Abstract
Objective.
To test the hypothesis that an altered gut microbiota (dysbiosis) plays a role in obesity-associated osteoarthritis (OA).
Methods.
Stool and blood samples were collected from 92 participants with BMI ≥ 30 kg/m2 recruited from the Johnston County Osteoarthritis Project. OA cases (n=50) had hand plus knee OA (Kellgren-Lawrence [KL] grade ≥2 or arthroplasty). Controls (N=42) had no hand OA and KL grade 0–1 knees. Compositional analysis of stool samples was carried out by 16S rRNA amplicon sequencing. Alpha and beta diversity and differences in taxa relative abundances were determined. Blood samples were used for multiplex cytokine analysis and measures of lipopolysaccharide (LPS) and LPS binding protein. Germ-free mice were gavaged with case or control pooled fecal samples and placed on a 40% fat, high sucrose diet for 40 weeks. Knee OA was evaluated histologically.
Results.
OA cases were slightly older with more females and higher BMI, WOMAC pain and KL grades than controls. There were no significant differences in alpha or beta diversity or genus level composition between cases and controls. Cases had higher plasma levels of osteopontin (p=0.01) and serum LPS (p<0.0001). Mice transplanted with case or control microbiota exhibited a significant difference in alpha diversity (p=0.02) and beta diversity but no differences in OA severity.
Conclusion.
The lack of differences in the gut microbiota yet increased serum LPS levels suggest the possibility that increased intestinal permeability allowing for greater absorption of LPS, rather than a dysbiotic microbiota, may contribute to development of OA associated with obesity.
Keywords: microbiome, osteoarthritis, obesity, inflammation, articular cartilage
INTRODUCTION
Major risk factors for osteoarthritis (OA) include age, obesity, genetics, and prior joint injury (1). Obesity is a risk factor not only for knee and hip OA, but also hand OA, suggesting that factors associated with obesity, in addition to increased joint loads, have a role in OA, including altered metabolism and low grade systemic inflammation (2, 3). A number of studies have shown that the composition of the gut microbiota plays a central role in mediating the effects of what we eat on the development of obesity and obesity-related conditions (4–7). Very few studies reported to date have examined the role of the gut microbiota in human OA and none has focused on OA associated with obesity.
Two forms of arthritis that are mechanistically distinct from each other in etiology, ankylosing spondylitis (8) and rheumatoid arthritis (9), have been linked to altered composition of the gut microbiota. Theoretically, development of OA could also be promoted by gut microbial factors, not only because of the strong links of OA to diet and obesity, but also due to findings in older adults (the group at highest risk of OA) that the composition of the gut microbiota and the presence of dysbiosis correlates with measures of physical function, systemic markers of inflammation, and the number of co-morbidities (10). Intestinal dysbiosis is a state of microbial imbalance that results from “a change in the structural and/or functional configuration of the microbiota that disrupts homeostasis between the host and the microbial community” (11). Dysbiosis can result in altered metabolism, obesity, malnutrition, and increased intestinal permeability resulting in increased systemic levels of lipopolysaccharide (LPS) and other inflammatory microbial products (4–7, 11).
A potential connection between obesity-associated OA and the microbiome was suggested by a metabolomics analysis performed using urine samples from overweight and obese adults with knee OA that were participants in the Intensive Diet and Exercise for Arthritis (IDEA) trial. Participants who exhibited radiographic progression of their knee OA over the course of the 18-month trial had increased levels of several metabolites that included hippurate (12). Hippurate is a mammalian-microbial “co-metabolite” well known to be affected by the composition of the gut microbiota (5) suggesting that the gut microbiota could be influencing OA progression in overweight and obese adults.
The purpose of the present study was to determine if an altered gut microbiota plays a causal role in obesity-associated OA by analyzing the microbial composition of fecal samples from OA cases and controls, as well as plasma levels of cytokines and serum levels of the microbial product LPS also known as endotoxin. To enrich the study population with individuals more likely to have obesity-associated OA, and its associated systemic metabolic changes, we recruited participants who were obese (BMI ≥ 30 kg/m2) and exhibited both hand and knee OA In order to test for causality, and microbial functional changes not detected by DNA sequence analysis, we measured knee OA severity after transplanting pooled human fecal microbiota from OA cases and controls to germ-free mice and then inducing obesity-associated OA by placing the mice on a high-fat and sucrose (“Western”) diet for 40 weeks.
PATIENTS AND METHODS
Study Participants.
Blood and fecal samples were collected from selected cases and controls recruited from the Johnston County Osteoarthritis Project (JoCo OA), which is a longitudinal, population-based, prospective study designed to investigate prevalence, incidence, and progression of OA and its risk factors. A detailed description of the study can be found elsewhere (13). In short, at the baseline visit (T0, 1991–1997), 3,187 adults were enrolled into the original cohort. Participants completed four follow-up visits approximately 5 years apart: T1 – T4. An enrichment cohort (T1*) entered the study in 2003–2004, and therefore T1* enrollees have shorter follow-up: only T2 - T4. For the present microbiome study, participants were recruited at the time of the most recent follow-up evaluation (T4, 2016–2018).
Ninety-two individuals were enrolled (50 cases and 42 controls) who: (1) had agreed to participate in T4 and to be contacted about future studies, (2) met eligibility criteria and consented to participate in this substudy (UNC IRB#15–1834), and (3) provided stool samples and completed a diet questionnaire in addition to the standard JoCo OA measures. To be eligible for recruitment as an OA “case”, the participant needed to be obese (BMI ≥ 30 kg/m2) and have clinical and/or radiographic hand OA defined as involvement of at least 3 joints across both hands (14) plus Kellgren-Lawrence (KL) grade 2–4 knee OA or knee arthroplasty for OA. Controls were also obese but did not have evidence of hand OA and had KL grade 0–1 knees based on a reading by one of the study investigators (RL) of the radiographs taken at the time of recruitment during the T4 visit. Radiographs were re-read by the study radiologist (JR), paired with prior knee radiographs, after all participants had been enrolled in the study. Participants were older than 45 years of age and there was an attempt to recruit controls to the study that were age- and sex-matched to the cases. Exclusion criteria were: recent (in the past 6 weeks) antibiotic and/or probiotics use, bowel surgery, a history of inflammatory bowel disease and/or celiac disease, or history of fecal microbiota transplantation (all self-reported). All individuals who met eligibility criteria completed consent and had a stool sample collection kit (EasySampler®, ALPCO Diagnostics, Salem, NH) delivered to their home along with instructions on how to collect and store the stool samples.
Sample Collection.
Details of the fecal and blood sample collection are provided in the Supplementary Methods.
Demographic and clinical measurements.
Data collected on participants as part of the JoCo OA T4 follow-up and used in the present study included demographic information and evaluation of knee pain using the Knee injury and Osteoarthritis Outcome Score (KOOS) subscale for pain for the left and right knees separately as described (15). The AUStralian CANadian Osteoarthritis Hand Index (AUSCAN) subscales were used to assess hand symptoms (16). For the present study, we also added a validated dietary questionnaire to evaluate the fat content of the diet at the time of stool collection (17). Non-steroidal anti-inflammatory drug (NSAID) use and nutraceutical use (e.g., glucosamine, chondroitin sulfate) was also recorded.
Fecal Microbiome Analysis.
Isolation of total DNA from stool samples and analysis by Illumina MiSeq 16S rRNA amplicon sequencing was performed in the UNC Microbiome Core as described (18, 19). Details are provided in the Supplementary Methods.
Cytokine and LPS measures.
Cytokine analysis was performed using G5 series arrays to profile 80 markers following the manufacturer’s instructions (RayBiotech Life, Peachtree Corners, GA) in plasma samples (n=73). Details are provided in the Supplementary Methods and results for all cytokines in Supplementary Table S1.
Serum samples (n=78) were used to measure LPS using the EndoZyme® Recombinant Factor C (rFC) Endotoxin Detection Assay from Hyglos (#890030) and LPS binding protein (LBP) using an ELISA kit for human LBP (Hycult Biotech) as previously described (20).
Fecal Transplant to germ-free mice and subsequent diet.
The use of mice for this study was approved by UNC Animal Care and Use Committee (#16–294). Fecal samples from 5 OA cases and 5 controls (Supplementary Table S2) were randomly selected to be used for fecal transplant to germ-free mice. Pooled fecal samples from each group of 5 were used to reduce heterogeneity within groups. Thirty-one 10- to 13-week-old male germ-free C57BL/6 mice were obtained from the UNC National Gnotobiotic Rodent Resource Center. Mice were randomly assigned to be gavaged with either the case (n=16 mice) or control (n=15 mice) fecal pools. The number of mice per group was based on our prior studies using the same mouse strain for histologic OA studies where ACS score was the primary outcome. These studies ((21, 22) have shown that an n=15 per group is sufficient to provide at least 90% power at α = 0.05 to detect a biologically meaningful difference of 50% between two groups. Details of the fecal transplantation are provided in the Supplementary Methods. For the first two weeks after fecal transplant, recipient mice were fed, ad libitum, standard chow to allow sufficient time for complete colonization (“humanization”). Then all mice were switched to sterilized “Western” diet chow (D12079B) from Research Diets (New Brunswick, NJ), which is a 40% fat and high sucrose (341gm%) diet and were maintained on the diet until the end of the study (40 weeks on diet).
The fecal gavage was repeated every 4 weeks to help maintain the humanized microbiome. Mice were weighed every other week. Stool pellets were collected from individual mice two weeks after each gavage and stored at −800C until the end of the mouse study period when they were submitted to the UNC Microbiome facility for analysis along with samples from the original OA case and control pools that had been used for transplant. Microbiome analysis was performed as described above. After 40 weeks on diet, body composition was measured by MRI in the UNC Nutrition Obesity Research Center Animal Metabolism Phenotyping Core and then the mice were sacrificed.
Histologic Evaluation.
Mouse stifle (knee joints) were collected, fixed and processed for histology as previously described (21, 23). Details of the grading are presented in the Supplementary Methods.
Statistical Analysis.
Primary analysis, 16SrRNA amplicon sequencing data.
We used QIIME 1.9.0 to generate measures of microbial diversity (24). Alpha (within-sample) diversity measures, including Shannon index (25), Chao1 (26), Phylogenetic diversity (PD whole tree), and observed species number metrics, were estimated at a rarefaction depth of 1,000 sequences per sample. Associations of alpha diversity (Shannon Index) with OA status, sex, age, and BMI were examined by univariate and multivariable linear regression models. Beta (between-sample) diversity estimates were calculated using weighted and unweighted Unifrac distances between samples at a subsampling depth of 1,000 (27, 28). Principal coordinate analysis (PCoA) was used to summarize these results. Permutation Multivariate Analysis of Variance (PERMANOVA) was used to test the null hypothesis that case and control microbial communities share the same distribution. The associations with OA status were examined for all participants and stratified by sex.
Further, comparisons between the two groups were performed individually for each taxon as follows. Taxa were summarized based at their taxonomic levels with QIIME’s summarize_taxa.py and the analysis was focused on genera. The Operational Taxonomic Unit (OTU) table for genus level was normalized and transformed to the table format with taxa as columns and individuals as rows using Python. We applied an arbitrary threshold and removed taxa that were absent in more than 75% samples. To identify the individual OTU ids that were significantly different between the case and control groups and between females and males, we applied the non-parametric Wilcoxon test following Benjamini and Hochberg adjustment for P-values in multiple comparisons (29) with a significance level of 0.05 implemented in r.adjust R function. The effect size (ES) was calculated as Z statistics divided by square root of the sample size as described (30). The interpretation of ES coincides with the one for Pearson’s correlation coefficient.
To ensure that the results were not a consequence of the QIIME algorithm, we repeated these steps using the ‘vegan’ R package. The ‘adonis’ function in the ‘vegan’ package was used to test differences of the Bray-Curtis dissimilarity between obese OA and obese non-OA participants. Multidimensional Scaling (MDS) ordination was performed using the ‘capscale’ function with Bray-Curtis dissimilarity.
Pain andmicrobial community composition.
We performed exploratory analysis (as detailed in the Supplementary Methods) to evaluate whether the gut microbiota was associated with knee and/or hand pain. In addition, we attempted to replicate findings from a recent study that found an association between the composition of the gut microbiota and WOMAC knee pain in the Rotterdam study (31). In that study, the abundance of the Streptococcus genus in the order Lactobacillales and class Bacilli was found to be significant. We considered these taxa to replicate if they reached a nominal significance of p=0.05 in our data.
Methods for analysis of LPS/LBP results, cytokine levels, and the mouse data are provided in the Supplementary Methods.
RESULTS
Demographics and clinical characteristics of the study participants.
A total of 92 individuals were recruited for this project (50 obese hand plus knee OA cases and 42 obese controls). The cases were slightly older, consisted of more females, and had a higher mean BMI compared to the controls (Table 1). As expected, the cases had higher WOMAC knee pain scores and AUSCAN hand pain scores. There were no differences between groups in the proportion of African Americans, dietary fat intake, glucosamine use or NSAID use. The cases had a predominance of severe knee OA with 70% of the participants having knees with either KL grade 4 (42%) or prior knee arthroplasty (28%). The controls had mainly KL grade 1 knees (76%) but 9 individuals in the control group (21%) had a knee with KL grade of 2 when the radiographs were graded by the study radiologist. These had all been read as KL grade 1 at the time of screening for recruitment.
Table 1.
Demographics and clinical characteristics
| Participant Characteristic | Cases (N=50) | Controls (N=42) | p-value |
|---|---|---|---|
| Age, years | 73.7 (6.9) | 70.8 (6.4) | 0.04 |
| Female, n (%) | 43 (86%) | 26 (62%) | 0.01 |
| African American, N (%) | 17 (34%) | 18 (43%) | 0.39 |
| BMI, kg/m2 | 36.3 (4.4) | 33.4 (3.1) | 0.001 |
| WOMAC pain score, left knee | 5.2 (5.1) | 2.2 (3.7) | 0.002 |
| WOMAC pain score, right knee | 5.3 (4.9) | 1.8 (3.3) | 0.0001 |
| AUSCAN hand pain score | 5.3 (6.1) | 2.4 (3.9) | 0.01 |
| Percent calories from fat, % | 33.1 (2.6) | 34.2 (4.7) | 0.20 |
| Glucosamine use, n (%) | 2 (0.04%) | 2* (0.05%) | 0.53 |
| NSAID use, n (%) | 33 (66%) | 27* (66%) | 0.99 |
| Maximum KL Grade | |||
| 0 | 0 | 1 (2.4%) | |
| 1 | 0 | 32 (76.2%) | |
| 2 | 2 (4.0%) | 9 (21.4%) | |
| 3 | 13 (26.0%) | 0 | |
| 4 | 21 (42.0%) | 0 | |
| Total knee replacement | 14 (28.0%) | 0 | |
Values are mean (SD) unless otherwise specified; BMI, body mass index; WOMAC, Western Ontario and McMaster Universities Arthritis Index (range 0–20); KL, Kellgren and Lawrence
missing data-glucosamine use (n=1)
Microbiome analysis.
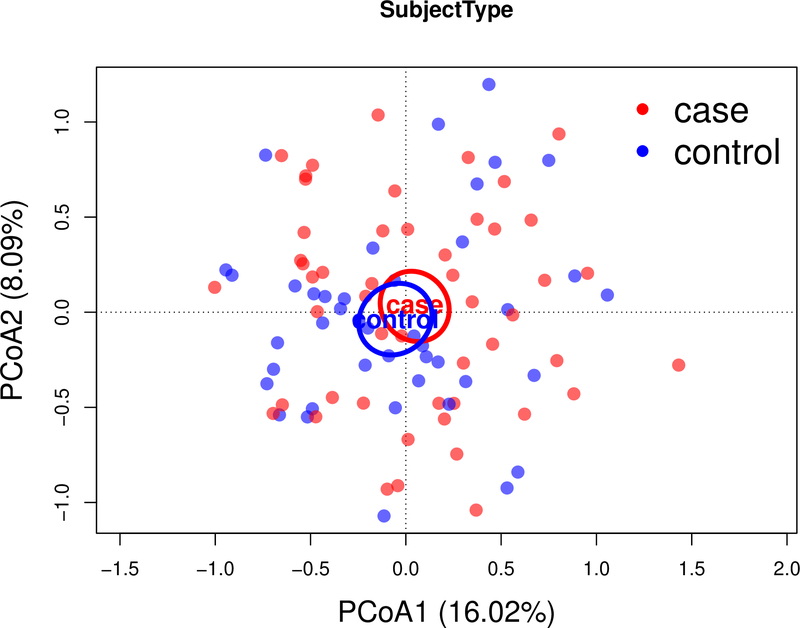
In univariate analyses, alpha diversity was inversely associated with BMI and not associated with OA status, sex, or age. In a multivariable linear regression model, none of these variables were significantly associated with the Shannon diversity Index (Supplementary Table S3). There was also no difference in beta diversity (p=0.73, PERMANOVA); a PCoA based on weighted UniFrac distance between samples showed no differences between the case and control groups (Figure 1). All results were consistent in the analysis using the vegan R package with Bray-Curtis dissimilarity (data not shown). We found that 3 taxa had nominally significant (p<0.05) associations with OA case status (ES ranges 0.23–0.33), but none of the associations remained significant with Benjamini and Hochberg adjusted p-values <0.1 (Table 2). There were no overall significant taxonomic differences between males and females, and no significant differences were found in the relative abundance of specific genera (Supplementary Table S4). No differences in alpha and beta diversity were seen when stratifying by sex. A sensitivity analysis was performed to account for the 9 participants in the control group with KL grade 2 but the results were not meaningfully different (data not shown).
Figure 1.
Microbial similarity biplot (Principal Coordinate Analysis (PCoA) axes) for study participants with knee and hand OA (red) and without OA (blue). Numbers in parentheses indicate the percent variation explained by the corresponding axis. Each point represents a single sample. The distance between 2 points shows how compositionally different the samples are. Ellipsoids illustrate the 95% CI for the mean location of each group.
Table 2.
Relative abundance of genera in JoCo participants with (cases) and without (controls) knee plus hand OA
| Taxon name | ALL | CASES | CONTROLS | |||||
|---|---|---|---|---|---|---|---|---|
| mean | SD | mean | SD | mean | SD | ES(P) | Q | |
| k_Bacteria.p_Cyanobacteria.c_4C0d.2.o_YS2.f_.g_ | 2.64E–03 | 1.24E–02 | 5.17E–04 | 1.96E–03 | 5.17E–03 | 1.80E–02 | 0.33 (0.001) | 0.13 |
| k_Bacteria.p_Firmicutes.c_Clostridia.o_Clostridiales. f_Christensenellaceae.g_Christensenella |
3.00E–05 | 6.21E–05 | 1.71E–05 | 4.83E–05 | 4.53E–05 | 7.30E–05 | 0.26 (0.01) | 0.56 |
| k_Bacteria.p_Lentisphaerae.c_.Lentisphaeria.o_Victivallales. f_Victivallaceae.g_ |
9.57E–04 | 4.86E–03 | 4.84E–04 | 1.70E–03 | 1.52E–03 | 6.96E–03 | 0.23 (0.03) | 0.68 |
| k_Bacteria.p_Firmicutes.c_Clostridia.o_Clostridiales. f_Eubacteriaceae.g_Anaerofustis |
3.01E–05 | 6.74E–05 | 2.52E–05 | 5.85E–05 | 3.58E–05 | 7.70E–05 | 0.20 (0.06) | 0.68 |
| k_Bacteria.p_Synergistetes.c_Synergistia.o_Synergistales. f_Dethiosulfovibrionaceae.g_Pyramidobacter |
1.24E–03 | 8.19E–03 | 1.43E–03 | 9.99E–03 | 1.02E–03 | 5.42E–03 | 0.20 (0.06) | 0.68 |
| k_Bacteria.p_Proteobacteria.c_Gammaproteobacteria. o_Enterobacteriales.f_Enterobacteriaceae.Other |
1.12E–04 | 2.74E–04 | 6.81E–05 | 1.48E–04 | 1.64E–04 | 3.68E–04 | 0.19 (0.07) | 0.68 |
| k_Bacteria.p_Firmicutes.c_Clostridia.o_Clostridiales. f_Peptococcaceae.g_rc4.4 |
1.74E–04 | 6.22E–04 | 5.04E–05 | 1.34E–04 | 3.20E–04 | 8.92E–04 | 0.19 (0.07) | 0.68 |
| k_Bacteria.p_Proteobacteria.c_Gammaproteobacteria. o_Pasteurellales.f_Pasteurellaceae.g_Haemophilus |
1.08E–03 | 3.98E–03 | 1.28E–03 | 4.93E–03 | 8.50E–04 | 2.44E–03 | 0.19 (0.07) | 0.68 |
| k_Bacteria.p_Firmicutes.c_Clostridia.o_Clostridiales. f_Lachnospiraceae.g_Roseburia |
1.30E–03 | 1.51E–03 | 1.09E–03 | 1.39E–03 | 1.54E–04 | 1.63E–03 | 0.18 (0.08) | 0.68 |
| k_Bacteria.p_Firmicutes.c_Clostridia.o_Clostridiales. f_Lachnospiraceae.g_Dorea |
5.78E–03 | 7.71E–03 | 5.55E–03 | 8.36E–03 | 6.06E–04 | 6.94E–03 | 0.18 (0.08) | 0.68 |
P, p-values from the non-parametric Wilcoxon test; ES, effect size; Q, false discovery rate adjusted p-values; SD, standard deviation
Association of gut microbiota composition and pain.
We conducted a secondary analysis to examine an association between the gut microbiota and WOMAC knee pain as well as AUSCAN hand pain in our cases and controls but did not find any significant differences after adjusting for multiple comparisons (Supplementary Tables S5 and S6). In addition, the associations of Lactobacillales or Streptococcus with WOMAC pain reported in the Rotterdam study (31) did not replicate in our study (p=0.44 and 0.64 respectively).
Differences in cytokines and LPS.
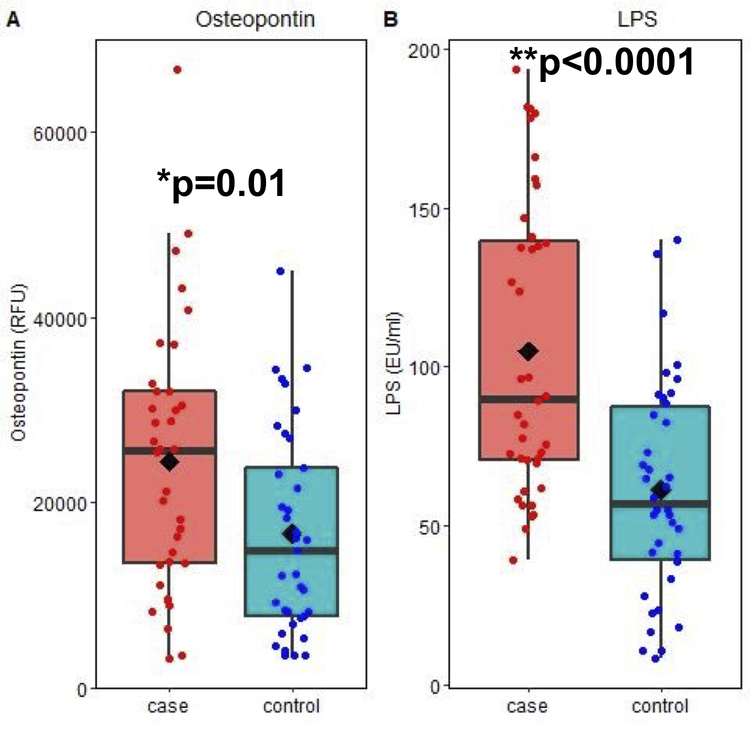
Plasma samples from cases (n=36) and controls (n=37) were analyzed for differences in 80 cytokines (Supplementary Table S1) that included 55 characterized as pro-inflammatory, 15 as anti-inflammatory and 10 as having pleotropic activity. Osteopontin, a pro-inflammatory cytokine (32, 33) was noted to be significantly higher in the cases (mean ± SD = 24380.6 ± 14315.9 RFU) than controls (16620.2 ± 10997.53 RFU), p=0.01) (Figure 2A). None of the other cytokines differed between the case and control groups. However, there were differences in several cytokines within either group between men and women, or African American versus Caucasian participants (Supplementary Table S1). Serum LPS levels measured in 40 cases (104.9 ± 45.8 EU/ml) and 38 controls (61.3 ±33.9 EU/ml) were also significantly higher in cases (p<0.0001, Figure 2B). There was no difference in serum LBP between cases (11.3 ± 5.0 μg/ml) and controls (13.3 ± 6.0 μg/ml). The osteopontin and LPS results remained significant when re-analyzed with the 9 participants in the control group with KL grade 2 treated as cases (data not shown). No correlation between osteopontin, LPS or LBP levels with knee or hand pain was noted (data not shown).
Figure 2.
Osteopontin and LPS levels in blood samples from OA cases and controls. Osteopontin was measured in plasma samples by a cytokine array and LPS was measured in serum using an endotoxin assay as detailed in the methods. The units for osteopontin are relative fluorescence units and for LPS are endotoxin units/ml. Data were analyzed using two-sided Student’s t tests.
Severity of osteoarthritis in mice after fecal transplant and treatment with a “Western” diet.
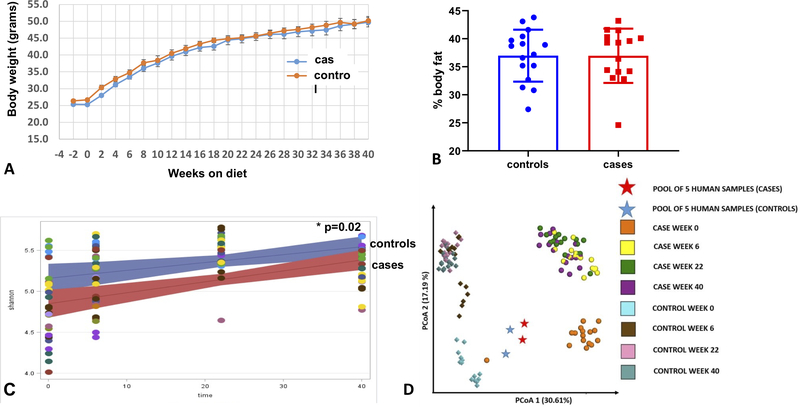
All 31 mice that received the fecal transplants and were treated with the high fat and high sucrose “Western” diet for 40 weeks completed the study. There were no differences between mice that received the case fecal pool compared to those that received the control fecal pool in weight gain over the course of the study (Figure 3A) or in percent body fat at sacrifice (Figure 3B). There were significant differences (p=0.02) in the Shannon (alpha) diversity between fecal microbiota in the two groups at baseline which remained over the course of the study (Figure 3C). A PCoA Emperor Plot for beta diversity revealed that one case mouse clustered with the control mice at baseline (time 0), which was 2 weeks after transplant when the diet was started, but at the subsequent time points the case and control mice clustered in their respective groups (Figure 3D). Individual clade analysis discovered 15 taxa that were different between groups at baseline (FDR-adjusted p-value < 0.05, Supplementary Table S8).
Figure 3.
Effects of a high fat and high sucrose diet after transfer of human OA case and control fecal samples to germ free mice. A, Body weight of mice starting at time of fecal transplant (−2 weeks) and then bi-weekly while on the high fat and high sucrose diet for 40 weeks. B, %body fat mesured by MRI after 40 weeks on diet. C, Linear mixed-effects model regression of Shannon index from fecal samples collected at the start of the diet (time 0 which is 2 weeks after fecal transplant) and after 6, 22, and 40 weeks on diet. Lines represent linear mixed-effects regression (and 95% confidence interval) of Shannon diversity over time. Each dot represents a mouse and those with the same color indicates they were caged together. D, PCoA Emperor Plot based on Bray-Curtis beta-diversity metric. Points represent samples from the two groups: OA cases (circle) and OA controls (diamond). Time points are colored coded as indicated on the graph. Stars indicate duplicate samples of the pooled human feces analyzed prior to transplant to mice. Numbers in parentheses indicate the percent variation explained by the corresponding axis. The distance between 2 points shows how compositionally different the samples are.
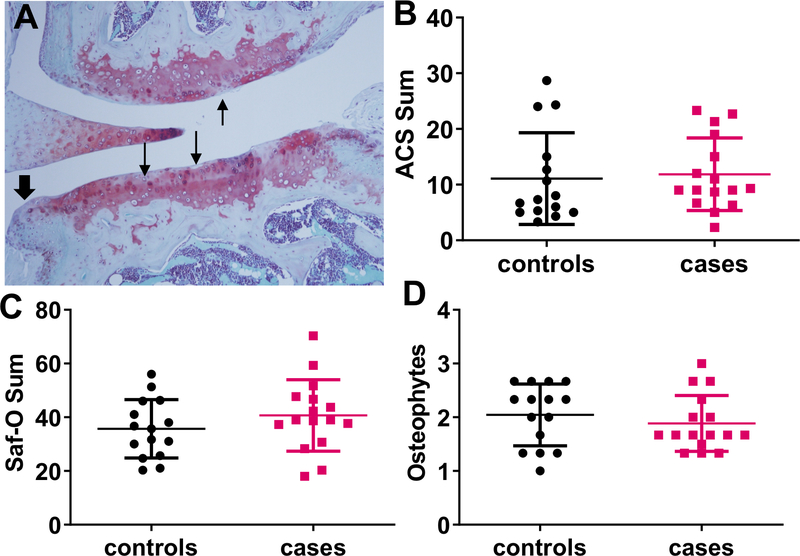
The “Western” diet induced histologic OA changes including cartilage degradation and matrix loss, measured using the ACS and Safranin-O scores, as well as osteophytes (Figure 4) but minimal synovitis (data not shown). There were no differences in any of the histologic measures between mice that received the fecal transplants from cases compared to the controls.
Figure 4.
Results of histologic analysis of knee joints from mice transplanted with case or control fecal pools and placed on a high fat and high sucrose diet for 40 weeks. A, representative images demonstrating histologic findings of OA including cartilage degradation with loss of matrix staining (arrows) and osteophyte formation (arrowhead). B-D, Results of histologic measures of OA including ACS scores (B), Saf-O scores (C) and osteophyte scores (D) presented as means and standard deviation. None of the measures were significantly different between cases and controls (Mann Whitney test). ACS, Articular cartilage structure, Saf-O, safranin-O.
DISCUSSION
This study did not demonstrate a difference in the fecal microbial communities between obese adults with hand OA and advanced knee OA when compared to obese controls without hand and knee OA. However, we did find serum levels of LPS were significantly elevated in the OA cases relative to controls. Since LPS is produced by enteric Gram-negative bacteria, including those in the gut, elevated serum levels suggest the presence of increased intestinal mucosal permeability, sometimes referred to as a “leaky gut”. This is consistent with a previous report of increased LPS in the serum and synovial fluid of adults with knee OA where the levels of serum LPS were associated with activated macrophages in the knee (20). Huang and Kraus proposed that LPS uptake from the gut due to an altered microbiome contributes to a low-grade inflammatory state that promotes the development of OA as a “second hit” when other OA risk factors are present, such as obesity or joint injury (34). In further work, they demonstrated an association of plasma LBP with radiographic progression of knee OA (35) but we were unable to show a difference in LBP between the cases and controls in the present study.
We also noted a significant increase in the pro-inflammatory mediator osteopontin in the OA cases. Although originally described as a bone matrix protein, subsequent studies have shown that osteopontin is a proinflammatory mediator and can serve as a Th1 cytokine (reviewed in (33). Osteoblasts from OA subchondral bone have increased expression of osteopontin (36) as do OA chondrocytes (37, 38) and, as we found with hand plus knee OA, systemic levels of osteopontin have been shown to be increased in association with knee OA (39).
Similar to our results in 92 JoCo OA participants, a study of 1400 participants in the Rotterdam study did not find an association between the fecal microbiome and the presence of radiographic knee OA (31). However, they did find an association of Streptococcus species with WOMAC pain scores, which they replicated in a second Dutch cohort of 867 individuals. We could not replicate this result but our sample size of 92 individuals may have been too small, although not even a trend was observed. Published studies on the association between the gut microbiota and OA in humans were lacking when we designed our study. We based our recruitment of 92 participants on prior work in RA (9) where highly significant microbial associations were observed in 44 new-onset rheumatoid arthritis patients compared to 28 healthy controls with multiple taxa displaying robust effect sizes (> 2). We attempted to enrich our OA cases with a phenotype expected to have a systemic component to their OA, potentially related to the gut microbiome, by recruiting obese individuals who exhibited both hand and knee OA. Our findings suggest that the fecal microbial communities found in obese adults with hand plus knee OA do not have as strong of an effect on the risk for OA as those seen in RA. Since our participants had advanced knee OA with the majority exhibiting KL grade 4 or prior knee arthroplasty, our findings do not rule out the possibility that a dysbiosis was present earlier in the disease course.
Rodent studies have suggested a role of the gut microbiota in OA. A study of diet-induced obesity in rats noted a link between the severity of cartilage damage and fat mass, serum LPS concentrations, and the increased presence of Methanobrevibacter species in fecal samples, while presence of Lactobacillus was negatively associated with cartilage damage (40). Treatment of rats (41) and mice (42) on a high fat diet with the pre-biotic fiber oligofructose restored the fecal microbiota to that seen with a normal chow diet. This restoration was associated with reduced serum LPS levels and, in the mouse study, less cartilage damage after OA was surgically induced. In a previous study, we noted less cartilage damage and fewer osteophytes in germ-free mice with surgically-induced OA compared to mice housed in a standard facility and the severity of cartilage damage correlated with serum levels of LBP (43).
In the present study, we transplanted germ-free mice with pools of fecal samples from human OA cases and controls and then placed the mice on a “Western” diet. Although there were no differences in the human case and control gut microbiome by 16S rRNA sequencing for the entire group of 92 participants, this method does not allow for the full taxonomic identification of taxa, down to the species level, and so strain-level differences or functional alterations could still be present. Therefore, lack of differences in composition by 16S rRNA sequencing do not necessarily translate into lack of differences in microbial metabolic activities or metagenomic differences that could contribute to the development of OA. Importantly, clade analysis did identify differences in the mouse fecal samples after transfer of human case and control fecal pools (Supplementary Table S8). This included higher levels of Eubacterium in the obese non-OA control group. Huang et al (44) reported that the presence of Eubacterium correlated with lower OARSI scores of histologic OA in mice that had OA induced by meniscal injury after receiving human fecal transplants. Higher levels of Akkemansia were noted in our obese OA group. In a rat study of diet-induced obesity reported by Rios et al (41), Akkemansia was positively correlated with severity of joint damage. We also found 3 taxa from the Ruminococcaceae family that were different between controls and OA samples with Ruminnococcus found to be more abundant in non-OA group while Anaerotruncus and Oscillospira were more abundant in the OA group. Ruminococcaceae are butyrate producers and butyrate is proposed to be a beneficial metabolite; a decreased abundance of Ruminococcaceae was associated with more severe OA in the Huang et al study (44).
Despite a difference in the microbiota between the two groups of mice after fecal transfer of human case and control fecal samples that included taxa that might be expected to influence the development of OA, there was no difference in the severity of histologic OA after 40 weeks of the “Western” diet. Although human fecal transfer to germ-free mice has been used to establish causality in other conditions including obesity (7), a limitation to this technique is that selective colonization of microbial community members during transfer from humans to germ-free mice can occur such that potential causative organisms may not be in the same abundance in the recipient mice as in the human donors (45.)
Our results, combined with the published studies noted above, indicate future studies are needed to further evaluate increased intestinal mucosal permeability as a potential contributing factor to the development of OA and to elucidate the underlying mechanisms. Although NSAIDs can cause increased intestinal permeability in some individuals, we did not find an association with LPS levels and NSAID use; both cases and controls reported similar use. Increased intestinal permeability, resulting in higher levels of systemic LPS, can occur independent of differences in the microbiome or from differences in just a few taxa that may not have been detected given the sample size available for this study. Causes of increased LPS also include impaired clearance by the liver, high fat intake, alterations in the endocannabinoid system, deceased intestinal motility and decreased physical activity (reviewed in (34). In addition to LPS, other factors produced by gut microbes that could enter the circulation through a leaky gut may be discovered that could promote OA. Intriguingly, recent reports that microbial DNA, including that from typical gut microbes, was present in OA cartilage (46) and synovial tissue (47), suggests that increased intestinal permeability could allow gut microbes access to joint tissues. Although there was no evidence that the microbial DNA was from viable bacteria or associated with joint infections, these studies taken together indicate that in OA, microbial products or microbes from the gut that enter the circulation may have a local effect on joints.
Supplementary Material
ACKOWLEDGEMENTS
We would like to thank the staff and participants in the Johnston County Osteoarthritis Project without whom this work would not be possible.
Supported by the Arthritis Foundation, the National Center for Advancing Translational Sciences (NCATS) (UL1TR002489), and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (P30AR072580). The Johnston County Osteoarthritis Project is funded in part by the Centers for Disease Control and Prevention (U01 DP006266). The UNC Microbiome and Gnotobiotic Cores are supported in part by the National Institute of Diabetes and Digestive and Kidney Diseases (P30 DK034987 and P30 DK056350) and the Office of the Director, NIH (P40 OD010995). The cytokine analysis was funded, in part, by the Nutrition Obesity Resource Core (P30DK056350).
Footnotes
Conflict of interest: No disclosures relevant to this manuscript were reported.
Publisher's Disclaimer: This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: 10.1002/ART.41955
REFERNCES
- 1.Johnson VL, Hunter DJ. The epidemiology of osteoarthritis. Best Pract Res Clin Rheumatol. 2014;28(1):5–15. [DOI] [PubMed] [Google Scholar]
- 2.Sellam J, Berenbaum F. Is osteoarthritis a metabolic disease? Joint Bone Spine. 2013;80(6):568–73. [DOI] [PubMed] [Google Scholar]
- 3.Visser AW, Ioan-Facsinay A, de Mutsert R, Widya RL, Loef M, de Roos A, et al. Adiposity and hand osteoarthritis: the Netherlands Epidemiology of Obesity study. Arthritis Res Ther. 2014;16(1):R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474(7351):327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–7. [DOI] [PubMed] [Google Scholar]
- 6.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costello ME, Elewaut D, Kenna TJ, Brown MA. Microbes, the gut and ankylosing spondylitis. Arthritis Res Ther. 2013;15(3):214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2013;2:e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488(7410):178–84. [DOI] [PubMed] [Google Scholar]
- 11.Gordon JI. Honor thy gut symbionts redux. Science. 2012;336(6086):1251–3. [DOI] [PubMed] [Google Scholar]
- 12.Loeser RF, Pathmasiri W, Sumner SJ, McRitchie S, Beavers D, Saxena P, et al. Association of urinary metabolites with radiographic progression of knee osteoarthritis in overweight and obese adults: an exploratory study. Osteoarthritis Cartilage. 2016;24(8):1479–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordan JM, Linder GF, Renner JB, Fryer JG. The impact of arthritis in rural populations. Arthritis Care Res. 1995;8(4):242–50. [DOI] [PubMed] [Google Scholar]
- 14.Scherzer ZA, Alvarez C, Renner JB, Murphy LB, Schwartz TA, Jordan JM, et al. Effects of comorbid cardiovascular disease and diabetes on hand osteoarthritis, pain, and functional state transitions: The Johnston County Osteoarthritis Project. J Rheumatol. 2020;47(10):1541–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roos H, Lauren M, Adalberth T, Roos EM, Jonsson K, Lohmander LS. Knee osteoarthritis after meniscectomy: prevalence of radiographic changes after twenty-one years, compared with matched controls. Arthritis Rheum. 1998;41(4):687–93. [DOI] [PubMed] [Google Scholar]
- 16.Bellamy N, Campbell J, Haraoui B, Gerecz-Simon E, Buchbinder R, Hobby K, et al. Clinimetric properties of the AUSCAN Osteoarthritis Hand Index: an evaluation of reliability, validity and responsiveness. Osteoarthritis Cartilage. 2002;10(11):863–9. [DOI] [PubMed] [Google Scholar]
- 17.Thompson FE, Midthune D, Williams GC, Yaroch AL, Hurley TG, Resnicow K, et al. Evaluation of a short dietary assessment instrument for percentage energy from fat in an intervention study. J Nutr. 2008;138(1):193S–9S. [DOI] [PubMed] [Google Scholar]
- 18.Allali I, Arnold JW, Roach J, Cadenas MB, Butz N, Hassan HM, et al. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC Microbiol. 2017;17(1):194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azcarate-Peril MA, Butz N, Cadenas MB, Koci M, Ballou A, Mendoza M, et al. An attenuated Salmonella enterica serovar typhimurium strain and galacto-oligosaccharides accelerate clearance of Salmonella infections in poultry through modifications to the gut microbiome. Appl Environ Microbiol. 2018;84(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Z, Stabler T, Pei F, Kraus VB. Both systemic and local lipopolysaccharide (LPS) burden are associated with knee OA severity and inflammation. Osteoarthritis Cartilage. 2016; 24(10):1769–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loeser RF, Olex A, McNulty MA, Carlson CS, Callahan M, Ferguson C, et al. Microarray analysis reveals age-related differences in gene expression during the development of osteoarthritis in mice. Arthritis Rheum. 2012;64(3):705–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loeser RF, Kelley KL, Armstrong A, Collins JA, Diekman BO, Carlson CS. Deletion of JNK enhances senescence in joint tissues and increases the severity of age-related osteoarthritis in mice. Arthritis Rheumatol. 2020;72(10):1679–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rowe MA, Harper LR, McNulty MA, Lau AG, Carlson CS, Leng L, et al. Deletion of macrophage migration inhibitory factor reduces severity of osteoarthritis in aged mice. Arthritis Rheumatol. 2017;69:352–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7(5):335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shannon CE. The mathematical theory of communication. 1963. MD Comput. 1997;14(4):306–17. [PubMed] [Google Scholar]
- 26.Chao A Nonparametric-estimation of the number of classes in a population. Scandinavian Journal of Statistics. 1984;11(4):265–70. [Google Scholar]
- 27.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57(1):289–300. [Google Scholar]
- 30.Fritz CO, Morris PE, Richler JJ. Effect size estimates: current use, calculations, and interpretation. J Exp Psychol Gen. 2012;141(1):2–18. [DOI] [PubMed] [Google Scholar]
- 31.Boer CG, Radjabzadeh D, Medina-Gomez C, Garmaeva S, Schiphof D, Arp P, et al. Intestinal microbiome composition and its relation to joint pain and inflammation. Nature Commun. 2019;10(1):4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sennels H, Sorensen S, Ostergaard M, Knudsen L, Hansen M, Skjodt H, et al. Circulating levels of osteopontin, osteoprotegerin, total soluble receptor activator of nuclear factor-kappa B ligand, and high-sensitivity C-reactive protein in patients with active rheumatoid arthritis randomized to etanercept alone or in combination with methotrexate. Scand J Rheumatol. 2008;37(4):241–7. [DOI] [PubMed] [Google Scholar]
- 33.Lund SA, Giachelli CM, Scatena M. The role of osteopontin in inflammatory processes. J Cell Commun Signal. 2009;3(3–4):311–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Z, Kraus VB. Does lipopolysaccharide-mediated inflammation have a role in OA? Nature Rev Rheumatol. 2016;12(2):123–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang ZY, Perry E, Huebner JL, Katz B, Li YJ, Kraus VB. Biomarkers of inflammation - LBP and TLR- predict progression of knee osteoarthritis in the DOXY clinical trial. Osteoarthritis Cartilage. 2018;26(12):1658–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez C, Deberg MA, Bellahcene A, Castronovo V, Msika P, Delcour JP, et al. Phenotypic characterization of osteoblasts from the sclerotic zones of osteoarthritic subchondral bone. Arthritis Rheum. 2008;58(2):442–55. [DOI] [PubMed] [Google Scholar]
- 37.Pullig O, Weseloh G, Gauer S, Swoboda B. Osteopontin is expressed by adult human osteoarthritic chondrocytes: protein and mRNA analysis of normal and osteoarthritic cartilage [In Process Citation]. Matrix Biol. 2000;19(3):245–55. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Xiao W, Sun M, Deng Z, Zeng C, Li H, et al. The expression of osteopontin and Wnt5a in articular cartilage of patients with knee osteoarthritis and its correlation with disease severity. BioMed Res Int. 2016;2016:9561058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Min S, Shi T, Han X, Chen D, Xu Z, Shi D, et al. Serum levels of leptin, osteopontin, and sclerostin in patients with and without knee osteoarthritis. Clin Rheumatol. 2021: 40(1): 287–294. [DOI] [PubMed] [Google Scholar]
- 40.Collins KH, Paul HA, Reimer RA, Seerattan RA, Hart DA, Herzog W. Relationship between inflammation, the gut microbiota, and metabolic osteoarthritis development: studies in a rat model. Osteoarthritis Cartilage. 2015;23(11):1989–98. [DOI] [PubMed] [Google Scholar]
- 41.Rios JL, Bomhof MR, Reimer RA, Hart DA, Collins KH, Herzog W. Protective effect of prebiotic and exercise intervention on knee health in a rat model of diet-induced obesity. Sci Rep. 2019;9(1):3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schott EM, Farnsworth CW, Grier A, Lillis JA, Soniwala S, Dadourian GH, et al. Targeting the gut microbiome to treat the osteoarthritis of obesity. JCI insight. 2018;3(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ulici V, Kelley KL, Azcarate-Peril MA, Cleveland RJ, Sartor RB, Schwartz TA, et al. Osteoarthritis induced by destabilization of the medial meniscus is reduced in germ-free mice. Osteoarthritis Cartilage. 2018;26:1098–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Z, Chen J, Li B, Zeng B, Chou CH, Zheng X, et al. Faecal microbiota transplantation from metabolically compromised human donors accelerates osteoarthritis in mice. Ann Rheum Dis. 2020;79(5):646–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fouladi F, Glenny EM, Bulik-Sullivan EC, Tsilimigras MCB, Sioda M, Thomas SA, et al. Sequence variant analysis reveals poor correlations in microbial taxonomic abundance between humans and mice after gnotobiotic transfer. ISME J. 2020;14(7):1809–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunn CM, Velasco C, Rivas A, Andrews M, Garman C, Jacob PB, et al. Identification of cartilage microbial DNA signatures and associations with knee and hip osteoarthritis. Arthritis Rheumatol. 2020;72(7):1111–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao Y, Chen B, Li S, Yang L, Zhu D, Wang Y, et al. Detection and characterization of bacterial nucleic acids in culture-negative synovial tissue and fluid samples from rheumatoid arthritis or osteoarthritis patients. Sci Rep. 2018;8(1):14305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.