replaced without dislodging the cells; however, it requires light-based manipulation of cells to isolate desired colonies that have a limited throughput of ∼10,000 cells per screen.
To address these issues, we have developed a hollow shell microparticle platform (PicoShells), which enables compartmentalization of growing colonies, continuous media exchange, phenotypic screening and sorting via FACS, and viable downstream recovery. The PicoShell particles are ∼90 µm in diameter, consist of a solid outer shell made of polymerized polyethylene glycol (PEG), and have a hollow inner cavity where microalgae can be encapsulated and cultured. More importantly, the solid PEG matrix is porous, allowing the PicoShells to be refreshed by external media, which would enable transport of nutrients into the compartment and facilitate potential communication between cells in nearby compartments or in surrounding media. We have also demonstrated that PicoShells can be placed into unique environments that have not been previously possible such as a stirring solution within a 100-mL beaker. Cell-containing PicoShells should, therefore, be compatible with culture in various bioreactors or other relevant environments, providing a production-similar environment for enclosed cells that is not possible to attain with any other nanoliter-scale compartments. As a result of these features, strains developed using PicoShells may be expected to exhibit their desired phenotypes in relevant scaled-up cultures, promising to save cell-line developers months or years of additional labor to reach a similar point. These PicoShell particles can be loaded with single cells such that those cells grow over a multiday period to form clonal colonies. Additionally, the pores in the outer hydrogel shell allow for encapsulated cells to be stained with common fluorescent tags such as 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene (BODIPY) and live/dead stains. Since these particles are stable in aqueous solution, they can be screened and sorted using standard FACS instruments, allowing colony-containing PicoShells to be sorted at throughputs >300 particles/s. Cells can be released from PicoShells via mechanical or chemical mechanisms, retaining viability such that selected cells can be recultured, further scaled, analyzed, and perhaps resorted (Fig. 1).
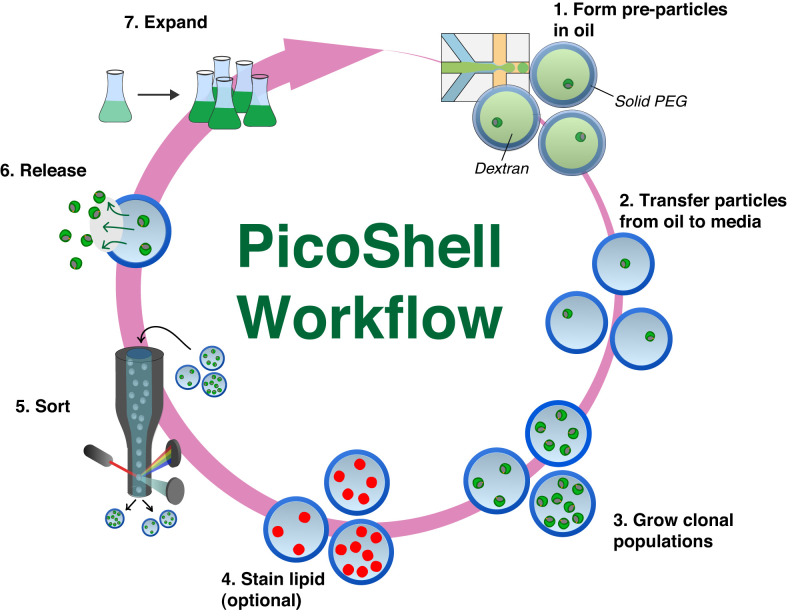
Fig. 1.
Workflow to enrich microalgae using PicoShell particles. 1) PicoShells are formed using droplet microfluidics, an aqueous two-phase system, and polymer chemistry. Particles are initially formed within an aqueous droplet surrounded by oil. Microalgae are within the dextran phase, which is surrounded by a solidifying PEG matrix. 2) Soon after particle formation, the particles are transferred into the algae’s native media. Pores in the solid outer shell allow for dextran to leak out and for continuous solution exchange. 3) Microalgae can divide within particles over multiple days to form clonal populations. 4) Pores in the solid matrix allow algal lipids to be fluorescently labeled. 5) High-performing populations can be sorted using FACS with scatter and/or fluorescence readouts. 6) Sorted particles can be broken down mechanically or by adding chemical reagents that degrade the PicoShell’s solid matrix, allowing associated cells to be released. 7) Released cells remain viable and can be recultured for further analysis and/or sorting.
Here, we compare growth of colonies in PicoShells with microfluidic droplet-in-oil approaches and demonstrate a proof-of-concept workflow for the selection of hyperperforming microalgae populations. In particular, we show that the accumulated growth of microalgae, yeast, and adherent Chinese hamster ovary (CHO) cell colonies is higher in PicoShells than in water-in-oil droplet emulsions. Also, we demonstrate that particles containing faster-growing algae colonies can be selected using FACS, that selected colonies can be released, and that the selected population can maintain a higher rate of accumulation of chlorophyll than the unselected population upon reculture. We anticipate that the PicoShell platform can play a key role in the selection of hyperproducing microalgae strains that translate to scaled-up culture environments as well as various other producer cell lines for a range of bioproducts.
Results
Fabrication of PicoShells.
PicoShell particles are made using a combination of microfluidic droplet technology (16–18), aqueous two-phase systems (19, 20), and PEG polymer chemistry (21) (SI Appendix, Fig. S1). When mixed together at certain concentrations, a PEG-rich and dextran-rich phase can form, with a degree of phase separation that is tunable by adjusting the relative concentrations of the PEG and dextran components (22, 23). Coalescence of the PEG-rich phase at different concentrations of PEG and dextran can lead to particles of unique shapes, owing to the unique interfacial tensions of the three-phase system (PEG-rich, dextran-rich, and oil phase) (24, 25). To determine the concentrations of PEG and dextran required to obtain PicoShell particles, we first obtained the binodal curve with the particular PEG and dextran used, a plot that defines the boundary between a completely mixed and phase-separated aqueous two-phase solution. The binodal is dependent on the molecular weights and chemical functionality of the materials (SI Appendix, Fig. S2). We found that regions close to the binodal but above and to the right of the boundary led to the formation of concentric phases. When droplets contain PEG/dextran concentrations within 1 to 2% into the phase separation region above and to the right of the binodal, the dextran-rich region orients in the center of the droplet with the PEG-rich region uniformly surrounding the dextran-rich region at the aqueous–oil interface (Movie S1). Complete stable phase separation occurs within ∼20 s following droplet formation. Cross-linking the PEG phase at these concentrations results in PEG hydrogel shells (i.e., PicoShell particles) that can remain stable when transferred out of oil and into aqueous solution (phosphate buffered saline [PBS], media, etc.). The molecular weight of the dextran is chosen such that it can diffuse out of the enclosed shell particle following the phase transfer (SI Appendix, Fig. S3). The mechanism to form such hollow shell or capsule particles using the methods we describe has been adapted from previous work (26, 27).
We identified a cell-compatible cross-linking chemistry to form PicoShell particles. While ultraviolet (UV)-dependent chemistries can be used to cross-link porous hydrogel particles of similar geometries (28), we chose a different approach as UV light and free radicals generated from photo initiators are likely to genetically or phenotypically affect cells being encapsulated, potentially introducing negative impacts on the assay/selection (29). We incorporated a biocompatible pH-induced cross-linking chemistry where gelation occurs within physiologically compatible pH ranges (pH ∼ 6 to 8) (30). However, pH-induced cross-linking introduces challenges as the mixing time of cross-linker and functionalized PEG within droplets along with solidification affects the final particle morphology, even at the same PEG/dextran concentrations (SI Appendix, Fig. S4). If the cross-linking time is too short, the PEG and dextran phases do not have enough time to fully phase separate, usually resulting in a nonuniform outer shell and/or undesired partial cross-linking in the cavity. In contrast, if the cross-linking reaction is too slow, the shift in the binodal resulting from the increasing molecular weight of the PEG phase as it starts to polymerize causes the formation of bowl-shaped particles instead. To obtain the ideal particle shape, we adjusted the cross-linking time by modulating the pH of the formed droplet. We found that repeatable, uniform shells could be formed by generating emulsions with in-droplet concentrations of 11% (wt/wt) 10-kDa dextran, 5% (wt/wt) 10-kDa four-arm polyethylene glycol maleimide (PEG-MAL), and 1.54 mg/mL dithiothreitol (DTT) cross-linker at pH 6.25. With this combination of reagents, we are able to form uniform particles (SI Appendix, Fig. S5) with an outer diameter of 91 µm and shell thickness of 13 µm (coefficient of variation-CVs of 1.7 and 6.9%, respectively) at a particle generation rate of 720 PicoShells/s.
Enhanced Growth in PicoShells vs. Droplets.
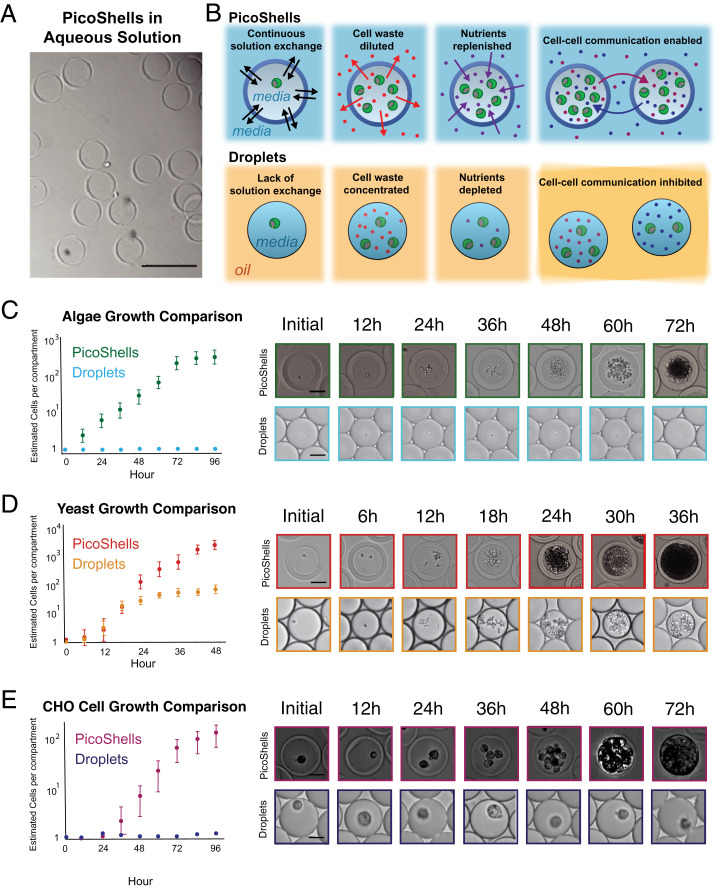
We found that encapsulated cells (Chlorella sp., Saccharomyces cerevisiae, and adherent CHO cells) grow more rapidly and to higher final densities in PicoShells than in microfluidic droplets in oil (Fig. 2 C–E). We found that cells tend to grow and settle to the bottom of the PicoShell rather than be suspended in the core (SI Appendix, Fig. S6).
Fig. 2.
Growth comparison between PicoShells and emulsion droplets. (A) PicoShells are solid spherical particles that contain a hollow inner cavity and a porous outer shell. (Scale bar: 200 µm.) (B) PicoShells allow for continuous solution exchange with the external environment such that cell waste can be diluted, nutrients can be replenished, and cell–cell communication factors can pass between adjacent PicoShells. (C) Chlorella were encapsulated into PicoShells and droplets to compare division rates in each compartment. Results show that the microalgae do not grow in droplets but grow readily in the particles; 5,000 to 6,000 cell–containing PicoShells/droplets were analyzed for each time point. Error bars represent the SD in the estimated number of cells per compartment at each time point. (Scale bars: 50 µm.) (D) S. cerevisiae were also encapsulated into PicoShells and droplets to compare growth rates. The yeasts initially grew at the same rate in both compartments, but growth eventually slowed down in droplets; 5,000 to 6,000 cell–containing PicoShells/droplets were analyzed for each time point. Error bars represent the SD in the estimated number of cells per compartment at each time point. (Scale bar: 50 µm.) (E) Adherent CHODP12 cells were also encapsulated into PicoShells and droplets to compare growth rates. CHO cells did not grow within droplets but grew readily in PicoShells; 400 to 500 cell–containing PicoShells/droplets were analyzed for each time point. Error bars represent the SD in the estimated number of cells per compartment at each time point. (Scale bars: 50 µm.)
Cells were encapsulated into PicoShells and droplets. Chlorella growth was tracked every 12 h over a 72-h period, and S. cerevisiae was tracked every 6 h over a 36-h period. The number of cells in each PicoShell and droplet was estimated by dividing the cell mean area within each compartment by the average area of a single cell in each population to obtain an estimated count (the code is provided in Dataset S1). Since the cells become very dense at longer times, counting individual cells was not feasible. Estimated count was, therefore, used to quantitatively compare relative differences in cell numbers between PicoShells and droplets rather than providing a precise measurement of cell count. Interestingly, we found that Chlorella grow rapidly in PicoShells (Movie S2) starting with the formation of a first generation of daughter cells following 12 h of incubation but did not grow when encapsulated in microfluidic droplets even over a 72-h period (Fig. 2C). Chlorella were encapsulated and incubated in autotrophic media, presumably making cells more susceptible to gas transport. Chlorella were found to double every 12.2 h and reached a carrying capacity within the 155-pL hollow cavity of a PicoShell of an estimated 250 cells (Fig. 2C). In parallel, we found that S. cerevisiae grow both in PicoShells and in water-in-oil droplet emulsions (Fig. 2D). However, while the growth rates of the yeast in both types of compartments were not statistically different before the first 18 h of culture (P = 0.28 at 12 h), the growth rate of droplet-encapsulated yeast became significantly slower than PicoShell-encapsulated yeast at later times (P < 0.001 at times >18 h). The average number of yeast cells in PicoShells is dramatically increased between 24 and 48 h after encapsulation to an estimated 2,900 cells/PicoShell, ∼20× higher than the carrying capacity in droplets (estimated to be ∼150 cells per droplet). We also found that adherent CHO cells can grow within PicoShells and not within droplets (Fig. 2E). CHO cells have a reduced growth rate for the first 24 h (doubling time = 178 h) and grow with a doubling time of 18 h per doubling starting at 24 h after encapsulation. CHO cells reach a carrying capacity of an estimated ∼100 cells/PicoShell 96 h after encapsulation. It is important to note that droplets containing CHO cells were covered with less dense mineral oil to reduce the evaporation of droplets kept at 37 °C during the study (SI Appendix, Fig. S7), which was not required for PicoShells. A number of factors, including the lack of adhesive surfaces to interact with or the reduction of gas transport from the mineral oil layer, may have resulted in the lack of growth by CHO cells in this assay. However, growing yeast cells in droplets with and without a mineral oil cap did not result in a significant difference in the growth rate over a 48-h period (SI Appendix, Fig. S8).
Differences in growth in PicoShells may be due to transport of nutrients and cytotoxic factors and/or increased cell–cell communication (Fig. 2B). The reduction in the growth rate of droplet-encapsulated yeast is likely due to the depletion of essential nutrients and/or accumulation of cytotoxic cellular waste. All nutrients present in both media types were below 200 Da. Such nutrients are expected to be freely exchanged through the outer membrane of the PicoShells given the molecular masses below ∼40 kDa (SI Appendix, Fig. S3). This result is in agreement with the enhanced growth rate of Escherichia coli observed when encapsulated in capsule particles compared with droplets in oil (27). To further explore these effects, we introduced a leucine-dependent yeast cell strain into PicoShells and transferred them into media with and without leucine after encapsulation. Yeast in PicoShells transferred to medium that contains leucine resulted in a statistically significant increase (P < 0.001) in the number of cell divisions over a 12-h period when compared with a leucine-free medium (SI Appendix, Fig. S9). This indicates that nutrients like the amino acid leucine transport across the PicoShells during incubation. We have also demonstrated that cell–cell communication factors have an effect on the behavior of cells within PicoShells by incubating encapsulated yeast in a solution with yeast cells at a concentration of 100 million cells/mL and another solution without yeast cells. We observed a statistically significant difference in the number of cell divisions over a 12-h period (SI Appendix, Fig. S10). Interestingly, we observed a reduced division rate when PicoShells are placed in a solution with a large background of yeast cells in stationary phase in the external environment, despite replenishing the nutrients just prior to the start of the study. We hypothesize that the quorum-sensing factors secreted by the yeast in the external solution were those normally secreted during stationary phase to inhibit growth (30).
Intriguingly, we also observed that S. cerevisiae and CHO cells do not stop dividing after they fully occupy the volume of the inner cavity of the particle, and additional cells actually cause the particle to stretch, increasing the overall diameter (SI Appendix, Figs. S11 and S12). The diameter of the PicoShells can actually expand from an initial diameter of ∼90 µm to a maximum size of ∼500 µm after 4 d, at which point the particle ruptures and releases the encapsulated cells (Movie S3). This phenomenon was not observed for encapsulated Chlorella colonies. Instead, the microalgae were observed to stop dividing when the colony reaches the particle boundary.
Sorting of Chlorella Based on Accumulated Growth.
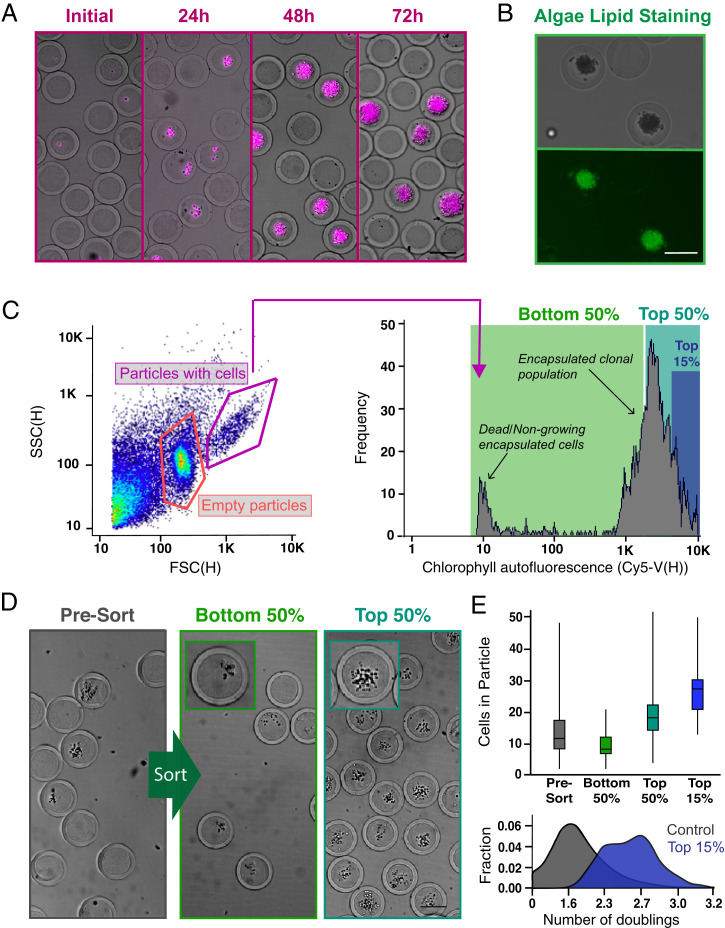
Chlorella colonies seeded and cultured in PicoShells were selected based on the accumulated growth of cells using an FACS instrument. We used the colony’s chlorophyll autofluorescence, appearing in the Cyanine5 (Cy5) channel (excitation [ex]: 620, emission [em]: 647), as a metric for growth and production of biological materials (Fig. 3A). Generally, colonies containing greater numbers of cells also contain higher amounts of chlorophyll, generating higher Cy5 fluorescence readouts, as we have demonstrated in a previous study (12). We demonstrated that lipids could also be stained through the PicoShells by mixing BODIPY with the colony-containing particles (Fig. 3B). However, to simplify the study design to focus on improving the engineering aspects of the workflow, we only sorted clonal colonies based on overall accumulation of chlorophyll and not lipid productivity. We encapsulated Chlorella at an average loading density, lambda of 0.1, which resulted in 91.7% of cell-containing particles with no more than a single cell.
Fig. 3.
Screening and sorting characterization of microalgae-containing PicoShells. (A) PicoShells were loaded with Chlorella at lambda = 0.1 and allowed to grow for 48 h. The growth of Chlorella can be characterized via the chlorophyll autofluorescence that appears in the Cy5 channel. (B) The lipids in encapsulated Chlorella cells were stained with the addition of BODIPY 505/515. Localization of the stain was observed in the FITC channel. (C) After allowing Chlorella to divide in PicoShells, the particles were screened using an On-Chip Biotechnologies Cell Sorter. Particles that contain colonies and cells can be distinguished from empty particles using scatter readouts. Colony-containing particles produce an observable Cy5 fluorescence distribution via the colony’s chlorophyll autofluorescence. SSC(H), side scatter height. (D) The colony-containing particles that produced the lowest 50%, highest 50%, and highest 15% of Cy5 fluorescence readouts were sorted with 94.0% purity and 72.7% yield; 400 particles were sorted in each sample. Insets show magnified views of a colony within a single PicoShell for the correspondingsort gate. (E) Selection of colony-containing PicoShells from different regions of the Cy5 distribution corresponds to particles containing different numbers of algal cells, with particles with higher Cy5 fluorescence readouts containing more cells than those with lower Cy5 fluorescent readouts. Particles sorted from the higher end of the Cy5 distribution contain colonies that have undergone more doublings and have divided more during the incubation period. The middle line within each of the boxes in the box and whisker plot represents the mean number of cells in the particle; the top and bottom of each box represent the first and third quartiles, respectively; and the top and bottom of the error bars represent the maximum and minimum values, respectively. In total, 350 to 400 PicoShells were counted in each sample. (Scale bars: 50 µm.)
Following culture for 48 h, PicoShells containing Chlorella colonies were sorted using the On-Chip Sort at an average event rate of 100 to 200 events per 1 s. We observed three distinct populations in the forward scatter (FSC) height vs. side scatter (SSC) height plot: one from colony-containing particles, one from empty particles, and one from debris (Fig. 3C). The debris population was confirmed to be from particulates naturally present in Chlorella media. If the media are filtered, a greatly reduced fraction of debris events is observed (SI Appendix, Fig. S13). As expected, ∼14.3% of detected particles contain cells, which correlates with the target loading fraction of 10%. In agreement with contrast observed in bright-field microscopy, PicoShells that contain microalgal colonies generally have increased forward and side scatter intensities. We verified that most of the colony-containing particles are within this high FSC/SSC gate by demonstrating that events in this gate also contained the highest Cy5 fluorescence (i.e., chlorophyll autofluorescence). A selected sample based on this gate had 94.0% purity for PicoShells that contained a cell colony (SI Appendix, Fig. S14).
Using the On-Chip Sort, we selected out PicoShell particles with the fastest-growing colonies by gating on chlorophyll autofluorescence. When we selected particles from different regions of the Cy5 distribution of colony-containing particles, we observed differing numbers of microalgae in the sorted colonies (Fig. 3D). PicoShells gated on the lowest 50% in the Cy5 channel and within the high scatter gate possessed on average 9.2 ± 3.7 cells. This was statistically different from colonies recovered when gating the highest 50% (19.5 ± 7.1 cells, P < 0.0001) and highest 15% (27.0 ± 7.2 cells) (Fig. 3E). Before sorting, colony-containing PicoShells contained on average 13.0 ± 7.7 cells. Overall, higher Cy5 fluorescence intensities corresponded to particles with a greater number of cells, and given our loading conditions favoring single cell–derived colonies, it is likely these particles contained microalgae subpopulations that have faster doubling times and/or increased production of chlorophyll (Fig. 3E).
Selection and Regrowth of a Hyperperforming Chlorella Subpopulation.
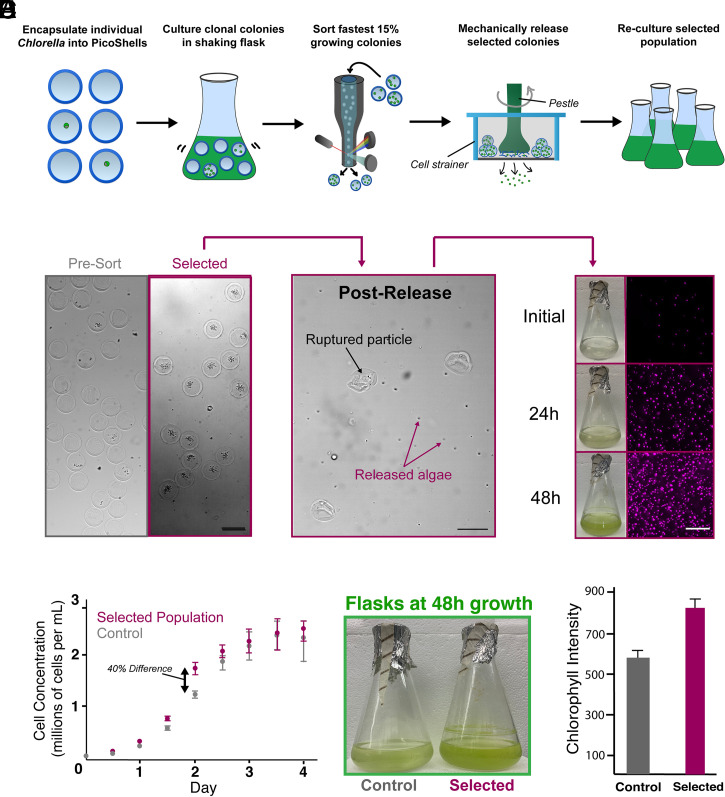
We used the workflow to select Chlorella colonies based on Cy5 fluorescence, released colonies from the particles, recultured, and verified after reculture that the selected subpopulation divides and accumulates chlorophyll faster than an unselected population (Fig. 4A). For these studies, we minimized PicoShells with more than one cell initially loaded by using lambda = 0.05, resulting in 3.2% of all particles containing colonies and ∼98.3% of cell-loaded particles containing no more than one cell. Following 48 h of growth in PicoShells immersed in Chlorella native media, we sorted colony-containing PicoShells gated to have the highest 11.1% of Cy5 fluorescence (425 events were selected from a population of 3,839 colony-containing particles) (SI Appendix, Fig. S13).
Fig. 4.
Selection of a hyperperforming Chlorella subpopulation based on division rate. (A) Single Chlorella were encapsulated into PicoShells and incubated under standard culturing conditions in a shaking flask to allow cells to produce greater numbers of cells . Colony-containing PicoShells from the top 15% of the Cy5 fluorescence distribution were selected by FACS and mechanically released from particles. Released cells were then recultured for further analysis. (B) From a particle population of 121,213 particles (3,839 containing colonies), 425 particles were selected. Selected particles were ruptured on top of a cell strainer, causing selected algae to be released into fresh culture media. This sample was regrown in an Erlenmeyer flask under standard culturing conditions for several days. (Scale bars: 100 µm.) (C) The selected population and an unselected population were seeded in separate flasks at the same concentration, and their cell concentrations were tracked for 4 d. The selected population had an 8% faster growth rate (10.2-h doubling times) than the unselected population (11.2-h doubling time) for the first 48 h after seeding before slowing down as the culture reached carrying capacity. Error bars represent the SD in the cell concentration at each time point between samples. (D) The largest difference in cell concentration was observed at 48 h after seeding (∼40% difference in cell concentration), a difference that can be visibly seen in the green color of the cultures. (E) The difference in growth was verified by measuring the chlorophyll density of each sample with a well-plate reader at 48 h after seeding. The selected population was measured to have a 27.6%-higher chlorophyll density (P < 0.05). Error bars represent the SD between the different wells used to measure the chlorophyll fluorescence at the 48-h time point. Ten wells were measured for each sample.
Selected colonies were released from PicoShells by applying mechanical shearing stress onto the particles, causing them to rupture (Movie S4). PicoShells were disrupted with mechanical shear, and released cells were recultured in a flask (Fig. 4B). We compared the division rate of the selected subpopulation during reculture with an unselected population by seeding each population at the same concentrations and tracking their cell concentrations over a 4-d period (Fig. 4C). We observed that the selected subpopulation had an ∼8% faster growth rate than the unselected population (doubling times of 10.2 and 11.2 h, respectively; P < 0.01) for the first 48 h of growth after seeding. This resulted in a 40% difference in the cell concentrations 48 h after seeding that can be visibly seen in the culture flask (Fig. 4D). We also measured the total chlorophyll autofluorescence within well-mixed aliquots from each culture at this time point (Fig. 4E), observing a 27.6% increase in chlorophyll autofluorescence for the selected subpopulation. As expected, the differences in cell number and overall chlorophyll accumulation between the two populations diminished after 48 h as the cultures reach carrying capacity.
Discussion
Advantages of PicoShells.
Several key aspects of the PicoShell workflow suggest that it can aid in the selection/evolution of cells and cell-based products, including the following. 1) Cell behavior and growth are significantly enhanced in PicoShells compared with water-in-oil droplet emulsions. 2) PicoShells containing desired cells/colonies can be selected using commercial fluorescence-activated cell sorters. 3) Selected cells/colonies can be successfully released from the PicoShells and recultured. 4) Selected populations maintained a high-growth phenotype postprocess at least for several generations. Importantly, PicoShells can be placed and remain stable in more production-relevant environments (e.g., a shaking culture flask, bioreactor, outdoor cultivation farms) that are not feasible with other high-throughput selection technologies (e.g., droplet technology, microwells, etc.) (SI Appendix, Fig. S11). The porous outer shell enables solution exchange with the external environment, likely allowing replenishment of nutrients; diffusive transport and dilution of cytotoxic cellular waste; access to quorum-sensing factors from external cells/colonies; and exposure to natural concentration, temperature, light, or physical gradients in the culture environment. As a result, PicoShell technology may provide a high-throughput screening tool that enables cell-line developers and researchers to select cells based on their behavior in production-relevant environments, making it much more likely that selected populations will exhibit the desired phenotypic properties when scaled up for real-world applications.
Transport across PicoShells is likely responsible for distinct growth phenotypes for encapsulated cells compared with droplets in oil. Previous studies demonstrated that droplet size affects the division of cells in microfluidic droplets surrounded by oil (31, 32). Another recent study has demonstrated that yeast cells cultured in large flasks, large droplets, and small droplets differed morphologically (14). There may be several reasons for this phenomenon. For example, smaller droplets have less nutrients, which could be depleted more rapidly when cells are placed within droplets and begin to grow and divide. Similar effects are expected with sealed nanowell arrays. With PicoShells, we have demonstrated that nutrients, such as the amino acid leucine, present in solution outside of the shell affect the growth and division of cells within (SI Appendix, Fig. S6). Based on these data, we conclude that leucine is able to transport across into the internal environment of the PicoShell, and we postulate that other factors and nutrients will be able to transport as well. In addition, it is well understood that cells release elements to the external environment that change its pH. For example, CHO cells release lactate that lowers the pH of the external media (33), and yeasts release alcohol that increases the pH of the external media (34). Charged species that modulate pH are expected to not easily transport out of droplets in oil or through solid barriers of nanowell arrays. Lastly, it is probable that proteins or other small molecules that are used for cell communication cannot pass through the walls of nanowell arrays or easily partition and transport through the oil phase of water-in-oil droplet emulsions. Since the outer shell of PicoShells is porous and has a molecular mass cutoff above 70 kDa, it is likely that these factors can pass through the outer matrix. This is supported by our data demonstrating that the presence of cells in the external solution can affect the growth behavior of cells within the cavity of the PicoShells (SI Appendix, Fig. S4). In addition, inclusion of a solid surface for adherent cell lines to attach to in PicoShells likely enhances the growth properties of adherent CHO cells in PicoShells compared with in droplets. It is important to note that the lack of adherent CHO cell growth in droplets that we observed may have resulted from reduced gas exchange through the mineral oil cap we placed on top of the droplets, which we use to reduce evaporation. However, we have demonstrated that S. cerevisiae do not have different growth properties within droplets capped or not capped with mineral oil (SI Appendix, Fig. S8).
Growth of other algae species in droplets has been previously shown (11, 12, 35, 36), making it intriguing why Chlorella in particular does not survive when encapsulated into water-in-oil droplet emulsions. While it is unclear exactly why this particular phenomenon occurs, we believe that the lack of cell survival is related to the restricted gas exchange across the oil barrier. This particular species is grown in autotrophic media and is very sensitive to gaseous CO2 concentrations. We have observed that bulk cultures of this particular species cannot grow when not cultured in an incubator that regulates CO2 or not cultured with media that are not supplemented with sodium bicarbonate (37). While previous studies have shown that gases can generally pass through fluorinated oil (38, 39), this diffusion may be limited or altered to an extent that sensitive species are greatly affected unlike more robust cell types (Chlamydomonas reinhardti, Euglena gracilis, etc.). Regardless of the root cause for the lack of growth in droplets, the results demonstrate that the environments in PicoShells and droplets are different enough that we can observe a noticeable effect on cell behavior, a result that is substantiated by the improved growth properties of S. cerevisiae in PicoShells.
Potential for Chemically Degradable PicoShells.
We have explored multiple mechanisms to chemically release cells from PicoShells by including chemically degradable motifs in the outer PEG shell. Currently, we can consistently fabricate PicoShells cross-linked with PEG-MAL and DTT. These are compatible with multiple cell types, including Chlorella, S. cerevisiae, and E. gracilis (SI Appendix, Fig. S15). PicoShells cross-linked with PEG-MAL and DTT can be broken down with the addition of sodium periodate (NaIO4) due to the presence of a diol in DTT. Unfortunately, NaIO4 can be toxic (40) and likely kills or has large negative impacts on many cell types. Previously, we have made hydrogel particles with degradable peptide cross-linkers (20), and similar incorporation of degradable cross-linkers could enable enzymatic or chemical degradation of particles to release selected cells/colonies. As an initial proof of concept of this approach, we developed PicoShells that contain disulfide linkages that can be degraded via the addition of DTT or Tris(2-carboxyethyl)phosphine (TCEP). S. cerevisiae encapsulated in these particles remain viable, grow, and can be chemically released (SI Appendix, Fig. S16 and Movie S5). Unfortunately, a chemical precursor we use to form these particles (four-arm polyethylene glycol ortho-pyridyldisulfide [PEG-OPSS]) is toxic to Chlorella (SI Appendix, Fig. S17), suggesting that the chemical formulation of the PicoShell should be matched to the cell type. We have also encapsulated and grown C. reinhardtii in PicoShells cross-linked with matrix metalloproteinase (MMP)–degradable peptides (SI Appendix, Fig. S18) that can be degraded with the addition of trypsin (Movie S6). Unfortunately, C. reinhardtii (and likely other cell types) naturally secrete MMPs that often prematurely break down the particles (41).
While the mechanical mechanism of release that we demonstrate works well for releasing bulk populations of selected particles, it is likely difficult to adapt the process to separately release individual colonies (e.g., a single particle in a single well). Such single-particle isolation is important if a researcher wishes to explore the different strategies for hyperperformance and the various underlying genetic mechanisms that result in such phenotypes. While it may be possible to engineer tools to mechanically break down a single particle, release of cells using these tools may be complicated and inefficient. In addition, there are studies that indicate that shear stress can negatively impact cells. For example, while shear stress may not induce cell death, such stress on microalgae or yeast may cause a decreased growth rate (42, 43). Shear stress can even reduce the expression of a recombinant protein in CHO cells (44). Hence, it may be necessary to fully develop PicoShells that are chemically degradable and compatible with several cell types. Although we have engineered disulfide cross-linked PicoShells that are compatible for yeast applications, we have also shown that it is difficult to discover chemistries that enable chemical degradation and maintain cell viability for more sensitive cell types.
Limitations on Throughput.
We have also found that there is a tendency for cross-linked material to stick to the walls near the droplet generation junction, causing a disruption in the flow (SI Appendix, Fig. S19). Since we use pH-induced gelation and the gellable materials (PEG-MAL and DTT) come into close proximity briefly before droplet generation, gelled material often forms at the junction, inducing jetting and disruption of particle formation ∼15 min after initial particle formation. As a result, the device needs to be replaced each time particle formation is halted, reducing the overall number of PicoShells that can be manufactured to 370,000 particles per device.
The jetting of reagents due to premature formation of gelled material that sticks to the walls of the droplet generator limits the overall throughput of PicoShell generation. One potential way to address this is to use a coaxial device geometry to reduce the amount of gelled material that sticks to the walls of the device (45). Use of UV-induced cross-linking mechanisms can also address this problem since gelation would occur downstream of droplet generation (29, 46, 47) unlike pH-induced mechanisms, where mixing of reagents immediately prior to droplet generation often results in gelled material forming in the droplet generation junction over time that disrupts the overall flow. UV cross-linking could also enable PicoShell manufacturing approaches with higher throughput (48). However, use of UV-induced cross-linking likely creates issues for particular cellular applications, as previously discussed. At the same time, UV-induced cross-linking may be used for workflows involving resilient cell types (e.g., bacteria) or workflows where cells are mutagenized prior to selection, and UV-induced mutations would be potentially beneficial. A summary of the different types of PicoShells that we can currently fabricate and their advantages and disadvantages is shown in Table 1.
Table 1.
Summary of current PicoShell variations
| Fabrication throughput (PicoShells/h) | Chemical release mechanism | Primary advantage | Primary disadvantage | |
| DTT cross-linked with UV | 2.5 million | NaIO4 | High fabrication throughput | Unclear how UV affects cells |
| DTT cross-linked via pH | 1.3 million | NaIO4 | Compatible with most cell types | Limited to mechanical degradation to viably release cells |
| Peptide cross-linked via pH | 1.3 million | MMPs or trypsin | Cells can be chemically released | Cells may prematurely release themselves via enzyme secretions |
| Disulfide cross-linked via pH | 1.3 million | DTT or TCEP | Cells can be chemically released | Only compatible with robust cell types such as bacteria and yeast |
Information is based on cells and chemistries explored in this study and previous studies.
Potential Future Applications.
Despite these solvable limitations, the experimental evidence we have presented shows that PicoShell technology has significant advantages. The workflow can be potentially used for directed evolution of cell populations (49) where mutagenized cells are placed under selection pressures to generate strains based on unique selection criteria that are time dependent (e.g., growth and production of pigments), at the colony level (multicellular construct formation), or require solution exchange steps (lipid staining, enzyme-linked immunosorbent assays [ELISAs]). For example, the technology may be used to produce microalgae strains that overperform in lipid accumulation rates without significantly reducing their rate of growth for biofuel applications. The technology may also be used to generate yeast strains that maintain a high growth rate at higher ethanol concentrations, potentially enhancing the overall production of ethanol biofuels (50, 51), plastics (52), materials (53), and alcoholic beverages (54).
Additionally, PicoShells have now enabled the ability to select single cells and/or clonal colonies based on their behavior in environments that have not been previously possible. For example, we have demonstrated that S. cerevisiae can grow in a bioreactor that has cells external to the PicoShells, with constant stirring, and temperature controls (SI Appendix, Fig. S20). Such a culture environment is not possible to achieve with other nanoliter-scale screening technologies, indicating the PicoShells may enable unique assays and applications that have previously been impossible. Given that PicoShells remain stable within our custom bioreactor and can be reisolated, it is probable that PicoShells can be placed into other commercial bioreactors, outdoor cultivation farms, and other unique environments and later isolated for screening and/or selection.
The outer shell’s PEG material is also able to be modified, enabling the technology to be potentially used for relevant mammalian cell applications. For example, affinity motifs such as antibodies and peptides can be added to the solid matrix that can capture cellular secretions (29). Antibody-conjugated PicoShells may be used to produce hypersecreting and hypergrowing CHO cell populations based on their behavior in bioreactors for scaled production of protein therapeutics. The pore size of the particles may also be modulated by changing the molecular weight (MW) of PEG used to cross-link the solid phase (55) or by including nonfunctionalized PEG (56), gelatin (57), or hyaluronic acid (58) in the PEG phase. Adherence motifs, such as arginine-glycine-aspartate (RGD) peptides, fibronectin, or poly-l-lysine, may be also added to the outer PEG matrix so that stem cells, adherent CHO cells, or other adherent cell types have a solid surface to adhere to, further expanding the potential applications of the PicoShell workflow.
In summary, we have shown that PicoShells may enable cell-line developers to develop cell populations based on their behavior in production environments. Unlike previously developed high-throughput screening tools, individual cells may be compartmentalized, placed into relevant environments such as bioreactors, exposed to natural stimuli, and selected based on their time-dependent behavior and growth in such environments via widely used flow cytometers. As a result, the technology has the exciting potential to rapidly accelerate the development of cell-derived bioproducts, such as biodiesel, materials, cell-derived agriculture, nutrient supplements, and protein therapeutics.
Materials and Methods
Bulk Culture of Cells.
Chlorella cells (CCMP1124 from the National Center for Marine Algae and Microbiota) used in the study were provided by Synthetic Genomics, Inc. (now called Viridos, Inc.). Chlorella populations were cultured in 500-mL Erlenmeyer flasks containing seawater-based medium with added vitamins, trace metals, nitrate, phosphate, and sodium bicarbonate (SM-NO3 medium) (59). SM-NO3 medium was also supplemented with penicillin-streptomycin (P/S; Thermo Fisher Scientific; 15140122). C. reinhardii (STR CC-4568) and E. gracilis Z (NIES-48) procured from the Microbial Culture Collection at the National Institute for Environmental Studies Japan were cultured in 500 mL using tris(hydroxymethyl)aminomethane-acetate-phosphate (TAP) medium (60) and Koren–Hunter (KH) medium at a pH of 5.5 (61), respectively. Flasks containing algae cultures were shaken continuously at 120 rpm with constant 150-µE light at room temperature. Algae cultures were kept at a concentration of 2 to 10 million cells/mL. Strains of S. cerevisiae were obtained from Sigma-Aldrich (STR YSC1). The yeasts were grown in yeast extract (1%; wt/vol), peptone (2%; wt/vol), glucose (2%; wt/vol; YPD) media supplemented with 50 mg/L ampicillin (Sigma-Aldrich; 69534). The strains were grown in 250-mL Erlenmeyer flasks containing 100 mL of YPD under aerobic conditions at 30 °C with agitation (300 rpm). Yeast cultures were kept at a concentration of 10 to 100 million cells/mL. Adherent CHODP12 cells (ATCC CRL-12445) were cultured in media containing Dulbecco's modified Eagle medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS), 1% P/S, 0.002 mg/mL recombinant human insulin (Sigma), 0.1% Trace Elements A (Fisher Scientific), 0.1% Trace Elements B (Fisher Scientific), and 200 nM methotrexate (Sigma). CHODP12 cells were also cultured in T75 flasks with vented caps that were placed into incubators at 37 °C and 5% CO2. CHODP12 cells were passaged and diluted at a 1:20 ratio using 3 mL 0.05% Trypsin-ethylenediaminetetraacetic acid (EDTA) (Thermo Fisher Scientific) after cells reached 70 to 90% confluency within the flask (approximately every 3 to 4 d).
PicoShell Fabrication.
Mechanically degradable particles demonstrated throughout the majority of the study were fabricated forming uniform water-in-oil droplet emulsions containing in-droplet concentrations of 5% (wt/wt) 10-kDa four-arm PEG-MAL (Laysan Bio), 11% (wt/wt) 10-kDa dextran (Sigma-Aldrich; D9260), and 1.54 mg/mL DTT (Sigma-Aldrich; 10197777001). Reagents were dissolved into SM-NO3 medium, YPD, TAP, or KH medium for the encapsulation of Chlorella, S. cerevisiae, C. reinhardtii, or E. gracilis, respectively (each at a pH of 6.25). Novec 7500 Engineered fluid (3M; 297730-92-9) with 0.5% Pico-Surf (Sphere Fluidics; C024) acting as surfactant was used as the continuous oil phase. Droplet emulsions were formed using a four-inlet microfluidic channel fabricated with polydimethylsiloxane (PDMS) using standard soft-lithography techniques (62). Reagents were loaded into separate syringes and pushed through the PDMS droplet generator using syringe pumps (Harvard Apparatus). In order to reduce the effects of functionalized PEG and cross-linker on cell growth during encapsulation in PicoShells, cells were suspended in the dextran phase such that the cells only interact with the PEG and DTT reagents for a short period of time (Movie S7). In-droplet concentrations of 3.25% (wt/wt) 20-kDa four-arm PEG-OPSS (Creative PEGWorks; PSB-459), 10% (wt/wt) 10-kDa dextran, and 0.80 mg/mL DTT were used to form disulfide-linked PicoShells. In-droplet concentrations of 5% (wt/wt) 10-kDa four-arm PEG-MAL, 11% (wt/wt) 10-kDa dextran, and 14.1 mg/mL dicysteine-modified MMP (Ac-GCRDGPQGIWGQDDRCG-NH2; Genscript) peptide substrate were used to form MMP-degradable PicoShells.
Following droplet generation, emulsions were stored at room temperature for 1 h to allow PicoShells to fully solidify. The PicoShells were demulsified by adding Pico-Break (Sphere Fluidics; C081) at a 1:1 volume ratio on top of the PicoShells. After Pico-Break had passed through all the PicoShells, the particles were transferred into aqueous solution (PBS or cell media) containing 10 µM N-ethylmaleimide (NEM; Sigma-Aldrich; E3876) at a pH of 6.5. The PicoShells were kept in NEM solution for 0.5 h to allow NEM to react to any free thiols on the particles to reduce clumping. PicoShells were then passed through a 100-µM cell strainer to remove any oversized or clumped particles and a 40-µM cell strainer to remove any free cells or small debris before being transferred into cell media to be used for the particular assay.
PicoShell vs. Droplet Emulsion Growth Comparison.
Chlorella and S. cerevisiae from the same respective initial culture were separately encapsulated into mechanically degradable PicoShells and microfluidically generated droplets in oil of approximately the same volume (155 pL) using the same droplet generator. PicoShells and droplets containing Chlorella were incubated in Eppendorf tubes with constant 150-µE light at room temperature (no shaking). Compartments containing S. cerevisiae were incubated in Eppendorf tubes at 30 °C (no shaking). Both PicoShells and droplets were not shaken since droplets tend to demulsify when shaken at speeds >100 rpm. PicoShells and droplets were imaged using an inverted microscope in brightfield (BF) and Cy5 (ex: 620 nm, em: 676 nm) fluorescence at equal time intervals over a multiday period to track the growth of cells in their respective compartments over time.
The same droplet generator design was used for both PicoShell fabrication and droplet generation for in-droplet growth assays (SI Appendix, Fig. S21). The devices were fabricated from PDMS (Ellsworth Adhesives) with a 1:10 curing agent to base ratio that was plasma bonded to a glass microscope slide (VWR). Holes for the inlets were punched using a 1.5-mm biopsy punch (Miltex). Reagents were loaded into 1-mL BD Luer-Lok syringes (Fisher Scientific) and were attached to the inlets of the device using flexible plastic tubing with a 0.02-in inner diameter (I.D.) and 0.06-in outer diameter (O.D.) (Tygon), 25-Ga Luer stubs (Fisher Scientific), and 0.02- × 1/32-in PEEK tubing (to connect Tygon tubing to the Luer stubs; IDEX Corporation). Syringes were loaded with the reagents required for PEG-MAL PicoShell generation in the appropriate media and at concentrations that generate in-droplet concentrations of PEG, dextran, and DTT detailed previously. Cells were placed into the dextran phase at a concentration of 2 million cells/mL. The dextran solution was connected into the middle bottom inlet (orientation is based on SI Appendix, Fig. S21). DTT and PEG solutions were connected to the left and right inlets (these inlets are interchangeable). For droplet generation in oil, three syringes were all loaded with the appropriate cell media, and the syringe connected to the middle bottom inlet was loaded with a cell concentration of 2 million cells/mL. For both PicoShell fabrication and droplet generation, Novec 7500 with 0.5% Pico-Surf was loaded into a 5-mL BD Luer-Lok syringe (Fisher Scientific) and connected to the top middle inlet the same way the 1-mL syringes were connected. Reagents were injected into the device using three separate Standard Infuse/Withdraw PHD 22/2000 Syringe Pumps from Harvard Apparatus (PEG/DTT or media with cells were loaded onto the same syringe pump, respectively). Cell-containing solutions for both PicoShell fabrication and droplet generation were injected into the device at a flow rate of 4 µL/min. Aqueous solutions without cells (PEG/DTT solutions for PicoShell fabrication) were each injected at a flow rate of 2 µL/min. Oil was injected into the device at a flow rate of 40 µL/min. PicoShells or droplets were flowed out to a 15-mL conical tube using flexible plastic tubing with a 0.02-in I.D. and 0.06-in O.D. for collection. PicoShells were phase transferred using previously discussed methods.
PicoShells for Chlorella and S. cerevisiae growth experiments were placed into 4 mL of their respective media at a concentration of 100,000 PicoShells/mL within 5-mL Eppendorf tubes (Fisher Scientific). Rather than using the standard caps to seal each tube, the caps were removed, and the top was sealed using a cut piece of sterile Kimwipes (Fisher Scientific). This enables a seal of the tube while still allowing gas exchange. The tubes were placed into an incubator with the appropriate temperature and lighting conditions for each cell type that were detailed in Bulk Culture of Cells.
PicoShells for CHODP12 growth experiments were placed into 8 mL of CHODP12 culture media (detailed previously) at a concentration of 100,000 PicoShells/mL within a T25 cell culture flask with vented cap (Fisher Scientific). The PicoShell-containing flask was placed into an incubator with the appropriate culture conditions detailed earlier (temperature, humidity, etc.).
For all cell types, cell-containing 500-µL volumes of droplets were placed into a 5-mL Eppendorf tube with 2.5 mL Novec 7500 with 0.5% Pico-Surf (1:5 ratio of droplets to oil). Droplets containing adherent CHO cells were covered with 1 mL of light mineral oil (Sigma-Aldrich) to reduce evaporation of droplets within an incubator at 37 °C. Mineral oil was not used for Chlorella sp. and S. cerevisiae droplet vs. PicoShells growth comparison studies. However, we also made a separate sample of S. cerevisiae containing droplets that were capped with mineral oil so that we could observe potential effects that covering droplets with mineral had on the growth of encapsulated cells. Multiple studies have covered cell-containing droplets to reduce evaporation (63, 64). The caps of each tube were removed, and the top was sealed using a cut piece of sterile Kimwipes (Fisher Scientific). Each tube was placed in an incubator with the appropriate culture conditions for each cell type.
A MATLAB code was used (Dataset S1) to count cells in PicoShells and droplets throughout the study. MATLAB’s FindCircles function was used to find PicoShells/droplets in the bright-field channel (referred to here as “compartment region”). A fluorescence overlay of the images was used to find the cells (Cy5 for microalgae and fluorescein isothiocyanate [FITC] for yeast) within each of these compartment regions, and the area for each cell cluster was obtained using blob analysis. The number of cells was calculated by taking the area of each blob and dividing it by the two-dimensional area of each cell within the compartment region. For later time points, the cell cluster became thick enough where a simple area calculation for each blob did not give the actual number of cells. To solve this, a secondary blob analysis was done on these high-density areas, and this added volume of cells was approximated to be a total of two times the amount of the low-density areas. This approximation was visually verified by testing the code on specific counted particles. Since CHODP12 cells do not provide a natural autofluorescence, these samples were counted manually. Several test clusters were used to verify that the number of cells computed by the code was not significantly different (P > 0.05) compared with visual counting (SI Appendix, Fig. S22).
Staining of Intracellular Lipids.
Following a 48-h culture of Chlorella in PicoShells, intracellular lipids were stained with BODIPY505/515. Stock BODIPY505/515 was prepared by dissolving 4,-difluoro-1,3,5,7-tetramethyl-4-bora-3a,4a-diaza-s-indacene (Life Technologies; D3921) powder into dimethyl sulfoxide at a concentration of 2.5 mg/mL and then diluted to 2.5 µg/mL using SM-NO3 media. Colony-containing PicoShells were placed at a concentration of 2 × 106 particles/mL in SM-NO3 media before being mixed at a volume ratio of 1:1 with 2.5 µg/mL BODIPY505/515 and incubated in the dark for 0.5 h. The PicoShells were washed three times with SM-NO3 before being imaged in the FITC channel (ex: 488 nm/em: 543 nm) using a fluorescence microscope.
Incubation and Flow Cytometric Sorting of PicoShells.
Chlorella were encapsulated into 90-µm-diameter PicoShells following Poisson loading with lambda = 0.1 for the initial sort and lambda = 0.05 for the full selection and placed into SM-NO3 medium at a particle to media volume ratio of 1:50. The particle-containing solution was then placed in a 250-mL Erlenmeyer flask shaking at 120 rpm and at room temperature under constant 150-µE light for 48 h to allow cells to divide.
Colony-containing PicoShells were screened and sorted using an On-Chip Sort (On-Chip Biotechnologies). The cytometer was equipped with both 488- and 561-nm excitation lasers and a PE-Cy5 (676/37-nm) filter. Events were triggered based on particle absorbance from the 488-nm laser. PicoShells were sorted based on their scatter readouts and thresholding desired intensity heights through the PE-Cy5 filter. PicoShell solutions were concentrated in fresh SM-NO3 media at a 1:10 particle to media volume ratio for screening and sorting. PicoShells within the selection gates were dispensed in a single collection reservoir. The sorted particles were then imaged using an inverted microscope, and the number of cells in each particle was counted using MATLAB code.
Release of Cells and Reculture of Selected Populations.
Postselection, Chlorella-containing PicoShells were placed onto a 37-µm cell strainer and placed over a 15-mL conical tube containing fresh SM-NO3 media supplemented with P/S. The PicoShells were then ruptured by “grinding” the particles with a pestle and washing with SM-NO3 media for ∼5 min, causing released cells to fall through the pores of the cell strainer and into the fresh media. Despite being able to be ruptured by direct mechanical shearing pressure, PicoShells remain stable in adverse indirect mechanical shearing pressures, such as mixing, vigorous pipetting, and vortexing. The solution containing released cells was then transferred into a 250-mL Erlenmeyer flask and put in standard bulk Chlorella culture conditions for 7 d to allow released cells to regrow to a concentration of 15 to 20 million cells/mL.
To test for maintenance of enhanced growth phenotypes in the selected population, we seeded the selected population and an unselected population into separate 250-mL Erlenmeyer flasks with SM-NO3 media supplemented with P/S at a concentration of 500,000 cells/mL The flasks were placed side by side under standard Chlorella culturing conditions for 4 d. The cell concentration was measured using a hemocytometer every 12 h. At 48 h of growth, we also measured growth by aliquoting several fractions from the selected and unselected samples into a well plate and measured the chlorophyll density (ex: 620 nm; em: 676 nm) using a well-plate reader at this time point.
Chemically Induced Degradation of PicoShells.
To chemically degrade the various types of PicoShells, we first diluted or concentrated PicoShells to a concentration of 1 × 106 particles/mL and added the following reagents at the indicated final concentration to degrade each PicoShell type: 10 µg/mL NaIO4 (Fisher Scientific; P120504) for particles cross-linked with four-arm PEG-MAL; DTT, 10 mg/mL DTT, or 3 mg/mL TCEP (Sigma-Aldrich; 646547) for particles cross-linked with four-arm PEG-OPSS and DTT; and 0.0025% Trypsin with EDTA (Thermo Fisher Scientific; 25300120) for particles cross-linked with four-arm PEG-MAL and dicysteine-modified MMP-degradable peptide.
Environmental Effects Studies.
To test the effects of the presence of leucine in the external solution on encapsulated cell behavior, we first encapsulated cdc3-mcherry::leu2 S. cerevisiae strains with an average loading lambda of 0.1 into PEG-MAL PicoShells using methods previously detailed. Half the sample was then placed into yeast media without any leucine at a concentration of 50,000 PicoShells/mL, and the other half of the sample was placed into yeast media supplemented with 76 mg/L leucine. The samples were allowed to incubate in their respective media within 5-mL Eppendorf tubes at 30 °C for 12 h. After the incubation period, each sample was imaged, and the number of cells within each PicoShell was counted visually.
To test the effects of the presence of cells in the external solution on encapsulated cell behavior, we first encapsulated cdc3-mcherry::leu2 S. cerevisiae strains with an average loading lambda of 0.1 into PEG-MAL PicoShells using methods previously detailed. Half the sample was then placed into yeast media without any cells at a concentration of 50,000 PicoShells/mL, and the other half of the sample was placed into yeast media containing cdc3-mcherry::leu2 S. cerevisiae at a concentration of 100 million cells/mL in stationary phase. The cell-containing media were prepared by pelleting a culture of cells and resuspending into fresh media within 5 min of mixing with PicoShells to make sure that the nutrients were fully replenished. The samples were allowed to incubate in their respective media within 5-mL Eppendorf tubes at 30 °C for 12 h. After the incubation period, PicoShells incubated in the media with external cells were isolated from this external population by running the sample through a CellTrics 20-µm cell strainer (Fisher Scientific). After PicoShells were isolated, the samples were imaged, and the number of cells within each PicoShell was counted visually.
To test the possibility of PicoShells being incubated in a bioreactor, we made a custom chamber with an autoclaved 100-mL glass beaker sealed with a rubber stopper with two holes (United States Plastic Corp.). Two 6-in-long 3-mm I.D. × 5-mm O.D. × 1-mm wall Excelon laboratory metric tubing (United States Plastic Corp) pieces were inserted into the two holes to allow gas to be exchanged with the external environment. The ends of the tubes outside of the beaker were sealed with sterile Kimwipes so that the environment could remain sterile while still allowing for gas exchange. The beaker was placed onto a hot plate set at 30 °C and contained a magnetic stir bar that rotated at 50 rpm. The beaker also contained 80 mL of yeast broth containing cdc3-mcherry::leu2 S. cerevisiae at a concentration of 2 million cells/mL at the start of the PicoShell incubation. cdc3-mcherry::leu2 S. cerevisiae yeast was encapsulated into PEG-MAL PicoShells with an average loading lambda of 0.1 using previously detailed methods and placed into the homemade bioreactor at a concentration of 50,000 PicoShells/mL for 12 h. PicoShells were isolated from the unencapsulated cells by running the entire solution through a pluriSelect 20-µm cell strainer three times. The isolated PicoShells were imaged, and the cells in each PicoShell were visually counted.
Supplementary Material
Acknowledgments
This work was supported by Presidential Early Career Award for Scientists and Engineers N00014-16-1-2997 and the California NanoSystems Institute at the University of California, Los Angeles Planning Award. We also thank the Yoshikazu Ohya laboratory for providing us with S. cerevisiae cell strains for the study.
Footnotes
Competing interest statement: M.v.Z., J.d.R., C.W., S.L., and D.D.C. are named inventors on a patent application by the University of California, Los Angeles.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109430119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
Change History
July 22, 2022: The editor’s affiliation has been updated to correct an error. Jens Nielsen’s affiliation is: BioInnovation Institute, DK2200 Copenhagen, Denmark.
References
- 1.Hood E. E., Plant-based biofuels. F1000Res. 5, F1000 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chisti Y., Biodiesel from microalgae. Biotechnol. Adv. 25, 294–306 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Sheehan J., Dunahay T., Benemann J., Roessler P., “A Look Back at the U.S. Department of Energy’s Aquatic Species Program: Biodiesel from Algae” (Close-Out Rep. NREL/TP-580-24190, National Renewable Energy Laboratory, Golden, CO, 1998).
- 4.Dismukes G. C., Carrieri D., Bennette N., Ananyev G. M., Posewitz M. C., Aquatic phototrophs: Efficient alternatives to land-based crops for biofuels. Curr. Opin. Biotechnol. 19, 235–240 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Radakovits R., Jinkerson R. E., Darzins A., Posewitz M. C., Genetic engineering of algae for enhanced biofuel production. Eukaryot. Cell 9, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan M. I., Shin J. H., Kim J. D., The promising future of microalgae: Current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb. Cell Fact. 17, 36 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen M., Mertiri T., Holland T., Basu A. S., Optical microplates for high-throughput screening of photosynthesis in lipid-producing algae. Lab Chip 12, 3870–3874 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Lindström S., Andersson-Svahn H., Single-cell culture in microwells. Methods Mol. Biol. 853, 41–52 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Chen B., et al. , High-throughput analysis and protein engineering using microcapillary arrays. Nat. Chem. Biol. 12, 76–81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu H., et al. , High throughput single cell counting in droplet-based microfluidics. Sci. Rep. 7, 1366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H. S., et al. , High-throughput droplet microfluidics screening platform for selecting fast-growing and high lipid-producing microalgae from a mutant library. Plant Direct 1, e00011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M., et al. , A gelatin microdroplet platform for high-throughput sorting of hyperproducing single-cell-derived microalgal clones. Small 14, e1803315 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Eun Y. J., Utada A. S., Copeland M. F., Takeuchi S., Weibel D. B., Encapsulating bacteria in agarose microparticles using microfluidics for high-throughput cell analysis and isolation. ACS Chem. Biol. 6, 260–266 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakagawa Y., et al. , Are droplets really suitable for single-cell analysis? A case study on yeast in droplets. Lab Chip 21, 3793–3803 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Mocciaro A., et al. , Light-activated cell identification and sorting (LACIS) for selection of edited clones on a nanofluidic device. Commun. Biol. 1, 41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muñoz H. E., et al. , Fractal LAMP: Label-free analysis of fractal precipitate for digital loop-mediated isothermal nucleic acid amplification. ACS Sens. 5, 385–394 (2020). [DOI] [PubMed] [Google Scholar]
- 17.de Rutte J., Koh J., Di Carlo D., Scalable high-throughput production of modular microgels for in situ assembly of microporous tissue scaffolds. Adv. Funct. Mater. 29, 1900071 (2019). [Google Scholar]
- 18.Kahkeshani S., Di Carlo D., Drop formation using ferrofluids driven magnetically in a step emulsification device. Lab Chip 16, 2474–2480 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Hatti-Kaul R., Aqueous two-phase systems. A general overview. Mol. Biotechnol. 19, 269–277 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Asenjo J. A., Andrews B. A., Aqueous two-phase systems for protein separation: Phase separation and applications. J. Chroma. 123, 1–10 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Griffin D. R., Weaver W. M., Scumpia P. O., Di Carlo D., Segura T., Accelerated wound healing by injectable microporous gel scaffolds assembled from annealed building blocks. Nat. Mater. 14, 737–744 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vijawakumar K., Gulati S., deMello A. J., Edel J. B., Rapid cell extraction in aqueous two-phase microdroplet systems. Chem. Sci. 1, 447–452 (2010). [Google Scholar]
- 23.Yasukawa M., Kamio E., Ono T., Monodisperse water-in-water-in-oil emulsion droplets, Yasukawa. ChemPhysChem 12, 263–266 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Ma S., et al. , Fabrication of microgel particles with complex shape via selective polymerization of aqueous two-phase systems. Small 8, 2356–2360 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Liu Q., et al. , Self-orienting hydrogel micro-buckets as novel cell carriers. Ange. Chem. Inter. Edit. 58, 547–551 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe T., Motohiro I., Ono T., Microfluidic formation of hydrogel microcapsules with a single aqueous core by spontaneous cross-linking in aqueous two-phase system droplets. Langmuir 35, 2358–2367 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Leonaviciene G., Leonavicius K., Meskys R., Mazutis L., Multi-step processing of single cells using semi-permeable capsules. Lab Chip 20, 4052–4062 (2020). [DOI] [PubMed] [Google Scholar]
- 28.de Rutte J., et al. , Massively parallel encapsulation of single cells with structured microparticles and section-based flow sorting. bioRxiv [Preprint] (2020). https://www.biorxiv.org/content/10.1101/2020.03.09.984245v2 (Accessed 13 November 2021).
- 29.McMillan T., et al. , Cellular effects of long wavelength UV light (UVA) in mammalian cells. J. Pharm. Pharmacol. 60, 969–976 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Phelps E. A., et al. , Maleimide cross-linked bioactive PEG hydrogel exhibits improved reaction kinetics and cross-linking for cell encapsulation and in situ delivery. Adv. Mater. 24, 64–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sprague G. F. Jr., Winans S. C., Eukaryotes learn how to count: Quorum sensing by yeast. Genes Dev. 20, 1045–1049 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Periyannan Rajeswari P. K., Joensson H. N., Andersson-Svahn H., Droplet size influences division of mammalian cell factories in droplet microfluidic cultivation. Electrophoresis 38, 305–310 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Dean J., Reddy P., Metabolic analysis of antibody producing CHO cells in fed-batch production. Biotechnol. Bioeng. 110, 1735–1747 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Casey G. P., Magnus C. A., Ingledew W. M., High-gravity brewing: Effects of nutrition on yeast composition, fermentative ability, and alcohol production. Appl. Environ. Microbiol. 48, 639–646 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carruthers D. N., Byun C. K., Cardinale B. J., Lin X. N., Demonstration of transgressive overyielding of algal mixed cultures in microdroplets. Integr. Biol. 9, 687–694 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Best R. J., et al. , Label-free analysis and sorting of microalgae and cyanobacteria in microdroplets by intrinsic chlorophyll fluorescence for the identification of fast growing strains. Anal. Chem. 88, 10445–10451 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Barahoei M., Hatamipour S., Afsharzadeh S., CO2 capturing by Chlorella vulgaris in a bubble column photo-bioreactor: Effect of bubble size on CO2 removal and growth rate. J. CO2 Utiliz. 37, 9–19 (2020). [Google Scholar]
- 38.Gruner P., et al. , Stabilisers for water-in-fluorinated oil dispersions: Key properties for microfluidic applications. Curr. Opin. Colloid Inter. Sci. 20, 183–191 (2015). [Google Scholar]
- 39.Hamza M., Serratrice G., Stebe M., Delpuech J., Solute-solvent interactions in perfluorocarbon solutions of oxygen. An NMR study. J. Am. Chem. Soc. 103, 3733–3738 (1981). [Google Scholar]
- 40.Lent E. M., Crouse L. C. B., Eck W. S., Acute and subacute oral toxicity of periodate salts in rats. Regul. Toxicol. Pharmacol. 83, 23–37 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Luxmi R., et al. , Proteases shape the Chlamydomonas secretome: Comparison to classical neuropeptide processing machinery. Proteomes 6, 36 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang C., Lan C. Q., Effects of shear stress on microalgae—a review. Biotechnol. Adv. 36, 986–1002 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Stafford R.A., “Yeast physical (shear) stress: The engineering perspective” in Brewing Yeast Fermentation Performance, Smart K., Ed. (Blackwell Science, Oxford, UK, ed. 2, 2003), 39–45. [Google Scholar]
- 44.Keane J. T., Ryan D., Gray P. P., Effect of shear stress on expression of a recombinant protein by Chinese hamster ovary cells. Biotechnol. Bioeng. 81, 211–220 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Morimoto Y., Tan W. H., Takeuchi S., Three-dimensional axisymmetric flow-focusing device using stereolithography. Biomed. Microdevices 11, 369–377 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Destgeer G., Ouyang M., Wu C. Y., Di Carlo D., Fabrication of 3D concentric amphiphilic microparticles to form uniform nanoliter reaction volumes for amplified affinity assays. Lab Chip 20, 3503–3514 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Wu C. Y., et al. , Monodisperse drops templated by 3D-structured microparticles. Sci. Adv. 6, eabb9023 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee S., de Rutte J., Dimatteo R., Koo D., Di Carlo D., Scalable fabrication and use of 3D structured microparticles spatially functionalized with biomolecules. ACS Nano, 10.1021/acsnano.1c05857 (2021). [DOI] [PMC free article] [PubMed]
- 49.Di Carlo D., Technologies for the directed evolution of cell therapies. SLAS Technol. 24, 359–372 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Zhou Y. J., et al. , Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat. Commun. 7, 11709 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petrovič U., Next-generation biofuels: A new challenge for yeast. Yeast 32, 583–593 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Lu W., et al. , Biosynthesis of monomers for plastics from renewable oils. J. Am. Chem. Soc. 132, 15451–15455 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Shen W., He Y., Zhang S., Li J., Fan W., Yeast-based microporous carbon materials for carbon dioxide capture. Chem. Sus. Shem. 5, 1274–1279 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Lodolo E. J., Kock J. L., Axcell B. C., Brooks M., The yeast Saccharomyces cerevisiae: The main character in beer brewing. FEMS Yeast Res. 8, 1018–1036 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Gorman D. S., Levine R. P., Cytochrome f and plastocyanin: Their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. U.S.A. 54, 1665–1669 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo Y., et al. , Effect of the molecular weight of polymer and diluent on the performance of hydrophilic poly(vinyl butyral) porous heddle via thermally induced phase separation. Mater. Chem. Phys. 261, 124227 (2021). [Google Scholar]
- 57.Pregibon D. C., Doyle P. S., Optimization of encoded hydrogel particles for nucleic acid quantification. Anal. Chem. 81, 4873–4881 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Witte R. P., Blake A. J., Palmer C., Kao W. J., Analysis of poly(ethylene glycol)-diacrylate macromer polymerization within a multicomponent semi-interpenetrating polymer network system. J. Biomed. Mater. Res. A 71, 508–518 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Broguiere N., et al. , Macroporous hydrogels derived from aqueous dynamic phase separation. Biomaterials 200, 56–65 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Verruto J., et al. , Unrestrained markerless trait stacking in Nannochloropsis gaditana through combined gene editing and marker recycling technologies. Proc. Natl. Acad. Sci. U.S.A. 115, 7015–7022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koren L. E., High-yield media for photosynthesizing Euglena gracilis. J. Eukaryot. Microbiol. 14, 17 (1967). [Google Scholar]
- 62.Qin D., Xia Y., Whitesides G. M., Soft lithography for micro- and nanoscale patterning. Nat. Protoc. 5, 491–502 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Isozaki A., et al. , Sequentially addressable dielectrophoretic array for high-throughput sorting of large-volume biological compartments. Sci. Adv. 6, eaba6712 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Langer K., Joensson H N.., Rapid production and recovery of cell spheroids by automated droplet microfluidics. SLAS Technol. 25, 111–122 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.