Abstract
Antibiotic persistence describes the presence of phenotypic variants within an isogenic bacterial population that are transiently tolerant to antibiotic treatment. Perturbations of metabolic homeostasis can promote antibiotic persistence, but the precise mechanisms are not well understood. Here, we use laboratory evolution, population-wide sequencing and biochemical characterizations to identify mutations in respiratory complex I and discover how they promote persistence in Escherichia coli. We show that persistence-inducing perturbations of metabolic homeostasis are associated with cytoplasmic acidification. Such cytoplasmic acidification is further strengthened by compromised proton pumping in the complex I mutants. While RpoS regulon activation induces persistence in the wild type, the aggravated cytoplasmic acidification in the complex I mutants leads to increased persistence via global shutdown of protein synthesis. Thus, we propose that cytoplasmic acidification, amplified by a compromised complex I, can act as a signaling hub for perturbed metabolic homeostasis in antibiotic persisters.
Subject terms: Ion channels, Antibacterial drug resistance, Experimental evolution, Antimicrobials
Antibiotic persisters are phenotypic variants within an isogenic bacterial population that are transiently tolerant to antibiotic treatment. Here, the authors provide evidence that cytoplasmic acidification, amplified by a compromised respiratory complex I, can act as a signaling hub for perturbed metabolic homeostasis in antibiotic persisters.
Introduction
Bacterial persistence describes the presence of phenotypic variants within an isogenic population that are transiently tolerant to otherwise lethal antibiotic therapy1–4. Such antibiotic-tolerant cells can endanger human health as they prolong therapy and can ultimately lead to relapsing infections1,3,5,6. In addition, tolerance levels are shown to increase during evolution under periodic antibiotic treatment7–10 and such increased tolerance levels were shown to proceed and accelerate the development of genetic resistance2,11–13.
Important advances towards understanding bacterial persistence have been made over the last years. First, toxin-antitoxin systems were found to be involved in persistence induction and awakening14–21. Second, the stringent response, a stress response that coordinates adaptation to stresses through altered levels of messenger nucleotides ppGpp and pppGpp (i.e. (p)ppGpp), has been linked to persistence via either increased levels of (p)ppGpp22–24 or a plethora of pathways including the induction of the RpoS regulon25,26, the inhibition of major processes such as DNA replication, transcription, and translation27,28 and the activation of toxin-antitoxin systems18,29,30.
A recently growing body of evidence suggests that also the ‘metabolic state’ is a highly important and general signal in the context of persistence31,32. First, a variety of metabolic genes affect the frequency of persister cells33–37. Second, metabolic perturbations like diauxic28 and rapid nutrient shifts26,38 and entry in stationary phase3,39,40 induce persister cell formation, where certain rapid carbon shifts can force almost an entire population into an antibiotic-tolerant state in a metabolic flux-dependent manner38. Other indications for metabolic perturbations driving persistence include the observed decrease in ATP concentration15,21,41–43 as well as other imbalances of the metabolic network such as very fast growth44, enzyme saturation45, or a perturbed TCA cycle and/or respiration40,46,47. Given the multitude of various metabolic perturbations leading to persistence, we hypothesize that one generic signal exists that triggers persistence upon perturbed metabolic homeostasis. Yet, it is unknown what this trigger is and how it initiates persister cell formation.
Here, we describe a mechanism for how different perturbations of metabolic homeostasis lead to persister cell formation. Through laboratory evolution of persistence and whole-genome, population-wide sequencing, we identified respiratory complex I as a key mutational target for increased persister formation upon entry into stationary phase, in model and pathogenic E. coli strains. Complex I mutants with increased persistence display compromised proton translocation leading to significant cytoplasmic acidification upon perturbations of metabolic homeostasis. We propose that acidification induces the persister state in two ways: through activation of the RpoS regulon and, upon more severe cytoplasmic acidification, through shutdown of protein synthesis. Our findings show how cytoplasmic acidification, amplified by a compromised complex I, acts as a central signaling hub to connect a perturbed metabolic homeostasis with persister cell formation.
Results
Evolution towards increased persistence leads to mutations in nuo
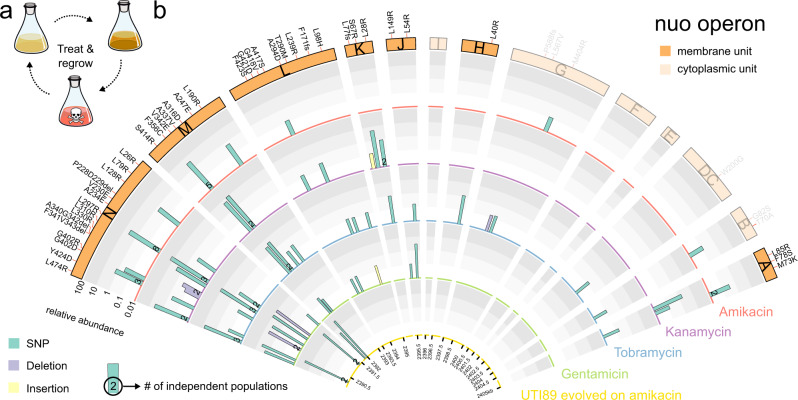
To discover genetic determinants of persister cell formation upon perturbations of metabolic homeostasis, we exploited laboratory evolution selecting for increased persistence in an environment that cycles between dilution and regrowth to stationary phase and antibiotic treatments (Fig. 1a)8,10,48. In this work, we investigated the genomic changes that occurred in 37 populations of an E. coli lab strain that were previously evolved under such cycling environment with daily exposure to amikacin (ten populations), kanamycin, tobramycin, or gentamicin (nine populations each)8. For comparison and generalization, we also included three populations of the uropathogen UTI89 that we evolved under daily exposure to amikacin in the current work, where we found 1000 to 10,000-fold increases in persistence, similar to the previously evolved populations of the lab strain (Supplementary Fig. 1a).
Fig. 1. As main target of evolution in E. coli towards increased persistence, the nuo operon is hit by mutations in genes primarily encoding membrane-spanning units.
a Scheme of experimental evolution alternating daily between dilution and regrowth to stationary phase to a density of 1–5 × 109 CFU ml−1 (respectively, light and dark orange) and antibiotic treatment (red), which selects for an increase in persistence. b A circos plot segment visualizes mutations that emerged in the evolved populations in genes of the nuo operon (orange rectangles with letters for each gene) which proved to be the main target of evolution (Supplementary Fig. 1). Genes encoding proteins that are part of the membrane-spanning arm of the complex and cytoplasmic domain are shown in bright and pale colors, respectively. Data of ten (amikacin), nine (kanamycin, tobramycin, and gentamicin) and three (UTI89) evolved populations were cumulated. Numbers in the bars show how many independently evolved populations contain that exact mutation (≥2). The concentric arcs show the relative frequency of mutations in populations evolved on different antibiotics (4 outer arcs; all treated for 5 h with 400 µg ml−1) and in populations of the uropathogenic strain UTI89 evolved on amikacin (inner arc; for 5 h with 400 µg ml−1). Target regions are rescaled for visual representation and different types of mutation are shown in different colors. The outer ring shows the resulting amino acid change at the protein level for each of the mutations. See also Supplementary Fig. 1, Supplementary Tables 1–2 and Supplementary Data 1–2. Source data are provided as a Source Data file.
To detect mutations in these 40 E. coli populations, we applied deep, whole-genome sequencing on population samples at the endpoint of evolution after 5–10 days. Initially focusing on mutations present at, or above, a frequency of 5% (see “Methods”), we identified on average 3.15 mutations per population amounting to a total of 128 mutations across all populations (Supplementary Data 1; Supplementary Fig. 1b, c). Statistical analyses indicate that the evolved populations consist entirely of mutants (Supplementary Fig. 1d, e). Confirming the low mutational complexity as inferred from population-wide sequencing, genome sequences of randomly picked clones at endpoints only contained one or two mutations (Supplementary Table 1). Mutations are significantly enriched in coding regions of 29 genes, specifically targeting genes coding for inner-membrane proteins (Supplementary Fig. 1f–h). Several targets are repeatedly hit by identical mutations in different populations, and/or emerge as multiple alleles (Supplementary Fig. 1b, j–l).
Mutations are strongly enriched in the nuo operon, which is hit 41 times by 19 unique alleles (Fig. 1b, Supplementary Fig. 1b, i–k), with eight alleles reoccurring identically in as many as ten populations (Supplementary Data 1; Supplementary Fig. 1l). When including mutations below our initial frequency cutoff of 5%, 237 additional mutations were uncovered in the 29 previously identified target genes (Supplementary Data 2), showing a similar enrichment in the nuo operon (52 hits) with 28 newly identified minor alleles (Fig. 1b). As overall 38.3% of each evolved population consists of nuo alleles (Supplementary Fig. 1m), we find that the nuo operon is the major target of evolution towards high persistence in our laboratory evolution experiment selecting for increased antibiotic survival upon perturbation of metabolic homeostasis through entry into stationary phase.
Mutations in nuo predominantly hit regions encoding transmembrane domains involved in proton translocation and are causal to high persistence
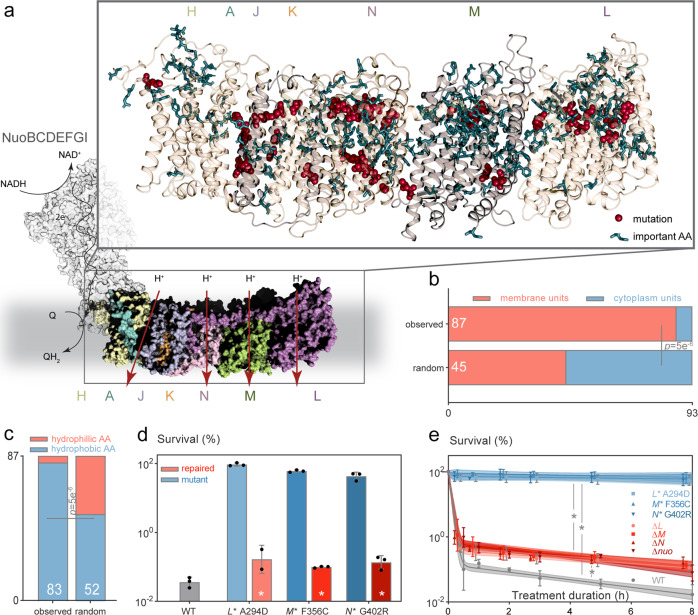
The E. coli nuo operon consists of 13 genes coding for NADH/ubiquinone oxidoreductase (respiratory complex I). The complex serves as an entry point for electrons from NADH into the electron transport chain (ETC) (Fig. 2a)49. 87 out of the 93 identified mutations and 41 unique alleles are present in the genes nuoAHJKLMN of complex I. Notably, the structurally and functionally distinct part of the complex, encoded by nuoBCDEFGI, contains significantly fewer mutations (only six) (Supplementary Fig. 2a, b). NuoBCDEFGI forms the cytoplasmic arm that catalyzes NADH oxidation and that transports the electrons via a series of iron-sulfur clusters to the ubiquinone reduction site, 100 Å apart50. NuoAHJKLMN builds the membrane arm that translocates protons across the membrane, thus contributing to the electrochemical membrane potential, i.e. proton motive force (PMF)51,52 (Fig. 2a). The mutations we found are primarily in the proton translocating units of the membrane arm and specifically hit codons of amino acids enriched in hydrophobic residues (83 times) and of those residues located in transmembrane helices (64 times) (Fig. 2c, Supplementary Fig. 2a, b). While many of the altered amino acids are in close proximity to regions involved in proton translocation (Fig. 2a inset, Supplementary Fig. 2c, d)51–53, the mutations do not directly affect amino acids reported to be crucial for proton translocation, such as amino acids involved in long-range structural changes along a central axis in the membrane arm, or in outlining the proton half-channels, or in the actual binding and export of protons (Fig. 2a). Based on these findings, we expected that the mutations do not abolish the overall complex I function, but rather induce smaller functional changes in complex I, likely connected with proton translocation, leading to the observed increase in persistence.
Fig. 2. High persistence-conferring nuo* mutations target hydrophobic amino acids in transmembrane subunits and do not cause a complete loss of function of the complex.
a Modeled structure of E. coli complex I (see Methods). The cytoplasmic subunits NuoBCDEFGI (gray surfaces) catalyze NADH oxidation and transfer two electrons over a series of iron-sulfur clusters (not shown) to the quinone reduction site. The quinol flows further down the electron transport chain in the membrane (gray background) to be re-oxidized by terminal oxidases (not shown). The hydrophobic subunits NuoAHJKLMN (in colored surfaces) translocate four H+ ions per molecule of NADH that is oxidized by the cytoplasmic subunits. The membrane part is magnified in the inset and is annotated with the positions of amino acid variants found in high-persistence mutants (red spheres) and with residues that are crucial for proton translocation (blue sticks, based on refs. 51–53). b, c High-persistence mutations are significantly enriched in b the membrane units and c predominantly target hydrophobic amino acids (Chi² comparison to random mutations, see Methods). d The nuo* mutations are causal for high persistence. Mutants with single mutations in each one of subunits L, M and N lose their high tolerance for amikacin (5h with 400 µg ml−1) when nuo* mutations are repaired (mean ±stdevs, n = 3; *p < 0.0001 for a within-strain comparison from a two-way ANOVA with Šidák’s posttest). e Killing dynamics with amikacin (400 µg ml−1) confirm the high persistence of nuo* point mutants in stationary phase and show that their effect cannot be mimicked by a single gene or operon knockout (in red). A model describing biphasic killing dynamics (lines ±95% CIs) was fitted to the data (means ±stdevs, n = 3; * fits are different based on AIC criterion). See also Supplementary Fig. 2. Source data are provided as a Source Data file.
To confirm causality of the mutations in nuo for persistence, we selected clones with single mutations in each one of the subunits L, M and N (called nuo* further on)—which are subunits that are all proposed to contain one distinct proton translocation path54—and restored the chromosomal wild-type alleles in these mutants by site-specific mutagenesis. Here we found that the strains again became susceptible towards amikacin, i.e. the antibiotic that imposed selective pressure during evolution (Fig. 2d). In addition, the minimal inhibitory concentration of amikacin remained unchanged in the nuo* mutants, excluding an increase in resistance as an explanation for their increased survival (Supplementary Fig. 2e). In fact, the nuo* mutants show clear cross-tolerance to other antibiotics in the stationary phase (Supplementary Fig. 2f–h), which is a known hallmark of persistence3. As already expected from the above-described characteristics of the mutations (e.g., that they are specific to the proton translocation arm and are not targeting crucial amino acids), the effect of the point mutations cannot simply be mimicked by destroying complex I functionality entirely, either by knocking out the individual subunits or the entire complex (Fig. 2e, Supplementary Fig. 2f, g). Restoring the wild-type alleles abrogates not only the amikacin tolerance as mentioned above, but also the multidrug tolerance of the point mutants for the related aminoglycoside kanamycin and for the unrelated fluoroquinolone ofloxacin (Supplementary Fig. 2i), demonstrating that nuo* mutations are causal to the high-persistence, multidrug-tolerant phenotype.
High-persistence-conferring variants of complex I oxidize NADH and transfer electrons but are impaired in proton translocation
To test whether the identified nuo* mutations indeed alter proton translocation, we produced and purified wild-type complex I and three variants, carrying a variation in either subunit L, M, or N. Despite its huge size (±550 kDa) and its multitude of cofactors, we were able to purify wild-type complex I and its variants with retained activity from E. coli membranes through several chromatographic steps (Supplementary Fig. 3a, b; see “Methods”). Yields of the preparations were similar between the samples (±0.2 mg protein g−1 cells) as were the banding patterns after SDS PAGE, both in line with previously obtained results55,56 (Supplementary Fig. 3c). Native gel electrophoresis confirmed stability and purity of the preparations, and in situ NADH oxidation demonstrated activity of the complexes (Supplementary Fig. 3d). The mutations did not introduce instability to the variants as judged from their “melting points”57 (Supplementary Fig. 3e, f). Thus, the purification yielded pure and active samples of the wild-type complex I and the three variants.
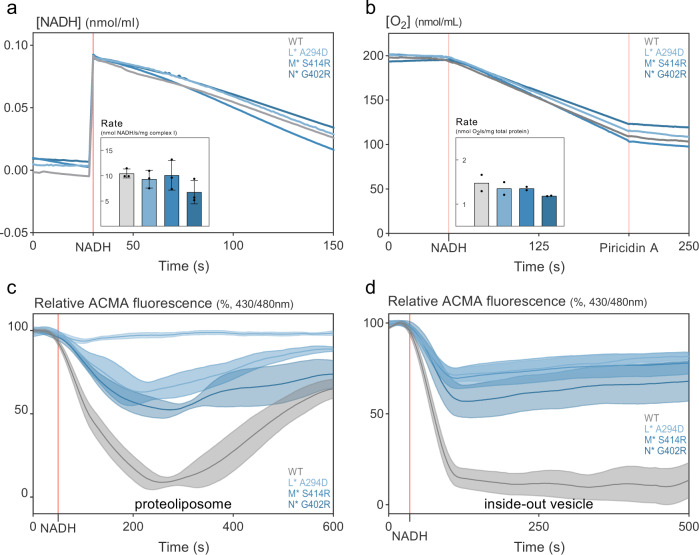
To profile the biochemical activities of the purified complexes, we made use 1) of proteoliposomes, i.e. vesicles made from E. coli polar lipids reconstituted with the individual preparations of complexes, of membrane fractions containing the wild-type complex I or the variants, and 2) of inside-out vesicles (ISOVs) prepared from E. coli membrane fractions (see “Methods”). Like proteoliposomes, the latter are closed compartments but additionally retain the full complexity of the in vivo membrane composition. NADH oxidation (and subsequent reduction of decyl ubiquinon in the membrane) was measured spectrophotometrically as a decrease in NADH absorption at 340 nm, where we found that the liposomes reconstituted with wild-type complex I and the variants oxidized NADH at a similar rate (Fig. 3a and inset). Electrons from NADH ultimately flow down the ETC via the quinone/quinol couple to the terminal oxidases, where in well-aerated conditions oxygen is reduced to water. Respirometry on membranes that contain wild-type complex I or its variants as sole NADH dehydrogenase (see “Methods”) revealed highly similar O2 reduction rates upon NADH addition (Fig. 3b and inset). Thus, NADH oxidation and electron transfer are both not affected by the nuo* mutations.
Fig. 3. High persistence-conferring nuo* variants are impaired in proton translocation.
a Complex I variants show similar NADH/decyl ubiquinone oxidoreductase activity as the wild-type complex. Complexes were reconstituted in liposomes and NADH concentration was monitored at 340 nm (one representative of n = 3). Average linear rates of NADH oxidation per mg complex I after NADH addition (inset, means ±stdev, n = 3) were not different (one-way ANOVA). Accounting for reconstitution efficiency, orientation, and activity of empty liposomes (see “Methods”), these rates amount to an overall NADH/decyl ubiquinone activity of 2.7 U mg−1, which is comparable to literature154. b The Nuo* variants and the wild-type complex transport electrons from NADH into the ETC with similar efficiency. Respirometry using a Clark-type electrode with membranes comprising complex I or its variants was used to measure the reduction of O2 by electrons which were released from NADH by complex I (one representative of n=2). Average linear rates of O2 consumption per mg protein after NADH addition (inset, means) were not different (one-way ANOVA) and amount to an overall NADH oxidase activity of 0.16 U mg−1, which is comparable to literature154. The second red vertical line indicates addition of piericidin A (10 µM), a potent complex I inhibitor, which indicates that O2 reduction indeed originates from NADH oxidation by complex I and that variants and wild-type complex I are equally sensitive to this inhibitor. c, d Proton translocation is impaired in all variants as estimated from the difference in fluorescence quenching of the pH-sensitive fluorophore ACMA in c proteoliposomes containing reconstituted complex I and in d ISOVs generated from membranes (means ±sems; n = 3). Relative fluorescent values were obtained by comparing the fluorescence to the start and through rescaling between 0 and 100% where 100% is the maximum value of each sample and 0% is the lowest value of the run (i.e. wild-type complex I which showed ±50% quenching, similar to values found in literature154). Proteoliposomes are leaky and therefore revert the ACMA quench over time. ISOVs, while additionally showing the activity of the terminal oxidases, are much tighter and show no reversion. In a, d, reactions were started by adding NADH (first red vertical line) and individual graphs are nudged horizontally and, in a, b also vertically to allow comparison. See also Supplementary Fig. 3. Source data are provided as a Source Data file.
To examine proton translation as third functionality of complex I, we made use of the fluorescence of 9-amino-6-chloro-2-methoxyacridine (ACMA) that can be quenched upon alkalization induced by proton translocation of complex I, allowing to investigate differences in proton pumping (Supplementary Fig. 3g, h). Compared to wild-type complex I, all variants showed, to varying degrees (50–75%), impaired proton translocation upon addition of NADH to the proteoliposomes (Fig. 3c). We confirmed this result by testing the activity of the variants in ISOVs (Fig. 3d). The ISOVs results furthermore quantitatively agreed with their more complex composition. As ISOVs also contain the terminal quinol oxidases, proton gradients measured with ACMA reflect the combined action of the quinol oxidases (1H+/e− for bd-type and 2H+/e− for bo3-type oxidase) and complex I (2H+/e−)58. ISOVs with complex I variants mainly translocate protons by the quinol oxidases and lack contribution from complex I (so 1–2 H+/e−), while complex I significantly contributes to proton translocation in wild-type ISOVs (so 3–4 H+/e−), which concurs with the 50–70% lower ACMA quenching in ISOVs with complex I variants compared to wild-type ISOVs (Fig. 3d). Overall, these results demonstrate that the identified nuo* mutations in complex I target proton translocation and uncouple translocation from electron transfer.
Neither impaired antibiotic uptake nor decreased energy levels fully explains complex I-dependent increased persistence
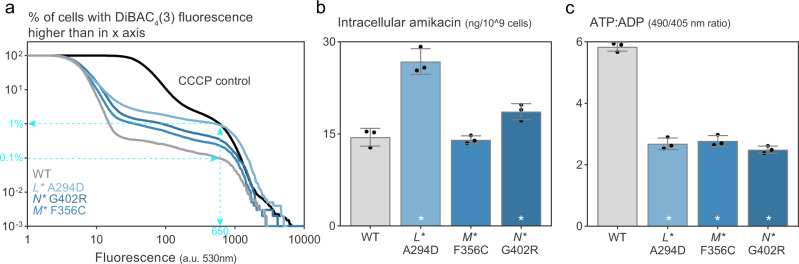
Given their compromised proton translocation, we hypothesized that the nuo* mutants could have a decreased proton motive force (PMF). Previously, persisters were shown to tolerate aminoglycosides by means of a decreased PMF leading to lower antibiotic uptake59. To test this hypothesis, we assessed the electrical gradient, i.e. the part of the PMF driving the uptake of charged aminoglycosides, using bis-(1,3-dibarbituric acid)-trimethine oxonol (DiBAC4(3)), a fluorescent dye whose uptake inversely correlates with the electrical potential across the membrane18. Using flow cytometry, we assessed the level of DiBAC4(3) of single cells to quantify their electrical gradient. Treatment with the dissipator carbonyl cyanide m-chlorophenyl hydrazone (CCCP) served as a depolarized control. The nuo* mutants showed a decreased electrical potential compared to the wild type (higher curves in Fig. 4a), resembling an expected consequence from the impaired proton translocation in complex I. However, the decrease in electrical gradient is minor—as expected, since the contribution of pH to the electrical gradient is usually only minor58,60—and does not agree with the completely depolarized control that received CCCP treatment. More importantly, under the assumption that depolarization fully explains the tolerant state, the differences in electrical gradient do not match with the differences in persister levels. For example, the 0.1% persister level of the wild type agrees with an electrical potential that is only present in 1% or less of the cells in the nuo* mutants (cyan lines in Fig. 4a) while these mutants have a persister level of ±50% (Fig. 2d, e, Supplementary Fig. 2f–i). Furthermore, and consistent with the small decrease in electrical gradient, nuo* mutants did not display decreased antibiotic uptake, as indicated by similar or even higher intracellular amikacin concentrations compared to the wild type (Fig. 4b). In addition, increased tolerance by a decreased antibiotic uptake as a result of a decreased electrical gradient would also not explain why the nuo* mutations also offer protection against ofloxacin (Supplementary Fig. 2g), a member of the fluoroquinolone antibiotics whose uptake does not dependent on an electrical gradient61–63 and which is also readily taken up by the nuo* mutants (Supplementary Fig. 4a). Altogether, these experiments show that a difference in antibiotic uptake does not explain the increased persister levels in the nuo* mutants.
Fig. 4. High-persistence nuo* mutants have no impaired antibiotic uptake and do not solely depend on a decreased ATP:ADP ratio.
a Single-cell fluorescence intensity distributions measured via cytometry where fluorescence corresponds to DiBAC4(3) uptake and serves a proxy for the extend of electrical depolarization. The data were plotted as “1-cumulative distributions”, showing the percentage of cells with an intensity above the fluorescence value specified in the x-axis to aid comparison between the strains. nuo* mutants in stationary phase show a minor decrease in electrical potential, indicated by the higher fraction of cells (higher curves) that have high DiBAC4(3) uptake (starting from a fluorescence value of ±10) (one representative of n = 2). Control samples treated with CCCP (in black, average across all repeats and strains), a protonophore and potent inducer of antibiotic tolerance70, show the expected increase in dye uptake in all cells for a depolarized sample. Cyan lines, arrows and numbers were added to put the results in biological context: the wild-type persister level (0.1% of the population, see Fig. 2e) corresponds to fluorescent level above 650. Only <1% of the nuo* mutants have such a high level of DiBAC4(3), whereas these strains carry on average 50% persister cells (Fig. 2d, e, Supplementary Fig. 2f–i). b Intracellular amikacin concentrations of stationary phase cultures of nuo* mutants measured in a bioassay are similar to the wild type or higher, confirming that antibiotic uptake is not decreased in the high-persistence mutants (means ±stdev, n = 3; *p < 0.01 for comparison to the wild type from a one-way ANOVA with Dunnett’s posttest). c In stationary phase (16–20 h after 1/100 dilution of an overnight culture), ATP:ADP ratios are significantly decreased as measured by the ratiometric GFP, Perceval (means ±stdev, n = 3; *p < 0.0001 for comparison to the wild type from a one-way ANOVA with Dunnett’s posttest). However, nuo* mutants show similar decreases in ATP:ADP ratios in exponential phase when they do not display increased persistence (Supplementary Figs. 4b, 2h). See also Supplementary Fig. 4. Source data are provided as a Source Data file.
Another consequence of the decreased proton translocation in the mutants could be a lowered ATP generation by the PMF-driven ATP synthase. Recently, a lower energy status has been linked to persistence15,21,41–43, while another study did not find decreased ATP levels in persisters26. We used a genetically encoded fluorescent reporter, Perceval64, to measure ATP:ADP ratios in stationary phase populations. The nuo* mutations led to a decreased ATP:ADP ratio compared to the wild type (Fig. 4c), which would be in line with the hypothesis of a de-energized cell status as cause of tolerance. However, the ATP:ADP ratios in the nuo* mutants are also decreased in the exponential phase (Supplementary Fig. 4e), a condition in which none of the mutants show increased tolerance (Supplementary Fig. 2h). While lower energy status might still contribute to the increased tolerance as an important prerequisite, we conclude that neither decreased antibiotic uptake nor a lower energy status by altered PMF can single-handedly explain the increased persister levels of the nuo* mutants, a result which we also confirmed for the evolved mutants of the uropathogenic strain (Supplementary Fig. 4c–g).
Increased cytoplasmic acidification in nuo* mutants is causal for increased persistence
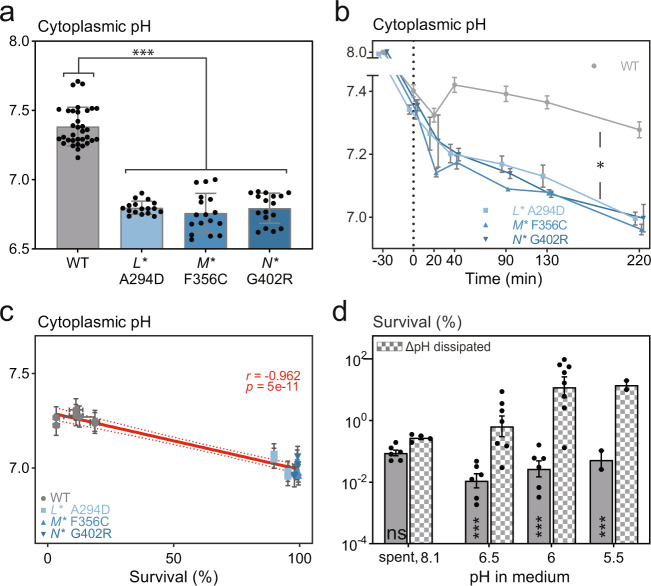
Next, we hypothesized that the impaired proton translocation of the complex I variants might influence pH homeostasis during metabolic perturbations, which in turn could lead to increased persistence. To test this hypothesis, we used the ratiometric fluorescent pH sensor pHluorin65 (Supplementary Fig. 4h) compared the cytoplasmic pH between the wild type and the high-persistence nuo* mutants. In exponential phase, a condition where we do not observe increased persistence in nuo* mutants (Supplementary Fig. 2h), the pH in the nuo* mutants did not significantly differ from the one in the wild type exhibiting an average pH value of 7.73 ±0.01 (Supplementary Fig. 4j). However, in stationary phase, all high-persistence nuo* mutants showed a significantly more acidified cytoplasm compared to the wild type (pHi = 6.79 ±0.04 versus pHi = 7.39 ±0.05; respectively, p < 0.0001) (Fig. 5a, Supplementary Fig. 4i). The increased persistence in the nuo* mutants correlates with stronger acidification of the cytoplasm (Supplementary Fig. 4k), likely being elicited by the perturbation in metabolic homeostasis during entry into stationary phase.
Fig. 5. Increased cytoplasmic acidification in nuo* mutants is causal for increased persistence.
a Cytoplasmic pH measured in cell populations using the ratiometric pHluorin sensor, is significantly decreased in the high-persistence nuo* mutants in comparison to the wild type in stationary phase, where the external pH is 8.1 ± 0.06 (means ±stdevs, n≥17; *** p<0.001 for a phenotype-level comparison from a linear mixed model). b Cytoplasmic pH of cells growing in exponential phase on M9 glucose medium decreases in all strains upon a shift to M9 fumarate medium (dotted line; pH = 7), known to induce persister formation. Dynamic measurement of cytoplasmic pH after a nutrient shift shows continued decreases over time only in the high-persistence nuo* mutants (means ±sems, n≥6; * slopes are different based on AIC criterion with the wild-type slope which is non-significantly different from 0 based on an F test). c Survival of amikacin treatment (for 4 h with 400 µg ml−1) assessed 30 min after the nutrient shift (in the same experiment as shown in b reveals a significant negative correlation between survival and cytoplasmic pH (means ±sems, n≥4). The linear regression line (±95% confidence interval) along with Pearson r and p values (top right corner) are shown in red. d Cytoplasmic acidification is causal for increased persister levels. Resuspending stationary phase cells in sterile spent medium buffered at different pH values increased the survival of amikacin treatment (for 5 h with 400 µg ml−1) from a pH of 6.5 and below in the wild type only when benzoate and methylamine are simultaneously added (ΔpH dissipators at 40 mM; boxed bars) to equilibrate cytoplasmic to extracellular pH. Adding this weak acid-base pair does not influence tolerance levels in unbuffered spent medium that has a pH of 8.1 ±0.06 (means ±stdevs, n ≥ 2; ***p < 0.001 and ns = non-significant for within-pH comparison from a linear mixed model with Šidák’s posttest). See also Supplementary Figs. 4–5. Source data are provided as a Source Data file.
To examine whether acidification of the cytoplasm is a general trait of metabolically induced persisters, we measured the cytoplasmic pH dynamically in time during well-controlled carbon nutrient shifts that are known to induce extremely high-persistence levels38. During exponential growth on glucose, we again found a highly similar intracellular pH of 7.97 ±0.02 in all strains (Fig. 5b), which dropped to 7.35 ±0.03 directly after the shift to fumarate in all strains. While in the wild type, the pH stabilized around this level, the cytoplasm of the high-persistence nuo* mutants continued to acidify to pH values around 7.07 ±0.13 (at 220 min after the nutrient shift) (Fig. 5b). Thus, also with a completely different way of perturbing metabolic homeostasis and inducing persistence, we find that the cytoplasm acidifies, and that this acidification is stronger in the nuo* mutants. Remarkably, also here, the decreased pH of the nuo* mutants correlated with an increased tolerance (Fig. 5c, Supplementary Fig. 5a), generalizing the findings of a lowered pH and increased tolerance to a different perturbation of metabolic homeostasis.
Intrigued by the correlation between the decreased cytoplasmic pH and increased tolerance, both in the stationary phase and upon the sudden shifts between glucose and fumarate, we asked whether the increased cytoplasmic acidification in the nuo* mutants is causal for the increased tolerance levels. To test this, we aimed to lower the cytoplasmic pH in the wild type to see whether this also results in increased antibiotic tolerance. To accomplish this, we resuspended stationary-phase cells in spent medium buffered at a range of pH values and added a membrane-permeable pair of a weak acid and base, potassium benzoate and ethylamine hydrochloride65, to equilibrate the cytoplasmic pH with the pH of the spent medium. Here, using the wild type, we found that acidification of the cytoplasm increases persistence levels (Fig. 5d, Supplementary Fig. 5b). Thus, the decreased cytoplasmic pH, which seems to be a general feature associated with persistence-inducing conditions involving metabolic perturbations, is further aggravated in the nuo* mutants, likely due to their compromised proton translocation, being causal for their increased tolerance.
RpoS contributes to increased persistence
Next, we asked how the lowered intracellular pH in the nuo* mutants could lead to increased tolerance. In our previous study, we found that the persisters generated through sudden nutrient shifts have an increased RpoS response with higher RpoS levels26. The stationary-phase specific nature of the increased persistence in the nuo* mutants also points to a role for RpoS, which is the ‘stationary phase’ sigma factor. As RpoS is posttranscriptionally and posttranslationally upregulated at low pH66–68, we hypothesized that the increased tolerance could be due to an increased pH-induced RpoS response.
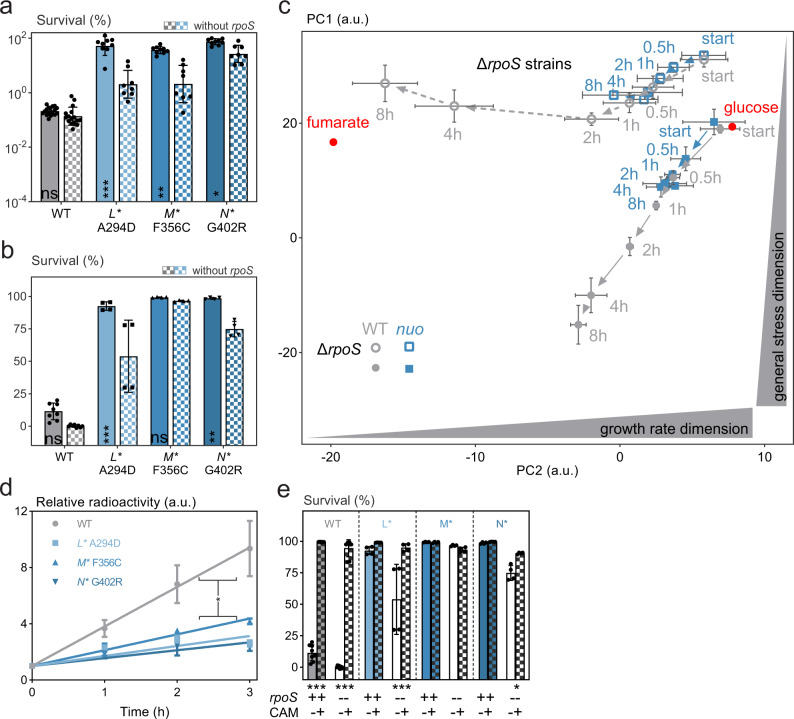
Indeed, supporting a role for RpoS, we found that deleting rpoS reduces persistence of the nuo* mutants to some extent in stationary phase conditions (Fig. 6a) and also upon the glucose-to-fumarate shift (Fig. 6b). A mechanism that might explain how cytoplasmic acidification leads to increased RpoS levels is via increased ppGpp levels through a pH-based inhibition of both the spontaneous and the SpoT-dependent ppGpp hydrolysis, which we found to occur at pH values characteristic for persisters (Supplementary Fig. 6; see “Methods”). Surprisingly, however, high-persistence nuo* mutants lacking both RelA and SpoT, which are incapable of producing (p)ppGpp, remain highly antibiotic-tolerant in stationary phase conditions (Supplementary Fig. 5c). Thus, alternative and/or redundant routes need to exist that connect pH to RpoS induction. Moreover, since the nuo* mutants lacking RpoS are still more antibiotic tolerant than the wild type (Fig. 6a, b) and since the correlation between cytoplasmic pH (Supplementary Fig. 5d, e) and tolerance remains significant in those strains (Supplementary Figs. 4k, l, 5a), a route additional to RpoS needs to connect the stronger decrease in cytoplasmic pH with the increased antibiotic tolerance.
Fig. 6. RpoS and decreased protein synthesis contribute to increased persistence in nuo* mutants.
a, b Increased tolerance of the nuo* mutants is partially RpoS-dependent in a stationary phase (amikacin for 5h with 400 µg ml−1) and b upon the glucose-to-fumarate shift (amikacin for 4h with 400 µg ml−1) (means ±stdevs, n≥4; * p<0.05, ** p<0.01, *** p<0.001 and ns = non-significant for within-strain comparison from on a linear mixed model with Šidák’s posttest). c Proteome changes after a shift from growth on glucose to fumarate (red dots) indicate that the proteome of high-persistence nuo* mutants is in a locked state (means ±stdevs, n≥2). The wild type with functional RpoS (closed gray symbols) shows an activation of the RpoS regulon as it moves down on the stress dimension of the PCA plot over time (see Methods), while removing rpoS (open gray symbols) leads to an adaptation to steady-state growth on fumarate and a migration along the growth rate dimension. Starting from 30 min after the shift to fumarate, the proteome of nuo* mutants remains unchanged, regardless of the presence of RpoS (in blue; average of nuoM* and nuoN* mutants). The nuoL* mutant was omitted as, in this particular experiment, it did not show strong and persistent acidification and hence, displayed a deviating proteome change (Supplementary Fig. 5f, g), which further substantiates our finding. d Translation activity is lower in nuo* mutants in stationary phase as measured by the incorporation of radioactive H3-leucine (means ±stdevs, n ≥ 2; *p < 0.05 for a phenotype-level comparison from a linear mixed model). e Chloramphenicol (CAM) supplementation (25 µg ml−1 at the time of the shift) induces tolerance towards amikacin (for 4h with 400 µg ml−1; 30 min after the shift) upon shifting from glucose to fumarate in the WT with/without rpoS and the nuoL* and nuoN* mutant lacking rpoS (means ±stdevs, n = 4; *p < 0.05, ***p < 0.001 for within-strain comparison between presence/absence of CAM from a linear mixed model with Šidák’s posttest). See also Supplementary Figs. 5, 6. Source data are provided as a Source Data file.
Decreased pH inhibits translation and causes increased persistence
To investigate how the strong pH decrease in the nuo* mutants could cause the increased tolerance, we subjected the wild type and the nuo* mutant to proteome analyses at different time points during the controlled shift from glucose to fumarate, where we had found that the nuo* mutants acidify more strongly than the wild type (Fig. 5b). We mapped the high-dimensional proteome data onto a two-dimensional space created by a previously performed principal component analysis of a similar dataset (Fig. 6c, Supplementary Fig. 5f; see “Methods”)26,69. One dimension reflects a “growth axis” along which proteomes from cultures growing at steady-state in different growth conditions cluster. The other dimension reflects a “stress axis”. It was previously found that the proteome of the wild type after the nutrient shift leading to persister formation, moved down the “stress” dimension, in a process that continued over >8 h26.
Here, agreeing with these previous results, we find that wild-type cells, when shifted from glucose to fumarate, display proteome dynamics moving down the stress dimension (closed, gray symbols in Fig. 6c), characterized by induction of the global stress response26. Secondly, we found that the proteome of the wild-type strain lacking rpoS (open, gray symbols in Fig. 6c) changes steadily over time towards a proteome of cells with normal steady-state growth on fumarate (red dot in Fig. 6c). This is in line with the previous finding that, in an ΔrpoS mutant, almost all cells resume growth after a rapid shift to fumarate26. Furthermore, this observation supports our earlier conjectured hypothesis32 that an ΔrpoS mutant strain invests resources in restoring metabolic homeostasis, and not in massive activation of stress responses, which is also corroborated by a somewhat restored pH homeostasis (i.e., reversal of the initial acidification) in the absence of RpoS in the wild type after a shift to fumarate (Supplementary Fig. 5d).
The proteome of nuo* mutants carrying a functional rpoS gene (closed, blue symbols in Fig. 6c) showed changes in the direction of an induced stress response for the first 30 min after the nutrient shift, similar to the changes of the wild-type proteome. Surprisingly, however, after 30 min the proteome stops changing abruptly (Fig. 6c), suggesting that the proteome might be locked in a frozen state with no further net turnover of proteins (i.e., either no or a balanced translation and degradation). Notably, it was also around 30 min that the cytoplasmic pH started to deviate between the wild type and the nuo* mutants (Fig. 5b) and that a correlation emerged between cytoplasmic pH and antibiotic tolerance (Fig. 5c, Supplementary Fig. 5a). Additionally, nuo* mutants lacking rpoS show a similar halt in proteome dynamics (Fig. 6c), which is in agreement with the only minor decreases in tolerance upon deletion of rpoS in these backgrounds. Thus, these data suggest that after about 30 min, which is when the pH in the nuo* mutants has dropped to pH values below 7.2, net protein turnover comes to a halt.
To test whether protein turnover is also halted in the stationary phase conditions on complex medium, we performed radioactive labeling experiments to follow protein synthesis. Here, we found that translation is indeed impaired in all nuo* mutants under the conditions that they were originally selected for (Fig. 6d), excluding the possibility that any net changes in the proteome were masked by a translation activity that balances out protein degradation. Thus, we conclude that the strong cytoplasmic acidification in the nuo* mutants halts changes in proteome turnover. We conjectured that halted protein translation, because of inactivity of amikacin’s antibiotic targets, could render the nuo* mutants more tolerant than the wild type.
To test whether the halted translation in the nuo* mutants is indeed responsible for increased persistence, we artificially blocked translation after shifting cells to fumarate by the addition of a sublethal dose of chloramphenicol and checked for tolerance against amikacin after 30 min. Here, we found that such a pretreatment, similar to what was found previously70, results in a strong induction of persistence in the wild type, comparable to the levels in the nuo* mutants without pretreatment (Fig. 6e). Additionally, such an artificial inhibition of translation also abolishes the need for rpoS both in the wild type and nuo* mutants. Thus, we conclude that the pH-induced halted translation in the nuo* mutants is indeed the main factor responsible for the observed increased persistence against amikacin, bypassing the need for a full-blown activation of stress responses by RpoS as mechanism for tolerance as is the case for the wild type.
Discussion
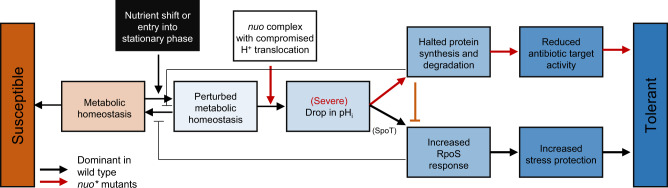
Here, we present a mechanistic model, which explains how persister cells are formed upon perturbations of metabolic homeostasis (Fig. 7). In this model, strong metabolic perturbations, such as the shift into stationary phase or abrupt nutrient shifts, lead to cytoplasmic acidification, likely through protons released through continued ATP consumption without ATP regeneration, which has been linked to acidification in other systems71–75. Mutations in complex I that compromise proton translocation result in a further decreased cytoplasmic pH upon metabolic perturbations. Part of the following induction of persistence, especially in the wild type, is conferred by the RpoS-dependent global stress response. In the nuo* mutants with increased cytoplasmic acidification, persistence is additionally conferred by a global halting of protein synthesis. At the system level, additional feedback and interaction mechanisms exist: an RpoS response inhibits restoration of metabolic homeostasis26 and the pH-induced halted protein synthesis further prevents restoration of homeostasis, while also halting a further activation of the RpoS response. Our newly identified mechanism also agrees with a growing body of recent research that suggest that bacterial metabolism, chemiosmotic homeostasis and membrane bioenergetics—in addition to drug uptake and activity of primary targets—dictates the lethality of bactericidal antibiotics76–78.
Fig. 7. Model connecting metabolic homeostasis, cytoplasmic acidification and two paths for persister cell formation.
A strong perturbation of metabolic homeostasis by nutrient shifts or entry in stationary phase acidifies the cytoplasm. Such acidification could occur due to an imbalance between ATP consumption and regeneration. In these conditions, complex I acts as a regulator of cytoplasmic pH: wild-type proton translocation activity counter-balances acidification; impaired proton translocation in the nuo* mutants enhances cytoplasmic acidification after ±30 min of perturbation of metabolic homeostasis. Mild cytoplasmic acidification leads to increased persistence by an increased RpoS response, possibly mediated by inhibition of SpoT-mediated hydrolysis of (p)ppGpp (dominant path in black arrows in the wild type). Stronger cytoplasmic acidification halts protein synthesis and renders antibiotic targets inactive leading to a second persistence mechanism (dominant path in red arrows in nuo* mutants with impaired complex I function.) At various points in this model, feedback loops are in place that affect persister formation. For instance, RpoS inhibits adaptation to maintain metabolic homeostasis, recovering from the initial pH drop. In turn, strong intracellular acidification via halted protein synthesis inhibits full RpoS activation.
Our work identifies complex I as an important persistence factor. Loss-of-function mutations in complex I (e.g., transposon or frameshift mutations) are long known to abrogate the growth advantage in long-term stationary-phase conditions of rpoS ‘GASP’ mutants in absence of antibiotics79. More recently, such mutations were shown to increase gentamicin tolerance in E. coli80 and aminoglycoside resistance in E. coli81–83 and Pseudomonas aeruginosa84 through a reduced PMF and antibiotic uptake. Such an effect could also be mimicked by artificially decreasing complex I activity85–88 and was recently proposed to underly the antibiotic tolerance of a point mutant in nuoN in P. aeruginosa89. Here, we describe a new way in which complex I alters antibiotic susceptibility, i.e. by changes that solely impair proton translocation activity of the complex. Mutations identified in this work specifically lie in the proximity of the three previously confirmed proton pathways in NuoL, M and N and were found at the interface of the NuoAHJK subunits, thereby corroborating a previously hypothesized fourth channel at this site51,53,90. Additionally, the nuo* mutants still take up antibiotics and show cross-tolerance towards other antibiotics, in contrast to the collateral hypersensitivity towards other antibiotics of (loss-of-function) mutations in nuo (and other genes) described in the context of aminoglycoside resistance due to reduced drug uptake83,91. Therefore, the nuo* mutations identified in the current study constitute a new mechanism in which complex I influences antibiotic survival in bacteria, i.e. by inducing persister formation through increased cytoplasmic acidification upon strong perturbations of metabolic homeostasis.
In this work, we uncovered that cytoplasmic acidification—induced by perturbations of metabolic homeostasis, i.e. upon nutrient shifts or entry into stationary phase—acts as a signal for persister formation in E. coli. In fact, a drop in cytoplasmic pH was observed in Salmonella upon macrophage engulfment and this acidification was furthermore important for the injection of modulating effector proteins that allow Salmonella to survive inside the vacuole92. At the same time, but without a connection to cytoplasmic acidification, Helaine and colleagues showed that macrophage internalization induced intracellular persister formation93. Indole, the inter-kingdom signal molecule that was previously connected to the persister state94–96, was shown to regulate intracellular acidification upon entry in stationary phase97 although very recently, the tryptophanase that catalyzes the formation of indole, not indole itself, was found to cause a lower pH in persisters98. In Mycobacterium smegmatis, bactericidal activity of antibiotics was linked to cytoplasmic alkalization during treatment99 while cytoplasmic acidification was recently found important for fully fledged resistance in marR E. coli mutants100. Beyond persisters and antibiotic survival, a drop in the cellular pH is more commonly associated with metabolic perturbations and dormancy. For instance, the pH of spores of Bacillus megaterium and Bacillus cereus is 1 to 1.3 pH units lower than the pH of growing cells, but rises rapidly upon germination101–103. In yeast, cytoplasmic pH was identified as a second messenger for glucose levels where low intracellular glucose induces acidification104. Even beyond microbial life, a pH drop seems to be a dormancy signal, as intracellular pH is a dormancy marker in strawberry plants105, and has been shown to regulate dormancy in shrimp embryos106. Thus, across different species and even kingdoms, cytoplasmic acidification might act as a ‘low-level’ signal, indicating strong perturbations of metabolic homeostasis, ultimately causing cells to enter ‘safe states’ such as dormancy or persistence.
Since RpoS is known to be upregulated at low pH66–68 its role in persistence after cytoplasmic acidification was somewhat expected. How cytoplasmic acidification influences the here uncovered halting of protein turnover as they occur in the nuo* mutants after 30 min (Fig. 5d), is less clear as mechanisms to downregulate protein synthesis during growth arrest are not fully understood107. The sudden and simultaneous stop of both protein production and degradation, as we inferred from both the sudden stop in proteome changes and the decreased translation activity (Fig. 6c, d), could result from cytoplasmic acidification leading to—as shown in yeast—a glass-like state with decreased intracellular dynamics that induces dormancy75, similar to the glassy cytoplasm connected with dormancy in other species108–111. Additional evidence for the significance of cytoplasm fluidity is the recently identified correlation of protein aggregate formation with persistence41,112–115, while, at the same time, widespread and pH-dependent aggregation of native-like proteins underlies the transition of the cytoplasm to a solid-like state75. Recently, mutations in translation-related genes were selected during laboratory evolution towards increased persistence and shown to affect translation more specifically116. Similarly, changes in cytoplasmic pH could also modulate ribosome activity more directly than via the induction of a global glass-like dormant cytoplasm, for example by unbalancing the ionic fluxes of other cations like K+, Ca2+, and Mg2, that were recently associated with ribosome functioning and/or antibiotic killing76,78,117. Therefore, a decrease in cytoplasmic pH could lead via a myriad of ways (e.g., cytoplasmic rigidity, ionic imbalances) to halted protein turnover and thus to increased tolerance.
With this work, we have unraveled the molecular basis of how metabolically induced persister cells are formed, where the dynamic nature of cytoplasmic pH, as a signal for strong metabolic perturbations, resembles a global connector of metabolism with persister formation. The generic nature of this mechanism, converging at cytoplasmic acidification, is consistent with the fact that numerous effects, including perturbations of many metabolic genes118,119, abrupt nutrient shifts, changes in extracellular pH93, sublethal concentrations of antibiotics and oxidative stress120 have all been observed to induce tolerance and could all converge in intracellular acidification.
Methods
Bacterial strains, media, and cultivation
All bacterial strains used in this study are listed in Supplementary Data 3, with the most important strains being SX43 or SX2513 (the ancestral lab strains used in most of our experiments as “wild type” or WT, which are close relatives to BW25113), and UTI89 (a uropathogenic E. coli strain). Genomic mutants were constructed with either P1vir phage transduction as described before121 or through homologous recombineering described before122. Where possible, mutants from an ordered gene knockout library in E. coli, the Keio collection123 were used as template to create new mutants and they were cured from their KmR-cassette by transformation with pCP20 expressing the FLP recombinase. At all steps, validity of mutants was checked by either PCR with gel electrophoresis or targeted Sanger sequencing or both using primers listed in Supplementary Data 4.
Experiments were performed using M9 minimal medium prepared as previously described38, Mueller Hinton Broth (MHB, Becton Dickinson; widely used in antibiotic sensitivity testing), lysogeny broth (LB) medium (10 g L−1 NaCl, 10 g L−1 trypton, 5 g L−1 yeast extract with/without 15 g L−1 agar), M63 minimum salts medium as previously described65 or ‘spent’ MHB medium. The carbon source in MHB and LB is an undefined mixture of peptides with minor traces of sugars. The spent MHB medium was prepared by removing wild-type cells from an overnight culture (37 °C, 200 rpm or 1g) by centrifugation and filtration through a 0.2 µm filter. For spent MHB medium buffered at a certain pH, the appropriate Good’s buffering agent124 was added (50 mM 2-(N-morpholino)ethanesulfonic acid for pH 5.5 and 6 and 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) for pH 6.5) with/without potassium benzoate (40 mM) and methylamine hydrochloride (40 mM) to dissipate ΔpH. Next, the pH was adjusted using HCl or KOH and the medium was filter sterilized. For minimal media, carbon source stock solutions were prepared in demineralized water, adjusted to pH 7.0 using NaOH or HCl, respectively, and sterile filtered through a 0.2 µm polyethersulfone (PES) filter. Glucose was added in a final concentration of 5 g L−1 and fumarate in a final concentration of 2 g L−1, respectively. Antibiotics, IPTG and arabinose were all prepared in demineralized water and sterile filtered through a 0.2 µm PES filter. Bacterial cultures were cultivated either in 50 ml medium in a 500 ml or 10 ml medium in a 100 ml Erlenmeyer flask closed with a 38 mm silicone sponge closure (Bellco Glass) at 37 °C, 300 rpm and 5 cm throw (5 × g) or in 100 ml medium in a 250 ml Erlenmeyer flask closed using cellulose stoppers (VWR) at 37 °C, 200 rpm and 2.5 cm throw (1 × g). Overnight cultures were diluted 1:100 in fresh medium and further diluted 1:100 as soon as OD600 of 0.5 is reached to keep cells in the exponential growth phase.
Assessment of the antibiotic sensitivity
MICs, survival levels, and killing curves were determined according to the methods described previously8,125,126. For MIC determination an overnight culture was diluted in fresh MHB to an inoculum of 1 × 106 colony-forming units per milliliter (CFU ml−1) and incubated in a range of two-fold antibiotic dilutions for 16–20 h. After incubation, the lowest antibiotic concentration where no growth was observed using the absorbance at 595 nm (BioTek), we defined as the MIC. Alternatively, commercially available MIC test strips were used (Liofilchem).
Antibiotic survival level in the stationary phase is obtained by taking the ratio CFU ml−1 after treatment (usually for 5h) and the total CFU ml−1 before antibiotic treatment (generally 1–5 × 109 CFU ml−1). CFU ml−1 were obtained by making serial dilutions, spiral plating, and semi-automatic quantification after 2 days of incubation at 37 °C (EddyJet and Flash & Grow). When using spent medium, stationary-phase cells were resuspended in an equal volume of the spent medium and treatment was started immediately.
The antibiotics that were used are all aminoglycosides or fluoroquinolones. We used these two classes as they are potent bactericidal drugs that furthermore can kill sensitive stationary phase cells and result in a biphasic killing pattern, indicative for the survival of persister cells. Furthermore, these antibiotics were used in our previous work which forms the basis of the current work8.
Assessment of antibiotic tolerance through cytometry upon nutrient shifts in M9 medium
Tolerance experiments upon nutrient shifts were always combined with ratiometric pH measurements. Cells were grown in M9 glucose medium containing ampicillin (100 µg ml−1) and arabinose (20 g L−1) with regular dilutions to maintain growth in the exponential phase. At OD600 0.5, pH was measured and persisters were induced by a switch to M9 fumarate medium. At multiple time points after the nutrient switch, samples were taken, and intracellular pH was determined as previously described. For determination of antibiotic tolerance, we treated the cells with amikacin (4 h at 400 µg ml −1) at 0.5 h after the switch. As a control we blocked translation using a low dose of chloramphenicol (25 µg ml −1) added direct after the switch to fumarate. After 4 h, amikacin treated cells were diluted 100-fold in LB and their ability to regrow was tracked using flow cytometry (for 3h, see refs. 26,127). Tolerant cells resumed growth, became bigger and lost their pHluorin fluorescent intensity whereas non‐growing cells retained their size and fluorescent intensity. The fraction of cells that did not resumed growth was determined by observing no cell size changes and no loss of fluorescence. Cells were measured every 30 min from 0 to 3 h after the transfer to LB medium. Fractions of non‐recovering cells from the period of 2.5 and 3 h after transfer to LB were used to calculate the fractions of tolerant cells by subtracting the nondividing cells from the total cells added to the culture at the start.
Evolution experiments
Experimental evolution using the uropathogenic E. coli strain UTI89 was performed as before8,48. Briefly, parallel cultures were grown overnight to stationary phase in MHB (±18 h), treated for 5 h with amikacin (400 µg ml−1; 100–200-fold MIC) to eliminate all non-persisters, washed three times in MgSO4 (10 mM) to remove antibiotics and diluted 1/100 into fresh MHB to allow another cycle of batch growth. A sigmoidal fit expected for the spread of a mutant in a haploid population was fitted to the data obtained under daily treatment:
with s the selective advantage of the assumed mutant and z and Za are the log-transformed persister fraction of the ancestor and mutant. A horizontal line was fitted to the control data without treatment and compared to a straight line with an extra sum of squares F test to confirm the absence of change of phenotype. All fitting was done in GraphPad Prism 8 using the least squares method.
Genome-wide next-generation sequencing
We detect mutations arising in evolved populations or clones by genome-wide sequencing using Illumina’s HiSeq platform. Genomic DNA was extracted by using the DNeasy Blood and Tissue Kit (Qiagen). Purity and concentration were assessed by Nanodrop, gel electrophoresis and Qubit. Average insert size of the prepared libraries was ±300 bp and sequencing was conducted at EMBL’s Genecore in Germany, the VIB Nucleomics core in Belgium, Eurofins, or the Genomics core of UZ Gasthuisberg in Belgium. Raw data files have been deposited in the NCBI SRA database with following accession IDs: PRJNA498891 (populations of the lab strain), PRJNA270307 (clonal data of the lab strain), PRJNA498717 (uropathogenic populations), PRJNA498708 (uropathogenic clones) and PRJNA768774 (to check the ΔrelAΔspoT deletion strains). The 100–150 bp paired-end output was analyzed using Qiagen’s CLC Genomics Workbench version 11.0. Full details on the used workflow and parameters are available upon request. Briefly, reads were mapped to reference sequences after quality control, read trimming and filtering (NC_000913.3 for lab strain, NC_007946.1 and NC_007941.1 for the uropathogen UTI89). Mutation lists were obtained with CLC’s low frequency variant detection tool. Further filtering of these lists was based on several quality features and prior experiences (scripts available upon request). Importantly, for clones, cutoff frequencies of 75% were used, while for population analysis, an initial cutoff frequency of 5% was used while later on, this cutoff was dropped for the regions of the target genes specifically and data were filtered on minimum coverage only (coverage >30, forward & reverse read count >0; script available upon request). Clonal results were furthermore confirmed using targeted Sanger sequencing with primers in Supplementary Data 4. While we did not specifically remove intergenic or synonymous mutations, only 3 and 0, respectively, were identified in the entire dataset at or above the cutoff frequency of 5%. The perl-based software package Circos was used (http://circos.ca/) to visualize data. Scripts and configuration files are available upon request. Mutations from the UTI background were added to figures, tables and data based on a pairwise alignment to the lab strain background.
For all the Chi² tests to examine for significant enrichment, we used 200,000 replicates in Monte Carlo simulations to compute p-values and computed the null hypothesis as the condition in which mutations would be hitting randomly in the genome (Supplementary Fig. 1f), in genes (Supplementary Fig. 1g), in genes coding for membrane proteins (Supplementary Fig. 1h), in genes coding for inner-membrane proteins (Supplementary Fig. 1i), in genes of the nuo operon (Fig. 2b), in nuo genes coding for the membrane-spanning subunit (Fig. 2c, Supplementary Fig. 3a, b). To do so, we used the genome-wide annotations in NC_000913: 4641652bp in total, 4089513bp of which are part of coding regions, 1040006bp of which code for membrane proteins, 822021bp of which code for inner-membrane proteins, 14673bp of which code for complex I of which 7110bp are for genes of the membrane subunit. For the membrane subunits, amino acids were scored to be part of either transmembrane helices or non-membrane-spanning loops based on predictions by Protter (wlab.ethz.ch/protter)128 and scored to be either hydrophobic or hydrophilic based on a scale computed before129 and a cutoff value of 0.6.
Genomically repairing nuo-mutation
P1vir phage transduction was used to genomically revert the identified mutations in nuoL, M and N by using the yfbP::KmR Keio mutant as donor strain as described elsewhere8,121.
Generating 3D structure of complex I from E. coli
No full protein structure exists for complex I of E. coli. To this end, we modeled the protein structure of the cytoplasmic domain (nuoBCDEFGI; uniprot IDs: P0AFC7, P33599, P0AFD1, P31979, P33602, P0AFD6) and membrane arm (nuoAHJKLMN; uniprot IDs: P0AFC3, P0AFD4, P0AFE0, P0AFE4, P33607, P0AFE8, P0AFF0) using the online SWISS modeling server (https://swissmodel.expasy.org/)130 with 6g2j and 4he8 PDB entries, respectively, as templates (Global Model Quality Estimation of, respectively, 0.56 and 0.67 with 35.8% and 38.5% sequence identity). Next, an overall structure of complex I of E. coli was obtained by aligning the modeled structures of the cytoplasmic and membrane domains to the structure of complex I of Thermus thermophilus (PDB entry 4hea)51. To this end, chains G and L were used as anchors by aligning them through the structural alignment of multiple proteins (STAMP) tool131 in Visual Molecular Dynamics (VMD)132 with standard settings. Images were generated by the rendering capabilities of PyMOL133.
Purification of complex I and its variants
Complex I transcribed from the pBADnuo nuoFHis6 was expressed in, and purified from an E. coli strain lacking complex I and the alternative NADH dehydrogenase as performed before by the Friedrich group55. Site-specific mutagenesis through PCR with Q5 polymerase (NEB) on this construct using primers nuoL_FW and nuoL_RV, nuoM_FW and nuoM RV and nuoN_FW and nuoN_RV (Supplementary Data 4) generated the variants carrying the point mutations of interest. Specifically, after 1:100 dilution in baffled flasks containing autoinduction medium, BW25113∆nuo∆ndh with pBADnuo nuoFHis6 was grown aerobically until late exponential phase (OD600 = 3–4). From that point onwards, purification steps were carried out at 4 °C. Cells were harvested through centrifugation at 3000g for 15 min. Cell sediments were suspended with a Teflon-in-glass homogenizer in a 5-fold volume of buffer A (50 mM NaCl, 50 mM MES/NaOH, pH 6.0) with 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and few grains of Dnase I. Cells were disrupted by 4 passes through an Avestin Emulsiflex at 110 MPa and cell debris was removed by low speed centrifugation at 9500g for 20 min. Next, membranes were separated from the supernatant by centrifugation at 257,000g for 1 h and the membrane sediment was suspended in an equal volume of buffer A* (buffer A + 5 mM MgCl2) with 0.1 mM PMSF. Membrane suspensions were either directly used or stored as sediments at −80 °C.
n-Dodecyl-β-D-maltopyranoside (DDM) was slowly added during 15 min to the membrane suspension until a final concentration of 2%. After gentle stirring for 30 min, insoluble material was removed at 250,000g for 15 min. Anion exchange chromatography on a Fractogel EMD TMAE Hicap (Merck) separated membrane proteins which were washed in buffer A* + 0.1% DDM and eluted using buffer B* (Buffer A* with 350 mM NaCl and 0.1% DDM). Fractions with NADH/ferricyanide oxidoreductase activity were pooled, corrected to contain 20 mM imidazole and applied on Ni-IDA material (Invitrogen) for affinity chromatography. After washing with binding buffer (500 mM NaCl, 50 mM MES/NaOH, 0.1% DDM and 20 mM imidazole (pH 6.3), fractions were eluted with 500 mM imidazole. Fractions with NADH/ferricyanide oxidoreductase activity were pooled, washed three times with buffer A* with 0.1% DDM, concentrated using ultrafiltration (Amicon Ultra-15, Millipore, 100 kDa MWCO) and stored in aliquots at −80 °C.
Protein concentration determination
Protein concentration was determined based on the biuret method using BSA as standard134. Concentration of purified complex I variants was measured by spectrometry (TIDAS II, J&M Analytik) subtracting the absorbance at 310 nm from that at 280 nm and with an ε of 763 mM−1 cm−1.
NADH/ferricyanide oxidoreductase activity
NADH/ferricyanide oxidoreductase activity of membranes, fractions or purified complex I was determined by following the decrease in absorbance of ferricyanide at 410 nm in buffer A with 0.2 mM NADH using an Ultrospec 1100 pro spectrophotometer (Pharmacia)56 and based on an ε of 1 mM−1 cm−1.
O2 reduction assay
Reduction of O2 by electrons released from NADH by complex I was determined in buffer A* on 5 µl of membranes with a Clark-type oxygen electrode at 30 °C (Hansatech). The reaction was started by adding 1.25 mM NADH while 10 µM piericidin A was used to inhibit the reaction.
Determination of purity and stability of complex I preparations
Initial production and stability of complex I were verified using polyacrylamide gel electrophoresis (PAGE). Under denaturing conditions, a sodium dodecyl sulfate (SDS) PAGE was run as described previously56 with a 3.9% stacking gel and a 10% separating gel. In addition, colorless native (CN) PAGE was performed as described previously135 with a 3.5% stacking gel and a 4–13% gradient separating gel (pH 6.0). For nitroblue tetrazolium (NBT) staining the gel was incubated for 5 min with 1 mg ml−1 NBT in 100 mM MOPS, pH 8 and the reaction was started by an addition of 100 μM NADH.
In addition to PAGE, stability of purified complex I was verified with thermal shift assays. ThermoFAD measures the intrinsic fluorescence of flavin, a cofactor of complex I, to determine the unfolding temperature of the complex136. Briefly, 25 µL of complex I (in buffer A, 1 µg µL−1) was heated in a CFX96 qPCR thermocycler (Bio-Rad) with fluorescence measurements at regular interval (ex. 470–500 nm, em. 523–543 nm). After subtracting blank data, a first derivative was calculated to obtain melting points as inflection points/maxima peaks. In addition, a Thermofluor-adapted assay137 made use of the increased fluorescent yield reaction of 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin (CPM) upon reaction with thiol groups that are released upon heating and unfolding. 300 pmol of complex I cysteines (1 µg µL−1 of complex I in buffer A) was mixed with a 5-fold excess of CPM, overlaid with silicone oil and assessed in a Perking Elmer LS-55 fluorescence spectrometer (ex. 384 nm, em. 470 nm). A dose-response equation was fitted to the data in GraphPad Prism 8 using the least squares method:
A first derivative was calculated from this fit to obtain melting points as inflection points/maxima peaks57.
Reconstitution of complex I into liposomes
Purified complex I was reconstituted in liposomes at 4 °C138. Briefly, complex I (2 mg ml−1) was mixed with a 4-fold excess of E. coli polar lipids (w/w; extract from Avanti) which were dissolved in lipid-buffer (5 mM MES/NaOH, pH 6.0) and DDM (20 mg ml−1) by sonication. The mixture was stirred gently for 10 min before BioBeads SM-2 (Bio-Rad) were added in an 8-fold excess, accounted for their binding capacity of and the presence of DDM in the sample and stirred gently for another 3 h. The proteoliposomes were sedimented by centrifugation for 45 min at 150,000g, resuspended in proteoliposome buffer (5 mM MES/NaOH, pH 6.0, 50 mM NaCl, 5 mM MgCl2) by gentle pipetting, extruded with 31 passes through a 0.1 µm polycarbonate membrane and used on the same day. The NADH/ferricyanide oxidoreductase activity before and after addition of 0.5% DDM showed no significant differences between variants and wild type. The ratio of both activities was with on average 0.75, indicating that 75% of the complex was oriented in the liposomes so that the NADH binding site was accessible from the buffer. Comparing to the NADH/FeCN oxidoreductase activity of purified proteins, the average protein reconstitution efficiency was 21%.
Preparation of ISOV vesicles
BW25113∆nuo∆ndh cells with pBADnuo nuoFHis6 were induced and harvested as described above. Inside-out vesicles (ISOVs) containing the complex I variants and endogenous lipids and other proteins; were prepared at 4 °C from frozen cells following a slightly modified, previously described procedure139. Cell sediments were suspended in equal amount of washing buffer (50 mM KH2PO4/KOH, 5 mM MgSO4, pH 7.5), sedimented (10 min, 4,500 g), suspended 6:1 in lysis buffer (washing buffer with 1 mM dithiothreitol, 0.1 mM PMSF and some grains of Dnase I) and disrupted by a single pass through a 40 ml French Pressure Cell at 55 MPa (SLM-Aminco). Centrifugation removed debris (20 min at 9500g). ISOVs were separated from the supernatant by two ultracentrifugation steps (70 min and 15 min at 257,000g) and resuspended in washing buffer. ISOV vesicles were kept on 4 °C and used on the same day.
Proton translocation and electron transfer activity
Proton translocation activity of complex I, either reconstituted in liposomes or in ISOVs, was measured by monitory quenching of fluorescence of 9-amino-6-chloro-2-methoxyacridine (ACMA)138. Briefly, proteoliposomes were incubated at 30 °C for 1 min with 0.2 µM ACMA and 60 µM decyl-ubiquinone in ACMA buffer (5 mM MES/NaOH, 50 mM KCl and 2 mM MgCl2 pH 6.0). The reaction was started by adding 100 µM NADH and ACMA fluorescence was followed on a Perking Elmer LS-55 fluorescence spectrometer through time (ex. 430 nm, em. 480 nm). For measurements of proton transport using ISOVs, no decyl-ubiquinone was added. Similarly, without ACMA, NADH/decyl ubiquinone oxidoreduction was followed by monitoring the decreasing NADH concentration by spectrometry at 340 nm with an ε of 6.3 mM−1 cm−1 (TIDAS II, J&M). As a control, the NADH/decyl ubiquinone oxidoreductase activity of empty liposomes was measured to be 15% of the activity of complex I-containing proteoliposomes.
Measurement of the electrical gradient in vivo
The electrical gradient across the membrane was determined using electrical-gradient dependent uptake of bis-(1,3-dibarbituric acid)-trimethine oxonol (DiBAC4(3))8. Stationary phase cultures were incubated for 10–20 min with 10 µg ml−1 DiBAC4(3). Single-cell uptake was measured on a BD influx cytometer (ex. 488 nm, em. 530/40 nm; 100,000 cells) and analyzed using FlowJo v10.3 (FlowJo, LLC). As control, samples were incubated for 1 h with 500 µM of the protonophore carbonyl cyanide m-chlorophenyl hydrazone (CCCP) prior to staining with DiBAC4(3).
Intracellular antibiotic accumulation measurement
To measure intracellular uptake of antibiotics, 50 ml stationary phase cultures were incubated with amikacin (100 µg ml−1) or ofloxacin (5 µg ml−1) for 60 min. Cells were separated from the extracellular solution by centrifugation through a 1:1.1 mixture of water-impermeable silicone oils barrier (AR 20 and AR 200) and frozen at −80 °C. For ofloxacin, cell sediments were suspended in 400 µl of 0.1 M glycine hydrochloride (pH 3) and lysed by incubating overnight at room temperature with light agitation. Cellular debris was removed by centrifugation (10 min at 14,000 rpm or 16,873g) and ofloxacin concentrations were analyzed by measuring fluorescence (ex. 292 nm, em. 496 nm) using a Synergy Mx Microplate Reader (BioTek). Ofloxacin concentrations were determined from a calibration curve for concentrations 0–300 ng ml−1. For amikacin, an agar well diffusion bioassay was used140. Cell sediments were suspended in 750 µl of PBS and lysed by incubation at 100 °C for 7 min. After centrifugation (10 min, 14,000 rpm or 16,873g) 100 µl supernatants was pipetted in 8 mm diameter holes on MHB agar plates inoculated with Bacillus subtilis ATCC 6051 as the indicator organism. Plates were incubated for 24 h at 37 °C after which the diameter of the inhibition zone was measured. Amikacin concentrations were deduced from a standard curve spanning a two-fold concentration range from 2 to 64 µg ml−1.
Ratiometric measurement of ATP:ADP ratios and cytoplasmic pH
ATP:ADP ratio was determined using the ratiometric GFP-based biosensor Perceval64. The pRsetB-his7-Perceval plasmid was obtained from Addgene (plasmid #20336) and the perceval gene was extracted by PCR using primer SPI-10577 and SPI-10578 (Supplementary Data 4). Next, Perceval was subcloned in pBAD/Myc-HisA, linearized through PstI and EcoRI digestion, using Gibson assembly (NEB)15. The 490/405 nm excitation fluorescence ratio (em. 530 nm) correlates with the ATP:ADP ratio and was determined using a Synergy MX Microplate reader (BioTek). Perceval’s sensitivity towards intracellular pH was countered by resuspending cells that were growing on MHB supplemented with 2 g L−1 arabinose and 100 mg L−1 ampicillin, in M63 medium buffered at pH 7 supplemented with potassium benzoate (40 mM) and methylamine hydrochloride (40 mM) before measurement.
Intracellular pH was measured with a GFP-based ratiometric sensor called pHluorin. We transformed the strains of interest with the pGFPR01 or pNTR-SD-pHluorin vector in which pHluorin is expressed from the arabinose-inducible or IPTG-inducible promoters, respectively, PBAD and Ptac65,141. To induce pHluorin expression, cells were either grown on MHB medium supplemented with 2 g L−1 arabinose or 1mM IPTG or on M9 medium supplemented with 5 g L−1 glucose and 20 g L−1 arabinose with ampicillin (100 mg L−1) to select for the maintenance of the plasmid. Note that no ampicillin and arabinose was added after nutrient shift. Ratiometric fluorescent measurements to determine pH were performed by either a Synergy MX Microplate reader (BioTek; ex. 410 and 470 nm ±20 nm; em. 530 nm ±20 nm) or a Spark plate reader (Tecan; ex. 380 and 470 nm ±20 nm; em. 530 nm ±20 nm). Cells were either resuspended in M63 minimal salts medium or spun through HPLC spin filters (BaseClear) and resuspended in fresh M9 medium before measurement. A calibration curve was generated by measuring fluorescence ratios of cultures resuspended for at least 5 min in a range of buffers with different pHs supplemented with 40 mM potassium benzoate and 40 mM methylamine hydrochloride to dissipate the transmembrane pH gradient (see Martinez et al., 2012). The relation between the fluorescence ratio and the intracellular pH was described by fitting a Boltzmann sigmoid:
Image analysis
Analysis of pictures of protein or DNA gels was done using Vilber’s VisionCapt software for quantification and to estimate sizes.
Dynamic proteomics
For the dynamic proteomics analysis, we followed previously published protocols with minimum modification26,142. Briefly, cells were grown in M9 glucose medium with regular dilutions to maintain growth in exponential phase. At OD600 0.45, a proteomics sample was collected, and persisters were generated by a switch to M9 fumarate medium. At multiple time points after the nutrient switch, samples were collected, all containing 3 × 108 cells. They were centrifuged and washed with phosphate buffered saline (2×) and then cell pellets were frozen in liquid nitrogen. Cell sediments were lysed in 2% sodium deoxycholate, 0.1 M ammonium bicarbonate and disrupted by two sonication cycles (Hielscher ultrasonicator). BCA assay (Thermo Fisher Scientific) determined the protein concentration. Sample preparation involved: reduction with 5 mM TCEP (10 min, 95 °C), alkylation with 10 mM iodoacetamide (30 min, in the dark at room temperature), quenching with 12.5 mM N-acetylcysteine, dilution with 0.1 M ammonium bicarbonate to a concentration of 1% sodium deoxycholate, digestion with trypsin (Promega; overnight with 50:1 protein:trypsin ratio), supplemented with 0.5% TFA and 50 mM HCl, removal of precipitated sodium deoxycholate (15 min at 4 °C at 21,000g), desalting of peptides (C18 reversed phase spin columns; Macrospin, Harvard Apparatus), drying under vacuum, and storage at −80 °C until further processing. Samples were run on a dual pressure LTQ-Orbitrap Velos mass spectrometer connected to an electrospray ion source with peptide separation through an EASY nLC-1000 system (all Thermo Fisher Scientific) equipped with a RP-HPLC column (75 µm × 45 cm) packed with C18 resin (ReproSil-Pur C18–AQ; Dr. Maisch GmbH) using a linear gradient from 95% solvent A (0.15% formic acid, 2% acetonitrile) to 28% solvent B (98% acetonitrile, 0.15% formic acid) over 90 min at 0.2 µl min−1. The acquisition mode obtained one high-resolution MS scan in the FT part at a resolution of 120,000 full width at half maximum (at m/z 400) followed by MS/MS scans in the linear ion trap of the 20 most intense ions. The charged state screening modus excluded unassigned and singly charged ions (dynamic exclusion duration: 20 s; ion accumulation time: 300 ms (MS) and 50 ms (MS/MS)). The raw files were imported into the Progenesis LC-MS software (Nonlinear Dynamics, Version 4.0) and analyzed using the default settings. MS/MS-data were exported in mgf format and searched against a decoy database of the forward and reverse sequences of the predicted proteome from E. coli (Uniprot, download date: 15/6/2012, total of 10,388 entries) using MASCOT. In the search criteria, full tryptic specificity was required (after lysine or arginine residues), three missed cleavages were allowed, carbamidomethylation (C) was set as fixed modification, oxidation (M) as variable modification and mass tolerance was 10 ppm for precursor ions and 0.6 Da for fragment ions. Results were imported into Progenesis, and the false discovery rate (FDR) was set to 1%. The final protein lists containing the cumulative peak areas of all identified peptides for each protein, respectively, were exported from Progenesis LC-MS and further statically analyzed using an in-house developed R script (SafeQuant)142. The raw mass spectrometry data is available through the ProteomeXchange Consortium (ID: PXD029006).
For the analysis, we normalized our data relative to a 2-fold difference in protein concentrations between all the analyzed conditions with the WT at start (before nutrient shift) as a reference. Next, we applied unscaled, two-dimensional principal component analysis using the FactoMineR package (R). For this, we used a previously published and analyzed dataset (from Schmidt et al.69 and Fig. 4a in Radzikowski et al.26, ProteomeXchange IDs: PXD000498 and PXD001968, respectively) to generate the two dimensions that constitute the PCA space. We plotted our data on this space to compare with previously found trends.
Radioactive assessment of translation activity
To probe protein synthesis, we opted to follow the incorporation of radioactive L-(4,5-3H)-leucine during stationary phase in complex MHB medium (±18 h after 1:100 dilution as done during the evolution experiment) instead of following induction of a fluorescent protein as done before8 as the latter might be prone to biased results. Specifically, L-(4,5-3H)-leucine was added to stationary phase cultures at 2.5 µCi ml−1 and further incubated shaking at 37 °C. As control, chloramphenicol was additionally added at 64 µg ml−1, which fully abrogated protein synthesis (data not shown). Over the course of 3h, samples were taken on an hourly basis and precipated in ice-cold trichloric acid (TCA; 10%). Precipitates were washed twice with ice-cold destilled water and added to scintillation liquid (Ultima-Flo M, Perking Elmer). Radioactive signal was measured as counts per minute with a Hidex 300SL scintillation counter.
HPLC-UV detection and quantification of ppGpp
Solutions containing ppGpp (Jena Biosciences) and products of its hydrolysis were analyzed using a HPLC-UV method measuring absorbance at 260 nm. A PL-SAX anion exchange column (PL-SAX 1000Å 8 µm, 50 × 4.6 mm, Agilent) was used for sample separation at 60 °C. Two methods were used: either (1) 0–2 min linear gradient from 80% to 70% A, 2–4 min linear gradient from 70% to 0% A, 4–9.5 min 0% A, at 9.5 min step change to 80% kept until 10 min; or (2) a 2.5 min (or 5 min, respectively) isocratic flow of B; A: 0.01 M K2HPO4, pH 2.6; B: 0.5 M K2HPO4, pH 3.5. The flow rate was 1 ml min−1 in (1) or 1.2 ml min−1 in (2).
Spontaneous ppGpp hydrolysis assays
Spontaneous (i.e. non-enzymatic) ppGpp hydrolysis can occur at low pH once the two distal phosphate groups of ppGpp are deprotonated and a positively charged divalent metal ion coordinates between the phosphate groups (Supplementary Fig. 6a). Spontaneous hydrolysis assays were performed in a buffer containing 0.05 M Tris, 0.09 M sodium formate and 0.025 M ammonium acetate, with adjusted to pH values and manganese concentrations as indicated. ppGpp (Jena Biosciences) was added in a final concentration of 1 mM sampled by HPLC at 37 °C. In order to prevent evaporation, the mixture was covered with PCR mineral oil. 2 µL of the reaction volume was injected at different points in time and the amount of ppGpp, as well as its hydrolysis products, were quantified by comparison to pure standards at different concentrations. Consistent with computational estimations of the pKa values of the two distal phosphate groups of ppGpp143 (i.e. 6.5 and 6.8), we found that spontaneous ppGpp hydrolysis is high at pH of 8, and absent at pH of 7, with a sharp change in velocity around pH 7.5, and only occurs in the presence of divalent metal cations like manganese (Supplementary Fig. 6b-d, g). Thus, at pH values found in persisters, spontaneous ppGpp hydrolysis is inhibited (Fig. 5a, b).
Overproduction and purification of 6His-SpoT
To identify whether an enzyme-based catalysis of ppGpp hydrolysis (potentially also pH inhibited) is partially responsible for the previously observed increase in ppGpp levels in persisters26, we focused on the enzymes synthesizing (RelA, SpoT) and hydrolyzing ppGpp (SpoT). As a deletion of relA had no effect on persister fractions, we had previously inferred that SpoT must be the player responsible for increased ppGpp levels in nutrient shift-induced persisters26. To further identify whether the ppGpp increase is due to an increased SpoT synthetase activity or an decreased SpoT hydrolysis activity, we initially analyzed a strain carrying a SpoT variant, which is defective in (p)ppGpp synthesis, but not in hydrolysis activity (SpoT-E319Q)144. After a glucose-to-fumarate nutrient shift, we found the same fraction of persisters as in the wild type (Supplementary Fig. 6e), suggesting that an inhibition of the hydrolase activity of SpoT must be responsible for the increased ppGpp levels in persisters formed upon a nutrient shift. To test whether, next to the spontaneous ppGpp hydrolysis, also the SpoT hydrolase activity is pH dependent, we aimed to purify SpoT, whose production and purification is notoriously difficult145,146. While the protein mostly resided in the insoluble protein fraction, we could purify minute amounts of soluble protein after a short induction time using either a Tris-based buffer (Supplementary Fig. 6h) or a cytosolic buffer (phosphate-based buffer, see below and Supplementary Fig. 6j).
SpoT was expressed with an N-terminal His-tag using plasmid pETM11-SpoT. pETM11-SpoT was generated by amplification of spoT using primer pair spoT_NcoI_s and spoT_NotI_stop_as (Supplementary Data 4) using genomic DNA from E. coli BW25113 as template. Subsequently the DNA fragment was cloned into pETM11 (EMBL Heidelberg) using NcoI and NotI restriction sites. Successful insertion was confirmed by restriction and sequencing analyses. The plasmid was transformed into E. coli BL21 (DE3) pLysS and selected on LB plates containing kanamycin and chloramphenicol.
E. coli strain BL21 (DE3) pLyS transformed with plasmid pETM11-SpoT was grown aerobically at 37 °C in 100 ml LB medium in a 1000 ml flask supplemented with kanamycin and chloramphenicol (both at 50 μg ml−1). Gene expression was induced at OD600 = 0.5 by adding 0.5 mM isopropylthio-β-galactoside (IPTG) and cells were grown for 1 h at 37 °C. Cells were harvested, washed with buffer (50 mM Tris/HCl pH 7.5, 10% glycerol) and disrupted by passage through a Cell disruptor (Constant Cell Disruption Systems, Northants, UK) at 25 bar and 4 °C in disruption buffer (50 mM Tris/HCl pH 7.7, 10% (v/v) glycerol, 300 mM NaCl, 50 mM KCl, 1 mM dithiothreitol, 0.5 mM PMSF, and 0.03 mg/ml (w/v) DNase). After removal of intact cells and cell debris by centrifugation (3000g, 10 min, 4 °C), the cytosol was incubated over night at 4 °C with pre-equilibrated Ni-Sepharose (with 50 mM Tris/HCl pH 7.7, 10% (v/v) glycerol, 300 mM NaCl, 10 mM MgCl2, 2 mM β-mercaptoethanol, 10 mM imidazole). Unbound protein was removed by washing with equilibration buffer and 6His-SpoT was eluted from the column by increasing imidazole concentrations up to 250 mM. Obtained purified protein degraded already at 4 °C (Supplementary Fig. 6i).
To obtain more stable protein, purification was attempted with cytosolic buffer. Cells were cultivated as described above and disrupted in cytosolic buffer pH 7.5 (6 mM KH2PO4, 14 mM KH2PO4, 140 mM KCl, 5.5% glucose, 10 mM NaCl) containing 0.03 mg/ml DNAse (w/v) and 0.5 mM PMSF. Removal of intact cells and purification of 6His-SpoT was performed as described above, but with use of cytosolic buffers containing imidazole. Note that pH of the buffers was re-adjusted after imidazole addition using phosphoric acid and KOH, respectively. For purification of SpoT at different pH, cells were disrupted in cytosolic buffer pH 7.5, bound to Ni-Sepharose and washed with buffer pH 7.5, and eluted with buffers adjusted to the corresponding pH and containing increasing imidazole concentrations. Protein content in samples was analyzed by SDS PAGE (12%) and Bradford protein assay147. The purified protein was slightly more stable than with the Tris-based buffer, but ppGpp hydrolysis activity declined strongly over time (Supplementary Fig. 6c, d). In addition, the amount of protein was rather low (0.3 mg ml−1) and attempts to concentrate the samples or exchange buffers remained unsuccessful, rendering systematic assessment of the pH dependency of SpoT hydrolysis impossible in this way.
Co-expression and purification of encapsulin and SpoT
To obtain purified and stable protein, we followed an alternative approach, where we expressed SpoT in nanocages, formed as icosahedral capsids of 60 monomers of a small bacterial protein (encapsulin), which can protect the cargo protein from aggregation and degradation148. Nanocages or encapsulins are proteins that form macromolecular assemblies, resembling viral capsids148. Such capsids usually contain DyP-peroxidase or ferritin-like proteins148,149. Efficient encapsulation in vivo is possible due to the presence of ~30 AA C-terminal signal peptide, which interacts with inner surface of encapsulin148. Genes encoding for encapsulin and cargo enzymes are found together in an operon, since encapsulin assembly and cargo loading is a concerted process148. Here, we used a newly discovered robust encapsulin from Mycolicibacterium hassiacum, for which it was demonstrated to increase the stability of foreign cargo enzymes150 and which enabled the characterization of the activity of the encapsulated and thus stabilized enzymes, as the nanocages contain three types of pores (5-9 Å), which allow small substrates to enter or leave the capsule148,149,151.
For co-expression of SpoT and encapsulin, spoT was amplified using forward and reverse primer (Supplementary Data 4). BsaI sites are underlined and specific overhangs are indicated in bold. spoT was cloned in a pENC vector based on pET28 with a changed origin of replication to p15A in order to be compatible with pBAD vector, and with an introduced BsaI restriction sites. These changes have been introduced using Gibson assembly. Briefly, the p15A ori was amplified from pEVOL-pAzF (Addgene plasmid #31186) and the pET-28a(+) vector was amplified excluding pBR322 ori. These two fragments were assembled using NEB’s Gibson assembly mix, with all the primers designed using Geneious software (Gibson cloning option; vector map available on request). The gene encoding SpoT was cloned into pENC in such way that, when expressed, it contains a N-terminal 6xHis-tag and also C-terminal fusion tag of 30 amino acids from DyP-peroxidase from the same operon as the original encapsulin gene150. EncMh encapsulin was provided by expression from pBAD-EncMh, obtained by cloning the codon-optimized gene encoding EncMh (GenScript) between the NdeI and HindIII restriction site of a pBAD-NK vector150. After sequencing, both the vectors were co-transformed into E. coli BL21-AI strain (ThermoFisher) and selected on LB plates containing ampicillin, kanamycin and tetracyclin.
E. coli strain BL21-AI transformed with plasmids pBAD-EncMh and pENC-spoT was grown aerobically at 37 °C in 200 ml LB containing ampicillin and kanamycin (both 50 μg ml−1) in a 1000 ml baffled flask. At OD600 = 0.5 expression of spoT was induced by 1 mM IPTG (final concentration) and expression of encapsulin by addition of 0.2% arabinose (final concentration) and cells were incubated for another 16–18 h at 30 °C. Cells were collected by centrifugation for 20 min at 6000 rpm (3099g) and 4 °C. Sonication was carried out in 50 mM TrisHCl pH 7.5 with 150 mM NaCl. As it can happen that cargo protein is expressed in higher amount than what can be encapsulated it is necessary to remove excess. Clarified cell-free extract was loaded on pre-equilibrated Ni-Sepharose column to remove excess of cargo protein which contains 6xHis-tag. Since encapsulin does not contain any 6xHis tag and due to its size, it comes in flow-through fraction, together with any cargo protein it may contain. Flow-through fraction was mixed on ice 1:0.75 with 10% PEG8000 in 100 mM Tris/HCl pH 7.5 with 2 M NaCl and left on a nutating shaker at 4 °C for 8 h. Precipitated proteins were collected by centrifugation for 30 min at 4000 rpm (1377g) and 4 °C. Sediment was resuspended in 20 ml of 50 mM TrisHCl pH 7.5 and left overnight on a nutating shaker at 4 °C to re-dissolve completely. Re-dissolved ENC-spoT was aliquoted, flash-frozen in liquid nitrogen and stored at −80 °C. Following this approach, we could obtain co-purified encapsulin with stable SpoT at 3 mg ml−1 (Supplementary Fig. 6m).
Enzymatic ppGpp hydrolysis assay
We used both the encapsulated SpoT and regularly purified SpoT to characterize the pH dependency of its ppGpp hydrolysis activity. While the product of the spontaneous hydrolysis reaction leads to GTP, experiments with the limited amounts of the regularly purified SpoT indicated that SpoT hydrolyzes ppGpp to GDP, and that manganese ions are necessary for hydrolysis (Supplementary Fig. 6g), consistent with earlier reported requirements of Rel/Spo homologues for hydrolysis152,153. Enzymatic ppGpp hydrolysis assays with 6His-SpoT and encapsulin SpoT were performed in cytosolic buffer containing 250 mM imidazole (and 0.1% triton-X100 in case of encapsulin SpoT) adjusted to the corresponding pH with a final protein concentration of 0.1 and 0.5 mg ml−1, respectively. Unless stated otherwise, manganese was added (1 mM) and the reaction was started by adding ppGpp (1 mM). Samples were incubated at 25 °C and, at different time points, aliquots were taken in which the reaction was stopped by adding EDTA (10 mM). Two microliters of each aliquot was used for HPLC analysis as described above. Here, similar to the results of the spontaneous hydrolysis, we found that the hydrolysis activity is strongly pH-dependent: hydrolysis starts at pH values higher than 7.0, sharply increases around pH 7.5 and reaches a maximum at around pH 8.3, above which the hydrolysis activity drops again likely due to enzyme denaturation (Supplementary Fig. 6f). A control experiment with an empty capsule sample at pH 8 revealed no ppGpp hydrolysis and few limited experiments with the minute amounts of regularly purified SpoT confirmed the pH dependency of hydrolysis (Supplementary Fig. 6f). Thus, also the ppGpp hydrolysis of SpoT is strongly pH-dependent, with hardly any activity around pH 7.0 and very much increased rates above pH 7.5. As persister cells have a cytoplasmic pH below 7.5 (Fig. 5a, b), we conjecture that the cytoplasmic pH, via inhibition of ppGpp hydrolysis, is a major regulator of ppGpp levels upon strong perturbation of metabolic homeostasis.
Statistics
Data were analyzed using either GraphPad Prism 8 or R in the RStudio environment. Relevant information regarding the statistical tests was added to each figure caption where appropriate. In Fig. 1b, c, Supplementary Figs. 1f–I and 2a, b, Chi² goodness-of-fit tests with 200,000 Monte Carlo simulations to compute p-values were used to compare the observed distribution of our mutations counts in various categories to the expected probabilities based on random occurring mutations. More detailed information can be found under the section explaining the genome-wide sequencing analyses and Chi² statistics are reported in Supplementary Data 4. For ANOVAs and linear (mixed) models, assumptions of homoscedasticity and normality of the residuals were checked visually comparing fitted value with residuals and comparing predicted with actual residuals. Where relevant, the Brown-Forsythe test was used to test whether SDs were equal, the Variance Inflation Factor was used to estimate the degree of multicollinearity and a Chi² test was performed to test the effectiveness of including random effects/matching. Post hoc tests (posttest) were performed to make and correct for multiple comparisons. All tests for which it applies were two-tailed. Throughout the manuscript, a significance level cutoff of alpha = 0.05 was used. For visual purposes, asterisks denote significance levels as defined in the figure captions and each exact p value can be found in Supplementary Data 4. In addition, test statistics and degrees of freedom are reported in Supplementary Data 4.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
The authors thank Susan Gottesman for the AB037 strain, Joan L. Slonczewski for the pHluorin plasmid, Steven B. Vik for the BA14 strain and Henri de Greve for the UTI89 strain. We further thank Ying Liu for help with cloning the plasmid for encapsulin-SpoT expression, Franziska Number for providing controls for the ACMA quenching, Alexandros Papagiannakis for valuable suggestions as well as for help with microscopy and Bert Poolman for valuable discussions. B.V.D.B. is recipient of fellowships from the Fund for Scientific Research, Flanders (FWO; 12O1917N, 12O1922N, V428917N, and 11C6812/4N), from the Federation of European Microbiological Societies (FEMS; RG-2016-0052), from the Belgian American Educational Foundation (BAEF; 2016-E083) and from the European Molecular Biology Organization (EMBO; ALTF 344-2017). J.E.M. was granted a fellowship by the Agency for Innovation by Science and Technology (IWT). This work was furthermore supported by grants from FWO (1528318N, G0B2515N and G055517N), KU Leuven (C16/17/006), the Flanders Institute for Biotechnology (VIB) (to J.M. and B.V.D.B), the Deutsche Forschungsgemeinschaft (278002225/RTG 2202) (to T.F.) and from the Dutch Research Council (NWO; VIDI grant 864.11.001) to M.H.
Source data
Author contributions
Conceptualization, B.V.D.B., H.S., J.L.R., M.F., T.F., J.M. and M.H.; Methodology, B.V.D.B., H.S., J.E.M., J.L.R., A. S., M.F., T.F., J.M. and M.H.; Investigation, B.V.D.B., H.S., J.E.M., T.E.P.K., J.L.R., T.M., N.L., J.S., S.V., L.D., A.S.; Writing—Original Draft, B.V.D.B., H.S., J.L.R., J.M. and M.H.; Writing—Review & Editing, B.V.D.B., H.S., J.M. and M.H.; Funding Acquisition, B.V.D.B., T.F., J.M. and M.H.; Resources, A.S.,T.F., J.M., and M.H.; Supervision, J.M. and M.H.
Peer review
Peer review information
Nature Communications thanks Gregory Cook and the other, anonymous, reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
The NGS Sequencing data generated in this study have been deposited in the SRA database under accession codes PRJNA498891, PRJNA498708, PRJNA498717, PRJNA270307, and PRJNA768774. Mass spectrometry raw data files have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository under dataset identifier PXD029006. The data underlying all the figures generated in this study are provided in the Source Data file. Mass spectrometry data used in this study are available via the ProteomeXchange Consortium via the PRIDE partner repository under the dataset identifiers PXD000498 and PXD001968. Sequence data used in this study are available in the NCBI nucleotide database (NC_000913.3, NC_007946.1, NC_007941.1), Uniprot database (P0AFC3, P0AFC7, P33599, P0AFD1, P31979, P33602, P0AFD4, P0AFD6, P0AFE0, P0AFE4, P33607, P0AFE8, P0AFF0), PDB database (6g2j, 4he8) and PubChem database (135398637). Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Jan Michiels, Matthias Heinemann.
Contributor Information
Jan Michiels, Email: jan.michiels@kuleuven.be.
Matthias Heinemann, Email: m.heinemann@rug.nl.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-28141-x.
References
- 1.Balaban NQ, et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019;17:441–448. doi: 10.1038/s41579-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen NR, Lobritz MA, Collins JJ. Microbial persistence and the road to drug resistance. Cell Host Microbe. 2013;13:632–642. doi: 10.1016/j.chom.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van den Bergh B, Fauvart M, Michiels J. Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol. Rev. 2017;41:1–33. doi: 10.1093/femsre/fux001. [DOI] [PubMed] [Google Scholar]
- 4.Dewachter L, Fauvart M, Michiels J. Bacterial heterogeneity and antibiotic survival: Understanding and combatting persistence and heteroresistance. Mol. Cell. 2019;76:255–267. doi: 10.1016/j.molcel.2019.09.028. [DOI] [PubMed] [Google Scholar]
- 5.Fisher RA, Gollan B, Helaine S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017;15:453–464. doi: 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- 6.Fauvart M, De Groote VN, Michiels J. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J. Med. Microbiol. 2011;60:699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- 7.Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature. 2014;99:1–9. doi: 10.1038/nature13469. [DOI] [PubMed] [Google Scholar]
- 8.Van den Bergh B, et al. Frequency of antibiotic application drives rapid evolutionary adaptation of Escherichia coli persistence. Nat. Microbiol. 2016;1:16020. doi: 10.1038/nmicrobiol.2016.20. [DOI] [PubMed] [Google Scholar]
- 9.Mechler L, et al. A novel point mutation promotes growth phase-dependent daptomycin tolerance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2015;59:5366–5376. doi: 10.1128/AAC.00643-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michiels JE, Van den Bergh B, Verstraeten NA, Fauvart M, Michiels J. In vitro emergence of high persistence upon periodic aminoglycoside challenge in the ESKAPE pathogens. Antimicrob. Agents Chemother. 2016;60:4630–4637. doi: 10.1128/AAC.00757-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levin-Reisman I, et al. Antibiotic tolerance facilitates the evolution of resistance. Science. 2017;355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 12.Windels EM, et al. Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J. 2019;13:1239–1251. doi: 10.1038/s41396-019-0344-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sebastian J, et al. De novo emergence of genetically resistant mutants of Mycobacterium tuberculosis from the persistence phase cells formed against antituberculosis drugs in vitro. Antimicrob. Agents Chemother. 2016;61:e01343–16. doi: 10.1128/AAC.01343-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rotem E, et al. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence suppl info. Proc Natl Acad Sci USA. 2010;107:1–5. doi: 10.1073/pnas.1004333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilmaerts D, et al. The persistence-inducing toxin HokB forms dynamic pores that cause ATP leakage. mBio. 2018;9:1–12. doi: 10.1128/mBio.00744-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheverton AM, et al. A Salmonella toxin promotes persister formation rhrough acetylation of tRNA. Mol. Cell. 2016;63:86–96. doi: 10.1016/j.molcel.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dörr T, Vulić M, Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verstraeten NA, et al. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol. Cell. 2015;59:9–21. doi: 10.1016/j.molcel.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 19.Schumacher MA, et al. HipBA–promoter structures reveal the basis of heritable multidrug tolerance. Nature. 2015;524:59–64. doi: 10.1038/nature14662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jurėnas D, et al. AtaT blocks translation initiation by N-acetylation of the initiator tRNAfMet. Nat. Chem. Biol. 2017;13:640–646. doi: 10.1038/nchembio.2346. [DOI] [PubMed] [Google Scholar]
- 21.Wilmaerts D, et al. HokB monomerization and membrane repolarization control persister awakening. Mol. Cell. 2019;75:1031–1042.e4. doi: 10.1016/j.molcel.2019.06.015. [DOI] [PubMed] [Google Scholar]
- 22.Fung DKC, Chan EWC, Chin ML, Chan RCY. Delineation of a bacterial starvation stress response network which can mediate antibiotic tolerance development. Antimicrob. Agents Chemother. 2010;54:1082–1093. doi: 10.1128/AAC.01218-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen D, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korch SB, Henderson TA, Hill TM. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 2003;50:1199–1213. doi: 10.1046/j.1365-2958.2003.03779.x. [DOI] [PubMed] [Google Scholar]
- 25.Gaca AO, Colomer-Winter C, Lemos JA. Many means to a common end: The intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J. Bacteriol. 2015;197:1146–1156. doi: 10.1128/JB.02577-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radzikowski JL, et al. Bacterial persistence is an active σS stress response to metabolic flux limitation. Mol. Syst. Biol. 2016;12:882. doi: 10.15252/msb.20166998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amato SM, Brynildsen MP. Persister heterogeneity arising from a single metabolic stress. Curr. Biol. 2015;25:2090–2098. doi: 10.1016/j.cub.2015.06.034. [DOI] [PubMed] [Google Scholar]
- 28.Amato SM, Orman MA, Brynildsen MP. Metabolic control of persister formation in Escherichia coli. Mol. Cell. 2013;50:475–487. doi: 10.1016/j.molcel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Germain E, Castro-Roa D, Zenkin N, Gerdes K. Molecular mechanism of bacterial persistence by HipA. Mol. Cell. 2013;52:248–254. doi: 10.1016/j.molcel.2013.08.045. [DOI] [PubMed] [Google Scholar]
- 30.Kaspy I, et al. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat. Commun. 2013;4:1–7. doi: 10.1038/ncomms4001. [DOI] [PubMed] [Google Scholar]
- 31.Stokes JM, Lopatkin AJ, Lobritz MA, Collins JJ. Bacterial metabolism and antibiotic efficacy. Cell Metab. 2019;30:251–259. doi: 10.1016/j.cmet.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radzikowski JL, Schramke H, Heinemann M. Bacterial persistence from a system-level perspective. Curr. Opin. Biotechnol. 2017;46:98–105. doi: 10.1016/j.copbio.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 33.Spoering AL, Vulić M, Lewis K. GlpD and PlsB participate in persister cell formation in Escherichia coli. J. Bacteriol. 2006;188:5136–5144. doi: 10.1128/JB.00369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Girgis HSH, Harris K, Tavazoie S. Large mutational target size for rapid emergence of bacterial persistence. Proc. Natl Acad. Sci. USA. 2012;109:12740–12745. doi: 10.1073/pnas.1205124109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen S, Lewis K, Vulić M. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob. Agents Chemother. 2008;52:2718–2726. doi: 10.1128/AAC.00144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, et al. Transposon mutagenesis identifies novel genes associated with Staphylococcus aureus persister formation. Front. Microbiol. 2015;6:1–9. doi: 10.3389/fmicb.2015.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma C, et al. Energy production genes sucB and ubiF are involved in persister survival and tolerance to multiple antibiotics and stresses in Escherichia coli. FEMS Microbiol. Lett. 2010;303:33–40. doi: 10.1111/j.1574-6968.2009.01857.x. [DOI] [PubMed] [Google Scholar]
- 38.Kotte O, Volkmer B, Radzikowski JL, Heinemann M. Phenotypic bistability in Escherichia coli’s central carbon metabolism. Mol. Syst. Biol. 2014;10:736–736. doi: 10.15252/msb.20135022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 40.Gutierrez A, et al. Understanding and sensitizing density-dependent persistence to quinolone antibiotics. Mol. Cell. 2017;68:1147–1154.e3. doi: 10.1016/j.molcel.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 41.Pu Y, et al. ATP-dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Mol. Cell. 2019;73:143–156.e4. doi: 10.1016/j.molcel.2018.10.022. [DOI] [PubMed] [Google Scholar]
- 42.Conlon BP, et al. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 2016;1:16051. doi: 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- 43.Shan Y, et al. ATP-dependent persister formation in Escherichia coli. mBio. 2017;8:e02267–16. doi: 10.1128/mBio.02267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orman MA, Brynildsen MP. Dormancy is not necessary or sufficient for bacterial persistence. Antimicrob. Agents Chemother. 2013;57:3230–3239. doi: 10.1128/AAC.00243-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ray JCJ, et al. Cellular growth arrest and persistence from enzyme saturation. PLoS Comput. Biol. 2016;12:1–21. doi: 10.1371/journal.pcbi.1004825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meylan S, et al. Carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem. Biol. 2017;24:1–12. doi: 10.1016/j.chembiol.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zalis EA, et al. Stochastic variation in expression of the tricarboxylic acid cycle produces persister cells. mBio. 2019;10:1–10. doi: 10.1128/mBio.01930-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van den Bergh, B., Michiels, J. E. & Michiels, J. In Bacterial Persistence: Methods and Protocols (eds Michiels, J. & Fauvart, M.) Vol. 1333, 131–143 (Springer, 2016).
- 49.Friedrich, T., Hellwig, P. & Einsle, O. in A Structural Perspective on Respiratory Complex I: Structure and Function of NADH:Ubiquinone Oxidoreductase (ed. Sazanov, L.) 46, 23–60 (Springer, 2012)..
- 50.Gnandt E, Dörner K, Strampraad MFJ, de Vries S, Friedrich T. The multitude of iron–sulfur clusters in respiratory complex I. Biochim. Biophys. Acta—Bioenerg. 2016;1857:1068–1072. doi: 10.1016/j.bbabio.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 51.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex I. Nature. 2013;494:25–30. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Efremov RG, Sazanov LA. Structure of the membrane domain of respiratory complex I. Nature. 2011;476:414–420. doi: 10.1038/nature10330. [DOI] [PubMed] [Google Scholar]
- 53.Di Luca A, et al. Symmetry-related proton transfer pathways in respiratory complex I. Proc. Natl. Acad. Sci. USA. 2017;114:E6314–E6321. doi: 10.1073/pnas.1706278114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Friedrich T, Dekovic DK, Burschel S. Assembly of the Escherichia coli NADH:ubiquinone oxidoreductase (respiratory complex I) Biochim. Biophys. Acta—Bioenerg. 2016;1857:214–223. doi: 10.1016/j.bbabio.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 55.Pohl T, Uhlmann M, Kaufenstein M, Friedrich T. Lambda Red-mediated mutagenesis and efficient large scale affinity purification of the Escherichia coli NADH:ubiquinone oxidoreductase (complex I) Biochemistry. 2007;46:10694–10702. doi: 10.1021/bi701057t. [DOI] [PubMed] [Google Scholar]
- 56.Steimle S, et al. Role of subunit NuoL for proton translocation by respiratory complex I. Biochemistry. 2011;50:3386–3393. doi: 10.1021/bi200264q. [DOI] [PubMed] [Google Scholar]
- 57.Hoeser J, Gnandt E, Friedrich T. Low cost, microcontroller based heating device for multi-wavelength differential scanning fluorimetry. Sci. Rep. 2018;8:1–10. doi: 10.1038/s41598-018-19702-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaila VRI, Wikström M. Architecture of bacterial respiratory chains. Nat. Rev. Microbiol. 2021;19:319–330. doi: 10.1038/s41579-020-00486-4. [DOI] [PubMed] [Google Scholar]
- 59.Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krulwich TA, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 2011;9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vesselinova N, Alexandrov BS, Wall ME. Dynamical model of drug accumulation in bacteria: Sensitivity analysis and experimentally testable predictions. PLoS ONE. 2016;11:1–20. doi: 10.1371/journal.pone.0165899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cramariuc O, et al. Mechanism for translocation of fluoroquinolones across lipid membranes. Biochim. Biophys. Acta—Biomembr. 2012;1818:2563–2571. doi: 10.1016/j.bbamem.2012.05.027. [DOI] [PubMed] [Google Scholar]
- 63.Aldred KJ, Kerns RJ, Osheroff N. Mechanism of quinolone action and resistance. Biochemistry. 2014;53:1565–1574. doi: 10.1021/bi5000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berg J, Hung YP, Yellen G. A genetically encoded fluorescent reporter of ATP:ADP ratio. Nat. Methods. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martinez KA, et al. Cytoplasmic pH response to acid stress in individual cells of Escherichia coli and Bacillus subtilis observed by fluorescence ratio imaging microscopy. Appl. Environ. Microbiol. 2012;78:3706–3714. doi: 10.1128/AEM.00354-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bearson SMD, et al. Acid shock induction of RpoS is mediated by the mouse virulence gene. Microbiology. 1996;178:2572–2579. doi: 10.1128/jb.178.9.2572-2579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bak G, Han K, Kim D, Lee Y. Roles of rpoS-activating small RNAs in pathways leading to acid resistance of Escherichia coli. Microbiologyopen. 2014;3:15–28. doi: 10.1002/mbo3.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmidt A, et al. The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 2016;34:104–110. doi: 10.1038/nbt.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kwan BW, et al. Arrested protein synthesis increases persister-like cell formation. Antimicrob. Agents Chemother. 2013;57:1468–1473. doi: 10.1128/AAC.02135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tannock IF, Rotin D. Acid pH in tumors and its potential for therapeutic exploitation. Cancer Res. 1989;49:4373–4384. [PubMed] [Google Scholar]
- 72.Urbach V, Hélix N, Renaudon B, Harvey BJ. Cellular mechanisms for apical ATP effects on intracellular pH in human bronchial epithelium. J. Physiol. 2002;543:13–21. doi: 10.1113/jphysiol.2001.015180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kanaporis G, Treinys R, Fischmeister R, Jurevičius J. Metabolic inhibition reduces cardiac L-type Ca2+ channel current due to acidification caused by ATP hydrolysis. PLoS ONE. 2017;12:1–17. doi: 10.1371/journal.pone.0184246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vroman R, et al. Extracellular ATP hydrolysis inhibits synaptic transmission by increasing pH buffering in the synaptic cleft. PLoS Biol. 2014;12:e1001864. doi: 10.1371/journal.pbio.1001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Munder MC, et al. A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. Elife. 2016;5:1–30. doi: 10.7554/eLife.09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bruni GN, Kralj JM. Membrane voltage dysregulation driven by metabolic dysfunction underlies bactericidal activity of aminoglycosides. Elife. 2020;9:1–25. doi: 10.7554/eLife.58706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Voskuil, M. I., Covey, C. R. & Walter, N. D. in Advances in Microbial Physiology Vol. 73, 77–122 (Elsevier Ltd., 2018). [DOI] [PubMed]
- 78.Lee DD, et al. Magnesium flux modulates ribosomes to increase bacterial survival. Cell. 2019;177:352–360.e13. doi: 10.1016/j.cell.2019.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zambrano MM, Kolter R. Escherichia coli mutants lacking NADH dehydrogenase I have a competitive disadvantage in stationary phase. J. Bacteriol. 1993;175:5642–5647. doi: 10.1128/jb.175.17.5642-5647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shan Y, Lazinski DW, Rowe SE, Camilli A, Lewis K. Genetic basis of persister tolerance to aminoglycosides in Escherichia coli. mBio. 2015;6:e00078–15. doi: 10.1128/mBio.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Girgis HSHHS, Hottes AKA, Tavazoie S. Genetic architecture of intrinsic antibiotic susceptibility. PLoS ONE. 2009;4:e5629. doi: 10.1371/journal.pone.0005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jahn LJ, Munck C, Ellabaan MMH, Sommer MOA. Adaptive laboratory evolution of antibiotic resistance using different selection regimes lead to similar phenotypes and genotypes. Front. Microbiol. 2017;8:1–14. doi: 10.3389/fmicb.2017.00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lázár V, et al. Bacterial evolution of antibiotic hypersensitivity. Mol. Syst. Biol. 2013;9:700. doi: 10.1038/msb.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schurek KN, et al. Novel genetic determinants of low-level aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2008;52:4213–4219. doi: 10.1128/AAC.00507-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ezraty B, et al. Fe-S cluster biosynthesis controls uptake of aminoglycosides in a ROS-less death pathway. Science. 2013;340:1583–1587. doi: 10.1126/science.1238328. [DOI] [PubMed] [Google Scholar]
- 86.Wong KS, et al. The MoxR ATPase RavA and its cofactor ViaA interact with the NADH:ubiquinone oxidoreductase I in Escherichia coli. PLoS One. 2014;9:e85529. doi: 10.1371/journal.pone.0085529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McCollister BD, Hoffman M, Husain M, Vázquez-Torres A. Nitric oxide protects bacteria from aminoglycosides by blocking the energy-dependent phases of drug uptake. Antimicrob. Agents Chemother. 2011;55:2189–2196. doi: 10.1128/AAC.01203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chareyre S, Barras F, Mandin P. A small RNA controls bacterial sensitivity to gentamicin during iron starvation. PLOS Genet. 2019;15:e1008078. doi: 10.1371/journal.pgen.1008078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Santi I, Manfredi P, Maffei E, Egli A, Jenal U. Evolution of antibiotic tolerance shapes resistance development in chronic Peudomonas aeruginosa infections. mBio. 2021;12:1–17. doi: 10.1128/mBio.03482-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mühlbauer ME, et al. Water-gated proton transfer dynamics in respiratory complex I. J. Am. Chem. Soc. 2020;142:13718–13728. doi: 10.1021/jacs.0c02789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Imamovic L, Sommer MOA. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci. Transl. Med. 2013;5:204ra132. doi: 10.1126/scitranslmed.3006609. [DOI] [PubMed] [Google Scholar]
- 92.Chakraborty S, Mizusaki H, Kenney LJ. A FRET-based DNA biosensor tracks OmpR-dependent acidification of Salmonella during macrophage infection. PLOS Biol. 2015;13:e1002116. doi: 10.1371/journal.pbio.1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Helaine S, et al. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vega NM, Allison KR, Khalil AS, Collins JJ. Signaling-mediated bacterial persister formation. Nat. Chem. Biol. 2012;8:431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vega NM, Allison KR, Samuels AN, Klempner MS, Collins JJ. Salmonella Typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc. Natl. Acad. Sci. USA. 2013;110:14420–14425. doi: 10.1073/pnas.1308085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kuczyńska-Wiśnik D, et al. Lack of intracellular trehalose affects formation of Escherichia coli persister cells. Microbiology. 2014;161:786–796. doi: 10.1099/mic.0.000012. [DOI] [PubMed] [Google Scholar]
- 97.Zarkan A, et al. Indole pulse signalling regulates the cytoplasmic pH of E. coli in a memory-like manner. Sci. Rep. 2019;9:1–10. doi: 10.1038/s41598-019-40560-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Goode, O. et al. Persister Escherichia coli cells have a lower intracellular pH than susceptible cells but maintain their pH in response to antibiotic treatment. mBio12, e0090921 (2021). [DOI] [PMC free article] [PubMed]
- 99.Bartek IL, et al. Antibiotic bactericidal activity is countered by maintaining pH homeostasis in Mycobacterium smegmatis. mSphere. 2016;1:1–18. doi: 10.1128/mSphere.00176-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reyes-Fernández EZ, Schuldiner S. Acidification of cytoplasm in Escherichia coli provides a strategy to cope with stress and facilitates development of antibiotic resistance. Sci. Rep. 2020;10:1–13. doi: 10.1038/s41598-020-66890-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Swerdlow, B. M., Setlow, B. & Setlow, P. Levels of H+ and other monovalent cations in dormant and germinating spores of Bacillus megaterium. J. Bacteriol. 148, 20–29 (1981). [DOI] [PMC free article] [PubMed]
- 102.Setlow B, Setlow P. Measurements of the pH within dormant and germinated bacterial spores. Proc. Natl. Acad. Sci. USA. 1980;77:2474–2476. doi: 10.1073/pnas.77.5.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Magill NG, Cowan AE, Koppel DE, Setlow P. The internal pH of the forespore compartment of Bacillus megaterium decreases by about 1 pH unit during sporulation. J. Bacteriol. 1994;176:2252–2258. doi: 10.1128/jb.176.8.2252-2258.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dechant RC, et al. Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase. EMBO J. 2010;29:2515–2526. doi: 10.1038/emboj.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Robert F, Gendraud M, Pétel G. Using intracellular pH to evaluate growth inhibition of strawberry plants. Plant Physiol. Biochem. 1999;37:155–160. [Google Scholar]
- 106.Busa W, Crowe J. Intracellular pH regulates transitions between dormancy and development of brine shrimp (Artemia salina) embryos. Science. 1983;221:366–368. doi: 10.1126/science.221.4608.366. [DOI] [PubMed] [Google Scholar]
- 107.Bergkessel M. Regulation of protein biosynthetic activity during growth arrest. Curr. Opin. Microbiol. 2020;57:62–69. doi: 10.1016/j.mib.2020.07.010. [DOI] [PubMed] [Google Scholar]
- 108.Dijksterhuis J, Nijsse J, Hoekstra FA, Golovina EA. High viscosity and anisotropy characterize the cytoplasm of fungal dormant stress-resistant spores. Eukaryot. Cell. 2007;6:157–170. doi: 10.1128/EC.00247-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cowan AE, Koppel DE, Setlow B, Setlow P. A soluble protein is immobile in dormant spores of Bacillus subtilis but is mobile in germinated spores: Implications for spore dormancy. Proc. Natl. Acad. Sci. USA. 2003;100:4209–4214. doi: 10.1073/pnas.0636762100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ablett S, Darke AH, Lillford PJ, Martin DR. Glass formation and dormancy in bacterial spores. Int. J. Food Sci. Technol. 1999;34:59–69. [Google Scholar]
- 111.Parry BR, et al. The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell. 2014;156:183–194. doi: 10.1016/j.cell.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Leszczyńska D, et al. The formation of persister cells in stationary-phase cultures of Escherichia coli is associated with the aggregation of endogenous proteins. PLoS ONE. 2013;8:e54737. doi: 10.1371/journal.pone.0054737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yu J, Liu Y, Yin H, Chang Z. Regrowth-delay body as a bacterial subcellular structure marking multidrug-tolerant persisters. Cell Discov. 2019;5:8. doi: 10.1038/s41421-019-0080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Goode, O. et al. Heterologous protein expression favors the formation of protein aggregates in persister and viable but nonculturable bacteria. ACS Infect. Dis. (2021). 10.1021/acsinfecdis.1c00154 (2021). [DOI] [PubMed]
- 115.Dewachter, L. et al. The dynamic transition of persistence towards the VBNC state during stationary phase is driven by protein aggregation. mBio12, e0070321 (2021). [DOI] [PMC free article] [PubMed]
- 116.Khare A, Tavazoie S. Extreme antibiotic persistence via heterogeneity-generating mutations targeting translation. mSystems. 2020;5:1–12. doi: 10.1128/mSystems.00847-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rozov A, et al. Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction. Nat. Commun. 2019;10:1–12. doi: 10.1038/s41467-019-10409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kaldalu N, Hauryliuk V, Tenson T. Persisters—as elusive as ever. Appl. Microbiol. Biotechnol. 2016;100:6545–6553. doi: 10.1007/s00253-016-7648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Amato SM, et al. The role of metabolism in bacterial persistence. Front. Microbiol. 2014;5:70. doi: 10.3389/fmicb.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wu Y, Vulić M, Keren I, Lewis K. Role of oxidative stress in persister tolerance. Antimicrob. Agents Chemother. 2012;56:4922–4926. doi: 10.1128/AAC.00921-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Thierauf A, Perez G, Maloy AS. Generalized transduction. Methods Mol. Biol. 2009;501:267–286. doi: 10.1007/978-1-60327-164-6_23. [DOI] [PubMed] [Google Scholar]
- 122.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Good NE, et al. Hydrogen ion buffers for biological research. Biochemistry. 1966;5:467–477. doi: 10.1021/bi00866a011. [DOI] [PubMed] [Google Scholar]
- 125.Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 126.Liebens V, et al. Identification and characterization of an anti-pseudomonal dichlorocarbazol derivative displaying anti-biofilm activity. Bioorg. Med. Chem. Lett. 2014;24:5404–5408. doi: 10.1016/j.bmcl.2014.10.039. [DOI] [PubMed] [Google Scholar]
- 127.Vedelaar, S., Radzikowski, J. L. & Heinemann, M. A robust method for generating, quantifying and testing large amounts of Escherichia coli persisters. (2020). 10.1101/2020.07.06.181891 (2020). [DOI] [PubMed]
- 128.Omasits U, Ahrens CH, Müller S, Wollscheid B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30:884–886. doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
- 129.Zhao G, London E. An amino acid “transmembrane tendency” scale that approaches the theoretical limit to accuracy for prediction of transmembrane helices: Relationship to biological hydrophobicity. Protein Sci. 2006;15:1987–2001. doi: 10.1110/ps.062286306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Waterhouse A, et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Russell RB, Barton GJ. Multiple protein sequence alignment from tertiary structure comparison: assignment of global and residue confidence levels. Proteins Struct. Funct. Bioinform. 1992;14:309–323. doi: 10.1002/prot.340140216. [DOI] [PubMed] [Google Scholar]
- 132.Humphrey W, Dalke A, Schulten K. VMD—visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 133.Schrödinger, L. The PyMOL molecular graphics system, Version~2.0. (2015).
- 134.Gornall AG, Bardawill CJ, David MM. Determination of serum proteins of the biuret reaction. J. Biol. Chem. 1949;177:751–766. [PubMed] [Google Scholar]
- 135.Schagger H, Cramer WA, Vonjagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- 136.Forneris F, Orru R, Bonivento D, Chiarelli LR, Mattevi A. ThermoFAD, a Thermofluor-adapted flavin ad hoc detection system for protein folding and ligand binding. FEBS J. 2009;276:2833–2840. doi: 10.1111/j.1742-4658.2009.07006.x. [DOI] [PubMed] [Google Scholar]
- 137.Alexandrov AI, Mileni M, Chien EYT, Hanson MA, Stevens RC. Microscale fluorescent thermal stability assay for membrane proteins. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 138.Stolpe S, Friedrich T. The Escherichia coli NADH:ubiquinone oxidoreductase (complex I) is a primary proton pump but may be capable of secondary sodium antiport. J. Biol. Chem. 2004;279:18377–18383. doi: 10.1074/jbc.M311242200. [DOI] [PubMed] [Google Scholar]
- 139.Kaback HR. Transport studies in bacterial membrane vesicles. Science. 1974;186:882–892. doi: 10.1126/science.186.4167.882. [DOI] [PubMed] [Google Scholar]
- 140.Tafin UF, et al. Gentamicin improves the activities of daptomycin and vancomycin against Enterococcus faecalis in vitro and in an experimental foreign-body infection model. Antimicrob. Agents Chemother. 2011;55:4821–4827. doi: 10.1128/AAC.00141-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Miesenböck G, Angelis DDE, Rothman J. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 142.Glatter T, et al. Large-scale quantitative assessment of different in-solution protein digestion protocols reveals superior cleavage efficiency of tandem Lys-C/trypsin proteolysis over trypsin digestion. J. Proteome Res. 2012;11:5145–5156. doi: 10.1021/pr300273g. [DOI] [PubMed] [Google Scholar]
- 143.Kanjee U, et al. Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase. EMBO J. 2011;30:931–944. doi: 10.1038/emboj.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bougdour A, Gottesman S. ppGpp regulation of RpoS degradation via anti-adaptor protein IraP. Proc. Natl. Acad. Sci. USA. 2007;104:12896–12901. doi: 10.1073/pnas.0705561104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lee J-W, Park Y-H, Seok Y-J. Rsd balances (p)ppGpp level by stimulating the hydrolase activity of SpoT during carbon source downshift in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2018;115:E6845–E6854. doi: 10.1073/pnas.1722514115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 2015;13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 148.Sutter M, et al. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat. Struct. Mol. Biol. 2008;15:939–947. doi: 10.1038/nsmb.1473. [DOI] [PubMed] [Google Scholar]
- 149.Contreras H, et al. Characterization of a Mycobacterium tuberculosis nanocompartment and its potential cargo proteins. J. Biol. Chem. 2014;289:18279–18289. doi: 10.1074/jbc.M114.570119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lončar N, Rozeboom HJ, Franken LE, Stuart MCA, Fraaije MW. Structure of a robust bacterial protein cage and its application as a versatile biocatalytic platform through enzyme encapsulation. Biochem. Biophys. Res. Commun. 2020;529:548–553. doi: 10.1016/j.bbrc.2020.06.059. [DOI] [PubMed] [Google Scholar]
- 151.Tamura A, et al. Packaging guest proteins into the encapsulin nanocompartment from Rhodococcus erythropolis N771. Biotechnol. Bioeng. 2015;112:13–20. doi: 10.1002/bit.25322. [DOI] [PubMed] [Google Scholar]
- 152.Heinemeyer E-A, Richter D. In vitro degradation of guanosine tetraphosphate (ppGpp) by an enzyme associated with the ribosomal fraction from Escherichia coli. FEBS Lett. 1977;84:357–361. doi: 10.1016/0014-5793(77)80724-2. [DOI] [PubMed] [Google Scholar]
- 153.An G, Justesen J, Watson RJ, Friesen JD. Cloning the spoT gene of Escherichia coli: Identification of the spoT gene product. J. Bacteriol. 1979;137:1100–1110. doi: 10.1128/jb.137.3.1100-1110.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gnandt, E. & Friedrich, T. The Mechanism of Nadh Oxidation by Respiratory Complex I With Focus on the Production of Reactive Oxygen Species (Universität, 2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The NGS Sequencing data generated in this study have been deposited in the SRA database under accession codes PRJNA498891, PRJNA498708, PRJNA498717, PRJNA270307, and PRJNA768774. Mass spectrometry raw data files have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository under dataset identifier PXD029006. The data underlying all the figures generated in this study are provided in the Source Data file. Mass spectrometry data used in this study are available via the ProteomeXchange Consortium via the PRIDE partner repository under the dataset identifiers PXD000498 and PXD001968. Sequence data used in this study are available in the NCBI nucleotide database (NC_000913.3, NC_007946.1, NC_007941.1), Uniprot database (P0AFC3, P0AFC7, P33599, P0AFD1, P31979, P33602, P0AFD4, P0AFD6, P0AFE0, P0AFE4, P33607, P0AFE8, P0AFF0), PDB database (6g2j, 4he8) and PubChem database (135398637). Source data are provided with this paper.