Abstract
The discriminatory power, speed, and interlaboratory reproducibility of tRNA intergenic length polymorphism analysis (tDNA-PCR) combined with capillary electrophoresis was evaluated for the identification of streptococci. This method was carried out in three different laboratories under highly standardized conditions for 54 strains belonging to 18 different species. It was concluded that interlaboratory reproducibility of tDNA fingerprints produced by means of capillary electrophoresis was sufficiently high to permit the exchange between different laboratories and the construction of common libraries which can be consulted for comparison with fingerprints obtained independently in separate laboratories. In a second step, 17 other species were included in the study and examined in one of the participating laboratories. All Streptococcus species studied, except S. mitis, S. oralis, S. parasanguinis, S. pneumoniae, S. thermophilus, and S. vestibularis, showed distinguishable tDNA fingerprints. A database of well-characterized strains was constructed to enable computer-aided identification of unknown streptococcal isolates.
Traditionally the clinically most important Streptococcus species have been identified by Lancefield carbohydrate antigen detection and the application of a few biochemical or physiological tests. Difficulties arise when less-prevalent species are to be dealt with. Lancefield groups are not species-specific (5, 10), and certain species groups (2) are notoriously difficult to differentiate phenotypically (8).
A number of genotypic methods have been evaluated for the identification of streptococci: amplified ribosomal DNA restriction analysis (7, 9), amplification of ddl genes (6), and sequencing of the MnSOD gene (13). tRNA intergenic length polymorphism analysis (tDNA-PCR) (15) has been used not only for the differentiation of streptococcal species (3, 12) but also for Acinetobacter (4, 16), staphylococci (11), Listeria (14), and enterococci (1). Thus far the interlaboratory reproducibility of this kind of genotypic identification technique has been ill studied although it is crucial with regard to the ability to compare fingerprints generated in different laboratories and with regard to the construction of publicly accessible DNA fingerprint data banks. Here we evaluated the interlaboratory reproducibility of tDNA-PCR in combination with capillary fluorescent electrophoresis and its suitability for identification in routine diagnostics.
MATERIALS AND METHODS
Bacterial strains.
Fifty-four BCCM-LMG culture collection strains (University of Ghent, K. L. Ledeganckstraat 35, B-9000 Ghent) belonging to 18 streptococcal species were used to standardize the method of tDNA-PCR and to evaluate its interlaboratory reproducibility (Table 1). The collection was extended with 47 strains of the BCCM-LMG culture collection belonging to 17 other streptococcal species (Table 2). Ten collection strains were subjected to blind testing in all three laboratories.
TABLE 1.
Strains used to standardize the tDNA-PCR method and to study the intra- and interlaboratory reproducibility
| Streptococcus species | Strain no. |
|---|---|
| S. agalactiae | LMG14694T, LMG14840,a LMG15081 |
| S. alactolyticus | LMG14617,a LMG14872, LMG14808T |
| S. anginosus | LMG14502T, LMG14696, LMG17666 |
| S. bovis | LMG15048,a LMG15055, LMG8518T |
| S. gallolyticus | LMG14618,a LMG15049, LMG16802T, LMG16880 |
| S. hyointestinalis | LMG14579T,a LMG14581, LMG14582 |
| S. hyovaginalis | LMG14710T, LMG14711, LMG14712 |
| S. mitis | LMG14553,a LMG14557T, LMG14552T |
| S. mutans | LMG14558T,a LMG14560 |
| S. oralis | LMG14532T, LMG14533,a LMG14534 |
| S. parasanguinis | LMG14537T,a LMG14538, LMG14539 |
| S. pyogenes | LMG14237, LMG14238, LMG14700T |
| S. salivarius | LMG13103, LMG13104, LMG11489T |
| S. sanguinis | LMG14656, LMG14657, LMG14702T |
| S. suis | LMG14181T, LMG14182, LMG14643 |
| S. thoraltensis | LMG13593, LMG14173, LMG14174 |
| S. uberis | LMG14375, LMG14892, LMG09465 |
| S. pneumoniae | LMG14545T, LMG15155a, LMG16738 |
Strains used in this study for blind identification of streptococci on the basis of their tDNA-PCR fingerprint pattern.
TABLE 2.
Strains used to expand our collection and the tDNA fingerprint database
| Streptococcus species | Strain no. |
|---|---|
| S. acidominimus | LMG 17755T |
| S. canis | LMG 14833, LMG 14834, LMG 15890T |
| S. constellatus | LMG 14503, LMG 14504, LMG 14507T |
| S. cricetus | LMG 14508T, LMG 14511 |
| S. crispatus | LMG 14512 LMG 16320T |
| S. downei | LMG 14514T |
| S. dysgalactiae | LMG 14602, LMG 14602, LMG 15832, LMG 15738, LMG 15747, LMG 16026T |
| S. equi subsp. zooepidemicus | LMG 15760, LMG 15888, LMG 16030T |
| S. gordonii | LMG 14515, LMG 14517, LMG 14518T |
| S. iniae | LMG 14520T, LMG 14521, LMG 15978 |
| S. intermedius | LMG 14548, LMG 14549, LMG 17840T |
| S. phocae | LMG 16735T, LMG 16736, LMG 16737 |
| S. porcinus | LMG 14615, LMG 14837, LMG 15980T |
| S. rattus | LMG 14559, LMG 14650T, LMG 14651 |
| S. pluranimalium | LMG 13649, LMG 14177, LMG 14385, LMG 14827 |
| S. thermophilus | LMG 6896T, LMG 7952, LMG 13564 |
| S. vestibularis | LMG 13516T, LMG 14645, LMG 14646 |
DNA preparation.
Bacterial cells were grown overnight on Columbia agar (Gibco Life Technologies, Paisley, Scotland) with 5% ovine blood for 24 h at 37°C in a 5% CO2-enriched environment and checked for purity. A 1-μl loopful of cells was suspended in 20 μl of lysis buffer (0.25% sodium dodecyl sulfate, 0.05 N NaOH) and heated at 95°C for 5 min. The cell lysate was spun down by brief centrifugation at 16,000 × g and neutralized by adding 180 μl of distilled water. The cell debris was removed by centrifugation at 16,000 × g for 5 min. Supernatants were used as the DNA in the PCR or were frozen at −20°C until further use.
tDNA-intergenic PCR.
PCR was carried out using outwardly directed tRNA gene consensus primers T5A (5′ AGTCCGGTGCTCTAACCAACTGAG) and T3B (5′ AGGTCGCGGGTTCGAATCC) as described by Welsh and McClelland (15). Reactions were carried out in a 10-μl volume containing 9.1 μl (dilution, 1.1) of High Fidelity Mix (Gibco Life Technologies). Primers were added to a final concentration of 0.1 μM. Primer T3B consisted of a mixture of one-fifth fluorescent TET-labeled oligonucleotides and four-fifths nonlabeled oligonucleotides (PE Biosystems, Nieuwerkerk a/d IJssel, The Netherlands). A volume of 0.7 μl of sample DNA was added (dilution, 1/15). After 2 min at 94°C, reaction mixtures were cycled 30 times in a Perkin-Elmer Cetus 9600 thermocycler with the following conditions: 30 s at 94°C, 1 min at 50°C, and 1 min at 72°C, without a final extension period. Reaction vials were then cooled to 10°C and kept on ice until used in electrophoresis.
Capillary electrophoresis.
Twelve microliters of deionized formamide was mixed with 0.5 μl of an internal size standard mixture containing 0.3 μl of the GS-400 high-density size standard and 0.2 μl of the GS-500 size standard, which both contain ROX-labeled fragments in the range of 50 to 500 bp. One microliter of tDNA-PCR product was added. The mixtures were denatured by heating at 95°C for 3 min and placed directly on ice for at least 15 min (according to the recommendations of the manufacturer).
Capillary electrophoresis was carried out using an ABI-Prism 310 genetic analyzer (Applied Biosystems) at 60°C, at a constant voltage of 1.5 kV, and at a more or less constant current of approximately 10 mA. Capillaries with a length of 47 cm and diameter of 50 μm were filled with performance-optimized polymer 4. Electropherograms were normalized using Genescan Analysis software, version 2.1 (Applied Biosystems).
Data analysis.
tDNA-PCR fingerprints were obtained as table files from the Genescan Analysis software and used in a software program developed at our laboratory (1). Using these sample files containing tDNA spacer fragment lengths (peak values) in base pairs, this program enabled us to construct manually a library which contains one entry for each species and whereby each entry consists of a number of numeric values representing the peak values in base pairs. The peak values in the library entries are the averages of the peak values obtained after testing different strains of each species, which are listed in Tables 1 and 2. The peaks retained for each entry are user selected, which means that the scientist takes the final decision about which peaks appear to be characteristic for each species. Negative values can be added to indicate that a certain peak must not be present in the fingerprint in order to be identified as a certain species. The similarity between the unknown fingerprint and a library entry is calculated with a coefficient, further referred to as dbp (differential base pairs): the number of fragments in common between the unknown fingerprint and the species entry, divided by the total number of fragments of the species entry in the library. A peak position tolerance of 0.7 bp was used.
A distance matrix was calculated with the in-house software. Clustering analysis was done with the Neighbor module of the Phylip software (http://evolution.genetics.washington.edu/phylip.html), using the unweighted pair group method using arithmetic averages (UPGMA) algorithm. Ten well-characterized strains were tested blindly in all three laboratories and identified on the basis of their tDNA-PCR fingerprint by using this software.
Reproducibility testing.
One S. agalactiae strain (LMG 14694T) was tested 135 times by tDNA-PCR in order to evaluate the variation caused by differences in PCR mixture preparation, PCR cycling, and electrophoresis runs at and between three different laboratories.
One 10-fold PCR mixture was made in each of the three laboratories and the DNA template was added. This mixture was divided into 10 equal volumes of 10 μl (samples 1 to 10). Samples 1 to 5 were cycled immediately, and samples 6 to 10 were kept at −70°C. On each of the following 5 days, a 10-μl PCR mixture was freshly prepared (samples 11 to 15) and this tube was cycled together with one of the samples 6 to 10. In total, this resulted for each laboratory in 15 tubes with tDNA-PCR product obtained from the same strain. The content of all 45 products was then divided over three tubes, exchanged between labs, and run on the ABI Prism 310 genetic analyzer at each laboratory. This resulted in 135 tDNA-PCR fingerprints of the same strain. For another S. agalactiae strain, LMG 14840, tDNA-PCR was performed on three other thermocyclers: the iCycler (Bio-Rad, Nazareth, Belgium), the PTC200 (MJ Research, Waltham, Mass.), and the Mastercycler (Eppendorf, Hamburg, Germany).
Reproducibility was evaluated by (i) calculation of the standard deviation of the peak values of the six predominantly present tDNA spacer fragment peaks and (ii) calculation of the similarity between all fingerprints by using the dbp coefficient and by clustering with the UPGMA algorithm, using the Phylip software.
RESULTS
Standardization.
In the three laboratories, different protocols were tested in order to assess the best fingerprint results. This revealed that the PCR conditions which produced the most reproducible and discriminatory tDNA fingerprints in each laboratory were obtained with the PCR mixture composition and the PCR cycling conditions as described in Materials and Methods.
Reproducibility.
Extensive testing of the reproducibility of tDNA-PCR was done by repeated amplification of one S. agalactiae strain (LMG 14694T) in different laboratories using different reaction mixtures, thermal cycling runs, and capillary electrophoresis runs. One of the 45 PCR mixtures and 10 of the 135 electrophoresis runs failed to produce a fingerprint. This resulted in 122 tDNA fingerprints available for the reproducibility studies.
In its tDNA-PCR fingerprint, the S. agalactiae strain showed six predominant peaks, of which the mean peak values, standard deviations, and percent standard deviations (SD/peak value × 100) for all 122 fingerprints are shown in Table 3. Only three samples (i.e., 2.5% of all cases), for all of which PCR was performed in one of the labs, lacked a single peak (of 241 bp).
TABLE 3.
Average values and standard deviations of six peak positions for all 122 fingerprints of S. agalactiae strain LMG 14694T
| Variable | Peak 1 | Peak 2 | Peak 3 | Peak 4 | Peak 5 | Peak 6 |
|---|---|---|---|---|---|---|
| Mean (bp) | 54.65 | 62.40 | 88.90 | 148.02 | 241.24 | 252.70 |
| SD (bp) | 0.27 | 0.20 | 0.15 | 0.21 | 0.37 | 0.33 |
| % SD | 0.49 | 0.31 | 0.17 | 0.14 | 0.16 | 0.13 |
| Minimum (bp) | 54.22 | 62.04 | 88.56 | 147.5 | 240.58 | 251.58 |
| Maximum (bp) | 55.29 | 62.73 | 89.19 | 148.56 | 242.04 | 253.38 |
Each of the 44 obtained PCR products was electrophoresed at the three different laboratories. For each PCR product, the ranges between maximal and minimal peak values obtained for the same DNA fragment were calculated. The lowest and highest ranges obtained for each of the six peaks are presented in Table 4. For the largest peak of 252.70 bp, a maximal range of 1.56 bp was observed.
TABLE 4.
Mean values and minimal and maximal range of the peak values for a total of 35 triplets and 9 doublets obtained from three different laboratories
| Variable | Mean values and range of peak values (bp)
|
|||||
|---|---|---|---|---|---|---|
| Peak 1 | Peak 2 | Peak 3 | Peak 4 | Peak 5 | Peak 6 | |
| Peak avg | 54.65 | 62.40 | 88.90 | 148.02 | 241.24 | 252.70 |
| Minimal range | 0.07 | 0.02 | 0.02 | 0.05 | 0.15 | 0.28 |
| Maximal range | 1.06 | 0.95 | 0.99 | 0.68 | 1.11 | 1.56 |
For all samples for which capillary electrophoresis was carried out in the same laboratory, the mean peak values, standard deviations, and percent standard deviations were calculated for the six peaks and are summarized in Table 5. This table also shows minimal and maximal peak positions obtained. In laboratory A (41 samples), the maximal standard deviation was 0.25 bp, for a mean peak value of 241.65 bp; in laboratory B (39 samples), it was 0.22 bp for a mean peak value of 252.56 bp; and in laboratory C (42 samples), it was 0.21 bp for a mean peak value of 240.97 bp.
TABLE 5.
Mean, standard deviation, and percent standard deviation of the six peaks in all fingerprints obtained in each laboratory
| CE results ina: | Peak 1 | Peak 2 | Peak 3 | Peak 4 | Peak 5 | Peak 6 |
|---|---|---|---|---|---|---|
| Lab A (n = 41) | ||||||
| Mean (bp) | 54.39 | 62.48 | 88.72 | 148.22 | 241.65 | 253.05 |
| SD (bp) | 0.09 | 0.20 | 0.08 | 0.16 | 0.25 | 0.22 |
| SD (%) | 0.16 | 0.31 | 0.09 | 0.11 | 0.10 | 0.09 |
| Minimal (bp) | 54.22 | 62.04 | 88.56 | 148.00 | 240.82 | 252.08 |
| Maximal (bp) | 54.62 | 62.73 | 88.85 | 148.56 | 242.04 | 253.34 |
| Lab B (n = 39) | ||||||
| Mean (bp) | 54.60 | 62.47 | 88.94 | 147.96 | 241.06 | 252.56 |
| SD (bp) | 0.13 | 0.17 | 0.07 | 0.14 | 0.16 | 0.22 |
| % SD | 0.23 | 0.27 | 0.08 | 0.09 | 0.07 | 0.09 |
| Minimal (bp) | 54.41 | 62.11 | 88.82 | 147.5 | 240.83 | 251.58 |
| Maximal (bp) | 54.88 | 62.68 | 89.06 | 148.29 | 241.74 | 253.38 |
| Lab C (n = 42) | ||||||
| Mean (bp) | 54.95 | 62.26 | 89.04 | 147.86 | 240.97 | 252.46 |
| SD (bp) | 0.18 | 0.14 | 0.14 | 0.12 | 0.21 | 0.18 |
| % SD | 0.33 | 0.22 | 0.16 | 0.08 | 0.09 | 0.07 |
| Minimal (bp) | 54.20 | 61.53 | 88.80 | 147.64 | 240.58 | 252.06 |
| Maximal (bp) | 55.29 | 62.46 | 89.61 | 148.19 | 241.44 | 252.85 |
CE, capillary electrophoresis. n, number of samples.
In addition to these standard deviation calculations, the similarity values of the 122 capillary electrophoresis runs of the same strain carried out in three laboratories with varying factors were also calculated, using the dbp coefficient and clustering with UPGMA. The lowest similarity obtained with all 122 fingerprints was 87.8%.
It was observed that the addition of a final extension period of half an hour at 72°C increased the reproducibility of the fingerprints. Without final extension, double peaks differing in length by one bp frequently occurred. With a final extension period, the double peaks mostly disappeared to be replaced by the larger of the two peaks only.
For 12 samples cycled on the iCycler, 6 samples on the Mastercycler, and 6 on the PTC200, the standard deviations were 0.18, 0.11, 0.22, 0.14, 0.20, and 0.16 bp for the six peaks.
All 35 streptococcal species tested gave a tDNA-PCR pattern which consisted of three to seven large and reproducibly present (i.e., present in 97.5 to 100% of all cases) peaks and several small (i.e., less than 20% of the average peak height in a fingerprint) nonreproducibly present peaks, which were considered as noise. All 54 strains used in the reproducibility study gave reproducible fingerprints regardless of the laboratory in which the assay was performed.
Discriminatory power.
Most species were easily distinguishable using tDNA-PCR (Table 6). The closely related species S. bovis, S. alactolyticus, and S. gallolyticus showed resembling but distinctive tDNA fingerprints. S. mutans and S. gordonii differed in one base pair of only one peak. The species S. anginosus, S. constellatus, and S. intermedius could be differentiated by the longer tRNA spacer fragments. S. canis and S. dysgalactiae fingerprints differed slightly in two peak values (Table 6). S. oralis, S. mitis, S. parasanguinis, and S. pneumoniae were not distinguishable on the basis of their tDNA pattern, nor were S. vestibularis and S. thermophilus. Some species were divided into two groups on the basis of different tDNA patterns. This was the case for S. iniae and S. porcinus.
TABLE 6.
tDNA-PCR library constructed manually and containing numeric values that represent lengths of amplified tDNA spacers that ought to be present or absent in the fingerprint of an unknown strain in order to be identified as a certain species
| Streptococcus species entry | Length of fragments considered (bp)a |
|---|---|
| S. acidominimus | 65.1, 75, 90.3, 103.9, 165.5, 261.5 |
| S. agalactiae | 54.8, 62.5, 89, 148, 241, 252.5 |
| S. alactolyticus | 55.8, 63.5, 154, −155, 253.8 |
| S. anginosus | 77, 88, 155.3 |
| S. bovis | 55.8, 63.5, 154, −155, −241, 255 |
| S. canis | 92, 164, 264.5 |
| S. constellatus | 88, 155.3 |
| S. cricetus | 72.6, 81, 165.5, 260.5 |
| S. crispatus | 56.3, 79.7, 90, 152, 251 |
| S. downei | 71.8, 166.5, 261.3 |
| S. dysgalactiae | 92, 163.4, 263.5 |
| S. equi subsp. zooepidemicus | 56.4, 63.5, 84.7, 92.8, 171.8, 234.6, 272.3 |
| S. gallolyticus | 55.8, 63.5, 155, −241, 255 |
| S. gordonii | 77, 88, 159.7 |
| S. hyointestinalis | 61.2, 69, −88.6, 159.6, 255.1 |
| S. hyovaginalis | −69, 88.6, 147.8, 160.2, 255.6 |
| S. iniae 1 | 130.5, 150, 167.5, 265.3 |
| S. iniae 2 | 130.5, 150, 169.5, 267.4 |
| S. intermedius | 77, 88, 155.3, 253.7 |
| S. mitis/oralis/parasanguinis/pneumoniae | 63.5, 84.5, 150 |
| S. mutans | 77, 88, 158.2 |
| S. parauberis | 66, 115.7, 165.5, 260.7, 290.5 |
| S. phocae | 56.3, 98.4, 165.3 |
| S. pluranimalium | 74.1, 99.5, 141, 162, 263 |
| S. porcinus 1 | 148.3, 171.5, 242, 266.5, 318.1 |
| S. porcinus 2 | 151.6, 168, 245, 263.3, 322.5 |
| S. pyogenes | 80.6, 92, 166.5, 266.5 |
| S. rattus | 56.3, 67.5, 261.6, 266.7 |
| S. sanguinis | 65.3, 92.7, 161.5, 258.6 |
| S. salivarius | 78.3, 148.3 |
| S. suis | 57.5, 65.5, 75.3, 92.8, 106.6, 162.2, 259.6 |
| S. thermophilus/vestibularis | 70.5, 79.5, 148.2, 247.5 |
| S. thoraltensis | 74, 83.2, 166.5, 258, 271.9 |
| S. uberis | 117.5, 133.5, 168.5, 263 |
Negative values indicate the length of tDNA spacers that should be absent in the fingerprint.
Use of the in-house software and a manually constructed library, which contained only the reproducible peak values, enabled straightforward differentiation between all of the species tested except between strains belonging to the species S. pneumoniae, S. mitis, S. oralis, and S. parasanguinis and between S. thermophilus and S. vestibularis strains. In three laboratories, identification of 10 well-characterized strains was attempted using tDNA-PCR and our software, without former knowledge of the species to which these strains belonged. All 10 strains were identified correctly in all laboratories.
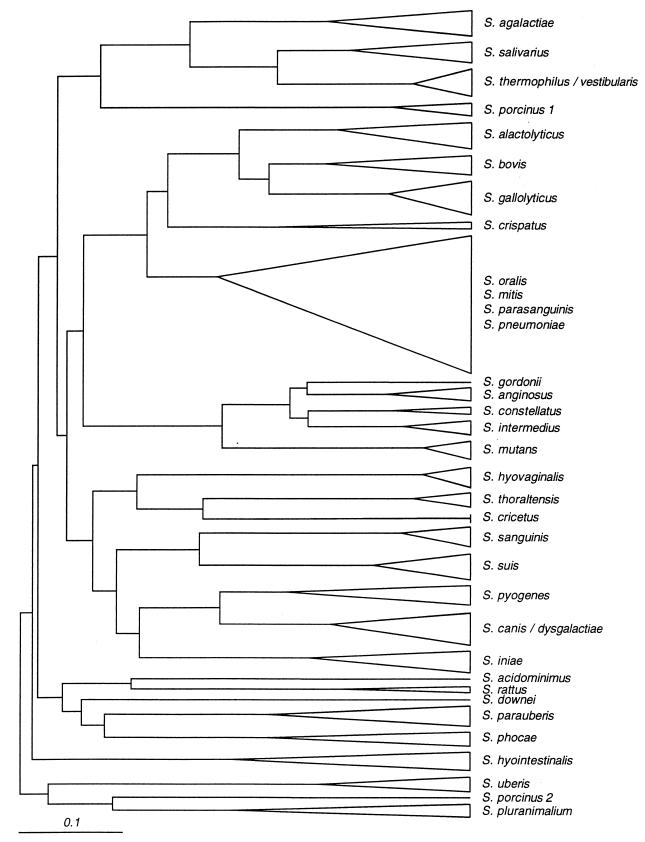
A dendrogram obtained with the tDNA-PCR patterns is shown in Fig. 1. All strains belonging to the same species clustered together. The strains belonging to S. mitis, S. oralis, S. parasanguinis, and S. pneumoniae were found in the same cluster, as was the case for strains belonging to S. thermophilus and S. vestibularis. Strains belonging to the species S. canis and S. dysgalactiae, and to S. anginosus, S. constellatus, and S. intermedius, which show resembling but still different tDNA patterns, clustered together.
FIG. 1.
Dendrogram obtained after similarity calculations between the tDNA fingerprints of streptococci. The bar represents a distance of 10%.
DISCUSSION
tDNA-PCR and capillary electrophoresis of the amplified DNA fragments already have been evaluated for the differentiation of Listeria species (14) and enterococci (1). To enable identification of a large number of strains, a software program which was described previously (1) has been developed at our laboratory. In the present study, the interlaboratory reproducibility of tDNA-PCR was evaluated in order to develop a fully exchangeable digital fingerprint database which can be consecutively extended with new fingerprints of species belonging to a wide array of genera.
For S. agalactiae strains, tDNA-PCR resulted in a fingerprint with six reproducibly present peaks. The standard deviation of the amplified tDNA spacer fragment lengths (peak values) was calculated for each of the six peaks obtained in 122 fingerprints of strain LMG14694T. The standard deviation of all samples ranged from 0.19 to 0.38 bp for peaks between 54 and 253 bp, which indicates that reproducibility with regard to peak values was extremely high.
The presence or absence of peaks must be caused by different PCR mixtures and/or PCR cycling runs and not by electrophoretic migration differences. The six DNA fragments which are most strongly amplified are reproducibly present in more than 97.5% of all cases. Therefore it can be concluded that peak presence reproducibility, i.e., PCR reproducibility, is high. Further, it becomes clear that the variation of peak values around the mean value is caused by electrophoretic run-to-run differences in and between laboratories (Table 4). The results show that electrophoresis of the same PCR product on another genetic analyzer can give peak position differences of more than 1 bp (the highest range found was 1.56 bp), which means that the difference is due to variation in migration and not to the addition or loss of a nucleotide during PCR. Therefore we conclude that the final variability between the obtained fingerprints was not caused by PCR mix preparation or by PCR cycling runs either in one laboratory or in different laboratories, but solely by differences in migration during capillary electrophoresis, whereby the largest difference occurred between ABI Prism 310 genetic analyzers in different laboratories. Nevertheless, the reproducibility was found to be very high and a peak position tolerance of less than 0.8 bp can be used to score corresponding fragments as identical.
In one laboratory, PCR mixtures were cycled on three other PCR cyclers and electrophoresis was carried out on the same genetic analyzer. The standard deviations for the six peaks ranged from 0.11 to 0.22 bp. This means that this PCR assay can be performed on the four PCR cyclers tested without the need for adjustment of the cycling conditions.
The observation that prolonged extension resulted in the disappearance of double peaks can be explained by the fact that this enables the Taq polymerase to add an extra A to most of the PCR fragments, causing these to outnumber the peaks without an additional A.
To be able to compare a large number of patterns, a software program was developed in our laboratory. It takes into account only peak values, not peak intensities. Importantly, extra peaks which are caused by electrophoretic impurities or other unknown factors are ignored by the approach used here and therefore cannot influence the identification results. Variability in peak positions due to electrophoretic differences can be compensated by enlarging the position tolerance in the software. Still, visual checking of the patterns to confirm the results is advised.
tDNA-PCR seems to be suited for the identification of most streptococcal species. However, S. mitis, S. oralis, S. parasanguinis, and S. pneumoniae, belonging to the clinically important viridans streptococci and to one phylogenetically closely related S. mitis species group (2), showed highly resembling patterns (see Table 6) and were not distinguishable. Recently, Degheldre et al. (3) have evaluated the discriminatory power of tDNA-PCR for the differentiation of viridans streptococci with separation of fragments on an ALFexpress DNA sequencer. Apparently, the patterns they found are not similar to those obtained in our study, because of the presence of more large fragments and fewer small ones. In their study, the large fragments enabled discrimination within the S. mitis group. The cause of this disagreement is not clear.
tDNA-PCR is very rapid and relatively easy to perform. In a PE 9600 thermocycler, 96 samples can be run at once. Starting from a single colony, DNA extraction takes about 3 h for 96 strains. The preparation of the tDNA-PCR mixtures in separate tubes and addition of the sample DNA takes about 1 h and the PCR run itself takes 2 h. During this run, the genetic analyzer ABI Prism 310 capillary electrophoresis apparatus can be prepared. Denaturation and preparation of the PCR products takes about 30 min. The electrophoresis run takes about half an hour per sample, but with the use of different dyes for labeling primers, three samples can be run at the same time. Quality control of the obtained profiles by means of GeneScan analysis and comparison of the tDNA-PCR profiles of the unknown strains with the database takes another hour. Summarized results for all 96 strains can be available within 25 h if the three-dye technology is used. Obviously, taking fewer samples at once will reduce the manipulation time, which makes it possible to have the first electrophoresis results within 8 h after colony picking.
The cost, including culture, DNA extraction, PCR, and capillary electrophoresis, was calculated as $2.50 per strain. Given the possibility for automation, the broad applicability of tDNA-PCR for species identification, and the interlaboratory exchange of data due to its high reproducibility, tDNA-PCR could be developed as a routinely applicable genotypic identification technique.
ACKNOWLEDGMENTS
This work was supported by the Research Fund of the University of Ghent, Ghent, Belgium, Codenr. BOF98/GOA/014. M.V. and M.G. are indebted to the Fund for Scientific Research—Flanders for an appointment as research associate (M.V.) and for research and personnel grants (M.G.).
We are grateful to R. Coopman, F. Grillaert, A. Vandekerckhove, and L. Van Simaey for their excellent technical assistance.
REFERENCES
- 1.Baele M, Baele P, Vaneechoutte M, Storms V, Butaye P, Devriese L A, Verschraegen G, Gillis M, Haesebrouck F. Application of tDNA-PCR for the identification of enterococci. J Clin Microbiol. 2000;38:4201–4207. doi: 10.1128/jcm.38.11.4201-4207.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley R W, Leigh J A, Collins M D. Intrageneric structure of Streptococcus based on comparative analysis of small-subunit rRNA sequences. Int J Syst Bacteriol. 1991;41:487–494. doi: 10.1099/00207713-41-4-487. [DOI] [PubMed] [Google Scholar]
- 3.Degheldre Y, Vandamme P, Goossens H, Struelens M. Identification of clinically relevant viridans streptococci by analysis of transfer DNA intergenic spacer length polymorphism. Int J Syst Evol Microbiol. 1999;49:1591–1598. doi: 10.1099/00207713-49-4-1591. [DOI] [PubMed] [Google Scholar]
- 4.Ehrenstein B, Bernards A T, Dijkshoorn L, Gerner S P, Towner K J, Bouvet P J M, Daschner F D, Grundmann H. Acinetobacter species identification by using tRNA spacer fingerprinting. J Clin Microbiol. 1996;34:2414–2420. doi: 10.1128/jcm.34.10.2414-2420.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farrow J A, Collins M D. Taxonomic studies on streptococci of serological groups C, G and L and possibly related taxa. Syst Appl Microbiol. 1984;5:483–493. [Google Scholar]
- 6.Garnier F, Gerbaud G, Courvalin P, Galimand M. Identification of clinically relevant viridans group streptococci to the species level by PCR. J Clin Microbiol. 1997;35:2337–2341. doi: 10.1128/jcm.35.9.2337-2341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillespie B E, Jayarao B M, Oliver S P. Identification of Streptococcus species by randomly amplified polymorphic deoxyribonucleic acid fingerprinting. J Dairy Sci. 1997;80:471–476. doi: 10.3168/jds.S0022-0302(97)75959-9. [DOI] [PubMed] [Google Scholar]
- 8.Hardie J M, Whiley R A. Recent developments in streptococcal taxonomy: their relation to infections. Rev Med Microbiol. 1994;5:151–162. [Google Scholar]
- 9.Jayarao B M, Doré J J E, Jr, Oliver S P. Restriction fragment length polymorphism analysis of 16S ribosomal DNA of Streptococcus and Enterococcus species of bovine origin. J Clin Microbiol. 1992;30:2235–2240. doi: 10.1128/jcm.30.9.2235-2240.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawrence J, Yajko D M, Hadley W K. Incidence and characterization of beta-hemolytic Streptococcus milleri and differentiation from S. pyogenes (group A), S. equisimilis (group C), and large-colony group G streptococci. J Clin Microbiol. 1985;22:772–777. doi: 10.1128/jcm.22.5.772-777.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maes N, De Gheldre Y, De Ryck R, Vaneechoutte M, Meugnier H, Etienne J, Struelens M J. Rapid and accurate identification of Staphylococcus species by tRNA intergenic spacer length polymorphism analysis. J Clin Microbiol. 1997;35:2477–2481. doi: 10.1128/jcm.35.10.2477-2481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McClelland M, Welsh J. Length polymorphisms in tRNA intergenic spacers detected by using the polymerase chain reaction can distinguish streptococcal strains and species. J Clin Microbiol. 1992;30:1499–1504. doi: 10.1128/jcm.30.6.1499-1504.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poyart C, Quesne G, Coulon S, Berche P, Trieu C P. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J Clin Microbiol. 1998;36:41–47. doi: 10.1128/jcm.36.1.41-47.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaneechoutte M, Boerlin P, Tichy H V, Bannerman E, Jäger B, Bille J. Comparison of PCR-based DNA fingerprinting techniques for the identification of Listeria species and their use for atypical Listeria isolates. Int J Syst Bacteriol. 1998;48:127–139. doi: 10.1099/00207713-48-1-127. [DOI] [PubMed] [Google Scholar]
- 15.Welsh J, McClelland M. Genomic fingerprints produced by PCR with consensus tRNA gene primers. Nucleic Acids Res. 1991;19:861–866. doi: 10.1093/nar/19.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiedmann-Al-Ahmad M, Tichy H V, Schön G. Characterization of Acinetobacter type strains and isolates obtained from wastewater treatment plants by PCR fingerprinting. Appl Environ Microbiol. 1994;60:4066–4071. doi: 10.1128/aem.60.11.4066-4071.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]