Significance
Biofilms are multicellular, soft microbial communities that are able to colonize synthetic surfaces as well as living organisms. To survive sudden environmental changes and efficiently share their common resources, cells in a biofilm divide into subgroups with distinct functions, leading to phenotypic heterogeneity. Here, by studying intact biofilms by synchrotron X-ray diffraction and fluorescence, we revealed correlations between biofilm macroscopic, architectural heterogeneity and the spatiotemporal distribution of extracellular matrix, spores, water, and metal ions. Our findings demonstrate that biofilm heterogeneity is not only affected by local genetic expression and cellular differentiation but also by passive effects resulting from the physicochemical properties of the molecules secreted by the cells, leading to differential distribution of nutrients that propagate through macroscopic length scales.
Keywords: biofilms, small angle X-ray scattering (SAXS)/wide angle X-ray scattering (WAXS), extracellular matrix, functional amyloid proteins, phenotypic heterogeneity
Abstract
Biofilms are multicellular microbial communities that encase themselves in an extracellular matrix (ECM) of secreted biopolymers and attach to surfaces and interfaces. Bacterial biofilms are detrimental in hospital and industrial settings, but they can be beneficial, for example, in agricultural as well as in food technology contexts. An essential property of biofilms that grants them with increased survival relative to planktonic cells is phenotypic heterogeneity, the division of the biofilm population into functionally distinct subgroups of cells. Phenotypic heterogeneity in biofilms can be traced to the cellular level; however, the molecular structures and elemental distribution across whole biofilms, as well as possible linkages between them, remain unexplored. Mapping X-ray diffraction across intact biofilms in time and space, we revealed the dominant structural features in Bacillus subtilis biofilms, stemming from matrix components, spores, and water. By simultaneously following the X-ray fluorescence signal of biofilms and isolated matrix components, we discovered that the ECM preferentially binds calcium ions over other metal ions, specifically, zinc, manganese, and iron. These ions, remaining free to flow below macroscopic wrinkles that act as water channels, eventually accumulate and may possibly lead to sporulation. The possible link between ECM properties, regulation of metal ion distribution, and sporulation across whole, intact biofilms unravels the importance of molecular-level heterogeneity in shaping biofilm physiology and development.
Biofilms are multicellular microbial communities composed of cells that encase themselves in a secreted network of biopolymers and attach to surfaces and interfaces (1–3). Bacterial biofilms may be detrimental in hospital and industrial settings, but they are beneficial in agricultural settings as well as in food technologies contexts. They grow on natural surfaces as diverse as rocks, plant roots, and teeth (4–6), but they also develop on synthetic surfaces, such as medical devices, drinking water distribution, and desalination systems (7, 8). Adopting a communal form of life, cells in biofilms organize in space and time into functionally distinct microenvironments, rendering biofilms with phenotypic heterogeneity. Phenotypic heterogeneity, the expression of specific genes in subpopulations of otherwise genetically identical cells (9–14), is related with chemical gradients arising from consumption and production processes of oxygen, nutrients, and pH changes (15), as well as local processes, such as cell death (16), presence of antibiotics (17), starvation (18), and even random activation of dedicated DNA regions (19). In recent years, it has become evident that phenotypic heterogeneity serves as a survival strategy both for gram-negative and gram-positive bacteria because it ensures the selection of at least a subpopulation in constantly changing environmental conditions (12, 19–21).
In Bacillus subtilis biofilms (B. subtilis), a model organism for biofilm formation, phenotypic heterogeneity is exhibited by the formation of subgroups of cells expressing genes related with motility, extracellular matrix (ECM) production, sporulation (10, 22), and cannibalism (23, 24). Specifically, differential expression of ECM genes leads to regions with different mechanical properties (25–27). Importantly, as also observed in Escherichia coli (28), Pseudomonas aeruginosa (29, 30), and Vibrio cholerae (31) biofilms, matrix gene expression in B. subtilis biofilms influences biofilm architecture, leading to local formation of wrinkles that are absent in matrix mutant biofilms (22, 25, 29, 32). In B. subtilis biofilms, these wrinkles act as water channels, laterally transporting water and nutrients between biofilm subgroups (33).
While phenotypic heterogeneity has been related with subgroup gene expression heterogeneity, a question that remains unanswered is whether spatiotemporal heterogeneity in biofilms is also observed at the molecular level through elemental and structural variations across the biofilms themselves. Although biochemical studies generated compositional and structural knowledge of ECM (34–40) and spore components (41, 42), these studies were mostly performed in vitro with isolated molecules out of the biofilm context.
To study molecular heterogeneities within intact biofilms, we used microfocus X-ray diffraction (XRD) simultaneously with X-ray fluorescence (XRF) to scan along intact B. subtilis biofilm samples of different ages. XRD revealed an internal, spatiotemporal division to subgroups of spores and ECM components that depended mainly on the location and hydration level of the biofilm. Simultaneous XRF measurement allowed parallel tracking of the variations in metal ion composition across biofilms. XRF measurements further exposed variable preference of biofilms, ECM components, and spores to metal ions, mainly calcium, magnesium, manganese, iron, and zinc. Surprisingly, the XRD signature of spores and water, as well as the XRF signals of Zn, Mn, and Fe, were colocalized at the biofilm wrinkles. These findings allowed us to portray a comprehensive, multiscale molecular view of multicellularity in biofilms, demonstrating a link between biofilm morphology, molecular structure, metal ions preference, and the presence of spores, that mirrors the division of labor of subgroups across whole biofilms in situ.
Results
XRD of Intact, Mature B. subtilis Biofilms Reveals Dominant Diffraction Pattern of Spore Components and Water Molecules.
Scanning XRD/XRF with a microfocused X-ray beam is a powerful tool that allows probing local molecular structure together with elemental mapping of heterogeneous samples with high spatial resolution. We scanned a 2.6 × 6 mm2 area of an intact, sealed, and naturally hydrated biofilm using a microfocused X-ray beam, as described in the schematic presented in Fig. 1. We chose a mature, wild-type (WT) biofilm sample (10 d old) in which large (millimeter scale) wrinkles were preserved during sample preparation (Fig. 1). For each position along the scanned region (X-ray beam cross-section = 50 × 50 µm2 and step size = 300 and 200 μm on x-axes and y-axes, respectively), we simultaneously obtained a two-dimensional (2D) diffraction pattern collected on an area detector and an XRF spectrum. Simultaneous collection of XRD and XRF signals enabled correlating structural (XRD) and elemental composition (XRF) variations across a biofilm sample. The 2D XRD patterns were mostly isotropic (see, for example, the 2D diffraction pattern in Fig. 2A), indicating that within the biofilm plane there was no preferred alignment of structural components. We therefore azimuthally integrated the 2D patterns (in the full range 0 to 360°) to obtain a one-dimensional (1D) profile of the intensity (I) versus scattering vector (q). Peaks on the 1D profile correspond to reflections from ordered structures in the biofilm, with characteristic d-spacings (units of length) in real space inversely related to q ().
Fig. 1.
Schematic representation of the experimental set-up. Intact biofilms (A) are cut into slices (B) that are placed between an X-ray source and a 2D area detector (C) in transmission geometry. (D) XRD signal is collected simultaneously with XRF using a fluorescence detector located almost perpendicular to the incoming beam. The X-ray beam scans the sample in preset step sizes along the x and y directions, defining pixels of Δx × Δy areas (see for example the black-colored pixel in B). Scanning over the large area provides structural and compositional information with spatial resolution that is determined by the size of the beam cross-section and the scanning step size. and are the direct beam and scattering vectors, respectively. is defined as the difference between and . The image is not drawn to scale, and the pictures used are for illustration purposes only. Random areas in the biofilm are mapped in each biofilm during an experiment.
Fig. 2.
XRD of an intact, hydrated, and fixed–dry WT biofilm. (A) Full detector image showing a characteristic 2D pattern obtained from a natively hydrated biofilm; the green arrowhead shows the water-scattering signal. (B) Zoom into the low-q range, showing the dominant doublet (marked with a blue arrowhead in A and B); the orange arrowhead marks the sample carrier (polyimide film) signal. White grid lines are gaps between detector units. (C) Representative, azimuthally integrated 1D diffraction profiles of a native, hydrated (dark and light blue curves taken from a wrinkle and bulk region, respectively), and fixed–dry (black curve) biofilms. The intensity (arbitrary units [Arb. U]) versus q (nm−1) profiles show a doublet peak in the low-q range (marked with rectangle and corresponding to the blue arrow in A and B) and broad signals in the high-q range (marked with green arrowhead in A). Dashed lines point at peak positions q = 13.8 nm−1, attributed to biofilm biopolymers, and q = 19 and 26 nm−1, attributed to water-scattering signal. (D) Intensity map across a 2.6 × 6 mm2 area of peak (ii) showing higher intensity along biofilm's wrinkles. (E) Mapping the intensity of the dominant peaks in the high-q range, q = 14 nm−1 and q = 19 nm−1 highlight the drying versus hydrated areas, respectively; water-scattering signal at 19 nm−1 is highest along the biofilm wrinkles, which serve as water channels (33). (F) Mapping of the q position of the low-q peak at ∼6.7 nm−1 [peak (ii), in C], corresponding to ∼9.4 Å d-spacing across a WT biofilm. (G) Zoom into the low-q range (marked with a rectangle in C), showing the average profiles in different areas in the biofilm sample (X, Y, and Z in F corresponding to the blue, red, green curves, respectively) and the average profiles in a fixed and dry biofilm (black curve) and in a spore (dry) sample (olive green curve). In fixed–dry samples of 72-h WT biofilms and in isolated spore samples (Fig. 2C), the d-spacings were the smallest.
The diffraction patterns of the intact, hydrated, 10-d-old, WT biofilm sample showed scattering signals in both low- (q = 5 to 8 nm−1) and high- (q = 10 to 29 nm−1) q ranges (Fig. 2 A–C). In the low-q range (marked with a blue arrowhead in Fig. 2 A and B and a rectangle in Fig. 2C), WT biofilms exhibited several reflections residing around ∼6 nm−1, corresponding to the structural order with d-spacing of the order of 1 nm in real space, which typically contained a sharp doublet superimposed on a broader hump (Fig. 2G and SI Appendix, Fig. S1 and Tables S1 and S2). The low-q XRD pattern of dry, WT biofilms largely corresponded to the XRD profile reported before for isolated spore samples (42) and confirmed by us (Fig. 2G and SI Appendix, Fig. S3).
To evaluate the variation in specific reflections across the sample, we fitted the dominant scattering signals in the 1D XRD profiles, yielding peak position, width, and amplitude in each of the measured pixels (position in the biofilm) (SI Appendix, Fig. S1 and Tables S1 and S2). Mapping the spatial variation of the dominant doublet reflections in the biofilm, the doublet appeared across the whole biofilm sample, but its intensity [peak (i) in SI Appendix, Fig. S2A and peak (ii) in Fig. 2D] was highest along large wrinkles in the biofilms relative to areas away from or between the wrinkles (Fig. 2D). The relative intensities of the two peaks comprising the doublet were similar across the biofilm sample (SI Appendix, Fig. S2C), implying that they originate from a single-spore component or from components with similar, relative abundance. The doublet peak positions were rather uniform across the measured area, ranging between 6.19 to 6.24 nm−1 [peak (i) in SI Appendix, Fig. S2B] and 6.63 to 6.70 nm−1 [peak (ii) in Fig. 2F], yet they exhibited a gradient, shifting to lower-q values (larger d-spacings) along the direction going from the top left corner of the biofilm-sampling area to the bottom right (Fig. 2F).
In the high-q range, the diffraction profiles of the hydrated sample showed typically broad peaks that were deconvoluted into three or four dominant reflections. Two reflections were fitted around 14 nm−1, and two reflections were centered around 19 and 26 nm−1 (Fig. 2C and SI Appendix, Fig. S4). The broad peak around 14 nm−1 was present in all the biofilms we tested, namely, fixed and dry (curve “dry bulk” in Fig. 2C), as well as natively hydrated biofilms (“hydrated bulk” and “hydrated wrinkle” in Fig. 2C). It includes reflections from biopolymers present in the ECM and in ordered structures inside the cells. For example, the sugar–phosphate backbone in DNA, which is present both inside and outside the cells in the biofilm (43), gives rise to a characteristic reflection at 15 nm−1 (44), and polysaccharides purified from biofilms showed typical scattering at q ∼13 nm−1 (SI Appendix, Fig. S5).
In addition to the contribution of biopolymers to the XRD profile, native, hydrated biofilms showed contributions around q = 19 nm−1 and q = 26 nm−1, corresponding to scattering from water (45, 46). In some regions, especially along wrinkles (curve hydrated wrinkle; Fig. 2C), fitting results showed that the relative amplitudes of these two peaks are comparable to those of pure liquid water, in which the scattering signal originates from tetrahedral packing of water, with typical distance between oxygen atoms around 2.8 Å (46, 47) (SI Appendix, Table S1 and Fig. S4). While free water gives rise to both reflections at 19 and 26 nm−1, bound hydration water layers in proteins lose the tetrahedral packing, and as a result, they give rise only to scattering signal around q = 19 nm−1 (45). Strikingly, in bulk areas away from the wrinkles (curve hydrated bulk), the free water signal at 26 nm−1 was diminished (Fig. 2C and SI Appendix, Fig. S4), leading us to conclude that the contribution at q ∼19 nm−1 results from water bound to ECM and cellular biopolymers. These findings stand in agreement with a previous study that showed that biofilm wrinkles serve as channels filled with water and essentially function to transfer nutrients across biofilms (33). In biofilms aged older than 6 d, these channels become closed tubes (33), which can explain the entrapment of free water that we observe with XRD in the 10-d-old biofilm section that was removed from its original substrate.
Interestingly, the large spatial variation in the intensity of the water signal (Fig. 2E) reflects dehydration during the measurement period. Mapping the intensity of the peak contributions in the high-q range across the sample (Fig. 2E) revealed a dehydration gradient spanning along the diagonal of the biofilm sample from the top left part of the sample, where the biopolymers reflection was dominant, to the bottom right part of the sample, where the water reflection at 19 nm−1 dominated. In the hydrated regions, the 14 nm−1 peak was still visible but partially obscured by the presence of the broad, intense 19 nm−1 peak (water signal) (SI Appendix, Fig. S4 and Table S1). The dehydration gradient provided an interesting insight into hydration/dehydration processes in biofilm spores, as the drying front coincided with shifting of the spore-related doublet peak position to smaller atomic separations (larger q values), as shown in Fig. 2 F and G. This strengthens the suggestion that the doublet signal is attributed to highly organized spore coat proteins (41, 42, 48, 49) and therefore sensitive to biofilm hydration. An intriguing hypothesis then arises that spore coat protein swelling may serve as humidity sensors for spore germination.
Loss of Spore-Associated Reflections Exposes Weak Cross–β-sheet Structure that Is Also Observed in TasA Fibers Formed In Vitro.
The dominant contribution of the spores to the XRD signal concealed a broader hump in the low-q region (SI Appendix, Fig. S1). We wondered whether this hump originates from larger d-spacings in ECM components, in addition to those we identified at ∼5 Å (corresponding to q ∼14 nm−1). Therefore, to uncover the biofilm matrix XRD signal, we performed a temporal analysis of the biofilm XRD at early time points, namely at 12 and 24 h, in which WT biofilms express matrix to a variable degree, but they do not yet produce spores (22). 1D XRD profiles of 24-h-old biofilms indeed showed that the spore-associated doublet was not yet present in young biofilms (Fig. 3A, green curve and SI Appendix, Table S1). Instead, at 24 h, broad peaks appeared at ∼6 and ∼14 nm−1, attesting the contribution of matrix and cells to the XRD signal. At 12 h, the broad hump around 6 nm−1 was only occasionally present, and some regions showed only scattering signal at high q (Fig. 3A, cyan and blue curves and SI Appendix, Table S1). Indeed, at 12 h matrix, expression is initiated but the matrix is not yet uniform across the whole biofilm area (22). Probing the samples at this time point allowed us to isolate the XRD signature of cells with little contribution of the matrix. The loss of the low-q hump in these biofilms indicated that it depends on the presence of matrix components and that the cells themselves do not contribute to the signal at low q. We, therefore, deduce that the ECM contribution to the scattering signal contains a low-q reflection at q ∼6 nm−1 and likely an additional component at ∼14 nm−1 that reflects the (intra)molecular arrangement of the polymers. The combination of such reflections typically corresponds to the canonic cross–β-structure attributed to a 10- (intersheet) and 4.7-Å (interstrand) spacings, respectively (50).
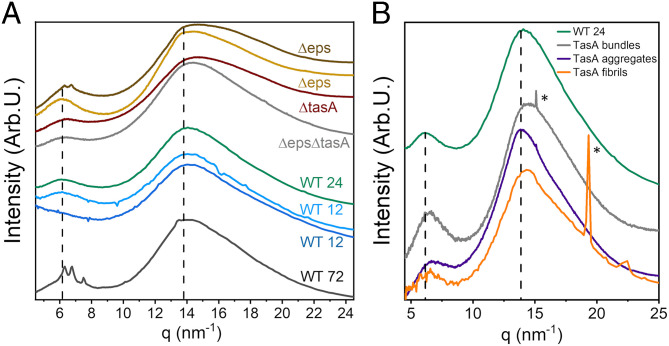
Fig. 3.
Comparison of the XRD of mature WT biofilms, young biofilms, and ECM mutant biofilms. (A) XRD of fixed, mature (72 h old) WT biofilms; fixed, mature biofilms made by matrix mutant cells Δeps and ΔtasA, double-mutant ΔepsΔtasA, as well as by fixed, young biofilms (aged 12 and 24 h), in which spores have not yet developed. The dashed line marks the position of the intersheet and interstrand peaks in cross β-sheet structures (q ∼6 and 14 nm−1). (B) XRD profiles of TasA aggregates (purple curve), fibrils (orange curve), and bundles (gray curve), along with the XRD profile of 24-h-old WT biofilms (green curve). The sharp peaks in the fibrils’ profile, marked with *, formed at high-NaCl concentration are attributed to salt crystal impurities.
TasA is the major matrix protein in B. subtilis biofilms and was reported to form amyloid fibers in vitro (34, 36, 51). We therefore also studied the XRD signal in matrix mutant biofilms, ΔtasA, as well as in the matrix mutant Δeps that lacks the gene responsible for the production of an exopolysaccharide and the double mutant, ΔepsΔtasA, that lack both matrix components. In addition, matrix mutant biofilms were reported to negligibly express sporulation genes (22), as we also verified using transmission electron microscopy (TEM) imaging (SI Appendix, Fig. S6). The TEM images in SI Appendix, Fig. S6 as well as a quantitative analysis of the cell types in different TEM images (SI Appendix, Fig. S6D) show clearly that the abundance of sporulating cells and spores in biofilms made by the ECM mutants, Δeps and ΔtasA, is negligible relative to their abundance in WT biofilms. The stark reduction in spore and the increased numbers of lysed cells in ECM mutant biofilms (SI Appendix, Fig. S6) suggest increased cannibalism in matrix mutant biofilms, as previously proposed (24).
In resemblance to the XRD profiles of young biofilms, the XRD profiles in tasA mutant biofilms (burgundy curve in Fig. 3A) as well as in the eps mutant biofilms (orange curve in Fig. 3A) showed loss of the spore-associated doublet. We note that some eps mutant biofilms also exhibited profiles in which the doublet was apparent as low-intensity peaks superimposed on top of a broad hump (brown curve in Fig. 3A).
We used curve fitting to estimate the contribution of the cross–β-structure, for example, intersheet and interstrand peaks at q ∼6 and 14 nm−1, respectively (see dashed lines in Fig. 3A), to the XRD profile of WT biofilms of different ages as well as of matrix mutant biofilms (SI Appendix, Tables S1 and S2 and Fig. S1). Whereas the 14-nm−1 reflection combines contributions from the 4.7-Å (interstrand) spacing in cross–β-structures and, additionally, from similar spacings originating from other biopolymers, the 6-nm−1 contribution originates solely form the cross–β-structures. Therefore, the intensity ratio (low q/high q) is indicative of the relative abundance of the cross–β-signature in the sample. Indeed, both reflections appeared in fixed, mature, WT biofilms and in matrix-mutated biofilms (Δeps, ΔtasA, and ΔepsΔtasA); however, the intensity ratio (SI Appendix, Table S1) was lower in mutants that lacked TasA (Fig. 3A, burgundy curve). We conclude, therefore, that intact biofilms harbor a cross–β-structure that results from matrix components, and although it is not exclusive to TasA, the latter is likely the major contributor to the cross–β-structure.
Interestingly, in eps mutant biofilms, wrinkles are absent despite the occurrence of structure in the remaining matrix components. This observation suggests that wrinkle formation depends on the cooccurrence of all the matrix components.
We were therefore encouraged to compare the structure of fibers formed in vitro from isolated TasA preparations with the general structural features of the ECM in intact biofilms. Recently, we have shown that TasA is polymorphic, as it forms fibers with different morphologies: aggregates in acidic solutions (termed “aggregates” henceforth), thin and long fibrils at high-salt concentrations (termed “fibrils” henceforth) and fiber bundles at high-protein and -salt concentration (termed “bundles” henceforth) (36, 52, 53). Fig. 3B shows the averaged, 1D, azimuthally integrated XRD signatures of isolated TasA that formed fibers in vitro under these three conditions. The XRD profiles of the three preparations of TasA fibers exhibited two broad reflections close to q ∼6 and ∼14 nm−1, corresponding to the canonic cross–β-sheet structure. Peak fitting shows that these peaks were slightly shifted to higher q, relative to those of 24-h-old, WT biofilms (marked by vertical dashed lines, see also SI Appendix, Table S1), indicating a more compact packing of TasA fibers formed in vitro relative to biopolymers within the biofilm. Of the three polymorphs, the position of the low-q peak in fibrils samples was closest to that observed in young biofilms in situ (6.25, 6.26, and 6.16 nm−1 for fibrils; 12-h WT biofilm; and 24-h WT biofilms respectively, SI Appendix, Table S1).
Diehl and coworkers reported XRD patterns of TasA fibers formed from recombinant protein in acidic conditions displaying narrow intersheet and interstrand reflections of a cross–β-sheet configuration (51), which, in our case, are very broad. While the combination of these two reflections is indicative of cross–β-sheet features, it is unlikely that they organize into a long-range, ordered amyloid structure in our case. This is consistent with our previous suggestions that TasA fibers form by oligomer-to-oligomer addition (53), but our data are insufficient to suggest a molecular model for the interactions between monomers and/or oligomers. We attribute the disparity in the molecular organization of TasA fibers between that reported here and that demonstrated for amyloid fibers to the fact that our fibrils are composed of full-length proteins, rather than short peptides (50, 54–56) or truncated (recombinant) sequences (51). Considering that NMR studies of TasA and its crystal structure demonstrated that it contains a core of β-strands arranged in a jelly roll fold, as well as alpha helices and loops (51), the cross–β-structure may arise from the internal core or from packing of intermolecular β-strands or -sheets (37). Importantly, the similarity between the XRD patterns of TasA fibers, formed in vitro, and the matrix XRD signal, obtained in situ, suggests that in intact biofilms TasA and other ECM components form ordered fibers that are not necessarily amyloidogenic.
Metal Ions Preferentially Accumulate along Biofilm Wrinkles as well as in Isolated TasA.
Following the discovery that macroscopic biofilm morphology is followed by structural heterogeneity at the molecular level, we wondered whether the elemental composition of the biofilms is as heterogeneous and whether it also tracks the morphology of biofilms. Previous work of us and others have shown that metal ions accumulate in biofilms (57–59); however, their spatial distribution across intact biofilms has not been addressed.
XRF signal results from the emission of X-ray photons following electron relaxation after its excitation by the X-ray beam, and it is indicative of the elemental composition of a sample. The XRF signal from biofilms was recorded concurrently with XRD to probe the distribution of accumulated metal ions across biofilms (Fig. 4). The fluorescence photon counts were then converted to relative atomic density, taking into account the energy-dependent absorption cross-section for each element (60–63). While Ca was the most abundant metal ion in all samples, and was rather homogenously distributed across the biofilms, we found an interesting spatial distribution for Zn, Fe, and Mn with increased signals along the biofilm wrinkles. A similar trend was observed for Mg (SI Appendix, Fig. S2D); however, the signal to noise in this measurement was relatively low because of the low-fluorescence emission energy of Mg, leading to significant absorption in air. For comparison, the distributions of Ca and K (Fig. 4 and SI Appendix, Fig. S2E, respectively) did not correlate with the wrinkle morphology. The uniform potassium distribution reported here is consistent with a previous report of extracellular potassium distribution and its involvement in electrical communication within B. subtilis biofilms (64).
Fig. 4.
Atomic density measured by XRF of intact, hydrated WT biofilm slice. XRF/elemental distribution maps are plotted for each metal ion. The Look Up Table (LUT) describes the XRF counts (values to the left of the scale) and the corrected fluorescence signals (values to the right of the scale) of Mn (blue), Zn (green), Fe (red), and Ca (magenta) of WT biofilms. The signal of Zn, Mn, and Fe, but not Ca, is stronger in the biofilm wrinkles, coinciding with water and spore accumulation along biofilm wrinkles.
The elevated signal of Zn, Mn, and Fe coincided with increased intensity of the spore-related doublet XRD signature and water scattering acquired simultaneously. A possible explanation of the increased XRD signal along the wrinkles could be that the biofilm is thicker along wrinkles; however, this effect alone would lead to an increase of the XRF signal of all the metal ions. The uniform Ca and K signals across the biofilms makes this possibility less probable.
While the XRF signal cannot be quantified into absolute values, after corrections for the absorption cross-section, the relative abundance of metal ions can be compared to obtain the general trends in ion abundance. We therefore evaluated the atomic abundance of Zn, Mn, and Fe ions relative to the most abundant Ca ion in areas positioned at the biofilm bulk (away from the wrinkles) and compared them to the relative concentrations of these metal ions in the medium (SI Appendix, Table S3).
We found that Ca accumulation is enhanced relative to the other metals ions, even after considering its higher abundance in the medium (SI Appendix, Table S3). This suggests that Ca is preferentially bound in the biofilm, thus maintaining a high-concentration gradient relative to the medium. Ca binding by the ECM was already shown before and suggested to stabilize the biofilm and inhibit biofilm dispersal (59). In contrast, Zn, Mn, and Fe ions are more free to diffuse and flow as solutes in water channels, and they accumulate along wrinkles, in which evaporation is enhanced (33). Strikingly, the relative abundance of Zn, Mn, and Fe, with respect to Ca, is similar in ECM mutant biofilms and in the bulk of WT biofilms (away from wrinkles), and yet sporulation is detained. We suggest that even though the metal ions are accumulated across the film, the lack of wrinkles and lateral water flow in the mutant films prevents localized accumulation of some metal ions with respect to Ca and that these altered Ca/metal ion ratios are crucial for sporulation. Interestingly, all these metal ions (Ca, Zn, Mn, and Fe) have been described to play a role in sporulation (59, 65, 66), and they also accumulated in isolated spore samples (SI Appendix, Fig. S7).
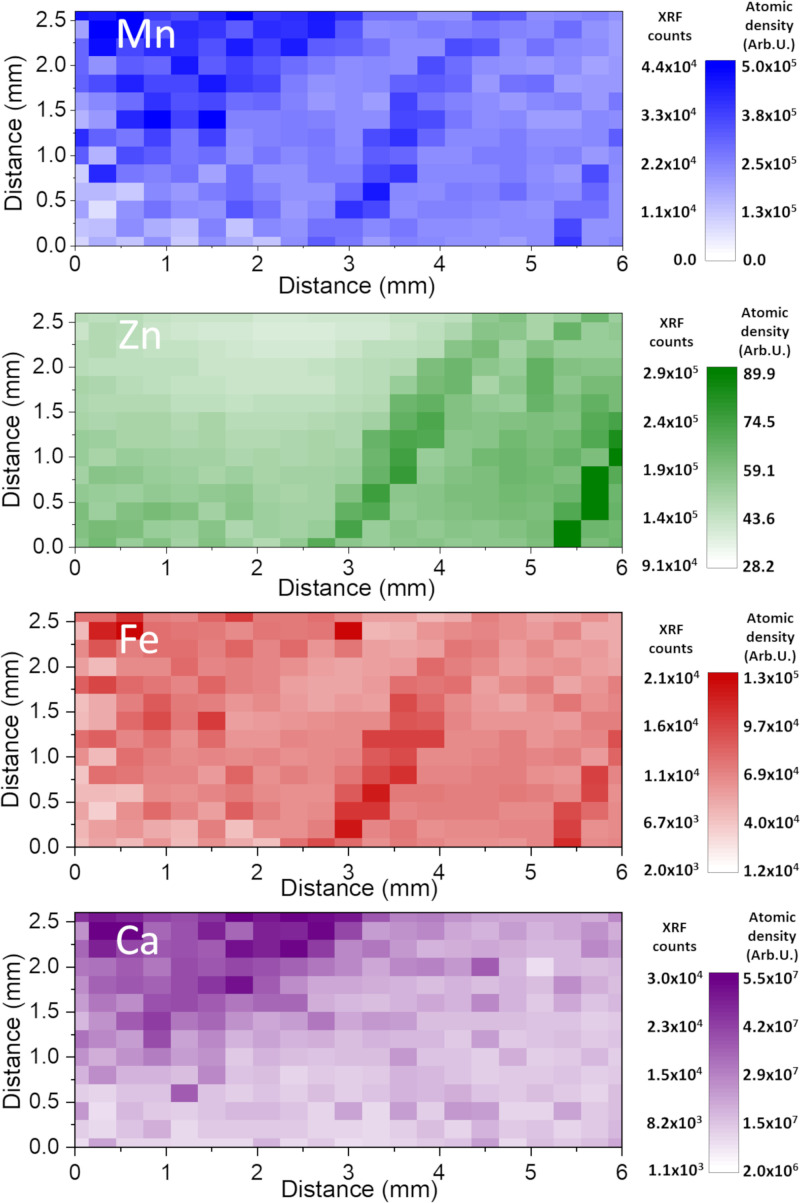
We speculated that the ECM binds Ca, in similarity with charged polyelectrolytes that often bind polyvalent ions (67, 68). To examine this possibility, we investigated protein fibers of isolated TasA preparations with XRF. XRF analysis revealed that TasA fibers contained all the metal ions: Ca, Zn, Fe, and, in some cases, Mn that were also probed in WT and mutant biofilms (Fig. 5). However, different TasA polymorphs showed different preference to these metal ions, as shown in an overlay of the XRF maps of these metal ions (Fig. 5 D–F and SI Appendix, Fig. S8). In particular, in aggregates that formed in an acidic environment, iron binding dominated over Ca, Mn, and Zn (Fig. 5D), whereas bundles that formed in high-salt (NaCl) and -protein concentrations accumulated more Ca relative to Zn, Mn, and Fe (Fig. 5F and SI Appendix, Fig. S8). The fibrils that formed at high-salt concentration were especially heterogeneous, as was reflected both in different morphologies and in metal ion–binding preferences residing in the same sample (Fig. 5E). These results suggest that some part of the metal ions in biofilms, especially Ca, are bound by TasA within the matrix. The variations in the XRF signal of TasA fibers, prepared under three different environmental conditions, is a further testimony of the protein’s polymorphism and its possible contribution to biofilm heterogeneity.
Fig. 5.
Morphology and metal ion binding of TasA fibers formed in different environments. (A–F) XRF of TasA fibers formed in acidic conditions (“TasA aggregates”), high-salt concentration (“TasA fibrils”), and high-protein and salt concentration (“TasA bundles”). XRF maps of Ca (A–C) are presented separately from the overlay of the XRF maps of Zn, Fe, and Mn (D–F) because of its higher abundance (atomic density) in all the samples. Color code: Ca (magenta), Zn (green), Mn (blue), and Fe (red); red and green = yellow. (Scale Bar, 300 µm.)
Discussion
Spatiotemporal analysis of molecular structures within B. subtilis biofilms reveals that cellular spatial differentiation by function also translates into structural and elemental heterogeneity across whole biofilms at the molecular level. Probing intact biofilms with XRD, we have found that B. subtilis biofilms convey three characteristic structural signatures originating from spores, ECM components, and water in bound and free state. Concomitant spatial elemental analysis by XRF showed that the metal ions, mainly, Ca, Zn, Mn, and Fe, known to play a crucial role in bacterial metabolism and sporulation (69), differentially accumulate in B. subtilis biofilms. While Ca and K are evenly distributed across the biofilms, other metal ions, mainly Zn, Mn, and Fe, accumulate along biofilm wrinkles.
Based on our observations, we suggest an inclusive view of biofilm development linking between macroscopic features, namely biofilm wrinkles, to elemental and structural heterogeneities in biofilms, as illustrated schematically in Fig. 6. We suggest a dual role for the ECM in biofilms. It provides structural support and leads to wrinkle formation, and it also serves as filter, selectively binding Ca over other metal ions. These two properties result with heterogeneous distribution of metal ions and spores, as detailed in the next paragraph.
Fig. 6.
Schematic representation of the relationships between biofilm structures, metal ion distribution, and their implications on biofilm physiology. (A) A side (cross-section) view of a WT B. subtilis biofilm with a central water-filled (blue) wrinkle. Bacterial cells are represented by brown ovals; spores are represented by circles with a dark brown circumference. The straight arrows represent water and nutrient uptake into the biofilm, and the curled arrows represent water evaporation from the biofilm surface. (B) The biofilm is composed of cells and ECM (represented by brown background and darker lines). (C) TasA fibrils in the ECM contain short cross β-sheet domains. (D) Zoom into a column across the biofilm wrinkle (black rectangle); water (blue background) and metal ions (we only refer to the metal ions that were observed in this study, Ca: purple and Fe/Zn/K/Mn: red and green) are free inside water channels (Bottom). The metals are adsorbed by the ECM (Middle), with preference to Ca that remains mostly bound to the ECM. Zn, Mn, and Fe are free to diffuse in the matrix, and they concentrate on the biofilm wrinkles (Top), in which water evaporation is the largest. Metal ion accumulation on biofilm wrinkles possibly leads to sporulation. This image is for illustration purposes, and it is not drawn to scale.
Metal ions, initially residing in the medium, accumulate in the ECM biopolymers with preferred Ca binding. This process is driven by water evaporation, which occurs throughout the film (curled blue arrows in Fig. 6). However, Wilking et al. (33) have shown that biofilm wrinkles act as water channels, in which water flow is driven by enhanced evaporation along the wrinkles. The water flow carries nutrients, but, as Ca is selectively bound by the matrix and therefore filtered out, the solution is relatively enriched with Zn, Mn, and Fe, which are only weakly bound by the matrix or taken up by cells. These ions eventually accumulate along the wrinkle because of water evaporation. The cooccurrence of these metal ions and spores along wrinkles is consistent with their essential role in sporulation (65, 70, 71) and points at the possibility that spores act as a drainage for metal ions, as a means to circumvent toxicity. Our study also suggests that, in the absence of wrinkles in ECM mutant biofilms, a Ca/metal ion ratio required for sporulation cannot be achieved, providing reason to the delayed sporulation in ECM mutant biofilms.
Our model therefore offers a functional link between ECM properties, macroscopic wrinkles, and sporulation via heterogenic metal ions distribution, showing that biofilm heterogeneity is not only affected by genetic expression and cellular differentiation but also by passive processes that stem from physicochemical properties of molecules secreted by the cells in the biofilm. These lead to a differential distribution of nutrients that propagates through macroscopic length scales. Multiscale approaches to biofilm internal structures and metal ion relationships may hold the key to understanding biofilm physiology and multicellularity and their relation to subpopulation survival in B. subtilis, as well as in other biofilms of single or mixed bacterial species.
Materials and Methods
XRD/XRF measurements of biofilms and proteins were performed at the mySpot beamline synchrotron BESSY II (Helmholtz Center) and at the ID13 beamline of the European Synchrotron Radiation Facility (ESRF) (Grenoble). Biofilm samples were mounted on aluminum frames overlaid with kapton films (13 μm thick from Goodfellow Cambridge limited or 25 μm thick from DuPont) or on a silicon nitride membrane (Silson Ltd., frame: 10.0 × 10.0 mm, 200 μm thick; membrane: 5.0 × 5.0 mm, 1 μm thick). Protein samples measured at ESRF were sandwiched between two 1-μm-thick silicon nitride membranes. Detailed information on the experimental methods can be found in SI Appendix.
Supplementary Material
Acknowledgments
Measurements were carried out at the mySpot beamline at the BESSY II electron storage ring operated by the Helmholtz-Zentrum Berlin für Materialien und Energie and at beamline ID13 at the European Synchrotron Radiation Facility. We thank the Helmholtz-Zentrum Berlin (HBZ) and the ESRF for provision of synchrotron radiation facilities and for financial support. We thank the Electron Microscopy Facility of Max Planck Institute of Molecular Cell Biology and Genetics (MPI-CBG) for support and use of equipment and reagents and Daniel Werner at the Max Planck Institute of Colloids and Interfaces (MPIKG) for X-ray measurements. We thank Jiliang Liu for ESRF ID13 beamline assistance at ESRF. Special thanks to Prof. Sigal Ben Yehuda for insightful discussions, Yosef Edery Aharony for his contribution with data sorting and image analysis, and Daniel Rosenblatt for assistance with the image analysis. D.N.A. acknowledges the support of the Kaete Klausner PhD scholarship and M.G. acknowledges the support of the Neubauer Foundation for the PhD fellowship.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2118107119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Romeo T., “Preface” in Bacterial Biofilms, Romeo T., Ed. (Springer-Verlag Berlin Heidelberg, ed. 1, 2008). [Google Scholar]
- 2.Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., Lappin-Scott H. M., Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745 (1995). [DOI] [PubMed] [Google Scholar]
- 3.O’Toole G., Kaplan H. B., Kolter R., Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Beauregard P. B., Chai Y., Vlamakis H., Losick R., Kolter R., Bacillus subtilis biofilm induction by plant polysaccharides. Proc. Natl. Acad. Sci. U.S.A. 110, E1621–E1630 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y., et al. , Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ. Microbiol. 15, 848–864 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall-Stoodley L., Costerton J. W., Stoodley P., Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Vickery K., Hu H., Jacombs A. S., Bradshaw D. A., Deva A. K., A review of bacterial biofilms and their role in device-associated infection. Healthc. Infect. 18, 61–66 (2013). [Google Scholar]
- 8.Prest E. I., Hammes F., van Loosdrecht M. C. M., Vrouwenvelder J. S., Biological stability of drinking water: Controlling factors, methods, and challenges. Front. Microbiol. 7, 45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence J. R., Korber D. R., Wolfaardt G. M., Heterogeneity of natural biofilm communities. Cells and Mater. 6, 175–191 (1996). [Google Scholar]
- 10.Dragoš A., et al. , Division of labor during biofilm matrix production. Curr. Biol. 28, 1903–1913.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jorth P., Spero M. A., Livingston J., Newman D. K., Quantitative visualization of gene expression in mucoid and nonmucoid Pseudomonas aeruginosa aggregates reveals localized peak expression of alginate in the hypoxic zone. MBio 10, e02622-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reyes Ruiz L. M., Williams C. L., Tamayo R., Enhancing bacterial survival through phenotypic heterogeneity. PLoS Pathog. 16, e1008439 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beebout C. J., et al. , Respiratory heterogeneity shapes biofilm formation and host colonization in uropathogenic Escherichia coli. MBio 10, e02400-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armbruster C. R., et al. , Heterogeneity in surface sensing suggests a division of labor in Pseudomonas aeruginosa populations. eLife 8, e45084 (2019). Correction in: eLife 9, e59154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart P. S., Franklin M. J., Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6, 199–210 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Aguilar B., et al. , Cell death as a trigger for morphogenesis. PLoS One 13, e0191089 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Díaz-Pascual F., et al. , Breakdown of Vibrio cholerae biofilm architecture induced by antibiotics disrupts community barrier function. Nat. Microbiol. 4, 2136–2145 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gingichashvili S., et al. , The adaptive morphology of Bacillus subtilis biofilms: A defense mechanism against bacterial starvation. Microorganisms 8, 62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang X., et al. , Invertible promoters mediate bacterial phase variation, antibiotic resistance, and host adaptation in the gut. Science 363, 181–187 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rainey P. B., Rainey K., Evolution of cooperation and conflict in experimental bacterial populations. Nature 425, 72–74 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Martin M., et al. , Cheaters shape the evolution of phenotypic heterogeneity in Bacillus subtilis biofilms. ISME J. 14, 2302–2312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vlamakis H., Aguilar C., Losick R., Kolter R., Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 22, 945–953 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López D., Vlamakis H., Losick R., Kolter R., Cannibalism enhances biofilm development in Bacillus subtilis. Mol. Microbiol. 74, 609–618 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Höfler C., et al. , Cannibalism stress response in Bacillus subtilis. Microbiology (Reading) 162, 164–176 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Otto S. B., et al. , Privatization of biofilm matrix in structurally heterogeneous biofilms. mSystems 5, e00425-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nadell C. D., Drescher K., Wingreen N. S., Bassler B. L., Extracellular matrix structure governs invasion resistance in bacterial biofilms. ISME J. 9, 1700–1709 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Besharova O., Suchanek V. M., Hartmann R., Drescher K., Sourjik V., Diversification of gene expression during formation of static submerged biofilms by Escherichia coli. Front. Microbiol. 7, 1568 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serra D. O., Richter A. M., Klauck G., Mika F., Hengge R., Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. MBio 4, e00103-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Branda S. S., Vik S., Friedman L., Kolter R., Biofilms: The matrix revisited. Trends Microbiol. 13, 20–26 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Sakuragi Y., Kolter R., Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J. Bacteriol. 189, 5383–5386 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fei C., et al. , Nonuniform growth and surface friction determine bacterial biofilm morphology on soft substrates. Proc. Nat. Acad. Sci. USA 117, 7622–7632 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trejo M., et al. , Elasticity and wrinkled morphology of Bacillus subtilis pellicles. Proc. Natl. Acad. Sci. U.S.A. 110, 2011–2016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilking J. N., et al. , Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U.S.A. 110, 848–852 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romero D., Aguilar C., Losick R., Kolter R., Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U.S.A. 107, 2230–2234 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romero D., Vlamakis H., Losick R., Kolter R., Functional analysis of the accessory protein TapA in Bacillus subtilis amyloid fiber assembly. J. Bacteriol. 196, 1505–1513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chai L., et al. , Isolation, characterization, and aggregation of a structured bacterial matrix precursor. J. Biol. Chem. 288, 17559–17568 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El Mammeri N., et al. , Molecular architecture of bacterial amyloids in Bacillus biofilms. FASEB J. 33, 12146–12163 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Dogsa I., Brloznik M., Stopar D., Mandic-Mulec I., Exopolymer diversity and the role of levan in Bacillus subtilis biofilms. PLoS One 8, e62044 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hobley L., et al. , BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm. Proc. Natl. Acad. Sci. U.S.A. 110, 13600–13605 (2013). Correction in: Proc. Natl. Acad. Sci. U.S.A. 112, E5371–E5375 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobayashi K., Iwano M., BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol. Microbiol. 85, 51–66 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Kadota H., Iijima K., The X-ray diffraction pattern of spores of Bacillus subtilis. Agric. Biol. Chem. 29, 80–81 (1965). [Google Scholar]
- 42.Qiu X., Setlow P., Structural and genetic analysis of X-ray scattering by spores of Bacillus subtilis. J. Bacteriol. 191, 7620–7622 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jakubovics N. S., Shields R. C., Rajarajan N., Burgess J. G., Life after death: The critical role of extracellular DNA in microbial biofilms. Lett. Appl. Microbiol. 57, 467–475 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Zuo X., et al. , X-ray diffraction “fingerprinting” of DNA structure in solution for quantitative evaluation of molecular dynamics simulation. Proc. Natl. Acad. Sci. U.S.A. 103, 3534–3539 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phan-Xuan T., et al. , Hydration-induced structural changes in the solid state of protein: A SAXS/WAXS study on lysozyme. Mol. Pharm. 17, 3246–3258 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skinner L. B., et al. , Benchmark oxygen-oxygen pair-distribution function of ambient water from x-ray diffraction measurements with a wide Q-range. J. Chem. Phys. 138, 074506 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Shi R., Tanaka H., Distinct signature of local tetrahedral ordering in the scattering function of covalent liquids and glasses. Sci. Adv. 5, eaav3194 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plomp M., Carroll A. M., Setlow P., Malkin A. J., Architecture and assembly of the Bacillus subtilis spore coat. PLoS One 9, e108560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bodík M., et al. , Diffraction pattern of Bacillus subtilis CotY spore coat protein 2D crystals. Colloids Surf. B Biointerfaces 197, 111425 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Eisenberg D. S., Sawaya M. R., Structural studies of amyloid proteins at the molecular level. Annu. Rev. Biochem. 86, 69–95 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Diehl A., et al. , Structural changes of TasA in biofilm formation of Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 115, 3237–3242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Azulay D. N., et al. , Colloidal-like aggregation of a functional amyloid protein. Phys. Chem. Chem. Phys. 22, 23286–23294 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Ghrayeb M., Hayet S., Lester-Zer N., Levi-Kalisman Y., Chai L., Fibrilar polymorphism of the bacterial extracellular matrix protein TasA. Microorganisms 9, 529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawaya M. R., et al. , Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 447, 453–457 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Li B., et al. , Cryo-EM of full-length α-synuclein reveals fibril polymorphs with a common structural kernel. Nat. Commun. 9, 3609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nelson R., et al. , Structure of the cross-beta spine of amyloid-like fibrils. Nature 435, 773–778 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ido N., et al. , Bacillus subtilis biofilms characterized as hydrogels. Insights on water uptake and water binding in biofilms. Soft Matter 16, 6180–6190 (2020). [DOI] [PubMed] [Google Scholar]
- 58.van Hullebusch E. D., Zandvoort M. H., Lens P. N. L., Metal immobilisation by biofilms: Mechanisms and analytical tools. Rev. Environ. Sci. Biotechnol. 2, 9–33 (2003). [Google Scholar]
- 59.Nishikawa M., Kobayashi K., Calcium prevents biofilm dispersion in Bacillus subtilis. J. Bacteriol. 203, e00114-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scofield J. H., Relativistic Hartree-Slater values for K and L X-ray emission rates. At. Data Nucl. Data Tables 14, 121–137 (1974). [Google Scholar]
- 61.Scofield J. H., “Theoretical photoionization cross sections from 1 to 1500 keV” (Tech. Rep. UCRL-51326, University of California Radiation Laboratory, Berkley, CA, 1973). [Google Scholar]
- 62.Onder P., Tursucu A., Demir D., K shell X-ray fluorescence parameters of some elements in the atomic range. Sci. Technol. Nucl. Install. 2013, 1–6 (2013). [Google Scholar]
- 63.Krause M. O., Atomic radiative and radiationless yields for K and L shells. J. Phys. Chem. Ref. Data 8, 307–327 (1979). [Google Scholar]
- 64.Prindle A., et al. , Ion channels enable electrical communication in bacterial communities. Nature 527, 59–63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kwak M.-K., Ryu H.-B., Song S.-H., Lee J.-W., Kang S.-O., Anti-σ factor YlaD regulates transcriptional activity of σ factor YlaC and sporulation via manganese-dependent redox-sensing molecular switch in Bacillus subtilis. Biochem. J. 475, 2127–2151 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Chung P. Y., Khanum R., Antimicrobial peptides as potential anti-biofilm agents against multidrug-resistant bacteria. J. Microbiol. Immunol. Infect. 50, 405–410 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Borkovec M., Koper G. J. M., Piguet C., Ion binding to polyelectrolytes. Curr. Opin. Colloid Interface Sci. 11, 280–289 (2006). [Google Scholar]
- 68.Zhu T., et al. , Metallo-polyelectrolytes as a class of ionic macromolecules for functional materials. Nat. Commun. 9, 4329 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chandrangsu P., Rensing C., Helmann J. D., Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 15, 338–350 (2017). Correction in: Nat. Rev. Microbiol. 15, 379 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chung L., Rajan K. S., Merdinger E., Grecz N., Coordinative binding of divalent cations with ligands related to bacterial spores. Equilibrium studies. Biophys. J. 11, 469–482 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Granger A. C., Gaidamakova E. K., Matrosova V. Y., Daly M. J., Setlow P., Effects of Mn and Fe levels on Bacillus subtilis spore resistance and effects of Mn2+, other divalent cations, orthophosphate, and dipicolinic acid on protein resistance to ionizing radiation. Appl. Environ. Microbiol. 77, 32–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.