Significance
The protovertebrate Ciona intestinalis is an ideal system to investigate both gene regulatory networks that underlie cell-type specification and how cell types have evolved. In this study, we use single-cell technology, experimental manipulations, and computational analyses to understand the role of the regulatory determinant POU IV—a homolog of Brn3 in vertebrates—in specifying various sensory cell types in Ciona. Surprisingly, the misexpression of POU IV throughout the epidermis led to the formation of hybrid sensory cell types, including those exhibiting properties of both palp sensory cells and bipolar tail neurons. These results demonstrate the interconnectedness of diverse sensory specification networks and give insights into the opportunities and challenges of reprogramming cell types through the targeted misexpression of cellular determinants.
Keywords: cell-type specification, evolutionary developmental biology, computational biology, cellular reprogramming
Abstract
The protovertebrate Ciona intestinalis type A (sometimes called Ciona robusta) contains a series of sensory cell types distributed across the head–tail axis of swimming tadpoles. They arise from lateral regions of the neural plate that exhibit properties of vertebrate placodes and neural crest. The sensory determinant POU IV/Brn3 is known to work in concert with regional determinants, such as Foxg and Neurogenin, to produce palp sensory cells (PSCs) and bipolar tail neurons (BTNs), in head and tail regions, respectively. A combination of single-cell RNA-sequencing (scRNA-seq) assays, computational analysis, and experimental manipulations suggests that misexpression of POU IV results in variable transformations of epidermal cells into hybrid sensory cell types, including those exhibiting properties of both PSCs and BTNs. Hybrid properties are due to coexpression of Foxg and Neurogenin that is triggered by an unexpected POU IV feedback loop. Hybrid cells were also found to express a synthetic gene battery that is not coexpressed in any known cell type. We discuss these results with respect to the opportunities and challenges of reprogramming cell types through the targeted misexpression of cellular determinants.
The specification of sensory neurons from lateral regions of the neural plate in protovertebrate Ciona intestinalis type A provides insights into the evolutionary origins of sensory placodes and the neural crest in vertebrates (1–10). Previous studies have identified early anteroposterior determinants that subdivide the lateral ectoderm into anterior, trunk, and tail regions (Foxc, Six1/2, and Msxb, respectively) (9). These trigger regulatory networks that produce related but distinct sensory neurons, including palp sensory cells (PSCs), anterior apical trunk epidermal neurons (aATENs), and bipolar tail neurons (BTNs). All of these cell types feature robust expression of POU IV (Fig. 1A), a key regulatory determinant of sensory neurons in vertebrates (Brn3) (11). These studies suggest that POU IV produces a sensory “ground state” that is modulated by additional determinants such as Six1/2 to specify distinctive neuronal cell types.
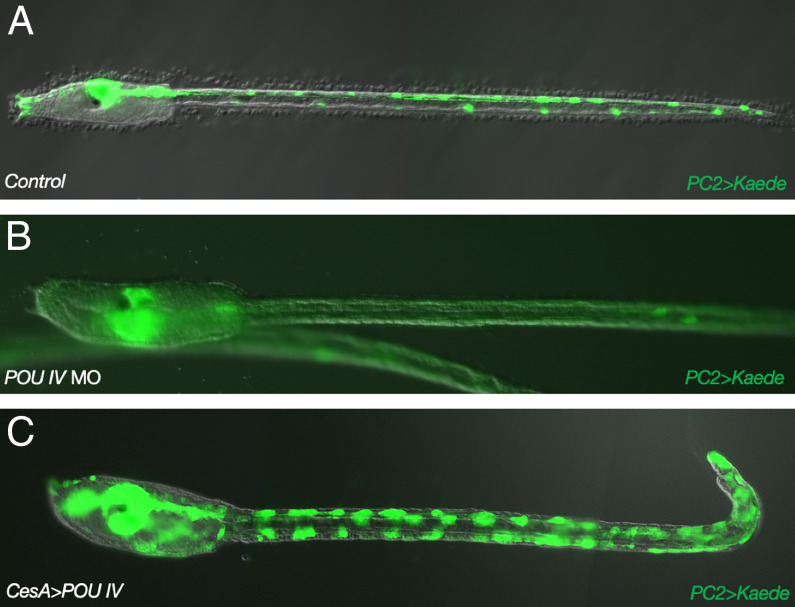
Fig. 1.
POU IV is necessary for the specification of epidermal sensory neurons. PC2>Kaede transgenic larvae. (A) Kaede fluorescence in the central nervous system (CNS) and epidermal sensory neurons in control larvae. (B) Same as A except that larvae were injected with a POU IV MO. (C) Same as A except that POU IV was misexpressed throughout the epidermis by a CesA 5′ regulatory sequence.
POU IV has also been implicated in the development of caudal epidermal sensory neurons (CESNs) (12, 13). These are arranged in single rows along the dorsal and ventral midlines of the Ciona tail, where they are thought to serve mechanosensory functions (14–16). Previous misexpression studies suggest that POU IV is sufficient to transform epidermal cells into supernumerary sensory neurons (13). Epidermal-specific cis-regulatory DNAs were used to drive expression of POU IV throughout the epidermis. This led to the surprising observation that the entire epidermis is neurogenic, not just the midline regions. Transformed epidermal cells have the appearance of supernumerary CESNs, suggesting that they might represent an ancestral sensory ground state. In this study, we employ single-cell RNA-sequencing (scRNA-seq) assays to examine their identities.
We used the CesA epidermal enhancer (17) to misexpress POU IV/Brn3 throughout the epidermis of developing tailbud embryos. As documented previously (13), there is a striking expansion of epidermal sensory neurons. However, single-cell transcriptome analyses are not consistent with a simple transformation of epidermis into CESNs. Instead, transformed cells express many of the genes associated with a BTN sensory identity. Different subclusters of transformed epidermal cells also express varying numbers of genes associated with other sensory identities, PSCs, aATENs, and CESNs. Moreover, transformed cells also express a set of genes that are not a specific transcriptome signature of any known cell type in the Ciona embryo or tadpole, which we will refer to as a “synthetic gene battery.” These results give insight into the opportunities and challenges of manipulating gene regulatory networks to reprogram cell types.
Results and Discussion
POU IV is a POU/homeobox gene that is expressed in all of the sensory neurons of the Ciona tadpole (Fig. 1A) (12). Homologous genes in vertebrates (Brn3) are essential for the development of a variety of sensory cell types, including visual, auditory/vestibular, and somatosensory neurons (18–20). Mutant embryos injected with a POU IV morpholino oligonucleotide (MO) at the one-cell stage resulted in the loss of expression of a pan-neuronal reporter gene (PC2>Kaede) in all sensory cell types at the larval stage (Fig. 1B), demonstrating that POU IV is an essential sensory determinant in Ciona. To determine whether it is sufficient for specification, POU IV was misexpressed throughout the epidermis of Ciona tadpoles using regulatory sequences from the CesA gene (Fig. 1C), which encodes a cellulase synthase that is essential for the elaboration of the protective test, a defining property of all urochordates (17). The CesA>POU IV transgene resulted in expanded expression of PC2>Kaede, suggesting transformation of epidermal cells into neurons (Fig. 1C).
To determine the nature of this transformation, we performed scRNA-seq of dissociated cells from transgenic tadpoles using the 10X Chromium System (9, 21–23). All wild-type sensory neurons, wild-type epidermal cells, and transformed epidermal cells were readily identified by their expression of a variety of known marker genes (Fig. 2A and SI Appendix, Tables S1 and S2). Transformed epidermal cells formed a heterogeneous population that exhibited differential expression of genes associated with each sensory cell type (Fig. 2A and SI Appendix, Fig. S1).
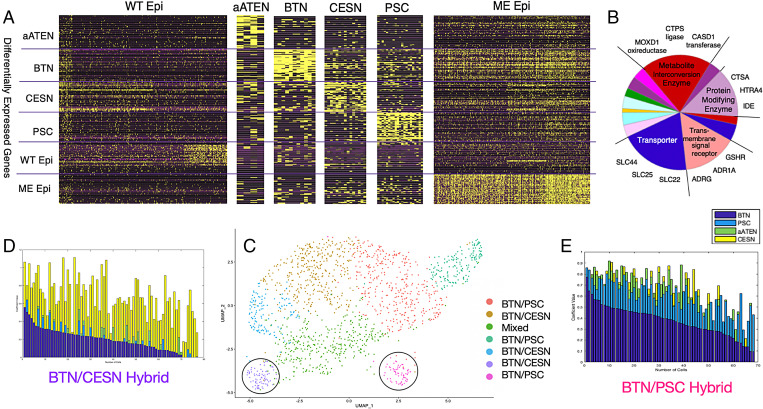
Fig. 2.
Quantitative characterization of wild-type and POU IV–misexpressed epidermis gene expression patterns. (A) Visualization of expression levels of 300 transcriptomic profile genes of wild-type epidermis and aATEN, BTN, CESN, PSC, and POU IV ME cell populations. Rows are the top 50 DEGs from each cell-type population (300 transcriptomic profile). Columns are individual cells. (B) Pie chart of the main PANTHER protein classes of ME DEGs and representative genes belonging to each class that define the various subclusters. More detailed information is provided in SI Appendix, Fig. S10. (C) UMAP visualization of seven subclusters generated from Louvain community–based clustering of ME cells. Circled purple and pink clusters contain representative BTN/CESN and BTN/PSC hybrid cells, respectively. (D and E) Stacked barplots of cell type–specific solved coefficients (Materials and Methods) for each cell in BTN/PSC and BTN/CESN subclusters, respectively. Solved aATEN, BTN, CESN, and PSC coefficients are represented by green, dark blue, yellow, and light blue bars, respectively. Cells are ordered by decreasing values of BTN solved coefficients, that is, “decreasing BTN character” and “increasing PSC/CESN character.”
This motivated us to subcluster the transformed cells into “misexpressed” (ME) clusters and model them as linear combinations of “average” wild-type cells based on their gene expression profiles (Materials and Methods, Fig. 2B, and SI Appendix, Fig. S2). We interpreted the coefficient values based on the extent to which an ME cell exhibited characteristics of the corresponding wild-type cell identity. In all subclusters, except for the BTN/CESN hybrid subcluster featured in Fig. 2D, BTN coefficients have the highest average values and right-skewed distributions, which implies that ME cells have the strongest bias toward a BTN-like identity. This is an unexpected result since previous studies suggested transformations of epidermal cells into CESNs (7, 9).
We also observed a distribution of coefficient values corresponding to all sensory cell types among ME cells, which implies “mixed” or “hybrid” sensory identities (SI Appendix, Fig. S3 and Table S3). For example, the purple and pink subclusters circled in Fig. 2C consist of ME cells with high average PSC and CESN coefficient values, respectively. We hereafter refer to these cells as “BTN/PSC” and “BTN/CESN” hybrids (Fig. 2 D and E). Moreover, ME cells concurrently express the PSC and BTN determinants Neurogenin and Foxg, respectively (7, 9, 10, 24) (SI Appendix, Fig. S4). This led us to take a particular interest in understanding the regulatory basis of the BTN/PSC hybrid identity.
While it is known that Foxg and Neurogenin directly bind and regulate POU IV expression (9), we explored the possibility of feedback regulation of both “upstream” determinants by POU IV. Thus, misexpression of POU IV could result in coactivation of both Foxg and Neurogenin in the transformed epidermis, a situation that is not encountered in wild-type sensory neurons. Computational analyses identified POU IV–binding motifs in the 5′ regulatory regions of both Foxg and Neurogenin (Materials and Methods and Fig. 3), and reporter genes containing these binding motifs exhibit specific expression in PSCs and BTNs, respectively (Fig. 3 A–D). Moreover, the Neurogenin enhancer contains a greater number of, and stronger, POU IV binding sites than Foxg. This correlates with altered activities of the Neurogenin and Foxg enhancers in mutant tailbud embryos injected with a POU IV MO at the one-cell stage (Fig. 4 A and B and SI Appendix, Fig. S5). Expression of the Neurogenin enhancer is abolished, while the Foxg enhancer is diminished.
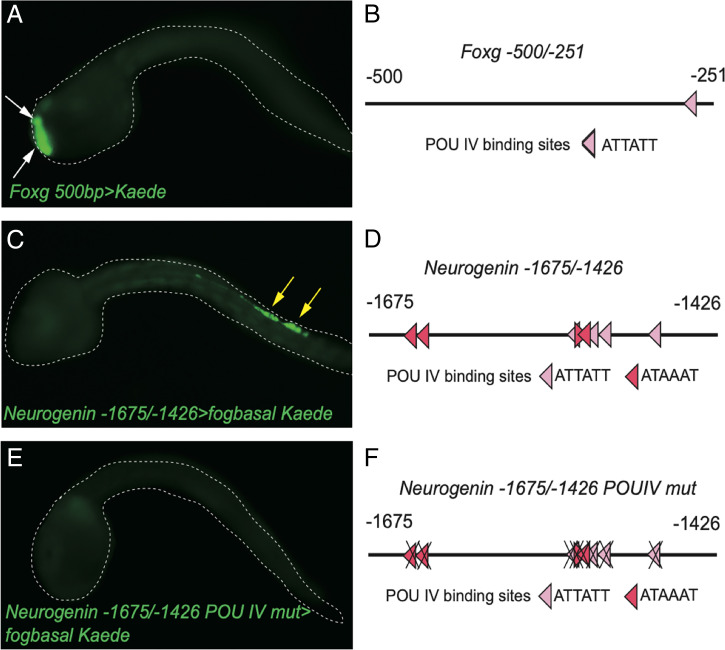
Fig. 3.
POU IV binding sites are present in the PSC minimal enhancer region of Foxg and BTN minimal enhancer region of Neurogenin and are necessary for the expression of Neurogenin in BTNs. (A) Kaede fluorescence in Foxg 500bp>Kaede–injected embryos. White arrows indicate Kaede fluorescence in PSCs. (B) Schematic diagram of -500/-251 bp of the PSC-specific Foxg minimal enhancer. One POU IV binding site is present. (C) Kaede fluorescence of Neurogenin -1675/-1472>Kaede–injected embryos. Yellow arrows indicate Kaede fluorescence in BTNs. (D) Schematic diagram of -1675/-1472 bp of the BTN-specific minimal enhancer. Eight POU IV binding sites are present. (E) Mutation analysis of eight POU IV binding sites showed that these POU IV binding sites are necessary for expression of Neurogenin in BTNs. (F) Schematic diagram of mutation analysis in E. Dark red binding sites are the higher-affinity POU IV binding sites.
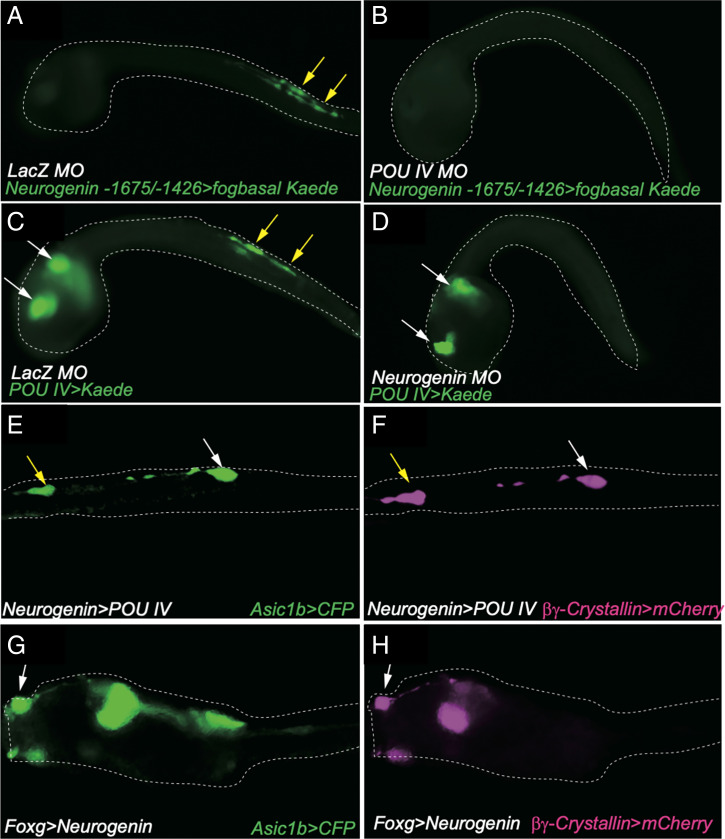
Fig. 4.
Positive feedback loop between POU IV and Neurogenin in BTN and synthetic cell-type specifications. (A and B) Kaede fluorescence of Neurogenin -1675/-1472bp>Kaede–injected embryos. (A) Kaede fluorescence in BTNs in LacZ MO–injected embryos. Yellow arrows indicate Kaede fluorescence in BTNs. (B) Same as A except that larvae were injected with the POU IV MO. (C and D) Kaede fluorescence of POU IV>Kaede–injected embryos. (C) Kaede fluorescence in BTNs and CNS in LacZ MO–injected embryos. Yellow and white arrows indicate Kaede fluorescence in BTNs and CNS, respectively. (D) Same as C except that larvae were injected with the Neurogenin MO. Kaede fluorescence disappears in BTNs but is maintained in the CNS. (E and F) Misexpression of POU IV in BTNs leads to expression of the PSC marker gene (βγ-Crystallin) in BTNs. Asic1b>CFP expression (E) and βγ-Crystallin>mCherry (F) expression in Neurogenin>POU IV–injected larvae. (G and H) Misexpression of Neurogenin in PSCs leads to expression of the BTN marker gene (Asic1b) in PSCs. Asic1b>CFP expression (G) and βγ-Crystallin>mCherry (H) expression in Foxg>Neurogenin–injected larvae.
There is a similar loss in activity of the BTN-specific minimal Neurogenin enhancer upon mutagenesis of POU IV binding sites (Fig. 3 E and F). Enhancer activity is also lost in tadpoles injected with either POU IV MO or Neurogenin MO (Fig. 4 A–D). Taken together, these studies suggest feedback regulation of Neurogenin, and to a lesser extent Foxg, by POU IV.
To determine whether this regulatory feedback results in the coactivation of Foxg and Neurogenin, we performed targeted misexpression of POU IV in BTNs using the minimal Neurogenin enhancer and targeted misexpression of Neurogenin in PSCs using the minimal Foxg enhancer. Both cases led to hybrid neuronal cell types that express both BTN (Asic1b) and PSC (βγ-crystallin) marker genes (Fig. 4 E–H) (25, 26). Similar results were obtained upon targeted misexpression of Foxc, which functions upstream of Foxg and POU IV (9, 10). These findings demonstrate interconnectedness of the Foxg–POU IV and Neurogenin–POU IV regulatory networks.
We next sought to understand two key aspects of ME cell identity—their strong BTN bias, and expression of a synthetic gene battery that is not observed in normal cell populations (ME Epi, Fig. 2 A and B). ME cells might exhibit a preponderance of BTN-like properties due to a more efficient induction of Neurogenin than Foxg by POU IV, since the Neurogenin enhancer is predicted to contain higher-affinity POU IV binding sites than its Foxg counterpart (Fig. 2). Consistent with this possibility is the finding of a more complete transformation of PSCs into BTNs upon targeted misexpression of both POU IV and Neurogenin (SI Appendix, Figs. S6 and S7). Interestingly, the combination of Brn3 and Neurogenin was found to be sufficient to reprogram human fibroblasts into different sensory cell types comprising dorsal root ganglia, a derivative of the neural crest (27). Our studies are therefore consistent with previous evidence that BTNs may correspond to derivatives of a “protoneural crest” in the Ciona lateral neural plate (7, 9).
To explore the characteristics of the synthetic gene battery that is deployed in hybrid sensory cell types, we performed both functional annotation and statistical overrepresentation Gene Ontology (GO) analyses of the human orthologs of these genes (Materials and Methods). Over 65% encode cell transporters, transmembrane signal receptors, protein-modifying enzymes, and metabolic enzymes. In addition, there is a >100-fold enrichment of various kinases, synthases, membrane transporters, and cell receptor functions (Fig. 2B and SI Appendix, Figs. S8–S12). Some of these genes are implicated in the biogenesis of lysosomes and exosomes, such as cathepsin. These are not signatures of cell stress, senescence, or death but instead suggest hyperactive growth. This surprising, emergent property of transformed ME cells is not predicted by the features of the regulatory networks underlying the specification of the different sensory cell types.
To understand what may have caused the induction of the synthetic gene battery, we examined up to 2 kb of their upstream regulatory regions and performed motif analysis (Materials and Methods). Our discovered motif (SI Appendix, Fig. S13) had significant matches to various Forkhead transcription factors in Ciona and their orthologs in mouse and human (SI Appendix, Fig. S14). Given this analysis and the up-regulation of Foxg expression throughout ME cells (SI Appendix, Fig. S1), we postulate that Foxg broadly associates with Forkhead binding sites to activate the synthetic gene battery. We also noted an up-regulation of other Forkhead transcription factors (SI Appendix, List S1), which might lead to further activation. These observations highlight the hazards and uncertainties created by targeted misexpression of cellular determinants such as POU IV.
Our study provides evidence for a POU IV feedback loop that maintains the identities of distinct sensory neurons, including PSCs and BTNs (Fig. 5). Misexpression of POU IV leads to ectopic activation of upstream regulators such as Foxg and Neurogenin via these feedback loops. Normally, these determinants are separately expressed in PSCs and BTNs but are coactivated upon misexpression of POU IV. This results in the development of hybrid sensory neurons exhibiting properties of both PSCs and BTNs. We consistently observe higher levels of expression of BTN identity genes as compared with PSC markers. In addition, these cells express genes not found in sensory cell types but are implicated in lysosomes, exosomes, and hyperactive growth. We postulate that they are fortuitously activated by the binding of Foxg and other Forkhead transcription factors.
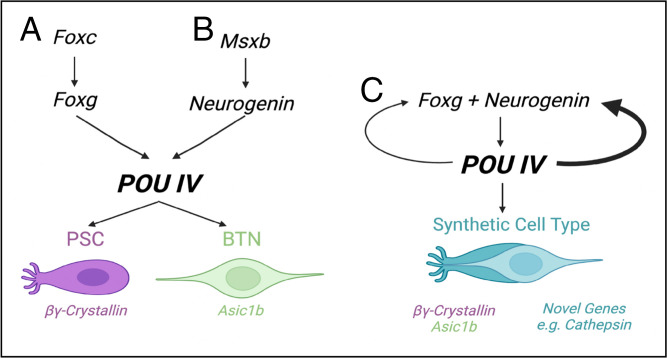
Fig. 5.
Summary of gene regulatory networks underlying PSC, BTN, and synthetic cell-type specifications. The schematics pertain to (A) PSCs, (B) BTNs, and (C) synthetic cell types found in POU IV–misexpressed embryos.
It has been suggested that the diversification of sensory cell types in vertebrates was facilitated by the duplication and divergence of POU IV/Brn3 paralogs (28). However, Ciona contains just a single POU IV ortholog but nonetheless produces diverse sensory cells. This study, along with several previous reports, suggests that this lone POU IV ortholog works in concert with regional determinants such as Foxg, Six1/2, and Neurogenin to specify distinct yet related sensory cell types (9). Misexpression of POU IV short circuits these cell-specific programs to produce a variety of hybrid cell types due to coexpression of Foxg, Neurogenin, and other regional determinants. In addition, we also observe fortuitous induction of a synthetic gene battery that is not coordinately expressed in any known natural cell type. It seems unlikely that they are induced by the combination of Foxg and Neurogenin. Instead, we found that their regulatory regions contain a significant overrepresentation of Forkhead-binding motifs. Thus, the success of future efforts to reprogram cell types with defined properties will depend on employing strategies to predict and diminish induction of unwanted secondary gene expression patterns. We believe that the strategy employed in our study—the iterative application of computational predictions and experimental manipulations of the underlying gene regulatory networks—will provide the precision required to achieve this goal.
Materials and Methods
Biological Materials.
Wild-type C. intestinalis type A (also called Ciona robusta) adults were obtained from M-REP and the National Bio-Resource Project for Ciona in Japan. Sperm and eggs were collected by dissecting the sperm and gonadal ducts. The PC2>Kaede transgenic lines (29, 30) were cultured and maintained in an island system (31).
Constructs.
Reporter genes were designed using previously published enhancer sequences: Asic1b (32), βγ-Crystallin (26), and CesA (17). The minimal enhancers of Foxg, Foxg -500/-201, and of Neurogenin, Neurogenin -1675/-1426, were isolated via PCR using sequence-specific oligonucleotide primers (SI Appendix, Table S4) and cloned into the XhoI/BamHI restriction site of the pSPfogbasalKaede vector. To generate pSPCiPOUIVcDNA, the coding sequence of Ci-POU IV was amplified by PCR with sequence-specific oligonucleotide primers (SI Appendix, Table S4). The PCR product was digested with NotI and inserted into the NotI and blunted EcoRI sites of pSPeGFP. To generate pSPCiCesACiPOUIVcDNA, the 5′ upstream region of Ci-CesA was inserted into the BamHI site of pSPCiPOUIVcDNA. To generate pSPCineurogeninPOUIVcDNA, the Neurogenin -1675/-1426+fogbasal promoter was inserted into the XhoI and NotI sites of pSCiPOUIVcDNA. To generate pSPFoxgPOUIV, Foxg 500bp was inserted into the XhoI and NotI sites of pSPCiPOUIVcDNA.
Microinjection of Antisense Morpholino Oligonucleotides.
MOs were obtained from Gene Tools. The antisense oligonucleotide sequence of the MO against Ci-POU IV is 5′-gcatgttagtaaacatatcgtatca-3′. MOs were dissolved in nuclease-free water (AM9930; Invitrogen) containing 1 mg/mL tetramethylrhodamine dextran (D1817; Invitrogen). The concentrations of MO and plasmid DNA in the injection medium were 0.5 mM and 2.5 to 10 ng/μL, respectively. Microinjections of MOs and reporter constructs were performed as described previously (9). All experiments were repeated at least twice with different batches of embryos.
Image Acquisition.
Images of transgenic larvae were obtained with a Zeiss AxioPlan, AX 10 epifluorescence microscope.
Single-Cell RNA-Seq Assays.
CesA>POU IV (2.5 ng/μL) + CesA>GFP (5.0 ng/μL) –injected eggs and control eggs were fertilized side by side and allowed to develop to the late-tailbud stage (13.5 h after fertilization at 18 °C). For each sample, 120 morphologically normal embryos were used for scRNA-seq assays. Dissociation of the embryos and scRNA-seq assays by the 10X Genomics Chromium System were done as described previously (9).
Computational Analysis Notes.
“OE cell” refers to a cell in POU IV–misexpressed epidermis. OEs and MEs are used interchangeably, and all code/lists use OE nomenclature. All code, analyses, and files can be accessed from the GitHub repository https://github.com/Singh-Lab/Pou4_Misexpression.
Single-Cell Data Analysis.
Data quality control and visualization.
According to the Cell Ranger 3.0 cell-calling algorithm based on the EmptyDrops algorithm (33), we removed cells using a cutoff of UMIs (unique molecular identifier) 5 SDs above the mean and those expressing fewer than 1,000 genes. This rendered 5,078 total cells derived from both control and POU IV–misexpressed embryos with 1,163 median genes per cell. All analyses were performed using Seurat version 2.3.4. Cells were further filtered so that only cells expressing at least 200 genes that are expressed in at least three cells were considered. For each cell, gene expression measurements were normalized by total gene expression, multiplied by a factor of 10,000, and then log-transformed. Gene expression values were regressed on UMI count data to remove unwanted variation, and residuals were then scaled and centered. These gene expression values were used for downstream analyses. Variable genes were outliers on a mean variability plot, calculated, and used as inputs for principal-component analysis (PCA) dimension reduction. For cells in the control embryos, 10 PCs were used as inputs to UMAP (Uniform Manifold Approximation and Projection) dimension reduction. For cells in POU IV–misexpressed embryos, 16 PCs were used. The resultant PCs were subjected to the unsupervised learning Louvain modularity community detection algorithm to cluster cells, which were then visualized in UMAP plots. Quality control metrics and figures are in SI Appendix, Figs. S15–S18.
Identification of cell populations.
Epidermal markers were used to identify 1,683 and 1,570 epidermal cell populations in both control and POU IV–misexpressed embryos, respectively. Four aATENs, 11 BTNs, and 21 PSCs were identified using their marker genes (SI Appendix, Table S1). We do not currently possess marker genes for CESNs, but 26 of them were identified as cells with the following code: POU IV+ Klf+ Neurogenin− β-Thymosine, in which + indicates expression and − indicates lack of expression (SI Appendix, Table S2). All genes in these tables and their corresponding gene IDs from the KH Assembly Model (34) (KHIDs) were obtained from the Aniseed database (35).
Calculation of differentially expressed genes and definition of transcriptomic profiles.
For each cell population, the edgeR differential expression analysis package (36) was used to calculate differentially expressed genes (DEGs) between itself and select cell populations. Backgrounds were decided based on our knowledge of similarity of gene expression patterns. For example, since CESNs and PSCs have very similar gene expression patterns, DEGs were calculated with respect to one another, to sufficiently distinguish them. aATEN DEGs were calculated with respect to CESNs and PSCs. BTN DEGs were calculated with respect to aATEN, CESN, PSC, and wild-type epidermis. Wild-type and ME epidermis DEGs were calculated with respect to all other cell-type populations. (Note: We refer to ME epidermis DEGs as “novel genes.”) All resultant DEGs were ordered by false discovery rate–corrected P values of less than 0.05, and then by nondecreasing log–fold change values. The top 50 most highly expressed DEGs for each cell population were identified, save for CESNs, which had 48 DEGs after this correction. We defined “transcriptomic profile” as a means of quantifying cell population identity. There were two such profiles. The first was the “300” transcriptomic profile, which consisted of the expression of all 298 DEGs. The other was the “250” transcriptomic profile, which consisted of the expression of the first 248 DEGs (all DEGs excluding those of ME cells).
Linear model to quantify OE cell identity with respect to wild-type cell population identities.
This model approximates the 250 transcriptomic profile of an ME cell as a linear combination of those of average wild-type cells. According to SI Appendix, Fig. S1, in the 250 × 5 matrix (wild-type matrix), rows are 250 transcriptome profile genes and columns are “average” wild-type vectors. A given entry in an average vector is the average expression level of a 250 transcriptome profile gene in all cells belonging to the same cell population. The 250 × 1 matrix (OE vector) contains the expression levels of the 250 transcriptome profile genes for a given OE cell. The entries in the 5 × 1 vector comprise a nonnegative linear least-squares solution to this system of equations. Each entry is a “cell-type specific solved coefficient.” Nonnegative linear least-squares solutions (solution vector) were calculated using the lsqnonneg function in MATLAB (version 9.2, release name R2017a). We then wanted to retain cells that were well-described by the linear modeling. To do this, the solution vector was multiplied by the wild-type matrix, and a Spearman correlation between this resultant vector and the OE vector was calculated using the corr function. If the Spearman correlation had a P value < 0.05, the corresponding solution matrix was retained for downstream analyses. Distributions of the coefficients of statistically significant solutions (cell-type specific solved coefficients) were plotted in histograms and stacked bar plots, as visualized in SI Appendix, Fig. S2.
Identification of POU IV binding sites in Foxg and Neurogenin minimal enhancer regions.
POU IV binding sites were found from Selex data for POU IV (KH2012:KH.C2.42) provided by the Aniseed database. “ATAAAT” has a higher binding affinity than “ATAATT.” Instances of these motifs were found in the sequences of the minimal enhancer regions by a simple string search.
Identification of putative 5′ cis-regulatory regions of novel genes.
Genomic sequences for novel genes were manually obtained from the Washington Genome Browser. Their KHIDs and human orthologs are given in "Final_OE_khids_orthologs.csv" and the genomic coordinates used are in "manual_oe_intervals.txt." Up to 2 kb sequence upstream of the transscriptional start site was analyzed. If the sequence overlapped a gene body of a neighboring gene, it was rejected. This rendered a total of 33 sequences.
Discovery of and matches to motifs in putative 5′ cis-regulatory regions of novel genes.
Putative cis-regulatory regions were used as inputs to MEME (37) with these parameters: motif site distribution: ANR; maximum number of motifs: 3; minimum motif length: 5; maximum motif length: 15 (SI Appendix, Fig. S13). One out of three discovered motifs had a significant match (q value < 0.05) to various Ciona Forkhead proteins. All motifs were used as inputs to TOMTOM (38) with the CIS-BP_2.00/Ciona_intestinalis motif database. Results are in SI Appendix, Fig. S14 A–E.
GO analyses of novel genes.
Human orthologs of 50 novel genes were used as input to the PANTHER Classification System (39), with “Homo sapiens” as the selected organism. Statistical overrepresentation test of all categories (GO biological process complete, GO cellular complete, etc.) was performed using human orthologs and “Homo sapiens genes” as inputs. Results are in SI Appendix, Figs. S10–S14.
Identification of significantly higher expressed Forkhead transcription factors in ME cells.
The Wilcoxon signed-rank test was performed on expression levels of each Forkhead transcription factor (given in "forkhead.csv") between wild-type and ME cells to find those that were significantly more highly expressed in ME cells.
Supplementary Material
Acknowledgments
We thank Kai Chen, Julia Buckles, Wei Wang, Lance Parsons, and all the members of the Lewis-Sigler Institute genome facility for technical support of the scRNA-seq assays and the M.S. laboratory for all computational analyses. This study was supported by a grant from the NIH (to M.L.) (NS076542) and by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science to T.H. (19H03204, 21K19249, 21H05239), T.G.K. (19H03213), and R.H. (20J00278, 21K15099). T.H. is supported by the Toray Science Foundation, Japan Science and Technology Agency FOREST Program (Grant JPMJFR2054) and Collaborative Research in Computational Neuroscience (CRCNS2021). R.H. is supported by the Kao Foundation for Arts and Sciences. T.G.K. is supported by a research grant from the Hirao Taro Foundation of KONAN GAKUEN for Academic Research. This study was further supported by the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science and Technology, Japan. P.P.C. is supported by a National Human Genome Research Institute T32 Training Grant.
Footnotes
Reviewers: I.A., Karolinska Institutet; and N.S., Okinawa Institute of Science and Technology Graduate University.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2118817119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix. All code, analyses, and files can be accessed from the GitHub repository https://github.com/Singh-Lab/Pou4_Misexpression. In addition, the data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Chacha et al., 2021) and are accessible through GEO Series accession number GSE192645 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE192645).
References
- 1.Manni L., et al. , Neurogenic and non-neurogenic placodes in ascidians. J. Exp. Zoolog. B Mol. Dev. Evol. 302, 483–504 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Mazet F., et al. , Molecular evidence from Ciona intestinalis for the evolutionary origin of vertebrate sensory placodes. Dev. Biol. 282, 494–508 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Patthey C., Schlosser G., Shimeld S. M., The evolutionary history of vertebrate cranial placodes—I: Cell type evolution. Dev. Biol. 389, 82–97 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Schlosser G., Patthey C., Shimeld S. M., The evolutionary history of vertebrate cranial placodes II. Evolution of ectodermal patterning. Dev. Biol. 389, 98–119 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Wagner E., Stolfi A., Gi Choi Y., Levine M., Islet is a key determinant of ascidian palp morphogenesis. Development 141, 3084–3092 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abitua P. B., et al. , The pre-vertebrate origins of neurogenic placodes. Nature 524, 462–465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stolfi A., Ryan K., Meinertzhagen I. A., Christiaen L., Migratory neuronal progenitors arise from the neural plate borders in tunicates. Nature 527, 371–374 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waki K., Imai K. S., Satou Y., Genetic pathways for differentiation of the peripheral nervous system in ascidians. Nat. Commun. 6, 8719 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horie R., et al. , Shared evolutionary origin of vertebrate neural crest and cranial placodes. Nature 560, 228–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu B., Satou Y., Foxg specifies sensory neurons in the anterior neural plate border of the ascidian embryo. Nat. Commun. 10, 4911 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McEvilly R. J., et al. , Requirement for Brn-3.0 in differentiation and survival of sensory and motor neurons. Nature 384, 574–577 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Candiani S., et al. , Ci-POU-IV expression identifies PNS neurons in embryos and larvae of the ascidian Ciona intestinalis. Dev. Genes Evol. 215, 41–45 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Tang W. J., Chen J. S., Zeller R. W., Transcriptional regulation of the peripheral nervous system in Ciona intestinalis. Dev. Biol. 378, 183–193 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Horie T., Kusakabe T., Tsuda M., Glutamatergic networks in the Ciona intestinalis larva. J. Comp. Neurol. 508, 249–263 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Pasini A., et al. , Formation of the ascidian epidermal sensory neurons: Insights into the origin of the chordate peripheral nervous system. PLoS Biol. 4, e225 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryan K., Lu Z., Meinertzhagen I. A., The peripheral nervous system of the ascidian tadpole larva: Types of neurons and their synaptic networks. J. Comp. Neurol. 526, 583–608 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Sasakura Y., et al. , Transposon-mediated insertional mutagenesis revealed the functions of animal cellulose synthase in the ascidian Ciona intestinalis. Proc. Natl. Acad. Sci. U.S.A. 102, 15134–15139 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gan L., Wang S. W., Huang Z., Klein W. H., POU domain factor Brn-3b is essential for retinal ganglion cell differentiation and survival but not for initial cell fate specification. Dev. Biol. 210, 469–480 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Murakami D. M., Erkman L., Hermanson O., Rosenfeld M. G., Fuller C. A., Evidence for vestibular regulation of autonomic functions in a mouse genetic model. Proc. Natl. Acad. Sci. U.S.A. 99, 17078–17082 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Badea T. C., et al. , Combinatorial expression of Brn3 transcription factors in somatosensory neurons: Genetic and morphologic analysis. J. Neurosci. 32, 995–1007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horie T., et al. , Regulatory cocktail for dopaminergic neurons in a protovertebrate identified by whole-embryo single-cell transcriptomics. Genes Dev. 32, 1297–1302 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao C., et al. , Comprehensive single-cell transcriptome lineages of a proto-vertebrate. Nature 571, 349–354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemaire L. A., Cao C., Yoon P. H., Long J., Levine M., The hypothalamus predates the origin of vertebrates. Sci. Adv. 7, 7452 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim K., et al. , Regulation of neurogenesis by FGF signaling and neurogenin in the invertebrate chordate Ciona. Front. Cell Dev. Biol. 8, 477 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., et al. , Conserved gene regulatory module specifies lateral neural borders across bilaterians. Proc. Natl. Acad. Sci. U.S.A. 114, E6352–E6360 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimeld S. M., et al. , Urochordate betagamma-crystallin and the evolutionary origin of the vertebrate eye lens. Curr. Biol. 15, 1684–1689 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Blanchard J. W., et al. , Selective conversion of fibroblasts into peripheral sensory neurons. Nat. Neurosci. 18, 25–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma N., et al. , The emergence of transcriptional identity in somatosensory neurons. Nature 577, 392–398 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osugi T., Sasakura Y., Satake H., The nervous system of the adult ascidian Ciona intestinalis type A (Ciona robusta): Insights from transgenic animal models. PLoS One 12, e0180227 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osugi T., Sasakura Y., Satake H., The ventral peptidergic system of the adult ascidian Ciona robusta (Ciona intestinalis type A) insights from a transgenic animal model. Sci. Rep. 10, 1892 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joly J. S., et al. , Culture of Ciona intestinalis in closed systems. Dev. Dyn. 236, 1832–1840 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Coric T., Passamaneck Y. J., Zhang P., Di Gregorio A., Canessa C. M., Simple chordates exhibit a proton-independent function of acid-sensing ion channels. FASEB J. 22, 1914–1923 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Lun A. T. L., et al. , Participants in the 1st Human Cell Atlas Jamboree, EmptyDrops: Distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satou Y., et al. , Improved genome assembly and evidence-based global gene model set for the chordate Ciona intestinalis: New insight into intron and operon populations. Genome Biol. 9, R152 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brozovic M., et al. , ANISEED 2015: A digital framework for the comparative developmental biology of ascidians. Nucleic Acids Res. 44, D808–D818 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bailey T. L., Elkan C., Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 (1994). [PubMed] [Google Scholar]
- 38.Gupta S., Stamatoyannopoulos J. A., Bailey T. L., Noble W. S., Quantifying similarity between motifs. Genome Biol. 8, R24 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mi H., et al. , Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 14, 703–721 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix. All code, analyses, and files can be accessed from the GitHub repository https://github.com/Singh-Lab/Pou4_Misexpression. In addition, the data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Chacha et al., 2021) and are accessible through GEO Series accession number GSE192645 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE192645).