Significance
Loss-of-function mutations in MECP2 cause the neurological disorder Rett syndrome (RTT), but the precise molecular mechanism driving pathogenesis remains unclear. Using an unbiased approach to identify proteins that interact with MeCP2, we identified the transcription factor 20 (TCF20) complex and discovered that RTT-causing mutations in MECP2 disrupt this interaction. Using biochemical, morphological, behavioral, and transcriptional studies, we examined the importance of this interaction for brain function and found that the TCF20 complex plays a direct role in MeCP2-dependent gene regulation and modifies MECP2-induced synaptic and behavioral deficits. Our data uncovered a previously unknown molecular aspect of MeCP2 function and revealed a converging molecular mechanism, whereby mutations of genes encoding several subunits in the same complex contribute to shared neurological symptoms.
Keywords: neurodevelopmental disorders, Rett syndrome, MeCP2, BioID, TCF20 complex
Abstract
MeCP2 is associated with Rett syndrome (RTT), MECP2 duplication syndrome, and a number of conditions with isolated features of these diseases, including autism, intellectual disability, and motor dysfunction. MeCP2 is known to broadly bind methylated DNA, but the precise molecular mechanism driving disease pathogenesis remains to be determined. Using proximity-dependent biotinylation (BioID), we identified a transcription factor 20 (TCF20) complex that interacts with MeCP2 at the chromatin interface. Importantly, RTT-causing mutations in MECP2 disrupt this interaction. TCF20 and MeCP2 are highly coexpressed in neurons and coregulate the expression of key neuronal genes. Reducing Tcf20 partially rescued the behavioral deficits caused by MECP2 overexpression, demonstrating a functional relationship between MeCP2 and TCF20 in MECP2 duplication syndrome pathogenesis. We identified a patient exhibiting RTT-like neurological features with a missense mutation in the PHF14 subunit of the TCF20 complex that abolishes the MeCP2–PHF14–TCF20 interaction. Our data demonstrate the critical role of the MeCP2–TCF20 complex for brain function.
Approximately 3 to 5% of children in the United States suffer from some form of neurodevelopmental disorder (NDD). Many of these disorders have overlapping features, with autistic behaviors, intellectual disability, motor incoordination, and seizures being among the most common (1). Large-scale sequencing studies have found that a given feature, such as autistic behavior, can be associated with mutations in any one of hundreds of different genes (2, 3). Bioinformatic approaches have revealed that diverse NDD-related proteins converge on shared molecular pathways through protein–protein interaction (PPI) networks (4), but this computational approach, typically based on high-throughput PPI and human genetic data, is often insufficient to uncover the specific molecular mechanism. Even when the genetic basis of a disease is known, the molecular pathogenesis can remain stubbornly elusive.
Such is the case with disorders associated with the X-linked methyl-CpG–binding protein 2 (MECP2). Rett syndrome (RTT) and MECP2 duplication syndrome (MDS), caused by loss-of-function (LoF) mutations and duplications of MECP2, respectively (5, 6), are complex conditions that affect cognition, motor function, mood, and multiple organ systems. The genotype–phenotype relationships in this subset of NDDs are surprisingly broad, as very mild MECP2 mutations, often in conjunction with favorably skewed X inactivation, can be associated with only a mild learning disability or mild autism in girls, and in males can cause anything from rapidly fatal neonatal encephalopathy to a syndrome that includes psychosis (7–9). Complicating the picture further is the fact that roughly 5% of clinically diagnosed RTT cases have no identifiable mutation in MECP2. Some of these individuals turn out to have mutations in other genes, such as CDKL5 or FOXG1, which are now considered distinct diseases thanks to more detailed phenotyping (7). Interestingly, a de novo deletion in transcription factor 20 (TCF20) was reported in a female with a clinical presentation that closely resembles RTT (10); LoF mutations in TCF20 have been identified to cause TCF20-associated NDD (TAND), which is associated with ataxia, hypotonia, autistic behaviors, and sleep disturbances (11–13). The overlapping clinical symptoms of these monogenic NDDs suggest there might be shared pathophysiological mechanisms that could provide greater insight into these diseases.
Such clues could lead to better understanding of RTT and MDS, because the precise molecular mechanism by which MeCP2 drives disease remains unclear. Transcriptomic analyses in mice lacking Mecp2 reveal changes in the expression levels of thousands of genes, suggesting that MeCP2 dysfunction broadly influences gene expression (14–16). Given that MeCP2 does not possess a transactivation or enzymatic domain, it is believed that it functions as a bridge between methylated DNA, which it binds via its methyl-CpG binding domain (MBD), and chromatin modifiers, which it recruits mainly via its transcriptional repression domain (TRD) (17). MeCP2 represses transcription by recruiting the histone deacetylase 3 (HDAC3) subunit of the NCoR1/2 complex (18, 19). RTT-causing mutations in the TRD of MeCP2 disrupt MeCP2 binding to the NCoR1/2 complex, suggesting this interaction plays a critical role in RTT pathogenesis (18). Yet this is clearly not the whole story, because in both humans and mice, several mutations within the TRD that completely abolish the interaction between MeCP2 and the NCoR1/2 complex cause milder forms of disease compared to mutations within the MBD (20–24). Furthermore, the inactivation of HDAC3 in the NCoR1/2 complex does not mitigate the deficits caused by MeCP2 overexpression (25). There must, then, be other MeCP2 interactors that mediate its molecular functions and pathogenesis.
To search for functional MeCP2 interactors in situ at the chromatin interface, we used proximity-dependent biotin identification followed by mass spectrometry (BioID-MS) (26) and identified a chromatin complex that physically interacts with MeCP2 in neurons. We examined these interactions in cells and in vivo to reveal an essential role for this complex in brain function and identify human patients with previously uncharacterized mutations in the interactors who share features of RTT.
Results
MeCP2 Interacts with the TCF20 Complex in the Mouse Brain.
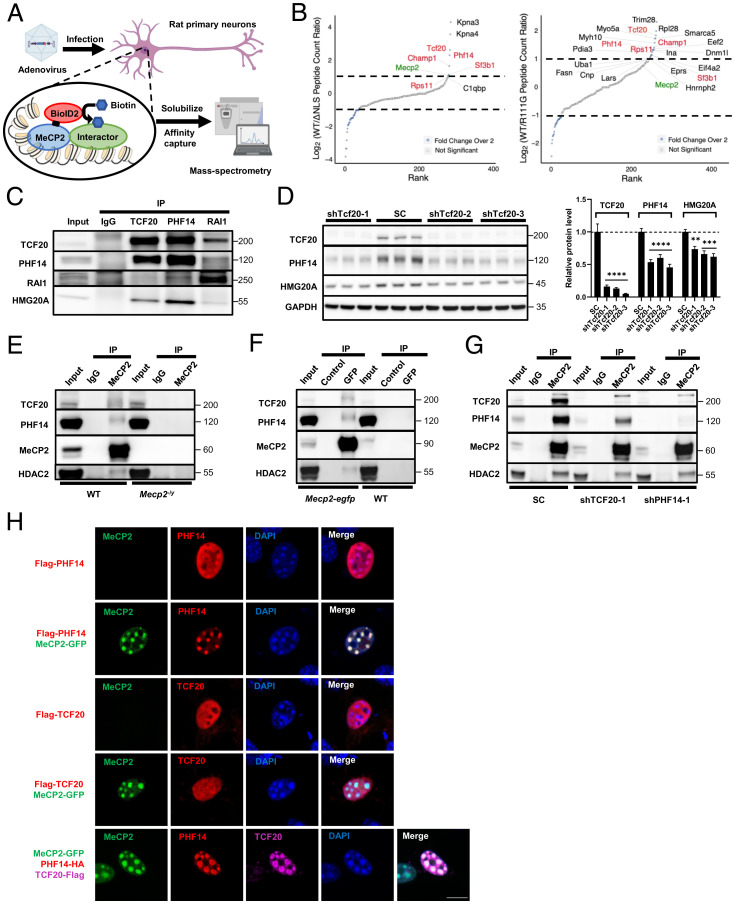
To identify novel MeCP2 interactors at the chromatin interface, we performed BioID-MS in cultured mature rat primary neurons with a biotin ligase called BioID2 tagged to MeCP2 at day in vitro (DIV)19 (Fig. 1A) (26). To home in on interactors specifically at the chromatin, we used two mutant alleles, MECP2R111G and MECP2ΔNLS, which disrupt DNA binding and nuclear localization of MeCP2, respectively, as negative controls (23, 27). Consistent with our previous work showing that Karyopherin-α3 (KPNA3) and KPNA4 bind to the nuclear localization sequence (NLS) of MeCP2 for nuclear import (28), we found a strong enrichment of KPNA3/4 with WT MeCP2 but not MeCP2ΔNLS (Fig. 1B). The other known interactors, TBL1X and TBLXR1, were enriched in twofold over one of the negative controls. We focused on interactors that were enriched more than twofold over both negative controls and identified five MeCP2 candidate interactors (Fig. 1B and SI Appendix, Fig. S1A). Of these, TCF20 and PHF14 function as subunits of a putative chromatin-binding complex that includes two other subunits, RAI1 and HMG20A (SI Appendix, Fig. S1B) (29). To verify the presence of this complex in the mouse brain, we performed coimmunoprecipitation (co-IP) of mouse cortical extracts using anti-TCF20, anti-PHF14, and anti-RAI1 antibodies. All three antibodies specifically coimmunoprecipitated TCF20, PHF14, RAI1, and HMG20A from cortical extracts, indicating that these proteins form a stable protein complex in vivo (Fig. 1C). Interestingly, knockdown of TCF20 in human 293T cells using short-hairpin RNA (shRNA) significantly reduced PHF14 and HMG20A protein levels without altering their mRNA levels (Fig. 1D and SI Appendix, Fig. S1C), whereas knockdown of PHF14 did not affect TCF20 or HMG20A protein levels (SI Appendix, Fig. S1D). These data suggest that PHF14 and HMG20A could be more stable by forming a complex with TCF20. For convenience, we refer to this protein complex as the TCF20 complex.
Fig. 1.
MeCP2 interacts with the TCF20 complex in vitro and in vivo. (A) The experimental strategy using BioID2-dependent proximity biotinylation to identify MeCP2 interactors. (B) Rank plots showing the enrichment of MeCP2-BioID2 peptide spectral matches (PSMs) over MeCP2ΔNLS-BioID2 (Left) and MeCP2R111G-BioID2 (Right) PSMs. BioID2-tagged protein (MeCP2) is indicated in green, proteins shown enrichment in both experiments are indicated in red. Only proteins with more than 10 peptide counts collected in the WT sample were plotted. Proteins with fewer than 10 peptide counts, such as RAI1, ATRX, and two components of the NCoR1/2 complex, TBL1XR1 and TBL1X, were not shown in the plots, although they were enriched for twofold or more compared to at least one negative controls (full lists of the peptides collected in the BioID MS is given in Dataset S1). (C) Representative immunoblot of TCF20, PHF14, RAI1, and HMG20A protein levels following IP of TCF20, PHF14 or RAI1 from mouse cortical lysates. (D) Representative immunoblot (Left) and quantification (Right) of TCF20, PHF14, and HMG20A protein levels following Tcf20 knockdown or scramble control (SC) in HEK293T cells (n = 3 per group; two-way ANOVA with post hoc Tukey’s tests). **P < 0.01, ***P < 0.001, ****P < 0.0001; data are mean ± SEM. (E and F) Representative immunoblots of TCF20, PHF14, and HDAC2 protein levels following IP of MeCP2 from WT and Mecp2-null cortical lysates (E) or GFP from Mecp2-egfp and WT cortical lysates (F). GFP-Trap Dynabeads (GFP) and control Dynabeads (Control) were used to IP GFP-tagged MeCP2 in F. (G) Representative immunoblot of TCF20, PHF14, and HDAC2 protein levels following IP of MeCP2 from HEK293T cells transduced with a nontargeting scramble (SC) lentivirus or lentiviral shRNA targeting TCF20 or PHF14. (H) Representative immunocytochemical images in mouse 3T3 fibroblasts showing the colocalization of Flag-tagged PHF14, Flag-tagged TCF20, or both HA-tagged PHF14 and Flag-tagged TCF20 relative to densely methylated heterochromatic foci (stained by DAPI) upon overexpression of MeCP2-GFP. (Scale bar, 10 µm.)
To determine whether these proteins interact with MeCP2 in vivo, we immunoprecipitated MeCP2 from mouse cortical lysate and probed for TCF20 and PHF14. Both proteins coimmunoprecipitated with MeCP2 in WT mice, but were not pulled down in Mecp2-null (Mecp2–/y) brain (Fig. 1E). This result was validated in the brains of Mecp2-enhanced green fluorescent protein (EGFP) mice by co-IP using an anti–green fluorescent protein (GFP) nanobody (Fig. 1F). As positive controls, we probed for two known MeCP2 interactors, HDAC2 and TBL1 (subunits of the SIN3A and NCoR1/2 complexes, respectively) (Fig. 1 E and F and SI Appendix, Fig. S1E) (18, 30). To determine which proteins mediate the interaction with MeCP2, we knocked down either TCF20 or PHF14 in HEK293T cells, and immunoprecipitated MeCP2 to probe for either TCF20 or PHF14. Knockdown of TCF20 slightly reduced the amount of PHF14 protein bound to MeCP2 and total PHF14 protein levels (Fig. 1G), possibly due to the destabilization of PHF14 in the absence of TCF20 (Fig. 1D). However, knockdown of PHF14 almost abolished the interaction of TCF20 with MeCP2 without altering TCF20 levels (Fig. 1G). These data indicate that MeCP2 binds TCF20 though PHF14 to form a stable complex in vivo.
MeCP2 overexpression in mouse embryonic fibroblast cells (NIH 3T3 cells) leads to the redistribution of its interactor TBL1 to mCpG-rich heterochromatic foci (18). To test whether MeCP2 similarly recruits TCF20 and PHF14 to heterochromatic foci, we overexpressed MeCP2-EGFP with PHF14-FLAG or TCF20-FLAG in NIH 3T3 cells. When MeCP2-EGFP and PHF14-FLAG were coexpressed, PHF14 relocated predominantly to heterochromatic foci (stained by DAPI) and colocalized with exogenous MeCP2. However, TCF20-FLAG failed to localize to heterochromatic foci when coexpressed with MECP2-EGFP (Fig. 1H). Strikingly, when all three proteins were coexpressed, both PHF14 and TCF20 relocated to nuclear foci (Fig. 1H). We conclude that MeCP2 recruits TCF20 to methylated heterochromatic foci by interacting with PHF14.
RTT-Causing Mutations Reduce MeCP2 Interaction with TCF20 and PHF14.
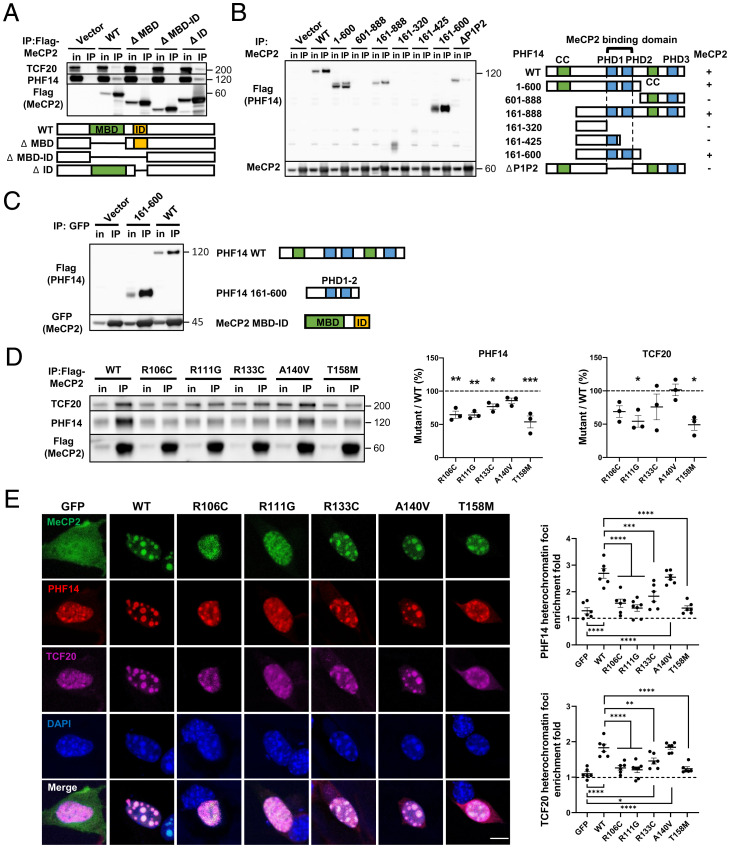
MeCP2 contains three major domains: the MBD, the TRD, and an intervening domain (ID) with unclear function (SI Appendix, Fig. S2A) (30). To determine which MeCP2 domain is required for TCF20 and PHF14 binding, we exogenously expressed a series of FLAG-tagged MeCP2 deletion fragments in HEK293T cells. By immunoprecipitating the FLAG-tagged MeCP2, we found that a region spanning the MBD and the ID in MeCP2 is necessary for binding TCF20 and PHF14 (SI Appendix, Fig. S2A). We then generated MeCP2 fragments lacking both the MBD and ID or lacking each domain separately (Fig. 2A) and tested their interaction with TCF20 and PHF14. Deleting the MBD and ID together abolished the interaction of MeCP2 with TCF20 and PHF14. Removing only the MBD partially impaired the interaction with TCF20 and PHF14, while removing the ID alone had no effect (Fig. 2A). These data suggest that the MBD–ID of MeCP2 serves as a critical interaction site for TCF20 and PHF14.
Fig. 2.
RTT-causing mutations in the MeCP2 MBD disrupt interactions with the TCF20 complex. (A) Representative immunoblot of TCF20 and PHF14 protein levels following IP of Flag-tagged WT and truncated MeCP2 in HEK293T cells. (B) Representative immunoblot (Left) and summary of the co-IP results (Right) of Flag-tagged WT and truncated PHF14 protein variants following IP of MeCP2 in HEK293T cells. The “+/−” denotes the presence (+) or absence (−) of MeCP2 interaction. (C) Representative immunoblot of Flag-tagged WT and truncated PHF14 (PHD1-2) proteins following IP of GFP-tagged truncated MeCP2 (MBD–ID) in HEK293T cells. (D) Representative immunoblot (Left) and quantification (Center and Right) of TCF20 and PHF14 protein levels following IP of Flag-MeCP2 variants in HEK293T cells. The abundance of the co-IP TCF20 and PHF14 was normalized to that of IP MeCP2 to quantify the interaction between TCF20/PHF14 and WT or mutant MeCP2. The ratio of normalized TCF20/PHF14 of each mutant to that of WT was calculated as the percentage of mutant/WT (n = 3 per group, one-way ANOVA with post hoc Tukey’s test). (E) Representative immunocytochemical images (Left) and quantification (Right) of HA-tagged PHF14 and Flag-tagged TCF20 enrichment to densely methylated heterochromatic foci (stained by DAPI) upon overexpression of WT and mutant MeCP2-GFP in mouse 3T3 fibroblasts. The heterochromatin foci enrichment fold was calculated as the ratio of mean gray value within foci area to mean gray value outside of foci area (n = 6 per group, one-way ANOVA with post hoc Tukey’s test). (Scale bar, 10 µm.) *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; data are mean ± SEM.
We next sought to identify the interacting domains of PHF14 and TCF20. Given that PHF14 mediates the interaction between MeCP2 and TCF20, we expressed a series of FLAG-tagged PHF14 deletion fragments in 293T cells to map the region of PHF14 that interacts with MeCP2. We found that the tandem plant homeodomains (PHDs) PHD1-2 in PHF14 were required to bind MeCP2 (Fig. 2B). Using co-IP, we also determined that the same tandem PHD1-2 on PHF14 and a C-terminal extended PHD (ePHD) on TCF20 were essential for the interaction between PHF14 and TCF20 (SI Appendix, Fig. S2 B and C). Finally, we found the MBD–ID on MeCP2 and the PHD1-2 on PHF14 are sufficient for MeCP2–PHF14 interaction (Fig. 2C). We conclude that MeCP2 interacts with the TCF20 complex through its MBD–ID region, PHF14 interacts with MeCP2 and TCF20 through its tandem PHD1-2 domains, and TCF20 interacts with PHF14 through its C-terminal ePHD.
Because the MBD–ID of MeCP2 is essential for its interaction with TCF20 and PHF14, we asked whether any of five RTT-causing mutations in this region (R106C, R111G, R133C, A140V, or T158M) might disrupt its binding to TCF20 and PHF14 (31). The abundance of the co-IP TCF20 and PHF14 was normalized to that of IP MeCP2 to quantify the interaction between TCF20/PHF14 and WT or mutant MeCP2. The R106C, R111G, R133C, and T158M mutations impaired the interaction with PHF14 or both PHF14 and TCF20 (Fig. 2D). In contrast, the A140V mutation did not impair MeCP2 interactions with either protein. Although the mutations that disrupt MeCP2 binding to TCF20 and PHF14 also interfere with DNA binding (7, 32, 33), it is unlikely that the protein interactions were mediated by DNA, because the co-IP experiments were performed in the presence of nuclease to digest all DNA and RNA (Materials and Methods). Finally, we were curious as to whether the most common RTT mutation within the TRD, R306C, would impair binding. We found that it abolished MeCP2 binding to the NCoR1/2 complex subunit HDAC3, as shown previously (18, 23), but did not affect binding to TCF20 and PHF14 (SI Appendix, Fig. S2D).
Results in NIH 3T3 cells were consistent with the co-IP data: PHF14 and TCF20 showed significantly less enrichment to methyl-CpG–rich heterochromatic foci when coexpressed with the MeCP2 R106C, R111G, R133C, or T158M mutants than with WT MeCP2 (Fig. 2E). These data suggest that mutations in the MBD of MeCP2 impair its interaction with the TCF20 complex, which in turn contributes to RTT pathogenesis.
Tcf20 Is Coexpressed with Mecp2 in Mouse Neurons and Regulates MECP2-Mediated Synapse Formation.
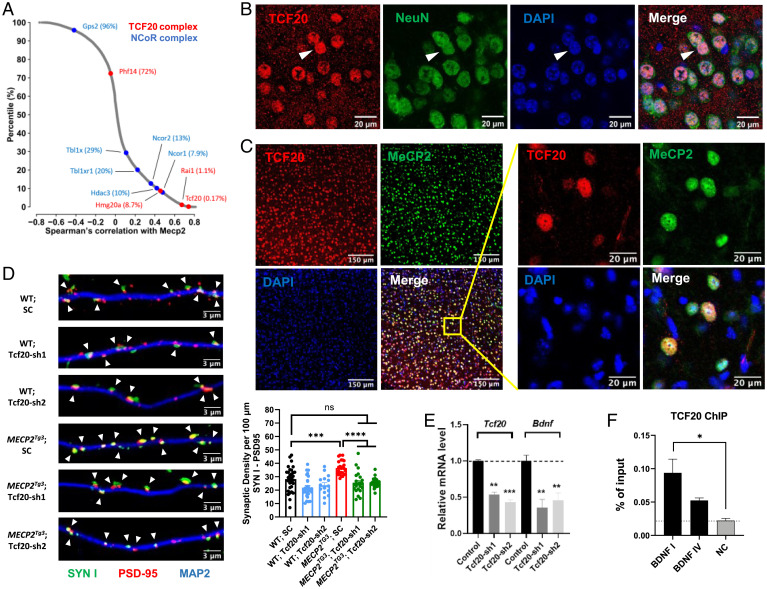
Having established that MeCP2 and the TCF20 complex physically interact, we next asked whether there is a functional association between these proteins. We reasoned that MeCP2 and the TCF20 complex should be expressed in the same cells in vivo to function together as interacting partners. Using single-cell RNA-sequencing (scRNA-seq) data of 565 cell types generated from the adult mouse brain (34), we calculated the correlation coefficient with Mecp2 for all the genes expressed in at least one cell type. Through this analysis we found that genes encoding the components of the TCF20 complex, including Tcf20, Rai1, and Hmg20a, are among the genes most highly coexpressed with Mecp2 (Fig. 3A). For comparison, the same analyses were performed for genes within the well-established MeCP2-interacting NCoR1/2 complex. As expected, Ncor1, Ncor2, and Hdac3 showed similar Mecp2 coexpression patterns. Interestingly, Phf14 and Tbl1x, the genes that encode the proteins mediating the interaction between MeCP2 and the two complexes, both showed relatively lower expression correlation with Mecp2 (Fig. 3A).
Fig. 3.
Tcf20 is coexpressed with Mecp2 in mouse neurons and regulates MECP2-mediated synapse formation. (A) Scatter plot shows the distribution of Spearman’s correlation with Mecp2 in adult mouse brain for all the genes expressed in at least one cell type. Red and blue dots denote the TCF20 complex and NCoR1/2 complex components, respectively, with top percentiles shown in parentheses. (B) Representative immunocytochemical images of mouse cortex showing TCF20 (red) is localized in the nucleus (blue, detected by DAPI) of NeuN+ (green) neurons, arrowheads. (C) Representative immunocytochemical images of mouse cortex showing TCF20 (red), MeCP2 (green), and DAPI (blue) proteins. (D) Representative images (Left) and quantification (Right bottom) of synaptic density marked by colocalization of Synapsin I (SYN I, green) and PSD-95 (red) puncta, arrowheads, with MAP2 (blue) in cultured mouse hippocampal neurons from WT and MECP2Tg3 mice infected with a nontargeting scramble (SC) AAV virus or AAV-shRNAs targeting TCF20 (n = 15 to 31, one-way ANOVA with post hoc Tukey’s tests). (E) Quantification of Tcf20 and Bdnf mRNA levels by qRT-PCR upon knockdown of Tcf20 in cultured hippocampal neurons from WT mice (n = 3, two-way ANOVA with post hoc Tukey’s tests). (F) ChIP with anti-TCF20 on cortex chromatin from WT mice. Three primer sets were used to amplify Bdnf promotor I, promoter IV, and an intergenic region as negative control (NC) (n = 3 per group, one-way ANOVA with post hoc Tukey’s tests). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; data are mean ± SEM.
Using the same scRNA-seq data, we next examined the cell type-specific expression of Tcf20, Phf14, and Mecp2 in the mouse brain. We found that Tcf20 and Mecp2 were preferentially expressed in neurons, whereas Phf14 was equally expressed in all brain cell types (SI Appendix, Fig. S3A). This may explain the poor expression correlation between Phf14 and Mecp2. By immunostaining, we further confirmed the specific expression of TCF20 protein in NeuN+ neurons in the adult mouse brain (Fig. 3B and SI Appendix, Fig. S3B), colocalizing with MeCP2 protein in the nucleus (Fig. 3C). In the developing mouse cortex, Tcf20 mRNA levels increased during pre- and postnatal development and then plateaued in adulthood, similar to the temporal expression pattern of Mecp2 (SI Appendix, Fig. S3C). These results agree with a previous high-throughput study showing that Tcf20 and Mecp2 were highly clustered in the same transcriptional wave of postmitotic neurons and potentially related to late-occurring processes, such as synaptogenesis during neuron maturation (35). Tcf20 and Mecp2 exhibit similar spatiotemporal expression patterns in the mouse brain, both being expressed predominantly in neurons and correlated with neuronal maturation.
Given the high spatiotemporal coexpression pattern of Tcf20 and Mecp2 in mouse neurons, we next examined the functional interaction between them. A previous study in cultured mouse primary neurons has shown that MeCP2 overexpression increases synapse number (36). To test whether Tcf20 contributes to this effect, we used shRNA to knockdown TCF20 protein (Fig. 3D and SI Appendix, Fig. S3D) in primary neurons derived from transgenic MeCP2 overexpression (MECP2Tg3) and WT mice (37). To quantify the number of synapses, we performed immunostaining and used synapsin-1 (SYN1) and postsynaptic density-95 (PSD95) as pre- and postsynaptic markers, respectively. Consistent with the previous study, we observed a 30% increase in synapse density, measured by colocalization of PSD95 and SYN1 puncta in MECP2Tg3 neurons compared to WT neurons (Fig. 3D). Although knockdown of Tcf20 mildly decreased PSD95-SYN1 puncta densities, we observed that the synaptic densities in MECP2Tg3 neurons reverted to WT levels upon Tcf20 knockdown (Fig. 3D), demonstrating that TCF20 modulates neuronal changes induced by MeCP2 overexpression.
The effect of MeCP2 on synapse density could be caused by dysregulation of the MeCP2 target gene Brain-derived neurotrophic factor (Bdnf), as overexpression of BDNF restores synapse density in Mecp2–/y neurons (38, 39). We therefore tested whether TCF20 regulates synapse density by modulating Bdnf expression levels. Remarkably, qRT-PCR analyses in cultured primary neurons showed that Bdnf expression is reduced by 60% upon Tcf20 knockdown (Fig. 3E). Furthermore, chromatin immunoprecipitation (ChIP) revealed that TCF20 binds to BDNF I and BDNF IV promoters (Fig. 3F). Given that the BDNF promoter is also bound by MeCP2 (39–42), these results suggest that TCF20 and MeCP2 coregulate Bdnf transcription in mouse neurons, which could subsequently affect synapse density.
Behavioral Deficits in Tcf20+/− Mice Overlap with RTT Mice.
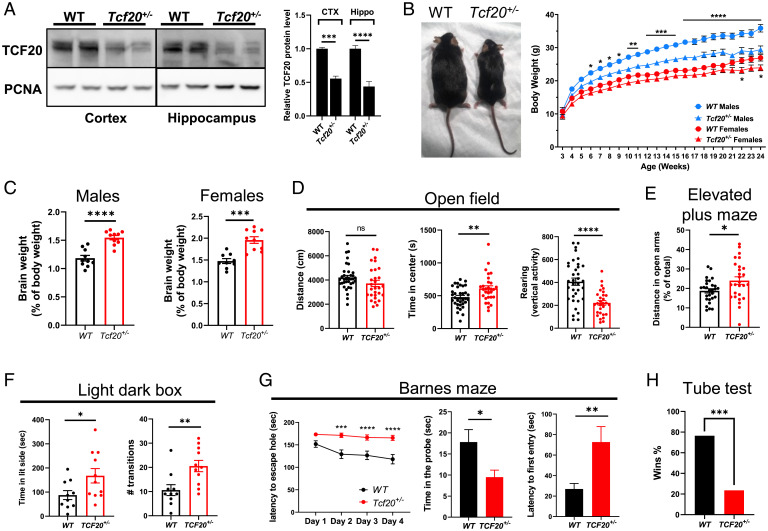
The overlapping clinical symptoms of TCF20-associated NDD and RTT inspired us to investigate whether mice deficient in Tcf20 phenocopy RTT mice. Tcf20 knockout (KO) mice were generated by deleting exon 2 using CRISPR/Cas9, which includes the translation start site and encodes 96% of the Tcf20 open-reading frame (SI Appendix, Fig. S4A). Most Tcf20 homozygous KO mice (Tcf20−/−) died 1 to 2 d after birth, and the few surviving animals were dramatically smaller than their WT littermates (SI Appendix, Fig. S4 B and C). In contrast, heterozygous mice (Tcf20+/−) were viable despite a 50% reduction of Tcf20 mRNA and protein levels in the brain compared to WT mice (Fig. 4A and SI Appendix, Fig. S4D). Tcf20+/− mice weighed less than WT, with the differences reaching significance at 6 wk of age in males and 22 wk in females (Fig. 4B). Despite their lower body weights, the brain weights of both male and female adult Tcf20+/− mice were greater than WT (Fig. 4C), consistent with the macrocephaly observed in TAND patients with Tcf20 mutations (12).
Fig. 4.
Haploinsufficiency in Tcf20 results in learning and memory deficits and autism-like phenotypes in mice. (A) Representative immunoblot (Left) and quantification (Right) of TCF20 protein levels in the cortex (CTX) and hippocampus (Hippo) from WT and Tcf20+/− mice (n = 3 per group, two-way ANOVA with post hoc Tukey’s tests). (B, Left) A photograph showing the body size of WT and Tcf20+/− mice at 8 mo of age. (Right) Body weights of WT and Tcf20+/− mice over the course of 24 wk. (For males, WT, n = 22, Tcf20+/−, n = 17; for females, WT, n = 21, Tcf20+/−, n = 14; two-way ANOVA with post hoc Tukey’s tests.) (C) Brain weights of adult WT and Tcf20+/− mice after being normalized to body weights. (For males, WT, n = 10, Tcf20+/−, n = 10; for females, WT, n = 9, Tcf20+/−, n = 10; unpaired two-tailed Student’s t test.) (D) Statistical analysis of open field test for WT and Tcf20+/− male mice. (Left) Total mouse movement in 30 min; (Center) time spent in the center area; (Right) time spent for vertical exploring (rearing) (WT, n = 35, Tcf20+/−, n = 29; unpaired two-tailed Student’s t test). (E) Statistical analysis of time spent in the open arm for WT and Tcf20+/− male mice in the elevated plus maze (WT, n = 28, Tcf20+/−, n = 27; unpaired two-tailed Student’s t test). (F) Statistical analysis of light–dark box test for WT and Tcf20+/− male mice. (Left) Time spent in the light side; (Right) the number of transitions between the two compartments (WT, n = 10, Tcf20+/−, n = 11; unpaired two-tailed Student’s t test). (G) Statistical analysis of Barnes maze for WT and Tcf20+/− male mice. (Left) Time spent to locate the escape hole during 4 d of training; (Center) time spent in the probe (escape hole) area during the test day; (Right) latency to first enter the escape hole during the test day (WT, n = 22, Tcf20+/−, n = 23, two-way ANOVA with post hoc Tukey’s tests for training, unpaired two-tailed Student’s t test for probe test). (H) Percentage of wins in test pairs between WT and Tcf20+/− male mice in the tube test (n = 51 matches, two-tailed binomial test). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; data are mean ± SEM.
We next examined the behavior of adult Tcf20+/− male mice. In the open field test, Tcf20+/− mice spent significantly more time in the center area and reared less than WT, which suggests less anxiety-like behavior and less exploratory activity (Fig. 4D). In the elevated-plus maze and light–dark box tests, Tcf20+/− mice spent significantly more time in the open arm and on the light side, respectively, which also indicates less anxiety-like behavior (Fig. 4E and F). In the Barnes maze, a test that requires the mouse to locate an escape tunnel in a brightly lit circular arena, the Tcf20+/− mice failed to learn the escape hole location during the training trials and spent significantly more time locating the probe hole and less time in the probe region, indicating impaired spatial learning and memory (Fig. 4G). In the tube test to measure social dominance between two unfamiliar mice (43), the Tcf20+/− mice lost most of the matches when tested against their WT opponents (Fig. 4H). Interestingly, similar to the milder difference in body weight changes (Fig. 4B), female Tcf20+/− mice showed milder behavioral deficits compared to male animals (SI Appendix, Fig. S4 E–H), perhaps because TCF20 negatively regulates estrogen receptor expression, which is associated with anxiety and depression-like behaviors of female mice (44, 45). These behavioral data suggest that Tcf20 haploinsufficiency in mice alters anxiety-like behavior, learning and memory, and social dominance. RTT mice also show reduced anxiety-like behavior in the open field, elevated-plus maze, and light–dark box tests, as well as impaired learning and memory in fear conditioning and Morris water maze (46, 47). The partial overlap of neurobehavioral deficits between the two mouse models suggests that TCF20 and MeCP2 share common downstream neuronal pathways.
We also generated Phf14 KO mice using CRISPR/Cas9 by deleting exon 3 (SI Appendix, Fig. S5A). Similar to Tcf20, complete KO of Phf14 in mice is embryonic-lethal, whereas heterozygous mice (Phf14+/−) with 50% reduction of Phf14 mRNA and protein levels appeared healthy with normal body weight (SI Appendix, Fig. S5 B–E), consistent with the previous study (48). We performed a battery of behavioral assays but found no differences between Phf14+/− and WT mice in the open field, elevated-plus maze, light–dark box, rotarod, or fear-conditioning tests (SI Appendix, Fig. S5 F–J). These data suggest that PHF14 protein is essential for prenatal development, but haploinsufficiency in Phf14 is tolerated pre- and postnatally.
MeCP2 and TCF20 Share Common Downstream Neuronal Genes and Pathways.
Our neuronal and behavioral data suggest that TCF20 and MeCP2 coregulate neuronal gene expression (Fig. 3 E and F). We therefore assessed global gene-expression changes in Tcf20+/− mice, exploring the extent to which these effects overlap with those observed in MeCP2 mutant mice. RNA-seq of Tcf20+/− mice prefrontal cortex (PFC) identified 145 up-regulated and 193 down-regulated genes (Padj < 0.05) (SI Appendix, Fig. S6A), consistent with previous studies showing TCF20 functions as both a transcriptional coactivator and corepressor (44, 49–51). Gene set enrichment analysis of these 338 dysregulated genes revealed enrichment of gene ontology terms related to behavioral regulation and neuron projection and signaling (SI Appendix, Fig. S6B), consistent with the behavioral and synaptic deficits in Tcf20-deficient mice and neurons (Figs. 3D and 4). Given that PHF14 mediates the interaction between MeCP2 and the TCF20 complex, we also performed RNA-seq in the PFC isolated from Phf14+/− mice. We identified only 22 up-regulated and 17 down-regulated genes (Padj < 0.05) in Phf14+/− mice, which is fewer than the differentially expressed genes (DEGs) found in Tcf20+/− mice (SI Appendix, Fig. S6C). This could be because PHF14 is not a transcription factor that directly regulates gene expression, and a 50% reduction of PHF14 is not sufficient to completely disrupt the function of the TCF20 complex in mice.
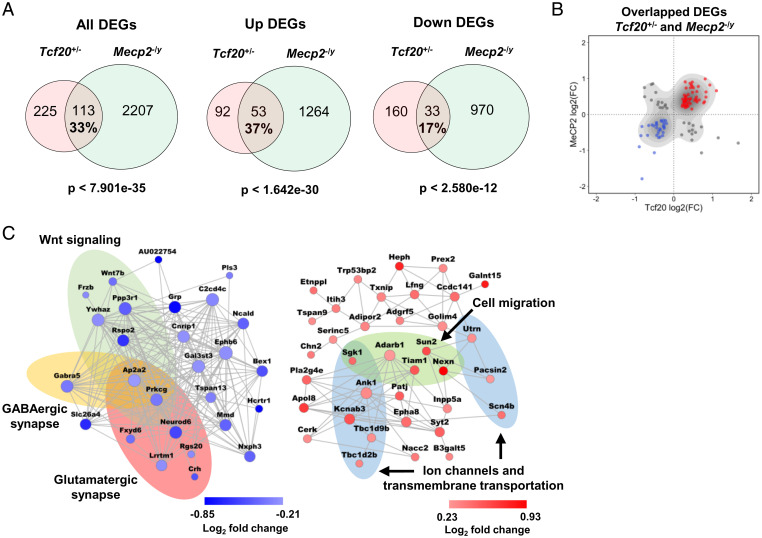
We next compared the Tcf20+/− DEGs to those in the PFC of Mecp2–/y mice. Interestingly, a large proportion (33%) of DEGs in Tcf20+/− mice were significantly altered in Mecp2–/y mice as well (Fig. 5A). The degree of overlap was somewhat higher when we considered only up-regulated DEGs. Comparing the log2 fold-change in gene expression between mouse models shows that a majority (72%) of the DEGs common to both models changed in the same direction and with similar magnitude (Fig. 5B). To assess the strength of the functional association of TCF20 and MeCP2 on gene expression, we performed the similar analysis using the same statistical criteria by overlapping the DEGs identified in Mecp2–/y mice with the DEGs found in mice carrying the NS-DAD mutation (NS-DADm) that abolishes HDAC3 activity in the NCoR1/2 complex (42, 52). We found the overall overlap between Mecp2–/y and NS-DADm DEGs was higher (47%) (SI Appendix, Fig. S6D), whereas less of the shared DEGs were altered in the same direction (33%) (SI Appendix, Fig. S6 D and E). Because the total number of DEGs in Mecp2–/y mice was much higher than that in the other two mouse models, only a small percentage of DEGs (∼5% each) in Mecp2–/y mice were significantly altered in Tcf20+/− or NS-DADm mice (Fig. 5A and SI Appendix, Fig. S6D).
Fig. 5.
MeCP2 and TCF20 share common downstream neuronal genes and pathways. (A) Venn diagrams showing DEGs common to Tcf20+/− and Mecp2–/y mouse models; (Left) all DEGs; (Center) up-regulated DEGs; (Right) down-regulated DEGs (full lists of Tcf20+/− and Mecp2–/y DEGs are given in Datasets S2 and S3, respectively). The percentage rate indicates the ratio of the number of overlapped DEGs to the number of Tcf20+/− DEGs. (B) Scatter plot showing log2 fold-change for overlapped DEGs in Tcf20+/− vs. Mecp2–/y mouse models. Majority of the DEGs common to Tcf20+/− and Mecp2–/y mouse models changed in the same direction. (C) Coexpression networks of the down- (Left, blue) and up-regulated (Right, red) overlapped genes between Tcf20+/− and Mecp2–/y mouse models. Colored ellipses indicate the enriched gene ontology terms or pathways in each network. Node size is proportional to coexpression degree. Node color reflects log2 fold-change in Tcf20+/− mice.
Further gene ontology coupled with coexpression network analyses of the DEGs shared between Tcf20+/− and Mecp2–/y mice identified significant enrichment in neuronal function among gene ontology terms. Pathways associated with ion channels and transmembrane transportation were enriched in the up-regulated DEGs, and pathways associated with GABAergic and glutamatergic synaptic function were enriched in the down-regulated DEGs (Fig. 5C). These results support the idea that TCF20 and MeCP2 coregulate the expression of neuronal genes and pathways.
Decreasing TCF20 Levels Improves Behavioral Deficits in MECP2 Duplication Mice.
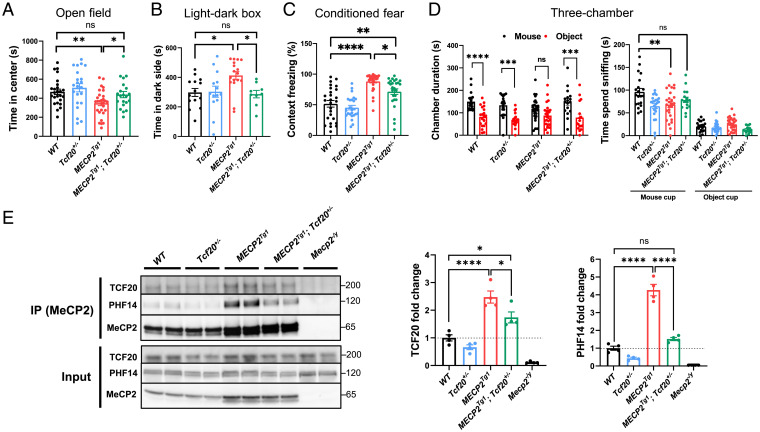
Several studies have shown that the behavioral abnormalities in mouse models of RTT and MDS can be reversed by correcting MeCP2 protein levels or reducing MeCP2 transcriptional targets (53–55), but not by manipulating MeCP2-interacting partners (25). Inspired by the ability of Tcf20 knockdown to reverse synapse density defects in MECP2Tg3 neurons (Fig. 3D), however, we decided to test whether genetically reducing Tcf20 could rescue the behavioral deficits in adult MDS mouse model (53).
We bred female MECP2Tg1 FVB mice to male Tcf20+/− C57BL/6J mice to genetically reduce the levels of TCF20 in MECP2Tg1 animals and performed a battery of behavioral tests on double-mutant mice (MECP2Tg1;Tcf20+/−) and their WT, MECP2Tg1, and Tcf20+/− littermates (SI Appendix, Fig. S7A). Consistent with previous studies (37, 53), MECP2Tg1 mice exhibited anxiety-like behavior, as demonstrated by less time spent in the center of the open field and more time spent on the dark side of the light–dark box (Fig. 6 A and B). These behaviors were rescued in MECP2Tg1;Tcf20+/− mice (Fig. 6 A and B). Moreover, MECP2Tg1;Tcf20+/− mice spent significantly less time freezing in the contextual fear assay relative to MECP2Tg1 mice, indicating partial rescue of this deficit (Fig. 6C). In the three-chamber social assay, MECP2Tg1 mice showed no preference for staying in the mouse chamber over the inanimate object chamber and spent significantly less time interacting with the mouse, but MECP2Tg1;Tcf20+/− animals were statistically indistinguishable from WT mice in this social assay (Fig. 6D). Reducing TCF20 levels did not improve the performance of MECP2Tg1;Tcf20+/− mice in the rotarod assay or elevated-plus maze (SI Appendix, Fig. S7 B and C). These data indicate that reducing TCF20 levels relieves some anxiety-like behavior, learning and memory deficits, and abnormal sociability observed in MECP2 duplication mice.
Fig. 6.
Genetic reduction of Tcf20 improves behavioral deficits in MECP2 duplication mice. (A) Statistical analysis of open field test showing time spent in the center area for mice with the indicated genotypes (n = 21 to 29 per genotype, one-way ANOVA with post hoc Tukey’s tests). (B) Statistical analysis of light–dark box test for mice with the indicated genotypes (n = 9 to 17 per genotype, one-way ANOVA with post hoc Tukey’s tests). (C) Altered learning and memory in conditioned fear test for mice with the indicated genotypes (n = 19 to 26 per genotype, one-way ANOVA with post hoc Tukey’s tests). (D) Statistical analysis of three-chamber test for mice with the indicated genotypes. (Left) Time the mice stay in chambers with the partner mouse or with the inanimate object; (Right) time the mice spent interacting with the partner mouse (n = 16 to 27 per genotype). (Left) Two-way ANOVA with post hoc Tukey’s tests; (Right) one-way ANOVA with post hoc Tukey’s tests. (E) Representative immunoblot (Left) and quantification (Right) of TCF20, PHF14 following IP using antibodies to MeCP2 from mouse cortex of each genotype. Mecp2–/y as negative controls. The ratio of co-IP TCF20 and PHF14 from each indicated genotypes to that of WT is calculated as fold-change (n = 4 per group, one-way ANOVA with post hoc Tukey’s tests) *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; data are mean ± SEM. ns, not significant.
Given the behavioral improvement seen in MECP2Tg1;Tcf20+/− mice, we hypothesized that neuronal dysfunction caused by MECP2 duplication results, in part, from abnormally increased recruitment of the TCF20 complex to the overexpressed MeCP2. To test this hypothesis, we immunoprecipitated MeCP2 from the cortex of WT, MECP2Tg1, Tcf20+/−, and MECP2Tg1;Tcf20+/− mice. Indeed, we found that more TCF20 and PHF14 were coimmunoprecipitated with MeCP2 in MECP2Tg1 brains (Fig. 6E), and significantly less TCF20 and PHF14 were coimmunoprecipitated in MECP2Tg1;Tcf20+/− brains (Fig. 6E), in agreement with the observed behavior results. Reduced PHF14 levels are likely due to the complex's instability upon TCF20 reduction (Fig. 1D). Taken together, these data suggest that some MECP2 duplication symptoms might be mediated by excessive recruitment of TCF20 and PHF14 and can be reversed through genetic reduction of Tcf20.
A Mutation That Disrupts the Interaction between MeCP2, PHF14, and TCF20 Is Associated with a Human Neurological Phenotype That Overlaps with RTT.
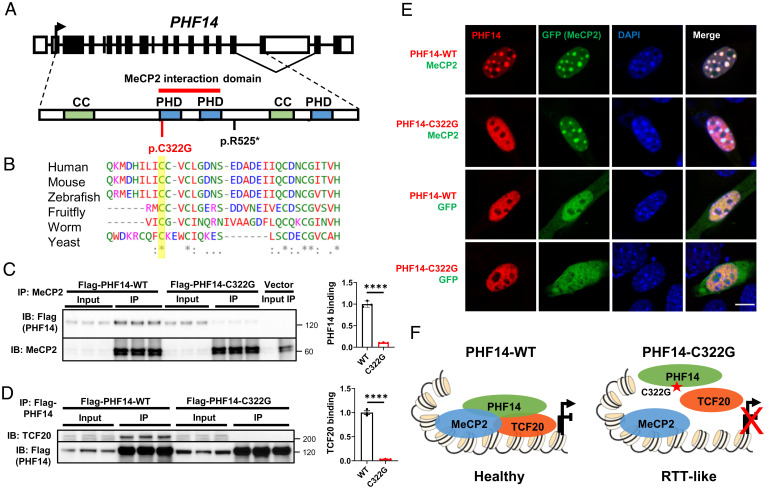
The functional interaction between MeCP2 and the TCF20 complex suggests they might contribute to a common etiology in a subset of NDDs. Prior to this study, microdeletion and duplication of PHF14 had been associated with Dandy-Walker malformation (56), in which the cerebellar vermis fails to form properly and the posterior fossa fill with cerebrospinal fluid; the syndrome is associated with hydrocephalus and seizures and a number of other organ defects. So far, however, PHF14 mutations have not been found in patients with NDDs. Given our data showing that PHF14 plays an important role in bridging the interaction between MeCP2 and the TCF20 complex, we asked whether mutations in PHF14 could also be associated with NDD.
We first searched the gnomAD database and found control individuals with LoF mutations in PHF14. Unlike MECP2 and TCF20, which have high LoF intolerance (pLI) scores of 0.89 and 1, respectively, PHF14 has a pLI score of 0.27, suggesting humans should be comparatively less affected by LoF PHF14 mutations. We mined whole-genome/exome sequencing through collaborative efforts and identified two individuals with de novo heterozygous PHF14 (NM_014660.4) variants without other known disease-causing mutations. The PHF14 variants in the affected individuals and their de novo status were confirmed by Sanger sequencing. One patient is a 5-y-old boy (patient GER01) with PHF14 nonsense variant (c.1573A > T, p.R525*) who presented with a neurological phenotype including autistic behavior (no eye contact), no speech, and sleep disturbance. The other patient is a 4-y-old girl who presented with clumsy gait, developmental delay/intellectual disability, speech delay, and an RTT-like regression in gross motor skills and balance. Her PHF14 missense variant (c.964T > G, p.C322G), which is absent in the control population (gnomAD database), is located in a highly conserved amino acid within the MeCP2-interacting domain (Fig. 7 A and B). To test whether this point mutation could disrupt MeCP2 interaction with PHF14, we transfected HEK293T cells with a Flag-tagged WT or PHF14-C322G mutant and measured their interaction with MeCP2 by co-IP. Strikingly, the PHF14 C322G mutation completely abolished PHF14 interactions with MeCP2 and TCF20 (Fig. 7C and D). Furthermore, immunostaining in NIH 3T3 cells showed that PHF14 carrying the C322G mutation cannot be recruited by MeCP2 to the heterochromatin foci (Fig. 7E). These human genetic and biochemical data raise the possibility that disruption of MeCP2–PHF14–TCF20 interaction could impair normal brain development (Fig. 7F).
Fig. 7.
A PHF14 mutation that disrupts the interaction of MeCP2 and TCF20 complex is associated with a human neurodevelopmental syndrome similar to RTT. (A) Diagram of PHF14 gene and PHF14 protein domains with the mutations currently identified. PHD, plant homeodomain finger domain; CC, Coiled-coil domain. A red bar above the protein structure indicates the domain on PHF14 that interacts with MeCP2. (B) ClustalW multispecies alignment obtained with the region containing C322G, yellow bar showing the high level of conservation of the mutated residue. (C) Representative immunoblot (Left) and quantification (Right) of Flag-tagged PHF14-WT and PHF14-C322G following IP of endogenous MeCP2 in HEK293T cells (n = 3 per group, unpaired two-tailed Student’s t test). (D) Representative immunoblot (Left) and quantification (Right) of TCF20 following IP of Flag-tagged PHF14-WT and PHF14-C322G in HEK293T cells (n = 3 per group, unpaired two-tailed Student’s t test). (E) Representative immunocytochemical images in mouse 3T3 fibroblasts showing the localization of Flag-tagged PHF14-WT and PHF14-C322G relative to densely methylated heterochromatic foci (stained by DAPI) upon overexpression of MeCP2-GFP and GFP only as controls. (Scale bar, 10 µm.) (F) Schematic overview of the proposed model of MeCP2 and the TCF20 complex interaction. In wild-type brains, MeCP2 recruits the TCF20 complexes by binding to PHF14 to coregulate gene expression. Patient mutation (C322G) in the PHD1-2 domain of PHF14 disrupted the interaction between MeCP2 and PHF14 and also prevent the assembly of the TCF20 complex, potentially leading to dysregulation of downstream genes and RTT-like symptoms in the patient. ****P < 0.0001; data are mean ± SEM.
Discussion
Previous PPI network analyses revealed that genes associated with heterogeneous NDDs could converge on interconnected molecular systems (4, 57). However, follow-up functional studies were needed to elucidate how different mutations can disturb the converging pathways. In this study, we uncovered an interaction between MeCP2 and the TCF20 complex in the mouse brain, demonstrating a shared molecular pathogenesis between RTT, MDS, TAND, and related conditions. The discovery of several pathological variants that disrupt this interaction supports the critical role of MeCP2–PHF14–TCF20 interaction for brain function. TCF20 regulates key neuronal pathways downstream of MeCP2 and modifies MECP2-induced synaptic and behavioral deficits, suggesting that mutations in MeCP2–TCF20 complex components could lead to certain neurological deficits through shared gene-expression changes. These findings shed light on the molecular function of MeCP2 and the mechanisms underlying RTT and these related disorders.
MeCP2 was found to predominately recruit the NCoR1/2 histone modification complexes through the TRD to repress transcription (18). Here, we showed that MeCP2 can also regulate gene expression by interacting with a transcriptional regulatory complex via its MBD and ID. This previously unknown aspect of MeCP2’s molecular function might explain why total loss of the gene cause a more severe RTT phenotype in both humans and mice than mutations in the TRD. Moreover, accumulating evidence suggests that MeCP2 recruits multiple protein complexes to carry out its function (14, 30, 33, 58–60). Besides the TCF20 complex, many of the MeCP2-interacting chromatin proteins, such as switch-insensitive 3 family member A (SIN3A) and several components of the NCoR1/2 complex, are also associated with neurodevelopmental phenotypes (52, 61, 62). These data therefore suggest that members of the protein-interaction network centered on MeCP2 could affect an even broader cluster of disorders.
We found that a portion of downstream genes shared by TCF20 and MeCP2 are enriched in key neuronal pathways. Certain critical neuronal genes, such as Bdnf and Crh, were significantly reduced in Tcf20+/− brains and contribute to anxiety-like behaviors and other disease features in MeCP2-related mouse models (54, 63). For example, knockdown of the critical ion channel gene Ankyrin 1 (Ank1), whose expression is altered in both Mecp2–/y and Tcf20+/− mouse models, resulted in reduced anxiety-like behavior in mice (64). These neural genes and pathways could mediate some of the overlapping deficits related to TCF20 and MeCP2 dysfunction. It is still unclear precisely how TCF20 modulates MeCP2’s function, as both increases and decreases in gene expression have been noted upon loss of Mecp2 and Tcf20. Future genome-wide chromatin binding studies of both TCF20 and MeCP2 coupled with the existing RNA-seq data could potentially address the mechanism of transcriptional regulation more directly.
Prior to the present study, it was unclear whether the TCF20 complex forms in the brain and what molecular function it plays. The presence of this complex in the brain, together with human genetic and mouse studies, suggests that it is essential to normal brain development (11–13). One subunit of the TCF20 complex is encoded by RAI1, the causal gene for Smith–Magenis Syndrome (SMS) (65). Rai1 mutant mouse models display many SMS-like symptoms including obesity, motor dysfunction, and learning and memory deficits (66–68). Similarly, our Tcf20+/− mice recapitulate the core TAND features, as does another Tcf20+/− allele (69). The clinical features of patients with PHF14 mutations—such as intellectual disability, autistic behaviors, speech delay or loss, sleep disturbance, motor incoordination, and regression—were also seen in TAND and RTT patients (SI Appendix, Fig. S8) (7, 12, 13). Our biochemical data on the PHF14 C322G mutation imply that its pathogenicity involves the loss of MeCP2 binding.
Interestingly, we did not observe significant behavioral abnormalities or transcriptional changes for Phf14 heterozygous mice (SI Appendix, Fig. S5), and there are apparently healthy individuals in gnomAD with LoF mutations in PHF14, suggesting that the effects of haploinsufficiency, whatever they may be, have low penetrance or late-life onset. We remain reserved about the pathogenicity of PHF14 variants, even though de novo LoF have been identified in patients with NDDs who lack other known disease-causing mutations. It is possible that patients who manifest phenotypes upon LoF of one PHF14 allele express slightly less PHF14 protein from their WT allele in the brain than those who do not manifest disease. Moreover, putative PHF14 orthologs, such as BRPF1 that also harbors a tandem PHD finger (PZP) domain (70), could functionally compensate for the loss of PHF14 in mice and humans. In fact, mutations in BRPF1, including a missense mutation within the PZP domain, also cause syndromic intellectual disability (71). Ultimately, the identification of additional patients with PHF14 mutations and a better assessment of genotype–phenotype relationships will be needed to confirm the contribution of PHF14 LoF to the neurodevelopmental phenotypes.
The clinical features of RTT and TAND patients overlap but are not identical (SI Appendix, Fig. S8). Just as it is likely that interactions with TCF20 do not account for all the functions of MeCP2, binding to MeCP2 does not account for all the functions of TCF20. Therefore, investigating the missense mutations that specifically disrupt MeCP2–TCF20 binding but retain other functions of the protein should shed light on those features that are most dependent on this interaction. Such an approach previously led to the discovery of several pathogenic missense mutations in TBL1XR1, the gene encodes the TBL1 subunit of the NCoR1/2 complexes that mediates the interaction with MeCP2 (61). These missense mutations were mapped within the MeCP2 interaction domain, suggesting that the impairment of TBL1 and MeCP2 interaction underlies pathogenesis. Clinical data are currently limited, however, and insufficient to evaluate the link between TBL1XR1 and RTT (61, 72). It will be important to identify more patients with missense mutations of PHF14 and TBL1XR1 that affect MeCP2 binding and compare their clinical phenotypes with those of RTT patients. Mouse models carrying these various missense mutations, such as the Phf14 C322G mutation, will be excellent tools to establish genotype–phenotype relationships.
In summary, our study demonstrates that it is feasible to identify shared molecular pathogenic mechanisms of NDDs by coupling protein interaction analysis with genotype–phenotype characterization in mouse models. Mapping the cause-and-effect relationships between mutations that disrupt certain protein interactions and the deficits seen in patients will shed light on the molecular pathology underlying both shared and distinct disease features. Identifying downstream points of convergence that mediate specific deficits could eventually lead to viable treatments for genetically distinct disorders.
Materials and Methods
Mecp2–/y, MECP2Tg1, MECP2Tg3, and MECP2-EGFP mice were previously described (23, 37, 73). The newly derived Tcf20 KO mice (MGI 6263655) were obtained from the Wellcome Trust Sanger Institute. Phf14 KO mice were generated by deleting exon 3 using two guide RNAs (5′-AGCTATTTGATATAAATCAG-3′ and 5′-AGATGCTAACTGGCAAAGAG-3′) through CRISPR/Cas9. Male Tcf20+/− and female MECP2Tg1 mice were bred to generate MECP2Tg1;Tcf20+/− mice. All KO and knockin mice were maintained on a C57BL/6 background and all transgenic mice were maintained on a FVB background in a 14-h:10-h light:dark cycle at 68 to 72 °F and 30 to 70% humidity, with standard mouse chow and water ad libitum. The Baylor College of Medicine Institutional Animal Care and Use Committee approved all research and animal-care procedures. Information on procedures involving BioID-MS, biochemical evaluation and immunostaining of brain tissues and cultured cells and primary neurons, behavior, RNA-seq and data analysis, genome/exome sequencing are detailed in SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We thank the families for their participation in the study and the clinical teams, particularly Jill A. Rosenfeld for her help in collecting some clinical data; Alexander Trostle and Ying-Wooi Wan for assistance in RNA-sequencing data processing; the Wellcome Trust Sanger Institute for providing the Tcf20 knockout mice; Vicky Brandt for critical review of the manuscript; and the Genetically Engineered Rodent Models Core led by Jason Heaney, which is partially supported by NIH Grant P30CA125123 at Baylor College of Medicine, for generating the Phf14 knockout mouse model. This work was supported by HHMI (H.Y.Z.), the Souki Foundation (H.Y.Z.), National Institute of Neurological Disorders and Stroke Grant 5R01NS057819 (to H.Y.Z.), National Institute of Child Health and Human Development Grant 1F32HD100048-01 (to S.S.B.), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (M.N.R. and A.L.B.), NIH Grant R01NS122073 (to M.N.R.), the US Department of Agriculture under Cooperative Agreement 58-3092-0-001 (to H.K.Y.), National Institute of General Medical Sciences Grant R01-GM120033 (to Z.L.), the Huffington Foundation (Z.L.), the Alabama Genomic Health Initiative (an Alabama-State earmarked project F170303004) through the University of Alabama in Birmingham (E.M.B. and M.L.T.), Deutsche Forschungsgemeinschaft Grants 418081722 and 433158657 (to T.B.H.), the Genomic and RNA Profiling Core at Baylor College of Medicine with funding from NIH Grant 1S10OD023469, and the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the NIH under Award P50HD103555 for use of the Neurovisualization and Neurobehavioral Core facilities. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Reviewers: G.M., Oregon Health and Science University; and C.W., Boston Children’s Hospital.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2119078119/-/DCSupplemental.
Data Availability
The data reported in this paper have been deposited in the Gene Expression Omnibus database (accession no. GSE179229). All other data are included in the manuscript and/or supporting information.
References
- 1.Zablotsky B., et al. , Prevalence and trends of developmental disabilities among children in the United States: 2009–2017. Pediatrics 144, e20190811 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Satterstrom F. K., et al. ; Autism Sequencing Consortium; iPSYCH-Broad Consortium, Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180, 568–584.e23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McRae J. F., et al. ; Deciphering Developmental Disorders Study, Prevalence and architecture of de novo mutations in developmental disorders. Nature 542, 433–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakai Y., et al. , Protein interactome reveals converging molecular pathways among autism disorders. Sci. Transl. Med. 3, 86ra49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amir R. E., et al. , Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23, 185–188 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Van Esch H., et al. , Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am. J. Hum. Genet. 77, 442–453 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandweiss A. J., Brandt V. L., Zoghbi H. Y., Advances in understanding of Rett syndrome and MECP2 duplication syndrome: Prospects for future therapies. Lancet Neurol. 19, 689–698 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Bienvenu T., Chelly J., Molecular genetics of Rett syndrome: When DNA methylation goes unrecognized. Nat. Rev. Genet. 7, 415–426 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Wen Z., et al. , Identification of autism-related MECP2 mutations by whole-exome sequencing and functional validation. Mol. Autism 8, 43 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sajan S. A., et al. , Enrichment of mutations in chromatin regulators in people with Rett syndrome lacking mutations in MECP2. Genet. Med. 19, 13–19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babbs C., et al. ; International Molecular Genetic Study of Autism Consortium (IMGSAC), De novo and rare inherited mutations implicate the transcriptional coregulator TCF20/SPBP in autism spectrum disorder. J. Med. Genet. 51, 737–747 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vetrini F., et al. ; DDD study, De novo and inherited TCF20 pathogenic variants are associated with intellectual disability, dysmorphic features, hypotonia, and neurological impairments with similarities to Smith-Magenis syndrome. Genome Med. 11, 12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torti E., et al. , Variants in TCF20 in neurodevelopmental disability: Description of 27 new patients and review of literature. Genet. Med. 21, 2036–2042 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chahrour M., et al. , MeCP2, a key contributor to neurological disease, activates and represses transcription. Science (80-.) 320, 1224–1229 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boxer L. D., et al. , MeCP2 represses the rate of transcriptional initiation of highly methylated long genes. Mol. Cell 77, 294–309.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clemens A. W., et al. , MeCP2 represses enhancers through chromosome topology-associated DNA methylation. Mol. Cell 77, 279–293.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyst M. J., Bird A., Rett syndrome: A complex disorder with simple roots. Nat. Rev. Genet. 16, 261–275 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Lyst M. J., et al. , Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat. Neurosci. 16, 898–902 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.You S. H., et al. , Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat. Struct. Mol. Biol. 20, 182–187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neul J. L., et al. , Specific mutations in methyl-CpG-binding protein 2 confer different severity in Rett syndrome. Neurology 70, 1313–1321 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cuddapah V. A., et al. , Methyl-CpG-binding protein 2 (MECP2) mutation type is associated with disease severity in Rett syndrome. J. Med. Genet. 51, 152–158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker S. A., et al. , An AT-hook domain in MeCP2 determines the clinical course of Rett syndrome and related disorders. Cell 152, 984–996 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heckman L. D., Chahrour M. H., Zoghbi H. Y., Rett-causing mutations reveal two domains critical for MeCP2 function and for toxicity in MECP2 duplication syndrome mice. eLife 3, e02676 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown K., et al. , The molecular basis of variable phenotypic severity among common missense mutations causing Rett syndrome. Hum. Mol. Genet. 25, 558–570 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koerner M. V., et al. , Toxicity of overexpressed MeCP2 is independent of HDAC3 activity. Genes Dev. 32, 1514–1524 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D. I., et al. , An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell 27, 1188–1196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nan X., Tate P., Li E., Bird A., DNA methylation specifies chromosomal localization of MeCP2. Mol. Cell. Biol. 16, 414–421 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baker S. A., Lombardi L. M., Zoghbi H. Y., Karyopherin α 3 and karyopherin α 4 proteins mediate the nuclear import of methyl-CpG binding protein 2. J. Biol. Chem. 290, 22485–22493 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eberl H. C., Spruijt C. G., Kelstrup C. D., Vermeulen M., Mann M., A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol. Cell 49, 368–378 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Nan X., et al. , Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393, 386–389 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Sheikh T. I., et al. , From function to phenotype: Impaired DNA binding and clustering correlates with clinical severity in males with missense mutations in MECP2. Sci. Rep. 6, 38590 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goffin D., et al. , Rett syndrome mutation MeCP2 T158A disrupts DNA binding, protein stability and ERP responses. Nat. Neurosci. 15, 274–283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nan X., et al. , Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc. Natl. Acad. Sci. U.S.A. 104, 2709–2714 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saunders A., et al. , Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174, 1015–1030.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Telley L., et al. , Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science 351, 1443–1446 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Chao H. T., Zoghbi H. Y., Rosenmund C., MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron 56, 58–65 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collins A. L., et al. , Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum. Mol. Genet. 13, 2679–2689 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Sampathkumar C., et al. , Loss of MeCP2 disrupts cell autonomous and autocrine BDNF signaling in mouse glutamatergic neurons. eLife 5, e19374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen W. G., et al. , Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302, 885–889 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Li H., Zhong X., Chau K. F., Williams E. C., Chang Q., Loss of activity-induced phosphorylation of MeCP2 enhances synaptogenesis, LTP and spatial memory. Nat. Neurosci. 14, 1001–1008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ito-Ishida A., et al. , Genome-wide distribution of linker histone H1.0 is independent of MeCP2. Nat. Neurosci. 21, 794–798 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen L., et al. , MeCP2 binds to non-CG methylated DNA as neurons mature, influencing transcription and the timing of onset for Rett syndrome. Proc. Natl. Acad. Sci. U.S.A. 112, 5509–5514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan Z., et al. , Using the tube test to measure social hierarchy in mice. Nat. Protoc. 14, 819–831 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Gburcik V., Bot N., Maggiolini M., Picard D., SPBP is a phosphoserine-specific repressor of estrogen receptor α. Mol. Cell. Biol. 25, 3421–3430 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walf A. A., Frye C. A., Estradiol reduces anxiety- and depression-like behavior of aged female mice. Physiol. Behav. 99, 169–174 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samaco R. C., et al. , Female Mecp2(+/-) mice display robust behavioral deficits on two different genetic backgrounds providing a framework for pre-clinical studies. Hum. Mol. Genet. 22, 96–109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Achilly N. P., Wang W., Zoghbi H. Y., Presymptomatic training mitigates functional deficits in a mouse model of Rett syndrome. Nature 592, 596–600 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang Q., et al. , Depletion of PHF14, a novel histone-binding protein gene, causes neonatal lethality in mice due to respiratory failure. Acta Biochim. Biophys. Sin. (Shanghai) 45, 622–633 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Lyngsø C., et al. , Interaction between the transcription factor SPBP and the positive cofactor RNF4. An interplay between protein binding zinc fingers. J. Biol. Chem. 275, 26144–26149 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Elvenes J., et al. , Pax6 represses androgen receptor-mediated transactivation by inhibiting recruitment of the coactivator SPBP. PLoS One 6, e24659 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Darvekar S. R., Elvenes J., Brenne H. B., Johansen T., Sjøttem E., SPBP is a sulforaphane induced transcriptional coactivator of NRF2 regulating expression of the autophagy receptor p62/SQSTM1. PLoS One 9, e85262 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou W., et al. ; DDD study, Loss of function of NCOR1 and NCOR2 impairs memory through a novel GABAergic hypothalamus-CA3 projection. Nat. Neurosci. 22, 205–217 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sztainberg Y., et al. , Reversal of phenotypes in MECP2 duplication mice using genetic rescue or antisense oligonucleotides. Nature 528, 123–126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samaco R. C., et al. , Crh and Oprm1 mediate anxiety-related behavior and social approach in a mouse model of MECP2 duplication syndrome. Nat. Genet. 44, 206–211 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shao Y., et al. , Antisense oligonucleotide therapy in a humanized mouse model of MECP2 duplication syndrome. Sci. Transl. Med. 13, eaaz7785 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao C., et al. , Prenatal diagnosis and molecular characterization of a novel locus for Dandy-Walker malformation on chromosome 7p21.3. Eur. J. Med. Genet. 55, 472–475 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Cristino A. S., et al. , Neurodevelopmental and neuropsychiatric disorders represent an interconnected molecular system. Mol. Psychiatry 19, 294–301 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Agarwal N., et al. , MeCP2 interacts with HP1 and modulates its heterochromatin association during myogenic differentiation. Nucleic Acids Res. 35, 5402–5408 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ballas N., Grunseich C., Lu D. D., Speh J. C., Mandel G., REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121, 645–657 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Della Ragione F., Vacca M., Fioriniello S., Pepe G., D’Esposito M., MECP2, a multi-talented modulator of chromatin architecture. Brief. Funct. Genomics 15, 420–431 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Zaghlula M., et al. , Current clinical evidence does not support a link between TBL1XR1 and Rett syndrome: Description of one patient with Rett features and a novel mutation in TBL1XR1, and a review of TBL1XR1 phenotypes. Am. J. Med. Genet. A. 176, 1683–1687 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Witteveen J. S., et al. , Haploinsufficiency of MeCP2-interacting transcriptional co-repressor SIN3A causes mild intellectual disability by affecting the development of cortical integrity. Nat. Genet. 48, 877–887 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Chang Q., Khare G., Dani V., Nelson S., Jaenisch R., The disease progression of Mecp2 mutant mice is affected by the level of BDNF expression. Neuron 49, 341–348 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Stevens S. R., et al. , Ankyrin-R regulates fast-spiking interneuron excitability through perineuronal nets and Kv3.1b K+ channels. eLife 10, e66491 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Slager R. E., Newton T. L., Vlangos C. N., Finucane B., Elsea S. H., Mutations in RAI1 associated with Smith-Magenis syndrome. Nat. Genet. 33, 466–468 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Huang W. H., et al. , Molecular and neural functions of Rai1, the causal gene for Smith-Magenis syndrome. Neuron 92, 392–406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bi W., et al. , Inactivation of Rai1 in mice recapitulates phenotypes observed in chromosome engineered mouse models for Smith-Magenis syndrome. Hum. Mol. Genet. 14, 983–995 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Bi W., et al. , Rai1 deficiency in mice causes learning impairment and motor dysfunction, whereas Rai1 heterozygous mice display minimal behavioral phenotypes. Hum. Mol. Genet. 16, 1802–1813 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Feng C., et al. , TCF 20 dysfunction leads to cortical neurogenesis defects and autistic‐like behaviors in mice. EMBO Rep. 21, e49239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng S., et al. , Molecular basis for bipartite recognition of histone H3 by the PZP domain of PHF14. Nucleic Acids Res. 49, 8961–8973 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yan K., et al. ; DDD Study; CAUSES Study, Mutations in the chromatin regulator gene BRPF1 cause syndromic intellectual disability and deficient histone acetylation. Am. J. Hum. Genet. 100, 91–104 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tillotson R., Bird A., The molecular basis of MeCP2 function in the brain. J. Mol. Biol. 432, 1602–1623 (2019). [DOI] [PubMed] [Google Scholar]
- 73.Guy J., Hendrich B., Holmes M., Martin J. E., Bird A., A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat. Genet. 27, 322–326 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper have been deposited in the Gene Expression Omnibus database (accession no. GSE179229). All other data are included in the manuscript and/or supporting information.