Abstract
A LightCycler-based PCR-hybridization gyrA mutation assay (GAMA) was developed to rapidly detect gyrA point mutations in multiresistant (MR) Salmonella enterica serotype Typhimurium DT104 with decreased susceptibility to ciprofloxacin (MIC, 0.25 to 1.0 mg/liter). Ninety-two isolates (49 human, 43 animal) were tested with three individual oligonucleotide probes directed against an Asp-87-to-Asn (GAC→AAC) mutation, an Asp-87-to-Gly (GAC→GGC) mutation, and a Ser-83-to-Phe (TCC→TTC) mutation. Strains homologous to the probes could be distinguished from strains that had different mutations by their probe-target melting temperatures. Thirty-seven human and 30 animal isolates had an Asp-87-to-Asn substitution, 6 human and 6 animal isolates had a Ser-83-to-Phe substitution, and 5 human and 2 animal isolates had an Asp-87-to-Gly substitution. The remaining six strains all had mismatches with the three probes and therefore different gyrA mutations. The sequencing of gyrA from these six isolates showed that one human strain and two animal strains had an Asp-87-to-Tyr (GAC→TAC) substitution and two animal strains had a Ser-83-to-Tyr (TCC→TAC) substitution. One animal strain had no gyrA mutation, suggesting that this isolate had a different mechanism of resistance. Fifty-eight of the strains tested were indistinguishable by several different typing methods including antibiograms, pulsed-field gel gel electrophoresis, and plasmid profiling, although they could be further subdivided according to gyrA mutation. This study confirmed that MR DT104 with decreased susceptibility to ciprofloxacin from humans and food animals in England and Wales may have arisen independently against a background of clonal spread of MR DT104.
Multiresistant (MR) Salmonella enterica serotype Typhimurium definitive phage type 104 (DT104) emerged during the 1990s as the most prevalent MR phage type in England and Wales. MR DT104 has been responsible for a substantial number of infections in humans and a wide range of food animals and is now recognized as a significant health problem in numerous European countries, North America, the Middle East, South Africa, and southeast Asia (38). MR DT104 is typically resistant to ampicillin, chloramphenicol, streptomycin-spectinomycin, sulfonamides, and tetracylines (resistance type [R type] ACSSpSuT; see Table 1 for definitions of individual elements of the R types). The ACSSpSuT resistance genes (blaCARB-2, cmlA, aadA2, sul1, and tetG) are all clustered together on a segment of the chromosome that is approximately 13 kb in length and includes two class 1 integrons (7, 8).
TABLE 1.
Antibiograms (R types) of 92 MR DT104 isolates from humans and animals
| Source | No. of isolates tested | No. of isolates with R typea:
|
|||
|---|---|---|---|---|---|
| ACSSuSpTNxCpL (A) | ACSSuSpTTmNxCpL (B) | ACNeKSSuSpTNxCpL (C) | SSuSpTNxCpL (D) | ||
| Humans | 49 | 46 | 3 | ||
| Turkeys | 14 | 14 | |||
| Cattle | 12 | 11 | 1 | ||
| Pigs | 8 | 7 | 1 | ||
| Sheep | 1 | 1 | |||
| Chickens | 2 | 1 | 1 | ||
| Dog | 1 | 1 | |||
| Cat | 1 | 1 | |||
| Rabbit | 1 | 1 | |||
| Pheasant | 1 | 1 | |||
| Partridge | 1 | 1 | |||
| Other wild birds | 1 | 1 | |||
Antimicrobial abbreviations: A, ampicillin; C, chloramphenicol; Ne, neomycin; K, kanamycin; S, streptomycin; Sp, spectinomycin; Su, sulfonamides; T, tetracyclines; Tm, trimethoprim; Nx, nalidixic acid; CpL, ciprofloxacin (low level). R type codes are in parentheses.
Since 1988 fluoroquinolones have become the antimicrobials of choice for extraintestinal salmonella infections. However, an increasing number of treatment failures have been associated with ciprofloxacin-resistant salmonella, including infections associated with strains with resistance levels below the designated MIC (27–29, 39, 40). Since 1993, the increasing number of MR DT104 isolates from humans and animals with additional resistance to nalidixic acid and decreased susceptibility to ciprofloxacin has become an important issue, particularly as this has followed the licensing of enrofloxacin for veterinary use. Moreover in a recent food-borne outbreak of MR DT104 with resistance to nalidixic acid and reduced susceptibility to ciprofloxacin in Denmark, four hospitalized patients did not respond to treatment with ciprofloxacin and there were two deaths (25). This clearly illustrates the potential risk that decreased susceptibility to ciprofloxacin can present in the effective antibiotic therapy of patients with serious salmonella infections, particularly when such strains are already resistant to several therapeutic antimicrobials.
Bacterial resistance to quinolones is chromosomal in origin and can be caused by alterations in target enzymes (DNA gyrase and topoisomerase IV), a decrease in drug permeability, or an active efflux mechanism (19, 31). Although quinolone resistance can involve a variety of different mechanisms, mutations within gyrA resulting in amino acid substitutions in the GyrA subunit of DNA gyrase play a major role in quinolone resistance in gram-negative bacteria including Salmonella (19). Point mutations in a region of the gyrA gene product, between amino acids 67 and 122, termed the quinolone resistance-determining region, have frequently been detected in salmonella serotypes that are resistant to nalidixic acid and that exhibit decreased susceptibility to ciprofloxacin (30, 42). Although several different GyrA mutations have been identified in Salmonella, Ser-83 and Asp-87 are most commonly mutated in clinically resistant isolates and in isolates with decreased susceptibility.
In this study we describe the adaptation of a rapid LightCycler assay, previously described by Gibson et al. (14), to detect the GyrA substitutions Ser-83 to Phe (TCC→TTC), Asp-87 to Gly (GAC→GGC), and Asp-87 to Asn (GAC→AAC) in strains of MR DT104 from humans and animals. This LightCycler gyrA mutation assay (GAMA) involves real-time PCR followed by the detection of mutations using oligonucleotide probes and thermal analysis. We have subsequently applied GAMA to investigate the type and frequency of gyrA mutations in 92 isolates from humans and animals that were resistant to nalidixic acid and that had reduced susceptibility to ciprofloxacin. These strains were additionally characterized using pulsed-field gel electrophoresis (PFGE), plasmid profiling, and a long-PCR assay to detect the ACSSpSuT resistance gene cluster.
MATERIALS AND METHODS
Bacterial strains.
Five MR S. enterica serotype Typhimurium DT104 strains with known gyrA mutations were selected as controls for the LightCycler GAMA protocol (33). These included P3801900 (Asp-87 to Asn), P4111690 (Asp-87 to Asn), P3424780 (Asp-87 to Gly), P4156400 (Asp-87 to Gly), and P3749380 (Ser-83 to Phe). P3343110 was included as a quinolone-sensitive DT104 strain with no gyrA mutations. Mutations in the gyrA gene in 49 human and 43 animal MR DT104 isolates were investigated. The human isolates were from patients in England and Wales in 1999 and were epidemiologically unrelated. The animal strains were isolated in 1999 and 2000 and came from a variety of sources (Table 1).
Phage typing and antimicrobial susceptibility tests.
Isolates were phage typed by the method of Callow (9) as extended by Anderson (2) and assigned to phage types in accordance with the scheme of Anderson et al. (3). Resistance to ampicillin, chloramphenicol, gentamicin, kanamycin, neomycin, streptomycin, spectinomycin, sulfonamides, tetracyclines, trimethoprim, low-level ciprofloxacin, high-level ciprofloxacin, nalidixic acid, and furazolidone was determined using a breakpoint method in Iso-Sensitest agar (12). The final concentrations (in micrograms per milliliter) of the antibacterial drugs were as follows: ampicillin, 8; chloramphenicol, 8; gentamicin, 4; kanamycin, 8; neomycin, 8; streptomycin, 16; spectinomycin, 64; sulfonamides, 64; tetracyclines, 8; trimethoprim, 2; low-level ciprofloxacin, 0.125; high-level ciprofloxacin, 1; nalidixic acid, 16; furazolidone, 8.
Detection of gyrA mutations using a LightCycler GAMA.
Briefly, GAMA involves amplification of a region of the gyrA gene (encompassing the nucleotides between codons 71 and 102) coupled with simultaneous detection of product formation by the LightCycler software (Roche Diagnostics Ltd., Lewes, United Kingdom), as the double-stranded DNA-specific fluorophore Sybr Green 1 (SG1) (BioGene, Cambridge, United Kingdom) is incorporated into the PCR amplicons. The level of SG1 fluorescence increases as the amount of PCR product doubles with each cycle and is measured by channel 1 (wavelength, 530 nm) of the LightCycler. The PCR product is then denatured, and a single gyrA mutation-specific oligonucleotide probe labeled with fluorophore Cy5 anneals to its target. Hybridization of the probe to the target DNA strand leads to an increase in Cy5 fluorescence as a result of fluorescence resonance energy transfer (FRET) between SG1 and Cy5 (14); this is measured by channel 3 (710 nm). The temperature is then increased, and the probe and target dissociate. Increasing the temperature stepwise to 94°C causes the Cy5 fluorescence to decrease as the probe and target dissociate and Cy5 and SG1 are no longer in close enough proximity for FRET to occur. If the specific mutation is not present, the mismatch of the probe with the target destabilizes the hybrid, so the decrease in fluorescence will occur at a melting temperature (Tm) lower than that for a hybrid where there are no mismatches.
A 96-bp fragment of gyrA was amplified with primers gyrAF (5′-GGTGACGTAATCGGTAAATA-3′) and gyrAR (5′-CAGCATGTAACGCAGCGA-3′), purchased from MWG Biotech UK Ltd. (Milton Keynes, United Kingdom) and based on the sequence of gyrA from S. enterica serotype Typhimurium NCTC 74 (17). The PCR and hybridization reaction were carried out in a 20-μl volume containing 1× buffer (10× buffer is 500 mM Tris-HCl, 20 mM MgCl2, and 5 μg of bovine serum albumin/ml), 20 ng of template DNA extracted using the DNeasy DNA extraction kit (Qiagen, Crawley, United Kingdom), 200 μM (each) deoxynucleoside triphosphate (Boehringer Mannheim, Lewes, United Kingdom), 0.5 μM forward primer (gyrAF), 0.2 μM reverse primer (gyrAR), 0.8 U of platinum Taq (Life Technologies, Paisley, United Kingdom), 1 mM MgCl2, a 1/10,000 dilution of SG1 (BioGene), and a 0.5 μM concentration of the probe complementary to either the Asp-87-to-Asn (GAC→AAC) mutation (gyrAPI [Cy5-5′-GGTGTTATACACTGCGGAAT-3′-biotin]), the Ser-83-to-Phe (TCC→TTC) mutation (gyrAPII [Cy5-5′-GGTGTCATACACTGCGAAAT-3′-biotin]), or the Asp-87-to-Gly (GAC→GGC) mutation (gyrAPIII [Cy5-5′-GGTGCCATACACTGCGGAAT-3′-biotin]) where bases complementary to specific point mutations are shown in bold. The labeled probes were also synthesized by MWG Biotech. The 3′ end of each probe is blocked with biotin to prevent the probes from behaving as primers. Forward and reverse primers were added in unequal concentrations in order to favor the amplification of the strand to which the probe bound. Table 2 lists the PCR amplification and probe melting conditions.
TABLE 2.
Amplification and melting cycles
| Step (no. of cycles) and target temp (°C) | Increment time (s) | Temp transition rate (°C/s) |
|---|---|---|
| Initial denaturation (1) | ||
| 94 | 10 | 20 |
| Amplification (50) | ||
| 92 | 0 | 20 |
| 50 | 0 | 20 |
| 55 | 0 | 3 |
| 74 | 10 | 20 |
| Melting (1) | ||
| 99 | 30 | 20 |
| 45 | 0 | 20 |
| 94 | 0 | 0.2 |
DNA sequencing.
DNA sequencing was performed by amplifying a fragment of the gyrA gene using the primers P1 (5′-TGTCCGAGATGGCCTGAAGC-3′) and P2 (5′-TACCGTCATAGTTATCCACG-3′), previously described by Griggs et al. (17). These primers surround the nucleotides between codons 37 and 151 of gyrA and therefore include all bases previously detected as mutation hot spots. PCR was performed in a 50-μl volume with the Hybaid (Ashford, United Kingdom). MultiBlock system. Reaction mixtures contained 1× MgCl2-free buffer (Life Technologies), 0.5 μM (each) primer, 2.5 U of Taq DNA polymerase (Life Technologies), 200 μM (each) deoxynucleoside triphosphate (Boehringer Mannheim), 1.5 mM MgCl2, and approximately 25 ng of template DNA. After an initial denaturation (3 min at 94°C), 25 cycles of amplification were performed as follows: denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and DNA extension at 72°C for 1 min. Finally DNA was extended at 72°C for 10 min for 1 cycle. The 347-bp products were purified using the QIAquick PCR purification kit (Qiagen), and the sequencing reactions were performed using the P1 and P2 primers and the CEQ dye terminator cycle sequencing kit (Beckman Coulter, High Wycombe, United Kingdom). The purified sequencing reaction samples were run on the CEQ 2000 DNA sequencer using the DTSC-2 method (Beckman Coulter DTCS sequencing protocol).
PFGE.
Chromosomal DNA was prepared by the method described by Powell et al. (32). DNA contained in the agarose plug was digested with 40 U of XbaI (New England BioLabs, Hitchin, United Kingdom). DNA fragments were resolved on 1% agarose gels (PFGE certified; Bio-Rad, Hemel Hempstead, United Kingdom) and run in 0.5% Tris-borate-EDTA on the contour-clamped homogeneous electric field DRII apparatus (Bio-Rad). The conditions for electrophoresis were 180 V for 44 h with a pulse time from 6 to 72 s. Gels were then stained with ethidium bromide (0.5 μg/ml) and visualized under UV light (312 nm). A mixture of lambda DNA HindIII fragments and lambda concatamers was used as a size standard (New England BioLabs).
Plasmid analysis.
Plasmid DNA was isolated by the method of Kado and Liu (22) and run on a 0.6% agarose gel with Escherichia coli K-12 strain 39R861 as the plasmid molecular mass marker (35). Plasmid sizes were determined with reference to plasmids carried in E. coli 39R861.
Long PCR of ACSSpSuT resistance gene cluster.
A 10,041-bp fragment of the ACSSpSuT resistance gene cluster was amplified using primers XLF (5′-TCAGAGGTGCTAAGCGTCATTGAG-3′) and XLR (5′-GCTTGATGCTCACTCCACACAACT-3′) based on the sequence of the antibiotic resistance gene cluster from S. enterica serotype Typhimurium DT104 isolate H3380 (8) and annealed to aadA2 and blaCARB-2, respectively. XLF and XLR were purchased from MWG Biotech. The reaction was performed on the Hybaid MultiBlock system in 100-μl volumes. The reaction mixtures contained 1× 3.3 XL buffer (PE Biosystems, Warrington, United Kingdom), 200 μM (each) deoxynucleoside triphosphate (Boehringer Mannheim), 30 pM (each) primer, 0.9 mM magnesium acetate, 4 U of rTth DNA polymerase XL (PE Biosystems), and 75 ng of template DNA. After an initial denaturation (1 min at 95°C), 30 cycles of amplification were performed as follows: denaturation at 95°C for 1 min, annealing at 59°C for 1 min, and DNA extension at 72°C for 10 min. Finally DNA was extended at 72°C for 10 min for 1 cycle.
RESULTS
Susceptibilities to antimicrobials.
All MR DT104 isolates were resistant to six or more antibiotics, including nalidixic acid (>16 mg/liter). All strains exhibited low-level resistance to ciprofloxacin (MIC, 0.25 to 1.0 mg/liter). The antibiograms are summarized in Table 1.
Type and frequency of gyrA mutations in MR DT104 using GAMA and DNA sequencing.
Table 3 shows the mean Tms of the three probes with the gyrA PCR products from MR DT104 quinolone-resistant control strains with known gyrA mutations and one quinolone-sensitive DT104 strain. The Tms of strains which had nucleotide mismatches with the three probes were consistently lower than the Tms of the strains where there was 100% homology between probe and target.
TABLE 3.
Tms of the three gyrA mutation probes (gyrAPI, gyrAPII, and gyrAPIII) with the PCR products from three MR DT104 control strains with characterized GyrA substitutions and one quinolone-sensitive DT104 straina
| Strain | GyrA mutation | Mean Tm (°C) (SD) with:
|
||
|---|---|---|---|---|
| gyrAPI (Asp-87→Asn) | gyrAPII (Asp-87→Gly) | gyrAPIII (Ser-83→Phe) | ||
| P3801900 | Asp-87→Asn | 64.0 (0.96) | 54.5 (0.86) | 52.7 (1.53) |
| P3424780 | Asp-87→Gly | 58.2 (0.41) | 64.3 (0.29) | 57.3 (0.64) |
| P3749380 | Ser-83→Phe | 55.4 (0.64) | 55.6 (0.77) | 64.5 (1.0) |
| P3343110 | No mutation | 58.8 (0.60) | 60.0 (0.73) | 60.4 (0.4) |
The results are the means of five samples.
The 92 MR DT104 isolates from humans and animals were assayed with three gyrA mutation probes. Probe-target Tm analysis showed that 37 human and 30 animal isolates had an Asp-87-to-Asn mutation. Six human and six animal isolates had a Ser-83-to-Phe mutation, and five human and two animal isolates had an Asp-87-to-Gly mutation. The remaining six strains were all found to have Tms lower than those of the positive controls when assayed with the different probes, which suggested that these strains had different gyrA mutations. Sequencing gyrA from these six strains showed that one human strain and two animal strains had an Asp-87-to-Tyr mutation, two animal strains had a Ser-83-to-Tyr mutation, and one animal strain had no gyrA mutation (Table 4).
TABLE 4.
Characteristics of 92 MR DT104 strains from humans and animals
| No. of isolates
|
R type codea | Plasmid profile (MDa) | PFGE profile | GyrA substitution | |
|---|---|---|---|---|---|
| Human (n = 49) | Animal (n = 43) | ||||
| 23 | 20 | A | 60 | XTm1 | Asp-87→Asn |
| 5 | 3 | A | 60 | XTm1 | Ser-83→Phe |
| 3 | 2 | A | 60 | XTm1 | Asp-87→Gly |
| 1 | 1 | A | 60 | XTm1 | Asp-87→Tyr |
| 1 | A | 60 | XTm46 | Ser-83→Tyr | |
| 2 | 4 | A | 60, 2.0 | XTm1 | Asp-87→Asn |
| 1 | A | 60, 2.0 | XTm1 | Asp-87→Gly | |
| 3 | A | 60, 1.0 | XTm1 | Asp-87→Asn | |
| 1 | A | 60, 1.0 | XTm1 | Asp-87→Gly | |
| 2 | 1 | A | 60, 2.5 | XTm1 | Asp-87→Asn |
| 1 | A | 60, 54 | XTm1 | Asp-87→Asn | |
| 1 | A | 60, 35 | XTm1 | Asp-87→Asn | |
| 1 | A | 52, 2.0 | XTm1 | Asp-87→Asn | |
| 1 | A | 70 | XTm1 | Asp-87→Asn | |
| 1 | A | 70, 3.0 | XTm1 | Asp-87→Asn | |
| 1 | A | 60, 3.0 | XTm1 | Asp-87→Asn | |
| 1 | A | 120, 60 | XTm1 | Ser-83→Phe | |
| 1 | A | 60, 4.0, 2.5 | XTm1 | Asp-87→Asn | |
| 1 | A | 60, 4.0, 2.0 | XTm47 | Ser-83→Phe | |
| 1 | A | 60, 3.0, 2.0 | XTm1 | Asp-87→Asn | |
| 1 | A | 60, 2.5, 2.0 | XTm1 | Asp-87→Tyr | |
| 1 | A | 60, 4.0, 2.0 | XTm18 | Asp-87→Asn | |
| 1 | B | 60, 4.6 | XTm1 | Asp-87→Asn | |
| 1 | B | 60, 4.6 | XTm1 | Ser-83→Phe | |
| 1 | B | 60, 40, 4.6 | XTm1 | Asp-87→Asn | |
| 1 | B | 60, 4.6 | XTm18 | Ser-83→Tyr | |
| 1 | B | 60, 4.6, 2.0 | XTm18 | No mutation | |
| 1 | C | 60, 4.1, 4, 3.6, 2.0 | XTm18 | Ser-83→Phe | |
| 1 | D | 60 | XTm1 | Asp-87→Asn | |
See Table 1 for the correlation of R type codes to R types.
Long PCR.
Ninety-one of the MR DT104 isolates (R-types: ACSSpSuTNxCpL, ACSSpSuTTmNxCpL, and ACNeKSSpSuTNxCpL) produced a 10,041-bp fragment and were therefore positive for the ACSSpSuT gene cassette. Only one animal isolate was negative for long PCR; this strain was of R type SSpSuTNxCp.
PFGE and plasmid profiling.
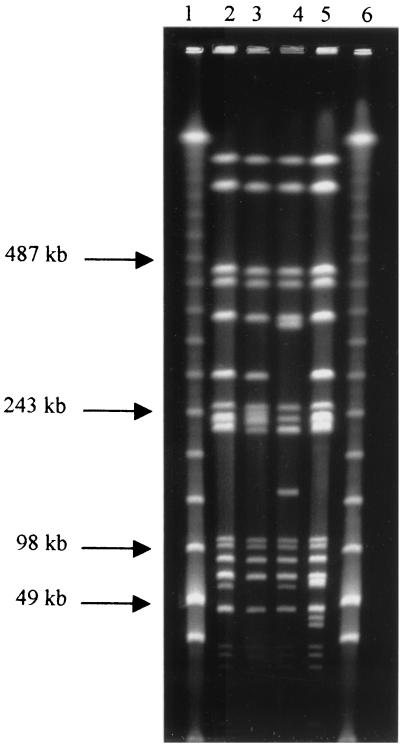
Four distinct patterns, which differed from each other by at least two band differences, were generated by PFGE (Fig. 1). The PFGE profiles of 49 human and 37 animal isolates were designated XTm-1, the predominant profile type for MR DT104 (33). The profiles of the remaining six animal isolates were designated either XTm18, XTm46, or XTm47 (Fig. 1; Table 4). Twenty plasmid profiles were identified (Table 4). Isolates contained between one and five plasmids ranging from 1.0 to 120 MDa. A 60-MDa plasmid was the most frequently identified plasmid type, present in 46 human and 43 animal isolates (Table 4).
FIG. 1.
PFGE of four profiles (XTm1, XTm18, XTm46, and XTm47) identified in 92 MR DT104 isolates. Lanes 1 and 6, size standard; lane 2, XTm1; lane 3, XTm18; lane 4, XTm46; lane 5, XTm47.
In total 58 isolates (32 human, 26 animal) had R type ACSSpSuTNxCpL and an XTm1 PFGE pattern, possessed a single 60-MDa plasmid (Table 4), and were positive by long PCR for the ACSSpSuT resistance gene cluster. These strains could, however, be further subdivided according to their GyrA substitution: Asp-87 to Asn for 43 strains, Ser-83 to Phe for 8, Asp-87 to Gly for 5, and Asp-87 to Tyr for 2 (Table 4). Seven strains that had R type ACSSpSuTNxCpL, that were positive by long PCR, that had the XTm1 PFGE pattern, and that possessed 60- and 2.0-MDa plasmids could also be further subdivided according to the GyrA mutation: six had an Asp-87-to-Asn mutation and one had an Asp-87-to-Gly mutation. Similarly four isolates that had an ACSSpSuTNxCpL R type, that were positive by long PCR, and that had an XTm1 profile and 60- and 1.0-MDa plasmids could be distinguished at the GyrA level: three had an Asp-87-to-Asn change and one had an Asp-87-to-Gly substitution. Finally two strains that had an ACSSuSpTTmNxCpL R type, that were positive by long PCR, that were characterized by an XTm1 PFGE profile, and that possessed 60- and 4.6-MDa plasmids could be distinguished by their GyrA substitution (Table 4).
DISCUSSION
In recent years the emergence of quinolone resistance in humans and food-producing animals has been a disturbing feature of salmonella infections in several European countries, including the United Kingdom, Germany, and Denmark (1, 18, 24, 36). In England and Wales fluoroquinolone resistance in human salmonella isolates was extremely rare up until 1994 (12, 36), despite the widespread use of ciprofloxacin in humans since 1987 (4). However, following the introduction of enrofloxacin for veterinary use in the United Kingdom in 1993 and its subsequent use in food-producing animals (28), decreased susceptibility to ciprofloxacin has rapidly developed, particularly among MR DT104 isolates of R type ACSSpSuT. In 1998 16% of human MR DT104 isolates were of R type ACSSpSuTNxCpL (37). A considerable increase in nalidixic acid resistance has also been observed among MR DT104 isolates from food-producing animals, especially turkeys (10). Studies in several other European countries have also shown that the incidence of quinolone-resistant salmonellae has increased substantially in the years following the approval of quinolones in livestock (1, 21, 23, 24).
In the present study we have described a LightCycler-based GAMA for the detection of three gyrA point mutations in MR DT104 with resistance to nalidixic acid and decreased susceptibility to ciprofloxacin. Eighty-six of 92 isolates possessed either an Asp-87-to-Asn, an Asp-87-to-Gly, or a Ser-83-to-Phe substitution when tested by GAMA. Although the method could not determine the gyrA mutation for the remaining six strains, it did dramatically reduce the requirement for DNA sequencing and the time taken to determine the presence of specific mutations. The procedure can be completed in less than 1 h following the extraction of DNA. Additionally, diluted cells can also be used in place of DNA (results not shown), thus decreasing further the length of time it takes to perform the test. Thus GAMA is a useful technique for rapidly screening large numbers of isolates in epidemiological investigations involving strains with decreased susceptibility to ciprofloxacin. GAMA has recently been applied to confirm the source and clonal identity of MR DT104 strains associated with a large outbreak involving unpasteurized milk (41). As the quinolone resistance-determining region of the gyrA gene appears to be highly conserved among salmonella serotypes (30), GAMA could also be used to investigate the frequency of different gyrA mutations in such organisms. Furthermore the method could be adapted to rapidly demonstrate gyrA mutations in a wide range of bacteria including Campylobacter spp. and E. coli (11, 34). In a clinical context GAMA could be applied to confirm decreased susceptibility to ciprofloxacin not detectable using the current susceptibility breakpoints of both the British Society for Antimicrobial Chemotherapy (5) and the National Committee for Clinical Laboratory Standards (26). Rapid identification of such strains may be useful in the management of serious salmonella infections.
In total 67 of 92 isolates (73%) had an Asp-87-to-Asn substitution. We therefore conclude that Asp-87 to Asn is the most common mutation giving rise to decreased susceptibility to ciprofloxacin in MR DT104 from both humans and animals. These findings are in agreement with two previous studies that have both identified Asp-87 to Asn as a common mutation among MR DT104 isolates (25, 33). The second-most-common mutation was at residue 83 and involved a Ser-to-Asp change. This mutation was identified in six human and six animal strains. The third-most-common mutation was an Asp-87-to-Gly substitution, detected in five human and two animal isolates. Partial sequencing of the gyrA gene from the remaining six strains showed that two human strains and one animal strain had an Asp-87-to-Tyr substitution, two animal strains had a Ser-83-to-Tyr substitution, and one animal strain had no gyrA mutation. It is possible that in the single strain with no gyrA mutation there may be a novel mutation outside of the sequenced region, although the sequenced area included all residues previously identified as mutation hot spots. These results suggest that other mechanisms and/or mutations are responsible for decreased susceptibility to ciprofloxacin in this strain. These could include a mutation in either gyrB, which encodes the B subunit of DNA gyrase, or parC, which codes for the ParC subunit of topoisomerase IV. However this may be unlikely, as gyrB and parC mutations are not normally associated with resistance to nalidixic acid and reduced susceptibility to ciprofloxacin in Salmonella (15, 42). Other alternative mechanisms of resistance could be a decrease in permeability to quinolones through alterations of the outer membrane proteins (30), modifications of lipopolysaccharide or an active efflux mechanism (16). The mechanism(s) of quinolone resistance in this particular strain is under further investigation.
Five different gyrA mutations were identified in this study. This variation can be used as an additional subtyping tool for MR DT104 with decreased susceptibility to ciprofloxacin and indistinguishable by several phenotypic and genotypic tests. For example 63% (58 of 92) of the strains tested were identical by R type, PFGE, and plasmid profiling and possessed the same 13-kb antibiotic resistance gene cluster but could be further subdivided according to their gyrA mutations. These results also demonstrate that in respect to their gyrA mutations these isolates were not all clonal in origin and may have arisen independently, over time, against a background of clonal spread of MR DT104.
The two most common gyrA mutations in the animal isolates were also those most common in the human strains. These findings confirm the association between quinolone-resistant human and animal isolates of MR DT104. However, although different gyrA mutations were identified in the animal isolates, there was no obvious link between gyrA mutation and isolate source. Most GyrA substitutions were detected in both animal and human isolates; the exception to this was a Ser-83-to-Tyr mutation, which was only identified in two animal strains. The finding that the Ser-83-to-Tyr mutation is only present in animals suggests that some of these mutants originate in animals before being widely distributed to the human population.
Fluoroquinolones are an important group of antibiotics for the treatment of potentially life-threatening extraintestinal salmonella infections in humans. It is therefore vital to limit their administration both in food-producing animals and humans so as to preserve their value for the treatment of not only humans but also sick animals. It is encouraging that in the United Kingdom the 1998 Codes of Practice recommended by the House of Lords Select Committee on Science and Technology for the use of fluoroquinolones in animal husbandry have been introduced by some pharmaceutical companies in attempts to reduce the unnecessary prophylactic use of such antimicrobials (20). The Advisory Committee on the Microbiological Safety of Food has also recently published guidelines aimed at reducing the use of veterinary antibiotics in the United Kingdom (6). However, probably the most significant recent news is the proposal by the Food and Drug Administration's Center for Veterinary Medicine to withdraw approval of enrofloxacin for use in poultry. This step has been taken as a result of the sharp increase in resistance to fluoroquinolones in Campylobacter spp. in the United States following the licensing of enrofloxacin for poultry production in 1996 (34). It is now hoped that these concerted actions will result in a reduction in resistance to fluoroquinolones, not only among MR DT104 and other nontyphoidal salmonella serotypes but also among Campylobacter spp. and other zoonotic bacterial pathogens.
ACKNOWLEDGMENTS
This work was funded by the Food Standards Agency, Code B10001 (formerly DH Code 240B), and the Ministry of Agriculture Fisheries and Food (MAFF Code FS3114).
We thank Mike Hampton for useful discussion.
REFERENCES
- 1.Aarestrup F M, Jensen N E, Jorsal S E, Nielsen T K. Emergence of resistance among bacteria causing infections in food animals in Denmark. Vet Rec. 2000;146:76–78. doi: 10.1136/vr.146.3.76. [DOI] [PubMed] [Google Scholar]
- 2.Anderson E S. The phage typing of salmonellae other than S. typhi, P. 89–110. In: van Oye E, editor. The world problem of salmonellosis. W. The Hague, The Netherlands: Junk; 1964. [Google Scholar]
- 3.Anderson E S, Ward L R, De Saxe M J, De Sa J D H. Bacteriophage-typing designations of Salmonella typhimurium. J Hyg. 1977;78:297–300. doi: 10.1017/s0022172400056187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angulo F J, Johnson K R, Tauxe R V, Cohen M L. Origins and consequences of antimicrobial-resistant nontyphoidal Salmonella: implications for the use of fluoroquinolones in food animals. Microb Drug Resist. 2000;6:77–83. doi: 10.1089/mdr.2000.6.77. [DOI] [PubMed] [Google Scholar]
- 5.Anonymous. Supplementary report on antibiotic sensitivity. J Antimicrob Chemother. 1996;38:1103–1105. [Google Scholar]
- 6.Anonymous. Report of the Advisory Committee on the Microbiological Safety of Food. London, United Kingdom: The Stationery Office; 1999. [Google Scholar]
- 7.Arcangoli M A, Leroy-Sétrin S, Martel J-L, Chaslus-Dancla E. A new chloramphenicol and florfenicol resistance gene flanked by two integron structures in Salmonella typhimurium DT104. FEMS Microbiol Lett. 1999;174:327–332. doi: 10.1111/j.1574-6968.1999.tb13586.x. [DOI] [PubMed] [Google Scholar]
- 8.Briggs C, Fratamico P F. Molecular characterization of the antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–849. doi: 10.1128/aac.43.4.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callow B R. A new phage-typing scheme for Salmonella typhimurium. J Hyg. 1959;57:346–359. doi: 10.1017/s0022172400020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies R H, Teale C J, Wray C, McLaren I M, Jones Y E, Chappell S, Kidd S. Nalidixic acid resistance in salmonellae isolated from turkeys and other livestock in Great Britain. Vet Rec. 1999;144:320–322. doi: 10.1136/vr.144.12.320. [DOI] [PubMed] [Google Scholar]
- 11.Everett M J, Jin Y F, Ricci V, Piddock L J V. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob Agents Chemother. 1996;40:2380–2386. doi: 10.1128/aac.40.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frost J A. Testing for resistance to antimicrobial drugs. In: Chart H, editor. Methods in practical bacteriology. Boca Raton, Fla: CRC Press; 1994. pp. 73–82. [Google Scholar]
- 13.Frost J A, Kelleher A, Rowe B. Increasing ciprofloxacin resistance in salmonellas in England and Wales 1991–1994. J Antimicrob Chemother. 1996;37:85–91. doi: 10.1093/jac/37.1.85. [DOI] [PubMed] [Google Scholar]
- 14.Gibson J R, Saunders N A, Burke B, Owen R J. Novel method for rapid determination of clarithromycin sensitivity in Helicobacter pylori. J Clin Microbiol. 1999;37:3746–3748. doi: 10.1128/jcm.37.11.3746-3748.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giraud E, Brisabois A, Martel J-L, Chaslus-Dancla E. Comparative studies of mutations in animal isolates and experimental in vitro- and in vivo-selected mutants of Salmonella spp. suggest a counterselection of highly fluoroquinolone-resistant strains in the field. Antimicrob Agents Chemother. 1999;43:2131–2137. doi: 10.1128/aac.43.9.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giraud E, Cloeckaert A, Kerboeuf D, Chaslus-Dancla E. Evidence for active efflux as the primary mechanism of resistance to ciprofloxacin in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother. 2000;44:1223–1228. doi: 10.1128/aac.44.5.1223-1228.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griggs D J, Gensberg K, Piddock L J V. Mutations in gyrA gene of quinolone-resistant salmonella serotypes isolated from humans and animals. Antimicrob Agents Chemother. 1996;40:1009–1013. doi: 10.1128/aac.40.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helmuth R, Protz D. How to modify conditions limiting resistance in bacteria in animals and other reservoirs. Clin Infect Dis. 1997;24:136–138. doi: 10.1093/clinids/24.supplement_1.s136. [DOI] [PubMed] [Google Scholar]
- 19.Hooper D C. Mechanisms of fluoroquinolone resistance. Drug Resist Updates. 1999;2:38–55. doi: 10.1054/drup.1998.0068. [DOI] [PubMed] [Google Scholar]
- 20.House of Lords Select Committee on Science and Technology. Resistance to antibiotics and other antimicrobial agents. London, United Kingdom: The Stationery Office; 1998. [Google Scholar]
- 21.Jacobs-Reitsma W F, Koenraad P M F, Bolder N M, Mulder R W A W. In vitro susceptibility of campylobacter and salmonella isolates from broilers to quinolones, ampicillin, tetracycline, and erythromycin. Vet Q. 1994;16:206–208. doi: 10.1080/01652176.1994.9694450. [DOI] [PubMed] [Google Scholar]
- 22.Kado C I, Liu S-T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kresken M, Hafner D, Mittermayer H, Verbist E, Bergogne-Bérézin E, Giamarellou H, Esposito S, van Klingeren B, Kayser F H, Reeves D S, Wiedemann B Study Group ‘Bacterial Resistance’ of the Paul-Ehrlich-Society for Chemotherapy. Prevalence of fluoroquinolone resistance in Europe. Infection. 1994;22:S90–S98. doi: 10.1007/BF01793572. [DOI] [PubMed] [Google Scholar]
- 24.Malorny B, Schroeter A, Helmuth R. Incidence of quinolone resistance over the period 1986 to 1998 in veterinary salmonella isolates from Germany. Antimicrob Agents Chemother. 1999;43:2278–2282. doi: 10.1128/aac.43.9.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mølbak K, Baggesen D-L, Aarestrup F M, Ebbesen J M, Engberg J, Frydendahl K, Gerner-Smidt P, Petersen A M, Wegener H C. An outbreak of multidrug resistant, quinolone resistant Salmonella enterica serotype Typhimurium DT104. N Engl J Med. 1999;341:1420–1425. doi: 10.1056/NEJM199911043411902. [DOI] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. 9th ed. 1999. Approved standard M-100-S9. National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 27.Pers C, Sogaard P, Pallensen L. Selection of multiple resistance in Salmonella enteritidis during treatment with ciprofloxacin. Scand J Infect Dis. 1996;28:529–531. doi: 10.3109/00365549609037954. [DOI] [PubMed] [Google Scholar]
- 28.Piddock L J V, Whale K, Wise R. Quinolone resistance in salmonella: clinical experience. Lancet. 1990;335:1459. doi: 10.1016/0140-6736(90)91484-r. [DOI] [PubMed] [Google Scholar]
- 29.Piddock L J V, Griggs D J, Hall M C, Yin Y F. Ciprofloxacin resistance in clinical isolates of Salmonella typhimurium obtained from two patients. Antimicrob Agents Chemother. 1993;37:662–666. doi: 10.1128/aac.37.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piddock L J V, Ricci V, McLaren I, Griggs D J. Role of mutation in the gyrA and parC genes of nalidixic-acid-resistant salmonella serotypes isolated from animals in the United Kingdom. J Antimicrob Chemother. 1998;41:635–641. doi: 10.1093/jac/41.6.635. [DOI] [PubMed] [Google Scholar]
- 31.Poole K. Efflux-mediated resistance to fluroquinolones in gram-negative bacteria. Antimicrob Agents Chemother. 2000;44:2233–2241. doi: 10.1128/aac.44.9.2233-2241.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Powell N G, Threlfall E J, Chart H, Rowe B. Subdivision of Salmonella enteritidis PT4 by pulsed field gel electrophoresis: potential for epidemiological surveillance. FEMS Microbiol Lett. 1994;119:193–198. doi: 10.1111/j.1574-6968.1994.tb06888.x. [DOI] [PubMed] [Google Scholar]
- 33.Ridley A, Threlfall E J. Molecular epidemiology of antibiotic resistance genes in multiresistant epidemic Salmonella typhimurium DT104. Microb Drug Resist. 1998;4:13–118. doi: 10.1089/mdr.1998.4.113. [DOI] [PubMed] [Google Scholar]
- 34.Smith K E, Besser J M, Hedberg C W, Leano F T, Bender J B, Wicklund J H, Johnson B P, Moore K A, Osterholm M T The Investigation Team. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992–1998. N Engl J Med. 1999;340:1525–1532. doi: 10.1056/NEJM199905203402001. [DOI] [PubMed] [Google Scholar]
- 35.Threlfall E J, Rowe B, Ward L R. Subdivision of Salmonella enteritidis phage types by plasmid profiling. Epidemiol Infect. 1989;102:459–465. doi: 10.1017/s095026880003017x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Threlfall E J, Ward L R, Rowe B. Increasing incidence of resistance to trimethoprim and ciprofloxacin in epidemic Salmonella typhimurium DT104 in England and Wales. Eurosurveillance. 1997;2:81–84. doi: 10.2807/esm.02.11.00187-en. [DOI] [PubMed] [Google Scholar]
- 37.Threlfall E J, Ward L R, Frost J A, Cheasty T, Willshaw G. The emergence and spread of antibiotic resistance in food-borne bacteria in the United Kingdom. Alliance Prudent Use Antibiot Newsl. 1999;17:1–4. [Google Scholar]
- 38.Threlfall E J. Epidemic Salmonella typhimurium DT 104—a truly international multiresistant clone. J Antimicrob Chemother. 2000;46:7–10. doi: 10.1093/jac/46.1.7. [DOI] [PubMed] [Google Scholar]
- 39.Vasallo F J, Martin-Rabadan P, Alcala L, Garcia-Lechuz J M, Rodiguez-Creixems M, Bouza E. Failure of ciprofloxacin therapy for invasive non-typhoidal salmonellosis. Clin Infect Dis. 1998;26:535–536. doi: 10.1086/517087. [DOI] [PubMed] [Google Scholar]
- 40.Wain J, Hoa N T T, Chinh N T, Vinh H, Everett M J, Diep T S, Day N P J, Solomon T, White N J, Piddock L J V, Parry C M. Quinolone-resistant Salmonella typhi in Viet Nam: molecular basis of resistance and clinical response to treatment. Clin Infect Dis. 1997;25:1404–1410. doi: 10.1086/516128. [DOI] [PubMed] [Google Scholar]
- 41.Walker R A, Lawson A J, Lindsay E A, Ward L R, Wright P A, Bolton F J, Wareing D R A, Corkish J D, Davies R H, Threlfall E J. Decreased susceptibility to ciprofloxacin in outbreak-associated multiresistant Salmonella typhimurium DT104. Vet Rec. 2000;147:395–396. doi: 10.1136/vr.147.14.395. [DOI] [PubMed] [Google Scholar]
- 42.Wiuff C, Madsen M, Baggesen D-L, Aarestrup F M. Quinolone resistance among Salmonella enterica from cattle, broilers and swine in Denmark. Microb Drug Resist. 2000;6:11–17. doi: 10.1089/mdr.2000.6.11. [DOI] [PubMed] [Google Scholar]