Abstract
Pulsed-field gel electrophoresis and antimicrobial sensitivity testing were used as tools to investigate the epidemiology of Streptococcus uberis mastitis in dairy cows. A total of 62 different strains were found among 138 isolates from the four herds investigated, and between 10 and 26 different strains were found in each herd. There was no strain common to all four herds. Identical strains of S. uberis were detected from different quarters of individual cows and from cows within the same herd, suggesting that transmission from quarter to quarter and cow to cow had occurred. Despite the great variation in S. uberis strains, persistent infection with the same strain within a lactation was observed in most cows. Predominant strains were present in two herds. Preliminary investigations could not clarify why these particular strains might predominate, but in one herd there was a significant difference between the prevalence of clinical mastitis in quarters infected with the predominant strain and that in quarters infected with other strains, suggesting the greater virulence of the predominant strain. The wide variety of S. uberis strains found is consistent with an environmental source of S. uberis. However, evidence of direct transmission, the persistence of infection, and the predominance of particular strains in some herds indicate that S. uberis infections are epidemiologically complex and that the relative importance of these factors in the occurrence of mastitis may differ between herds.
Implementation of mastitis control programs based on improved milking practices, postmilking teat disinfection, therapeutic and prophylactic antimicrobial administration, and culling of persistently infected animals has effectively controlled intramammary infections caused by contagious pathogens. However, these measures have been less effective against environmental pathogens. Consequently, environmental mastitis has become a major problem, particularly in well-managed herds that have successfully controlled contagious pathogens. It has been hypothesized that the niche vacated by contagious mastitis pathogens becomes occupied by environmental mastitis pathogens, resulting in an increased prevalence of intramammary infection by environmental pathogens (16). Streptococcus uberis is one of the more important environmental patho- gens implicated in bovine mastitis, accounting for a significant proportion of subclinical and clinical intramammary infections in both lactating and nonlactating cows.
An increasing prevalence of S. uberis mastitis has been reported throughout the world. Approximately 14 to 26% of clinical mastitis cases in Canada, the United States, The Netherlands, and the United Kingdom are caused by S. uberis (13). S. uberis is also one of the most significant causes of bovine mastitis in New Zealand and Australia, where the dairy industry is pasture based (21, 29). Despite its high prevalence, the epidemiology of S. uberis mastitis is incompletely understood. S. uberis has been isolated from many sites on the cow including the skin surface, genital tract, intestinal tract, and tonsils (6, 22, 23). It can also be isolated in large numbers from bedding material (4), which is thought to be a major source for intramammary infections in housed cattle. Although many potential reservoirs have been identified, their significance and their association with the occurrence of mastitis in a herd are still unclear. More information on reservoirs and modes of transmission is needed for development of better control programs for this pathogen.
The ability to identify specific strains of a causative bacterial species is an essential tool for epidemiological investigations. A number of typing techniques for differentiation of S. uberis strains have been investigated. The conventional typing methods based on phage typing, serotyping, bacteriocin-like inhibitory substance fingerprinting, and antibiograms have low typeability for closely related strains of S. uberis (5, 11, 17, 24). DNA-based methods have shown potential for typing S. uberis (9, 12, 15, 17, 28). DNA macrorestriction analysis by pulsed-field gel electrophoresis (PFGE) appears to be a simple, reliable, and highly discriminatory method. It produces distinct patterns that are easy to interpret and is highly reproducible. The reproducibility of PFGE makes it useful for comparison of strains between laboratories, and it has been used successfully to investigate genomic diversity among strains of S. uberis (2, 7, 27).
The aim of this study was to explore the epidemiology of mastitis caused by S. uberis in dairy cattle by using PFGE and antibiograms to investigate the persistence of strains and the relative importance of new infections from the environment and from cow-to-cow spread.
MATERIALS AND METHODS
Cattle herds and sample collection.
Milk samples from 38 cows infected with S. uberis were obtained from four pasture-based dairy herds located in Kyabram, Victoria, Australia. Each herd had a history of a high prevalence of S. uberis infection. Milk samples were collected aseptically from each quarter of most of these cows four times over two successive lactations, from 2 months prior to drying off to 2 months after the subsequent parturition. The samples were collected at 4- to 6-week intervals in each lactation. The cows were dried off in June and July and calved in August and September. Blanket dry-cow therapy with cefalonium dihydrate (Cepravin DC; Schering-Plough Animal Health, Baulkham Hills, New South Wales, Australia) was used by all four herds during the trial. Clinical mastitis and treatment data were available from herd 4.
Bacteriological methods.
Milk samples were mixed thoroughly, and 10 μl was plated onto Edward's medium. S. uberis isolates were identified to the species level by conventional tests and by PCR. Conventional tests included Gram straining, catalase production, esculin hydrolysis, the CAMP test, and the sodium hippurate test. PCR amplification of spacer regions between 16S and 23S rRNA of S. uberis was used to confirm identification (8).
Antimicrobial susceptibility testing.
The calibrated dichotomous sensitivity test of Bell (3) was used to assess the antimicrobial sensitivity of S. uberis isolates. Antimicrobial disks (Oxoid, Basingstoke, Hampshire, England) used included penicillin G (0.5 μg), cloxacillin (5 μg), erythromycin (5 μg), gentamicin (10 μg), trimethroprim (5 μg), sulfamethoxazole (300 μg), tetracycline (30 μg), vancomycin (5 μg), cephalexin (100 μg), neomycin (30 μg), novobiocin (30 μg), and kanamycin (50 μg). The diameter of the inhibition zone was measured after 18 to 24 h of incubation at 37°C. A standard strain of Staphylococcus aureus was used as a control strain.
S. uberis concentrations in milk from infected quarters.
The numbers of S. uberis colonies isolated from 38 milk samples collected from herds 3 and 4 were determined. Each sample was mixed thoroughly, and 50 μl from each sample was spread onto separate Edward's medium agar plates. The inoculated plates were then incubated at 37°C for 24 h. S. uberis colonies were observed using UV light (Wood's lamp), the colonies were counted, and each sample was allocated to a category depending on whether it yielded less than 50, 51 to 100, 101 to 200, 201 to 300, 301 to 400, or more than 400 colonies per 50-μl sample. The distribution of samples containing strain C2 into these categories was compared to the distribution of samples from herd 3 containing other strains, and the distribution of samples containing strain D2 was compared to the distribution of samples from herd 4 containing other strains using chi-square tests.
Preparation of genomic DNA in agarose blocks.
S. uberis isolates were grown in Todd-Hewitt broth (Oxoid) at 37°C overnight. Cells from 1 ml of culture were collected by centrifugation at 13,000 × g for 1 min, washed three times with 1 M NaCl–10 mM Tris-HCl (pH 7.6), and resuspended in 300 μl of the same solution. The cell suspension was mixed with an equal volume of molten 2% (wt/vol) low-melting-temperature agarose (SeaPlaque; FMC Bioproducts, Rockland, Maine) and dispensed into 100-μl molds. When solidified, blocks were incubated in lysis buffer (6 mM Tris-HCl [pH 7.6], 1 M NaCl, 100 mM EDTA [pH 7.6], 1% Sarkosyl, 1 mg of lysozyme/ml) for 18 h at 37°C. The lysis buffer was then replaced with ESP buffer (0.5 M EDTA [pH 9.2], 1% Sarkosyl, 1 mg of proteinase K/ml), and the blocks were incubated at 50°C for 72 h. Gel blocks were stored in 0.5 M EDTA (pH 8) until used.
Restriction endonuclease digestion of genomic DNA.
Before digestion blocks were equilibrated with TE (10 mM Tris-HCl [pH 8], 1 mM EDTA [pH 8]). Slices 1 to 2 mm thick were then cut from each block and incubated in 100 μl of 1× restriction endonuclease buffer as supplied by the manufacturer. Digestions were performed with 10 U of SmaI (Boehringer GmbH, Mannheim, Germany) at 25°C for 18 h or 20 U of ApaI (Boehringer GmbH) at 30°C for 18 h.
PFGE.
DNA fragments were separated by clamped homogeneous electric field (CHEF) electrophoresis using a CHEF DRIII (Bio-Rad Laboratories, Richmond, Calif.). Electrophoresis of digested samples was performed through 1% (wt/vol) agarose gels in 0.5× TBE (1× TBE is 89 mM Tris-HCl, 89 mM boric acid, and 2 mM EDTA, pH 8.3) at 6 V/cm for 20 h at 14°C, with the pulse time increasing linearly from 1 to 20 s for SmaI-digested samples and from 1 to 17 s for ApaI-digested samples. Gels were stained with 0.5 mg of ethidium bromide/liter, and DNA was visualized by UV transillumination. Lambda phage concatamers (Bio-Rad) and HindIII digests of lambda phage DNA were used as molecular size standards.
Strain classification.
Strains were defined using the criteria proposed by Tenover et al. (25). Isolates were designated different strains if the PFGE patterns differed by more than three bands and different subtypes of the same strain if patterns differed by one to three bands. Isolates that had similar PFGE patterns were considered to be genetically related isolates and to be derived from a common parent, as differences in two or three bands were most likely the result of a single genetic change such as a point mutation or an insertion or deletion of DNA. The discrimination of PFGE and antibiogram typing in this population were determined using Simpson's index of diversity (14), assuming that all isolates obtained from the same quarter in the same lactation were not independent strains if they belonged to the same subtype as determined by PFGE of SmaI-digested chromosomal DNA.
RESULTS
Discrimination and reproducibility of PFGE typing.
PFGE of chromosomal DNA digested with SmaI yielded 8 to 14 fragments in the 23- to 340-kb size range, and ApaI digests yielded patterns of 10 to 16 fragments of 23 to 340 kb. The stability of SmaI and ApaI PFGE patterns was examined by selecting one isolate (strain D2b) and looking at the PFGE patterns before and after 10 passages in Todd-Hewitt broth. The reproducibility of the PFGE patterns was also examined by repeated testing of the same isolate on separate occasions on different gels. In both cases identical patterns were obtained, indicating the stability and reproducibility of PFGE.
To increase the discriminatory power of PFGE and confirm the results obtained from SmaI digestion, ApaI digestion was used to examine isolates that had identical or similar SmaI restriction patterns. Identical ApaI patterns were seen in all isolates that had identical SmaI patterns. However, ApaI digestion was found to be less discriminatory than SmaI digestion for closely related isolates. Isolates that belonged to subtypes C2a and C2c as determined by SmaI digestion were found to have identical ApaI PFGE patterns. Isolates of subtypes D2a and D2b also had the same ApaI PFGE patterns. The SmaI and ApaI restriction patterns revealed a marked clonal diversity of S. uberis. A total of 62 different strains were obtained from 138 S. uberis isolates from the four herds (Table 1), indicating that the epidemiology of S. uberis infection could be very complex.
TABLE 1.
Strains of S. uberis isolated from individual quarters of dairy cows on four occasions over two lactations
| Herd | Cow | Strainsa detected in indicated sampleb from:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Left forequarter
|
Right forequarter
|
Left hindquarter
|
Right hindquarter
|
||||||||||||||
| S1 | S2 | S3 | S4 | S1 | S2 | S3 | S4 | S1 | S2 | S3 | S4 | S1 | S2 | S3 | S4 | ||
| 1 | 1 | A1 | ∗ | A2 | A2 | A1 | ∗ | — | — | A3 | ∗ | — | — | — | ∗ | — | — |
| 2 | — | — | ∗ | ∗ | A4 | A4 | ∗ | ∗ | — | — | ∗ | ∗ | — | — | ∗ | ∗ | |
| 3 | — | ∗ | ∗ | — | — | ∗ | ∗ | — | — | ∗ | ∗ | A5 | A6 | ∗ | ∗ | A7 | |
| 4 | A8 | ∗ | — | — | — | ∗ | — | — | — | ∗ | — | — | — | ∗ | — | — | |
| 5 | — | ∗ | — | — | — | ∗ | — | — | — | ∗ | — | — | A9 | ∗ | — | — | |
| 6 | — | — | — | — | — | — | — | — | A10 | — | — | — | — | — | — | — | |
| 2 | 1 | B1 | B1 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 2 | B2 | B2 | ∗ | — | B2 | B2 | ∗ | — | B3 | B3 | ∗ | — | — | — | ∗ | — | |
| 3 | — | — | B4 | — | B5 | B5 | — | — | — | — | B4 | — | — | — | — | — | |
| 4 | — | — | ∗ | — | — | — | ∗ | — | B6 | B6 | ∗ | — | — | — | ∗ | — | |
| 5 | — | — | — | — | — | — | — | — | B7 | B8 | — | — | — | — | — | — | |
| 6 | — | — | — | — | — | — | B9 | — | — | — | — | — | B10 | — | — | B9 | |
| 7 | — | B11 | ∗ | ∗ | — | — | ∗ | ∗ | — | — | ∗ | ∗ | B12 | B12 | ∗ | ∗ | |
| 3 | 1 | — | — | — | — | C1 | — | — | — | — | — | — | — | — | — | — | — |
| 2 | C2a | C2b | ∗ | ∗ | — | — | ∗ | ∗ | C2a | C2b | ∗ | ∗ | — | — | ∗ | ∗ | |
| 3 | C2c | C2c | — | — | C2c | C2c | — | — | — | C2a | — | — | C3 | — | — | — | |
| 4 | — | — | — | — | — | — | — | — | C2b | C2b | — | — | — | C2b | — | — | |
| 5 | — | — | ∗ | — | — | — | ∗ | — | C2c | — | ∗ | — | — | — | ∗ | — | |
| 6 | — | — | C4 | C4 | C2a | C2a | C5 | C6 | — | — | — | C7a | — | — | — | C6 | |
| 7 | — | — | C8 | C8 | C9 | C9 | C10 | C10 | C11 | C2b | C12 | C12 | C2d | C2d | — | — | |
| 8 | — | — | — | — | — | — | C13 | — | — | — | — | — | C14a | C14a | — | — | |
| 9 | — | — | ∗ | — | C15 | C15 | ∗ | C16 | C14b | C14b | ∗ | C17 | C18 | C18 | ∗ | — | |
| 10 | C19 | C19 | — | — | C19 | C19 | — | — | — | — | — | — | — | C20 | — | — | |
| 11 | C21 | C21 | — | ∗ | — | — | — | ∗ | — | — | — | ∗ | — | — | — | ∗ | |
| 12 | — | — | — | ∗ | — | — | — | ∗ | C22 | C22 | — | ∗ | C23 | C23 | — | ∗ | |
| 13 | — | — | — | — | — | — | — | C24 | C7b | — | — | — | C25 | C25 | C26 | C26 | |
| 4 | 1 | D1 | D1 | — | — | — | — | — | — | — | D1 | — | D2a | — | — | — | — |
| 2 | — | — | D2a | D2a | — | — | — | — | — | — | D2a | D2a | D2b | D2b | — | — | |
| 3 | — | — | D2a | D2a | — | — | — | — | — | — | — | — | D3 | D3 | — | — | |
| 4 | — | — | — | — | — | — | — | — | D2b | — | D2c | D2c | — | — | — | — | |
| 5 | — | — | — | — | — | — | — | — | D2b | — | — | — | — | — | — | — | |
| 6 | — | D4 | D5a | — | — | D6 | — | — | — | D4 | — | — | D2d | D2b | — | — | |
| 7 | — | — | — | — | D7 | — | — | D8 | — | — | — | — | — | — | — | D5b | |
| 8 | — | — | — | — | — | — | — | — | D9 | — | — | — | — | — | — | — | |
| 9 | D10 | D10 | ∗ | ∗ | — | D11 | ∗ | ∗ | — | D12 | ∗ | ∗ | — | D13 | ∗ | ∗ | |
| 10 | — | — | ∗ | ∗ | D14 | D14 | ∗ | ∗ | — | — | ∗ | ∗ | D15 | D15 | ∗ | ∗ | |
| 11 | — | — | ∗ | ∗ | — | — | ∗ | ∗ | D16 | D16 | ∗ | ∗ | — | — | ∗ | ∗ | |
| 12 | ∗ | ∗ | ∗ | ∗ | — | — | — | — | — | — | — | — | D17 | — | — | — | |
—, S. uberis not isolated from sample; ∗, sample not collected.
S1 to S4, samples 1 to 4. Collection dates (all in 1999) for herds 1 to 4 were, respectively, as follows: sample 1, 14 May, 17 May, 30 April, and 19 April; sample 2, 8 June, 14 July, 10 June, and 3 June; sample 3, 14 October, 8 October, 8 October, and 14 October; sample 4, 11 November, 10 November, 10 November, and 10 November.
Discrimination of antibiogram patterns.
Antimicrobial susceptibility testing revealed that all isolates were susceptible to vancomycin and cephalexin and resistant to gentamicin, sulfamethoxazole, neomycin, and kanamycin. A total of 11 different antibiogram patterns were obtained (Table 2). The majority of S. uberis isolates (91 of 138, 66%) were antibiogram type B or D (Tables 1 and 2). Although the discriminatory power of the antibiogram technique was relatively low, the results of antibiograms mostly correlated with the results obtained by PFGE (Fig. 1). S. uberis isolates designated as the same strains or subtypes of the same strains by PFGE had common antibiogram patterns. However, some isolates from different herds that shared PFGE patterns were found to have different antibiogram patterns. Two isolates (strain B12) from herd 2 and four isolates (strain C19) from herd 3 had the same PFGE pattern but had antibiogram types F and K, respectively, and two isolates (strain A4) from herd 1 and five isolates (strain D2b) from herd 4 that had the same PFGE pattern had antibiogram types B and D, respectively.
TABLE 2.
Antibiogram patterns of S. uberis isolates and corresponding PFGE strains
| Antibiogram pattern | Strainsa | Susceptibility tob:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | OB | E | SF | N | W | NV | CN | TE | CL | VA | K | ||
| A | B2, B4, B7 | S | S | S | R | R | S | S | R | S | S | S | R |
| B | A1, A3, A4, A5, A6, A8, A10, B6, B8, B9, B10, C9, C17, C18, C20, C21, C23, C25, D8 | S | S | S | R | R | R | S | R | S | S | S | R |
| C | B3, B5, C15 | S | S | R | R | R | S | S | R | S | S | S | R |
| D | A7, A9, B1, C1, C2a, C2b, C2c, C2d, C3, C5, C14a, C14b, C16, C22, C26, D1, D2a, D2b, D2c, D2d, D4, D5a, D5b, D7, D9, D10, D12, D16, D17 | S | S | R | R | R | R | S | R | S | S | S | R |
| E | C11 | S | S | S | R | R | R | R | R | S | S | S | R |
| F | A2, B12, D3, D13, D14 | R | R | S | R | R | R | S | R | S | S | S | R |
| G | C4, C7a, C7b, C13 | S | S | R | R | R | R | R | R | S | S | S | R |
| H | B11, C6, C24 | S | S | R | R | R | R | S | R | R | S | S | R |
| I | C8, C10, C12, D6 | R | R | R | R | R | R | S | R | S | S | S | R |
| J | D11 | R | R | S | R | R | R | R | R | S | S | S | R |
| K | C19, D15 | R | R | R | R | R | R | R | R | S | S | S | R |
Strains as determined by PFGE of chromosomal DNA digested with SmaI.
Abbreviations: P, penicillin; OB, cloxacillin; E, erythromycin: SF, sulfamethoxazole; N, neomycin; W, trimethoprim; NV, novobiocin; CN, gentamicin; TE, tetracycline, CL, cephalexin; VA, vancomycin; K, kanamycin; S, susceptible; R, resistant.
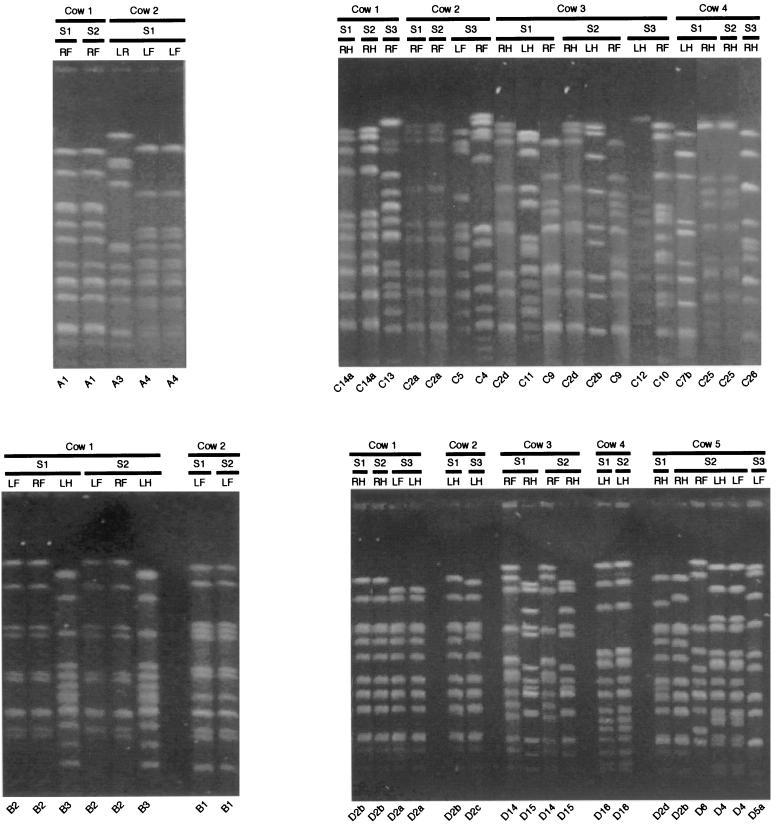
FIG. 1.
PFGE of SmaI-digested chromosomal DNA from some S. uberis isolates from each herd over two successive lactations. The cow from which each sample was obtained is indicated. S1, S2, and S3, different sampling times as shown in Table 1, with S3 obtained in the subsequent lactation; LF, RF, LH, and RH, isolates obtained from the left fore-, right fore-, left hind-, and right hindquarters, respectively. The strain designation of each isolate is indicated at the bottom. Each panel shows isolates obtained from a single herd.
The index of discrimination for PFGE examination of SmaI-digested chromosomal DNA was 0.989, while that for antibiogram typing was 0.731, indicating the much higher discriminatory power of PFGE typing.
Comparison of S. uberis isolates from the same quarters.
To determine if multiple strains were present in one infected quarter, milk samples from 12 S. uberis-infected quarters were evaluated. Three S. uberis colonies isolated from each of 12 milk samples were selected for PFGE. The 36 S. uberis isolates included 11 strains, but all isolates from the same quarter were the same strain, suggesting that infection with multiple strains of S. uberis within the same quarter was not common.
Comparison of S. uberis isolates from different quarters of individual cows.
Strains of S. uberis isolated from different quarters of the same cow were analyzed to determine whether different quarters were infected with different strains. Of 11 cows that had two quarters infected with S. uberis, 5 cows were infected with the same strain in both quarters. Two of the six cows that had three quarters infected had different S. uberis strains in each quarter, and the other four had the same strain in two of the three infected quarters. One cow was found to have different strains of S. uberis in all four quarters, and two cows had the same strain in two of four infected quarters. In 5 of 11 cows, the two quarters infected with the same strain were the left forequarter and left hindquarter, and in 4 of 11 cows the left and right forequarters were infected with the same strain. In no cow were diagonally opposite quarters infected with the same strain.
Comparison of S. uberis strains within and among herds.
A great degree of genetic variation was found in S. uberis within the same herd and among herds. Between 10 and 26 different S. uberis strains were present in each herd (Table 1). Within herds 1 and 2, no common strains were found. However, predominant strains were found in herds 3 and 4. Strain C2 was the most prevalent in herd 3 during the lactation prior to drying off, with 40% of isolates belonging to this strain. Strain D2 was the predominant strain found in herd 4 over both lactations, accounting for 21.4 and 75% of S. uberis isolates in the two successive lactations. Only two to six isolates in each herd were found to be identical to isolates from other herds. Two isolates (strain B12) from herd 2 and four isolates (strain C19) from herd 3 appeared to be identical, two isolates (strain A4) from herd 1 had the same pattern as five isolates (strain D2b) from herd 4, and two isolates that belonged to strain D5a and D5b from herd 4 were identical to six isolates (strain C2b) and five isolates (strain C2a), respectively, from herd 3. There was no strain found in all four herds.
Comparison of concentrations of predominant strains and other strains in milk from infected quarters.
The concentrations of S. uberis strains C2 and D2, which were the predominant strains in herds 3 and 4, respectively, and other strains from herds 3 and 4 in milk from infected quarters were examined by colony counting. There was no significant difference between the concentrations of strains C2 (P = 0.29; χ2 = 4.95) or D2 (P = 0.609; χ2 = 2.7) and the other strains in infected quarters in herds 3 and 4, respectively.
Incidence of clinical mastitis caused by strain D2 and the other strains in herd 4.
There was a significant difference between the prevalence of clinical mastitis in quarters infected with strain D2 and the prevalence of clinical mastitis in quarters infected with other strains (P = 0.002). Only 2 of the 18 quarters infected with the other strains developed clinical mastitis during the trial. Of eight quarters infected with strain D2, six quarters developed clinical mastitis, and four out of the six quarters were still infected with the same strain after treatment with a combination of oxytetracycline hydrochloride, oleandomycin, and neomycin (Mastalone Blue; Pfizer Animal Health, Highett, Victoria, Australia).
New and persistent S. uberis infections during a lactation and over lactations.
The 47 paired isolates of S. uberis recovered from the same quarters over a 4- to 6-week interval during a single lactation were compared. In 41 pairs strains of S. uberis were the same, and members of three pairs were different subtypes of the same strain. Different strains of S. uberis were detected in only three paired samples. The persistence of S. uberis in infected quarters over lactations was also investigated by comparing strains isolated in one lactation with strains found in the next lactation. The prevalence of S. uberis infection decreased significantly from one lactation to the next in all herds. The total number of infected quarters was reduced from 64 in one lactation to 27 in the next lactation, with only 13 quarters infected with S. uberis in both lactations. Of these 13 quarters, different S. uberis strains were detected in each lactation in 12, suggesting that new infections had occurred, and 1 quarter was found to have different subtypes of the same strain in each lactation.
DISCUSSION
PFGE and antibiogram analysis were used to examine strains of S. uberis to investigate the epidemiology of S. uberis mastitis in dairy cattle. As shown in previous studies (2, 7, 27) PFGE was proven to be a highly discriminatory method. It was able to resolve many isolates that were indistinguishable by antimicrobial susceptibility testing. Antibiograms alone were found to be of limited value in differentiating closely related strains, but agreement between antibiograms and PFGE was observed for most isolates. However, it was found that some isolates from different herds that were defined as the same strain by PFGE had different antibiogram patterns. This could be due to differences in management and treatment practices among the herds, particularly differences in the type and frequency of antibiotic use, and thus of exposure to antibiotics, among herds.
A number of S. uberis strains were isolated from cows in this study, demonstrating that a wide variety of strains are able to cause mastitis. This is consistent with the dogma that quarters are infected by this pathogen more frequently from their environment than from other infected quarters. Strains of S. uberis recovered from clinically affected quarters were also found in subclinically infected quarters. This finding agrees with the previous report of Jayarao et al. (18), who showed that most S. uberis strains isolated from clinical mastitis were also isolated from cows with subclinical mastitis.
The demonstration of identical strains of S. uberis in different quarters of individual cows and from some cows within the same herd suggests that direct transmission from an infected quarter to uninfected quarters and from cow to cow occurs, presumably during the milking process. Improper milking hygiene and machine function could therefore contribute to the incidence of S. uberis infection. However, direct contact with the same environmental source, although unlikely, cannot be discounted entirely. As only a few strains were isolated from more than one herd, it is unlikely that there is an environmental source of S. uberis common to different farms.
Despite the large number of different strains found within each herd, in two herds there was a predominant strain. Only one herd had the same strain predominating over two lactations. It is possible that this strain was widespread in the environment and relatively stable over time or, alternatively, that it persisted in a chronically infected quarter between lactations. The spread of this strain within this herd could be influenced by management factors, especially milking management. It is also possible that this strain is more transmissible or has a greater capacity to adhere to the mammary epithelium than the others. Evidence of differing abilities to establish intramammary infection among strains of S. uberis has been reported previously (10, 19). We were unable to demonstrate differences in the concentrations of these predominant strains compared to those of the other strains in milk from infected quarters.
It is possible that predominant strain D2 was more virulent than the other strains, as there were significant differences in the prevalence of clinical mastitis between quarters infected with strain D2 and those infected with the other strains. Strain D2 was also more resistant to treatment than other strains. The antibiograms showed that this strain was sensitive to the antibiotic used, and therefore resistance to the antibiotic could not be the reason for the poor response to therapy. Resistance of this strain to treatment might be due to a greater ability to adhere to and invade mammary epithelial cells and persist intracellularly, where it is difficult for the drugs to reach MICs. Alternatively this strain may cause more severe inflammation and fibrosis, preventing the antibiotic from reaching the infected areas of the gland.
Most S. uberis organisms isolated at different times from the same quarters in one lactation belonged to the same strain, indicating the persistence of these infections. It is unlikely that this results from reinfection with the same genotype of S. uberis from the surroundings of the cow given the amount of genomic variation of S. uberis within each herd. Such persistence of infection by S. uberis has been suggested by previous studies using PFGE, but the use of composite milk samples from each cow, rather than individual quarter samples, in these earlier studies precluded definitive conclusions about the relative significance of persistent infection with different strains in different quarters compared to that of new infections (27). The localization of S. uberis in the udder was not investigated in our study, but it could be hypothesized that S. uberis persisted intracellularly. The ability of S. uberis to adhere to and to invade mammary epithelium cells has been reported (1, 20, 26). Internalization of S. uberis could explain chronic infections, as this could protect it from the defence mechanisms of the mammary gland and from antimicrobial action. Chronically infected quarters could play a role as one of the sources of infection in a herd.
Different subtypes of the same strains were observed in some isolates from the same quarter during a lactation. The difference is likely to be due to genetic changes in persisting isolates between the times of sampling rather than being indicative of new infections. The greatest variations were observed among isolates of strains C2 and D2, which were the predominant strains in herd 3 and herd 4, respectively, suggesting that these isolates may have originated from a common clone.
It was found that the prevalence of S. uberis infections decreased significantly from one lactation to the next in all herds. Different strains of S. uberis were detected in all but one infected quarter, indicating that most cases in the subsequent lactation resulted from new infections, as would be expected of successful dry-cow therapy with an antibiotic effective against all the strains isolated. One quarter was found to be infected with different subtypes of the same strain in two lactations. This was most likely due to chronic infection with the same strain, with mutation of the strain over time. However, it could possibly be due to a new infection, as strain D2 was also the predominant strain in the second lactation. Previous studies using PFGE to type S. uberis have not compared strains isolated from the same cows in successive lactations (27).
In the present study, a wide variety of strains were shown to be able to infect the mammary gland, suggesting that the majority of cases of mastitis due to S. uberis are due to contamination of the gland from the environment. Nevertheless, the finding of persistent infection and predominant strains in some herds indicates the epidemiological complexity of S. uberis mastitis. Chronically infected quarters could be an important source of intramammary infection within the herd. The finding of identical strains in different quarters of the same cow and predominant strains in some herds suggests that direct transmission between quarters and cows may be an important means of spread of this pathogen. Further investigation is necessary to determine whether some strains are more virulent than others and to clarify the relative importance of chronically infected quarters, particular strains, and other factors such as herd management practices in the occurrence of S. uberis mastitis in dairy herds.
ACKNOWLEDGMENT
P.P. was supported by an Australian International Development Agency Scholarship.
REFERENCES
- 1.Almeida R A, Luther D A, Kumar S J, Calvinho L F, Bronze M S, Oliver S P. Adherence of Streptococcus uberis to bovine mammary epithelial cells and to extracellular matrix proteins. Zentbl Vetmed Reihe B. 1996;43:385–392. doi: 10.1111/j.1439-0450.1996.tb00330.x. [DOI] [PubMed] [Google Scholar]
- 2.Baseggio N, Mansell P D, Browning J W, Browning G F. Strain differentiation of isolates of streptococci from bovine mastitis by pulsed-field gel electrophoresis. Mol Cell Probes. 1997;11:349–354. doi: 10.1006/mcpr.1997.0126. [DOI] [PubMed] [Google Scholar]
- 3.Bell S M. Antibiotic sensitivity testing by the CDS method. In: Hartwig N, editor. Clinical Microbiology Update Program—1984. Sydney, Australia: NSW Branch of the Australian Society for Microbiology; 1984. [Google Scholar]
- 4.Bramley A J. Sources of Streptococcus uberis in the dairy herd. I. Isolation from bovine faeces and from straw bedding of cattle. J Dairy Res. 1982;49:369–373. doi: 10.1017/s0022029900022500. [DOI] [PubMed] [Google Scholar]
- 5.Buddle B M, Tagg J R, Ralston M J. Use of an inhibitor typing scheme to study the epidemiology of Streptococcus uberis mastitis. N Z Vet J. 1988;36:115–119. doi: 10.1080/00480169.1988.35504. [DOI] [PubMed] [Google Scholar]
- 6.Cullen G A, Little T W. Isolation of Streptococcus uberis from the rumen of cows and from sell. Vet Rec. 1969;85:115–118. doi: 10.1136/vr.85.5.115. [DOI] [PubMed] [Google Scholar]
- 7.Douglas V L, Fenwick S G, Pfeiffer D U, Williamson N B, Holmes C W. Genomic typing of Streptococcus uberis isolates from cases of mastitis, in New Zealand dairy cows, using pulsed-field gel electrophoresis. Vet Microbiol. 2000;75:27–41. doi: 10.1016/s0378-1135(00)00184-x. [DOI] [PubMed] [Google Scholar]
- 8.Forsman P, Tilsalatimisjarvi A, Alatossava T. Identification of staphylococcal and streptococcal causes of bovine mastitis using 16s–23s rRNA spacer regions. Microbiology. 1997;143:3491–3500. doi: 10.1099/00221287-143-11-3491. [DOI] [PubMed] [Google Scholar]
- 9.Gillespie B E, Jayarao B M, Pankey J W, Oliver S P. Subtyping of Streptococcus dysgalactiae and Streptococcus uberis isolated from bovine mammary secretions by DNA fingerprinting. Zentbl Vetmed Reihe B. 1998;45:585–593. doi: 10.1111/j.1439-0450.1998.tb00831.x. [DOI] [PubMed] [Google Scholar]
- 10.Hill A W. Pathogenicity of two strains of Streptococcus uberis infused into lactating and non-lactating bovine mammary glands. Res Vet Sci. 1988;45:400–404. [PubMed] [Google Scholar]
- 11.Hill A W, Brady C A. A note on the isolation and propagation of lytic phages from Streptococcus uberis and their potential for strain typing. J Appl Bacteriol. 1989;67:425–431. doi: 10.1111/j.1365-2672.1989.tb02513.x. [DOI] [PubMed] [Google Scholar]
- 12.Hill A W, Leigh J A. DNA fingerprinting of Streptococcus uberis: a useful tool for epidemiology of bovine mastitis. Epidemiol Infect. 1989;103:165–171. doi: 10.1017/s0950268800030466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogan J S, Smith K L. Proceedings of the Symposium on Udder Health Management for Environmental Streptococci—1997. Arlington, Tex: National Mastitis Council Inc; 1997. Occurrence of clinical and sub-clinical environmental streptococcal mastitis; pp. 59–75. [Google Scholar]
- 14.Hunter P R, Gaston M A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jayarao B M, Bassam B J, Caetano-Anolles G, Gresshoff P M, Oliver S P. Subtyping of Streptococcus uberis by DNA amplification fingerprinting. J Clin Microbiol. 1992;30:1347–1350. doi: 10.1128/jcm.30.5.1347-1350.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayarao B M, Gillespie B E, Lewis M J, Dowlen H H, Oliver S P. Epidemiology of Streptococcus uberis intramammary infections in a dairy herd. Zentbl Vetmed Reihe B. 1999;46:433–442. doi: 10.1046/j.1439-0450.1999.00254.x. [DOI] [PubMed] [Google Scholar]
- 17.Jayarao B M, Oliver S P, Tagg J R, Matthews K R. Genotypic and phenotypic analysis of Streptococcus uberis isolated from bovine mammary secretions. Epidemiol Infect. 1991;107:543–555. doi: 10.1017/s0950268800049244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jayarao B M, Schilling E E, Oliver S P. Genomic deoxyribonucleic acid restriction fragment length polymorphism of Streptococcus uberis: evidence of clonal diversity. J Dairy Sci. 1993;76:468–474. doi: 10.3168/jds.S0022-0302(93)77367-1. [DOI] [PubMed] [Google Scholar]
- 19.Leigh J A, Field T R, Williams M R. Two strains of Streptococcus uberis, of differing ability to cause clinical mastitis, differ in their ability to resist some host defence factors. Res Vet Sci. 1990;49:85–87. [PubMed] [Google Scholar]
- 20.Matthews K R, Almeida R A, Oliver S P. Bovine mammary epithelial cell invasion by Streptococcus uberis. Infect Immun. 1994;62:5641–5646. doi: 10.1128/iai.62.12.5641-5646.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pankey J W, Pankey P B, Barker R M, Williamson J H, Woolford M W. The prevalence of mastitis in primiparous heifers in eleven Waikato dairy herds. N Z Vet J. 1996;44:41–44. doi: 10.1080/00480169.1996.35932. [DOI] [PubMed] [Google Scholar]
- 22.Razavi-Rohani M, Bramley A J. A study of the frequency and distribution of Streptococcus uberis contamination on the body of lactating and non-lactating cows. Indian Vet J. 1981;58:804–811. [Google Scholar]
- 23.Sharma R M, Packer R A. Occurrence and ecologic features of Streptococcus uberis in the dairy cow. Am J Vet Res. 1970;31:1197–1202. [PubMed] [Google Scholar]
- 24.Tagg J R, Vugler L G. An inhibitor typing scheme for Streptococcus uberis. J Dairy Res. 1986;53:451–456. doi: 10.1017/s0022029900025061. [DOI] [PubMed] [Google Scholar]
- 25.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas L H, Haider W, Hill A W, Cook R S. Pathologic findings of experimentally induced Streptococcus uberis infection in the mammary gland of cows. Am J Vet Res. 1994;55:1723–1728. [PubMed] [Google Scholar]
- 27.Wang S M, Deighton M A, Capstick J A, Gerraty N. Epidemiological typing of bovine streptococci by pulsed-field gel electrophoresis. Epidemiol Infect. 1999;123:317–324. doi: 10.1017/s0950268899002745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams A M, Collins M D. DNA fingerprinting of Streptococcus uberis based on polymorphism of DNA encoding rRNA. Lett Appl Microbiol. 1991;12:23–28. doi: 10.1111/j.1472-765x.1991.tb00493.x. [DOI] [PubMed] [Google Scholar]
- 29.Williamson J H, Woolford M W, Day A M. The prophylactic effect of a dry-cow antibiotic against Streptococcus uberis. N Z Vet J. 1995;43:228–234. doi: 10.1080/00480169.1995.35898. [DOI] [PubMed] [Google Scholar]