Significance
Staphylococcus aureus invariably acquires resistance mechanisms against new antibiotics. The persistent colonization with S. aureus is the key risk factor for invasive disease and a driver for the evolution of antibiotic resistant isolates. Anti-S. aureus antibodies that could promote decolonization, prevent infection, or treat disease would alleviate the selection for drug resistance. The successful development of such antibodies is complicated by Staphylococcal protein A (SpA) in the envelope of S. aureus. SpA captures immunoglobulins via their constant region, preventing antibodies from initiating anti-staphylococcal activities. Here, we demonstrate that therapeutic anti-S. aureus antibodies can be engineered to avoid sequestration by SpA. Such antibodies display extended half-lives and improve bacterial uptake and killing by immune cells.
Keywords: engineered antibody, C1q, Staphylococcus aureus, FcRn, immune evasion
Abstract
Gram-positive organisms with their thick envelope cannot be lysed by complement alone. Nonetheless, antibody-binding on the surface can recruit complement and mark these invaders for uptake and killing by phagocytes, a process known as opsonophagocytosis. The crystallizable fragment of immunoglobulins (Fcγ) is key for complement recruitment. The cell surface of S. aureus is coated with Staphylococcal protein A (SpA). SpA captures the Fcγ domain of IgG and interferes with opsonization by anti-S. aureus antibodies. In principle, the Fcγ domain of therapeutic antibodies could be engineered to avoid the inhibitory activity of SpA. However, the SpA-binding site on Fcγ overlaps with that of the neonatal Fc receptor (FcRn), an interaction that is critical for prolonging the half-life of serum IgG. This evolutionary adaptation poses a challenge for the exploration of Fcγ mutants that can both weaken SpA–IgG interactions and retain stability. Here, we use both wild-type and transgenic human FcRn mice to identify antibodies with enhanced half-life and increased opsonophagocytic killing in models of S. aureus infection and demonstrate that antibody-based immunotherapy can be improved by modifying Fcγ. Our experiments also show that by competing for FcRn-binding, staphylococci effectively reduce the half-life of antibodies during infection. These observations may have profound impact in treating cancer, autoimmune, and asthma patients colonized or infected with S. aureus and undergoing monoclonal antibody treatment.
Immunoglobulin G (IgG) accounts for about 75% of serum antibodies in humans and consists of four subclasses, IgG1 (66%), IgG2 (23%), IgG3 (7%), and IgG4 (4%) (1). The effector functions of IgG are contributed by the crystallizable fragment (Fcγ). Antibody engagement with Fcγ receptors (FcγRs) on the surface of professional leukocytes enhances phagocytic uptake of marked pathogens, while Fc recruitment of complement component C1q initiates the activation of the classical complement pathway (2). IgG1 and IgG3 display the highest affinity toward FcγRs; IgG3 displays the strongest interaction with C1q followed by IgG1 (1). Human IgG1 (hIgG1) is often preferred for the development of therapeutic antibodies over hIgG3 owing to its stability (1). Following pinocytosis, IgG1 binding to the neonatal Fc receptor (FcRn) at the slightly acidic pH of endosomes allows for its recycling to the cell surface and its release from FcRn at the neutral extracellular pH (1). IgG3 fails to escape lysosomal degradation because of altered binding to FcRn and competition with IgG1 (3).
C1q is the recognition module of C1 complement complex that includes the serine proteases C1r and C1s. The weak affinity of C1q for monomeric IgG increases when its six globular domains interact with IgG hexamers (4–6), a clustering that is facilitated by IgG binding to cell surface antigens. The ensuing activation of C1r and C1s results in the generation of C3 convertases that catalyze the covalent modification of cell-surface determinants with opsonins such as C3b (2, 7). Most cells marked with C3-derived opsonins can be lysed by the membrane attack complex (MAC) (i.e., lytic pores made of five different complement proteins) (7). Because of their thick peptidoglycan, gram-positive bacteria cannot be lysed by MAC (8) but can be taken up in a complement receptor (CR)–dependent manner by phagocytes (7). Bacteria have evolved immune evasion strategies to escape opsonization by antibodies and complement and uptake by phagocytes (9). The conserved Staphylococcal protein A (SpA) and Staphylococcal binder of immunoglobulin (Sbi) contain five and two immunoglobulin-binding domains (IgBDs), respectively (10). The IgBDs fold into triple-helical bundles that associate with the Fcγ domain of human and vertebrate immunoglobulins (11, 12). The extensive tethering of SpA to peptidoglycan results in the capture and inhibition of IgG at the cell surface (13). SpA interaction with Fcγ also interferes with FcRn-binding (3, 14). However, the physiological implications of this interaction are not known. Lastly, SpA binds with the variant heavy chain of VH3-IgM, -IgG, -IgD, and -IgE (10). When released from the bacterial envelope (15), SpA binding to VH3-IgM B cell receptors promotes B-cell expansion and the transient flooding of nonspecific VH3-IgM and VH3-IgG (16–18).
Humans and vertebrates develop antibodies against molecular determinants of Staphylococcus aureus yet are unable to generate antibodies that bind to and neutralize the IgBDs of SpA and Sbi (17, 19, 20). By immunizing animals with a SpA variant that cannot bind IgG (19), we isolated earlier the mouse monoclonal antibody 3F6 with SpA-specific complementarity determining region (CDR) and introduced these sequences into hIgG1 to yield recombinant 3F6-hIgG1 (21). Here, we introduce amino acid substitutions in the Fcγ domain of 3F6-hIgG1 to block SpA- and Sbi-binding and generalize this approach using Tefibazumab (Tefi), a humanized antibody with specificity to the surface virulence factor, ClfA (22). The Fcγ-engineered antibodies were evaluated for their ability to bind C1q and FcRn in vitro and tested for blood half-life and opsonophagocytic activity of S. aureus in several models of disease.
Results
Engineering the Fcγ Domain of 3F6-hIgG1 to Escape SpA- and Sbi-Binding.
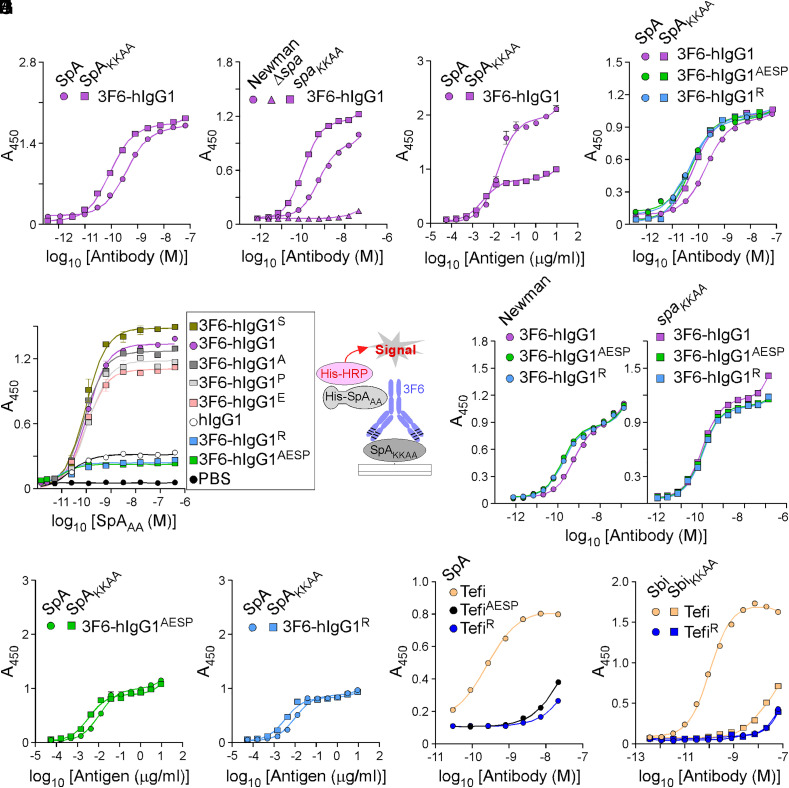
Although 3F6-hIgG1 can neutralize SpA via its 3F6 paratope (23), its Fcγ is still a ligand for SpA and Sbi. Each IgBD of SpA contains one 3F6 epitope and one Fcγ-binding site. Thus, full-length SpA contains 10 possible binding sites for 3F6 with two distinct binding domains (23). Half-maximal effective concentration (EC50) values measured by enzyme-linked immunosorbent assay (ELISA) represent the sum of all these interactions. The EC50 value of 3F6-hIgG1 for SpA was 4.11 times lower than for SpAKKAA (Fig. 1A and SI Appendix, Table S1). In this study, SpA and SpAKKAA refer to antigens composed of the five IgBDs of SpA. SpAKKAA has been modified with 20 amino acid substitutions to disrupt Fcγ and VH3 antigen-binding fragment (Fab) binding; in the conventional IgBD nomenclature, these residues correspond to Q9,Q10 changed to K9,K10 and D36,D37 changed to A36,A37 (19). When offered to wild-type S. aureus strain Newman (Newman) or the isogenic mutant strain with the spaKKAA sequence replacing the wild-type spa gene on the chromosome (spaKKAA), lower concentrations of 3F6-hIgG1 were required to saturate binding sites on spaKKAA bacteria as compared to Newman (Fig. 1B and SI Appendix, Table S1). Of note, Newman and the spaKKAA variant strain produce the same amount of protein A (13). No binding was observed with the Δspa control strain deleted for the spa gene (Fig. 1B). When 3F6-hIgG1 was coated to ELISA plates, SpA and SpAKKAA displayed distinct binding saturation (relative Bmax for SpA 1.911 versus SpAKKAA 0.771) (Fig. 1C and SI Appendix, Table S1); this is in agreement with the notion that SpA contains twice as many binding sites for 3F6 as compared to SpAKKAA. The Fc interaction with SpA can be gleaned from the atomic structure of SpA fragments bound to human Fcγ (11, 12). Four hydrogen bonds promote interactions between residues Q9, Q10, N11, and Y14 of SpA IgBD-B [also conserved in Sbi (23)] and residues S254, Q311, N434, and L432 of hIgG1, respectively. Single and combined amino acid substitutions (S254A, Q311E, L432S, or N434P) were generated yielding antibody variants 3F6-hIgG1A, 3F6-hIgG1E, 3F6-hIgG1S, 3F6-hIgG1P, and 3F6-hIgG1AESP. A sixth variant 3F6-hIgG1R with amino acid substitution H435R was generated, because the common IgG3 allotype with arginine at position 435 avoids SpA-binding (3, 24, 25). Of note, the same allotype also accounts for altered binding to FcRn (3). The substitutions (S254A, Q311E, L432S, N434P, and H435R) did not affect CDR-binding to SpAKKAA (SI Appendix, Fig. S1A). Next, immobilized SpAKKAA was incubated with test antibodies to form immune complexes (IC), and histidine-tagged SpAAA (His-SpAAA, a variant that can only bind hIgG1-Fc) (19) was added to preformed ICs. Bound His-SpAAA was detected with a nickel-activated derivative of horseradish peroxidase (HRP). SpAAA could no longer bind to ICs formed between 3F6-hIgG1AESP or 3F6-hIgG1R (Fig. 1D). Unlike 3F6-hIgG1, 3F6-hIgG1AESP and 3F6-hIgG1R interacted similarly with SpA and SpAKKAA (Fig. 1E and SI Appendix, Table S1), with Sbi and SbiKKAA (SI Appendix, Fig. S1B), and with Newman and spaKKAA bacteria (Fig. 1F and SI Appendix, Table S1). As a control, test antibodies did not interact with Δspa bacteria (SI Appendix, Fig. S1C). When 3F6-hIgG1AESP and 3F6-hIgG1R were coated to ELISA plates, the binding curves for both SpA and SpAKKAA exhibited similar saturation patterns (relative Bmax 0.969 versus 0.870) and similar EC50 values (Fig. 1 G and H and SI Appendix, Table S1) in contrast to 3F6-hIgG1 (Fig. 1C). KD measurements were also obtained using surface plasmon resonance (SPR) to compare antigen (SpAKKAA)–antibody interactions (SI Appendix, Fig. S1D). None of the substitutions in the Fc domain interfered with epitope binding; KD values of 45, 38, and 29 pM were obtained for 3F6-hIgG1, 3F6-hIgG1AESP, and 3F6-hIgG1R, respectively, and were in agreement with EC50 values obtained by ELISA (SI Appendix, Table S1). Together, the experiments show that SpA associates with both the Fc and CDR of 3F6-hIgG1 and that the antibody can be engineered to redirect SpA binding to CDR exclusively. To corroborate these results, the AESP and R substitutions were introduced in the sequence of Tefi, a hIgG1 monoclonal antibody that binds the A domain of Clumping factor A (ClfA-A) of S. aureus. The Fc substitutions did not significantly alter the binding to ClfA-A (SI Appendix, Fig. S1E), and Spa interacted only marginally with the modified TefiAESP and TefiR antibodies possibly through VH3 residues (Fig. 1I and SI Appendix, Table S1). Similarly, Sbi interacted marginally with TefiR (Fig. 1J and SI Appendix, Table S1).
Fig. 1.
hIgG1-Fc variants that escape SpA- and Sbi-binding. Antigen–antibody interactions were measured by ELISA. ELISA plates were coated as indicated on the figure panels: recombinant histidine-tagged SpA/SpAKKAA/Sbi/SbiKKAA proteins (A, E, I, and J); Newman/spaKKAA bacteria (B and F); 3F6 or its variants (C, G, and H); or ICs that were preformed between antigen (SpAKKAA) and antibody variants (with single or quadruple substitutions in the Fc region) as shown in the drawing (D). Bound antibodies and proteins were detected with HRP-conjugated secondary antibody and His-HRP, respectively, and absorbances were recorded at 450 nm (A450). Data are presented as mean ± SEM (n = 3 assays) and are representative of at least two independent experiments.
Engineering the Fcγ Domain of 3F6-hIgG1 to Restore Complement Activation.
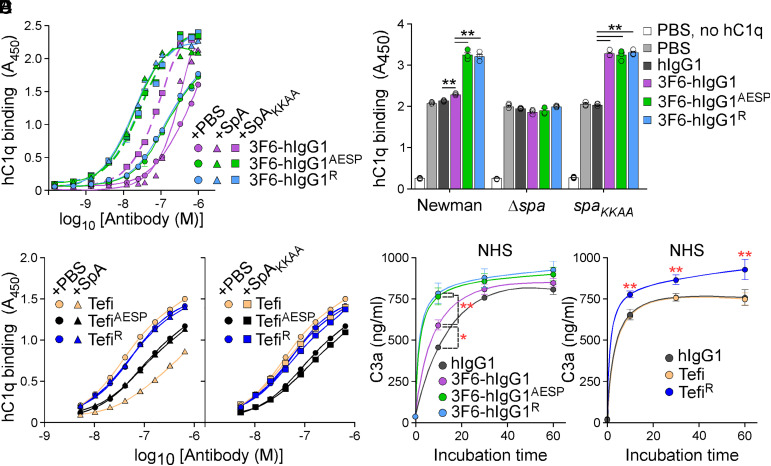
SpA binding to Fcγ prevents Fc–Fc association and, consequently, the formation of antibody hexamers needed for the recruitment of C1q to the bacterial surface (26). This was reflected by a 23-time-higher EC50 value for 3F6-hIgG1 interaction with hC1q in the presence of SpA as compared to SpAKKAA (Fig. 2A and SI Appendix, Table S2). Similarly, SpA but not SpAKKAA reduced interactions between the mouse IgG2a antibody, 3F6-mIgG2a, and mouse C1q (SI Appendix, Fig. S2A and Table S2). As predicted, SpA no longer blocked the association of hC1q with Fcγ-engineered 3F6-hIgG1AESP and 3F6-hIgG1R; in fact, hC1q binding to the modified AESP/R antibodies increased in the presence of SpA or SpAKKAA, as compared to hC1q binding to the unmodified 3F6-hIgG1 in complex with SpAKKAA (Fig. 2A and SI Appendix, Table S2). When tested with staphylococci, recruitment of purified hC1q was dramatically increased following opsonization of Newman cells with 3F6-hIgG1AESP and 3F6-hIgG1R; only a modest hC1q increase was observed in the presence of 3F6-hIgG1 as compared to hIgG1 (Fig. 2B). All three antibodies (3F6-hIgG1, 3F6-hIgG1AESP, and 3F6-hIgG1R) recruited C1q equally well when incubated with spaKKAA cells. As a control, hC1q binding was reduced to background levels for Δspa bacteria (Fig. 2B). Similar results were obtained when normal human serum (NHS) was used as the source of C1q (SI Appendix, Fig. S2B). hC1q binding to Tefi was also reduced in the presence of SpA, but not of SpAKKAA (Fig. 2C and SI Appendix, Table S2). SpA did not block hC1q binding to TefiAESP and TefiR; of note, TefiAESP establishes weaker interactions with hC1q as compared to Tefi/TefiR (Fig. 2C). To evaluate whether C1q recruitment correlated with complement activation, rates of C3a and C4d production were recorded (Fig. 2 D and E and SI Appendix, Fig. S2 C and D). Upon incubation with bacteria, the rate of C3a production increased in the presence of 3F6-hIgG1 versus control hIgG1 and was further accelerated in the presence of 3F6-hIgG1AESP and 3F6-hIgG1R (Fig. 2D). C4d levels increased similarly within the first 10 min of interaction with staphylococci (SI Appendix, Fig. S2C). C3a production occurred at the same rates when using C1q-depleted serum (SI Appendix, Fig. S2D). C3a production was also increased in the presence of TefiR; in this assay, unmodified Tefi behaved like the isotype control, hIgG1 (Fig. 2E).
Fig. 2.
hIgG1-Fc variants that restore C1q-binding in the presence of SpA. (A–C) Antibody interactions with purified (human) hC1q were assessed in the presence of mock (PBS), SpA, or SpAKKAA (A and C) or in the presence of bacteria: wild-type Newman or its variants, Δspa or spaKKAA (B). ELISA plates were coated with serially diluted test antibodies (A and C) or bacteria (B). Bound hC1q was detected with HRP-conjugated secondary antibody, and absorbances were recorded at 450 nm (A450). (D and E) Quantification of C3a production over time following incubation with bacteria (Newman 1 × 108 CFU/mL) and 3F6 (D) or Tefi (E) test antibodies. NHS was used as the source of opsonins. In all panels, data are presented as mean ± SEM (n = 3 assays) and differences identified in B, D, and E by two-way ANOVA with Bonferroni posttests (**P < 0.01; *P < 0.05). One representative of at least two independent experiments is shown.
Next, we examined antibody interactions with FcγRs. IgG-binding sites for SpA and FcγRs (at least FcγI and FcγRIIa) in Fc do not overlap (14, 27). Thus, unlike with C1q, SpA is not thought to affect interactions between human FcγRs (hFcγRs) and IgG. This was confirmed for six hFcγRs (SI Appendix, Fig. S2E). In fact, hFcγRs incubated with antibodies, including the non-VH3 control IgG1, interacted with bacterial cells in a SpA-dependent manner (SI Appendix, Fig. S2 E and F). All engineered antibodies interacted similarly with FcγRs in this assay (SI Appendix, Fig. S2E). In conclusion, SpA blocks hC1q but not hFcγR interactions with ICs. hC1q recruitment can be restored by introducing defined amino acid substitutions in the Fc region that do not affect interactions with antigen and hFcγRs.
Engineering the Fcγ Domain of 3F6-hIgG1 Improves Its Antistaphylococcal Activity.
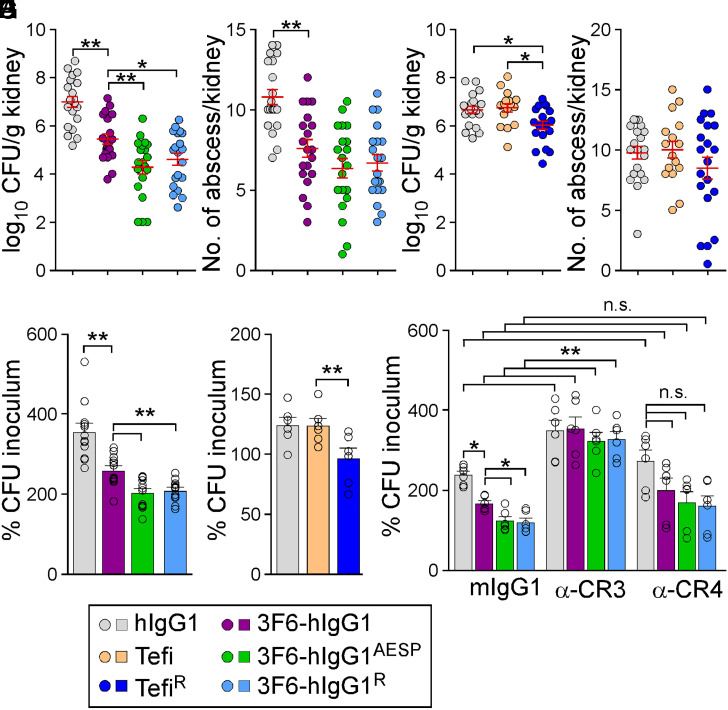
The therapeutic potential of antibodies can be evaluated in a mouse model of bloodstream infection (28). Consistent with previous results (29), animals that received 3F6-hIgG1 harbored reduced bacterial burdens (Fig. 3A) and fewer abscess lesions (Fig. 3B) in kidneys following challenge with methicillin-resistant Staphylococcus aureus (MRSA) strain MW2, as compared to animals that received hIgG1. Total bacterial loads were further reduced in animals treated with 3F6-hIgG1R and 3F6-hIgG1AESP (Fig. 3 A and B). At day 15 postinfection, serum concentrations of 3F6-hIgG1R were significantly higher than that of 3F6-hIgG1 (SI Appendix, Fig. S3A). Administration of Tefi in this animal model afforded no protection; nonetheless, the single H435R substitution in Tefi-Fc boosted the activity of this antibody as reflected by a small but significant reduction of bacterial loads in kidney tissues (Fig. 3 C and D). Serum concentrations of TefiR were significantly higher than that of Tefi 15 d post infection (SI Appendix, Fig. S3B). To assess the opsonophagocytic killing (OPK) potential of test antibodies in humans, we developed earlier a whole-blood killing assay using freshly drawn anticoagulated blood from healthy volunteers. The canonical HL60-based OPK assay lacks the hemostasis factors (prothrombin and fibrinogen) that are exploited by S. aureus to agglutinate and avoid phagocytic uptake (30). Addition of 3F6-hIgG1AESP and 3F6-hIgG1R to anticoagulated human blood resulted in increased killing of MRSA isolates MW2 and USA300 as compared to addition of 3F6-hIgG1 (Fig. 3E and SI Appendix, Fig. S3C). As observed with animals (Fig. 3C), Tefi displayed no antistaphylococcal activity in human blood, but significant OPK activity was observed with TefiR (Fig. 3F). Clearly, the lack of OPK activity correlated with the presence of protein A in the envelope of bacteria since Tefi promoted the killing of spaKKAA but not of Newman (spa+) in blood (SI Appendix, Fig. S3D). As a control, Newman and its spaKKAA variant displayed similar amounts of ClfA in their envelope (SI Appendix, Fig. S3E).
Fig. 3.
Antistaphylococcal activity of 3F6 and Tefi variants. (A and B) Enumeration of bacterial loads in kidney tissues (CFU) (A) and of tissue surface abscesses following 15 d infection with S. aureus MW2 of BALB/c mice (n = 20 animals, from two independent experiments) pretreated with antibody hIgG1, 3F6-hIgG1, 3F6-hIgG1R, or 3F6-hIgG1AESP at 5 mg.kg (body weight). (C and D) Enumeration of bacterial loads (C) and kidney surface abscesses (D) as described in A and B in animals pre-treated with antibody hIgG1, Tefi, or TefiR (n = 16 to 20 mice from two independent experiments). Data in A–D are presented as mean ± SEM (E–G) The OPK activity of antibodies toward S. aureus MW2 was analyzed using anticoagulated freshly drawn human blood. Test antibodies in E included hIgG1, 3F6-hIgG1, 3F6-hIgG1R, and 3F6-hIgG1AESP (n = 12 donors) and, in F, hIgG1, Tefi, and TefiR (n = 6 donors). In G, OPK activities of 3F6-hIgG1, 3F6-hIgG1R, and 3F6-hIgG1AESP in human blood were tested in the presence of control mIgG1, and anti-CR3 (α-CR3) or anti-CR4 (α-CR4) (n = 6 donors). Data were plotted as the average ± SEM of CFUs after 60-min incubation in blood compared to CFUs of inoculum (set as 100%). (H) Color code for test antibodies used in this figure. Significant differences were identified in A–F by one-way ANOVA with Tukey’s multiple-comparison test and in G by two-way ANOVA with Bonferroni posttests (*P < 0.05; **P < 0.01).
The whole-blood killing assay was repeated in the presence of anti-CR3 or anti-CR4 blocking antibodies. Myeloid cells produce various amounts of CR3 and CR4, and both share the ligand iC3b, the activation product of C3 on opsonized targets (7). Blocking CR4 had little effect on bacterial replication in general (Fig. 3G). Blocking CR3 resulted in increased bacterial replication in mock (hIgG1) samples, in agreement with the notion that CR3 is critical for the clearance of complement-opsonized S. aureus by effector cells (31–33). The OPK activity of all 3F6 antibodies was lost in the presence of anti-CR3, suggesting that these antibodies operate by enhancing the natural defenses of the host (Fig. 3G).
SpA Affects the Half-life of Antibodies In Vivo.
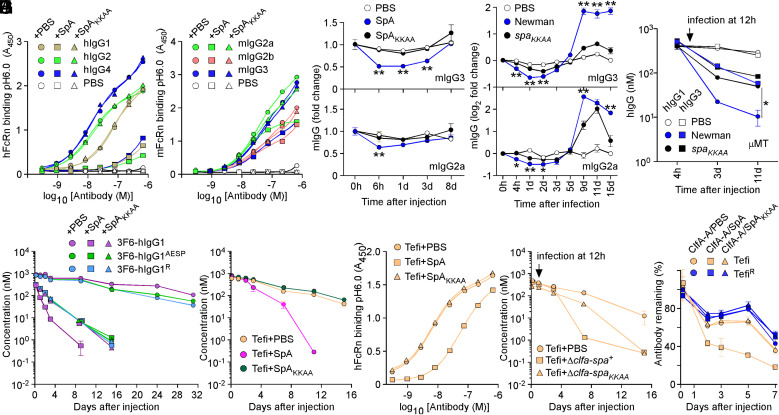
Atomic structures of protein complexes revealed overlapping binding sites for SpA and FcRn with IgG-Fc, and in vitro binding studies demonstrated that SpA and FcRn are competitive ligands for IgG-Fc (3, 11, 14, 34). Nonetheless, the physiological significance of this competitive interaction is not known but implies that SpA could affect the half-life of antibodies during infection (SI Appendix, Fig. S3 A and B). SpA but not SpAKKAA blocked interactions between hIgG1, hIgG2, and hIgG4 and hFcRn at pH 6.0 (Fig. 4A and SI Appendix, Table S2); hIgG3 that does not bind SpA was omitted from the analysis (SI Appendix, Fig. S4A). SpA also blocked interactions between mouse IgG (mIgG) and mouse FcRn (mFcRn) at pH 6.0 (Fig. 4B and SI Appendix, Table S2). The inhibitory activity correlated with SpA affinity for mIgG3 > mIgG2a > mIgG2b (SpA does not bind mIgG1) (SI Appendix, Fig. S4B and Table S1). Next, mice (n = 4) were injected with purified proteins (SpA/SpAKKAA) or bacteria (Newman/spaKKAA), the latter of which mimicked the bloodstream infection displayed in Fig. 3A. A single injection of SpA in the bloodstream but not of SpAKKAA resulted in the transient but noticeable reduction of mIgG2a and mIgG3 (Fig. 4C). Similarly, levels of mIgG2a and mIgG3 dropped in animals infected with Newman but not with spaKKAA (Fig. 4D). Levels of mIgG1 and mIgG2b that are poor ligands for SpA were not affected (SI Appendix, Fig. S4 C and D). Rising levels of mIgG after day 5 of infection with Newman (Fig. 4D and SI Appendix, Fig. S4D) were attributed to the B-cell superantigen activity of SpA (i.e., the expansion of VH3 antibodies as measured using antigen SpAKK) (SI Appendix, Fig. S4E) (18). To study the effect of SpA on human antibodies, μ chain gene mutation (μMT) mice that lack mature B cells and the ability to produce antibodies (35) were injected with 100 μg hIgG1 or hIgG3, 12 h prior to intravenous challenge with phosphate buffered saline (PBS) (mock), Newman, or spaKKAA (Fig. 4E). As compared to mock treatment, serum concentrations of hIgG1 and hIgG3 decreased slightly upon infection, presumably because S. aureus secretes the V8 protease that cleaves all hIgG (36, 37). Nonetheless, hIgG1 levels decreased much more rapidly in animals infected with Newman as compared to spaKKAA (Fig. 4E). Such a difference was not observed for IgG3 since hIgG3-Fc is not a ligand for SpA (Fig. 4E). Next, we asked whether SpA alters the half-life of therapeutic antibodies through Fc interactions. 3F6-hIgG1 and the engineered variants AESP and R were mixed with PBS, SpA, or SpAKKAA and injected in wild-type mice (Fig. 4F). Serum concentrations of all three antibodies remained stable when injected in PBS (Fig. 4F). In contrast, serum concentrations of 3F6-hIgG1 decreased precipitously when injected with SpA (Fig. 4F). When SpA–Fc interactions were circumvented, such as when 3F6-hIgG1 was coinjected with SpAKKAA or engineered variants (AESP/R) were coinjected with SpA/SpAKKAA, antibody clearance was less pronounced (Fig. 4F). We attribute this slower turnover to the uptake of productive antigen–antibody IC by phagocytes. This notion is supported by the increased antibody responses to 3F6-hIgG1 and its variants (SI Appendix, Fig. S4F). Injection of SpA with 3F6-hIgG1 that results in both productive (paratope-bound) and illegitimate (Fc-bound) complexes dampened antibody responses to 3F6-hIgG1 (SI Appendix, Fig. S4F). To further validate these observations, we used the therapeutic antibody Tefi that interacts with SpA though hIgG1-Fc exclusively. Coinjection of Tefi with SpA in animals resulted in enhanced clearance of the antibody; coinjection with SpAKKAA had no effect on the half-life of Tefi (Fig. 4G). Animals developed antibodies against Tefi injected with SpA but not with SpAKKAA (SI Appendix, Fig. S4G). When tested in vitro, Tefi interaction with hFcRn at pH 6.0 was inhibited by SpA but not by SpAKKAA (Fig. 4H and SI Appendix, Table S2). To examine the impact of infection, animals treated with Tefi were challenged 12 h later with the isogenic ΔclfA-spa+ and ΔclfA-spaKKAA variants or PBS (mock control). The clfA gene was deleted to eliminate antibody turnover caused by productive Tefi-ClfA ICs. Serum levels of Tefi dropped much more rapidly in animals infected with the spa+ (ΔclfA-spa+) strain as compared to the spa defective strain (ΔclfA-spaKKAA) (Fig. 4I). ICs formed between the ClfA-A antigen and Tefi or TefiR were coinjected in animals with SpA, SpAKKAA, or PBS. The fastest clearance was observed for the ClfA-A–Tefi complex when injected with SpA (Fig. 4J), further emphasizing that Fc–SpA interactions reduce antibodies’ half-life even when the antibody is bound to cognate antigen (Fig. 4J).
Fig. 4.
SpA influences the stability of antibodies in vivo. (A and B) Human and mouse hFcRn/mFcRn binding to human (A) and mouse (B) IgG at pH 6.0 was measured in the presence of PBS, SpA, or SpAKKAA by ELISA (n = 3 assays). ELISA plates were coated with hIgG, mIgG, or mock (PBS). Bound biotinylated FcRn was detected with HRP-conjugated streptavidin, and absorbances were recorded at 450 nm (A450). (C–D) Fold change over time of the serum concentration of mIgG2a and mIgG3 following injection of PBS, SpA, or SpAKKAA (C) and infection with mock (PBS), Newman, or its spaKKAA variant in BALB/c mice (D) (n = 4 animals per group). (E) hIgG1 and hIgG3 concentrations over time in μMT mice infected with mock (PBS), Newman, or spaKKAA 12 h after administration of hIgG1/3 (n = 4 mice per group). (F and G) Serum concentrations over time of test antibodies following injection of PBS, SpA, or SpAKKAA in BALB/c mice (n = 4 mice per group). (H) hFcRn binding to Tefi at pH 6.0 was measured in the presence of PBS, SpA, or SpAKKAA by ELISA (n = 3 assays). Bound FcRn was detected as in A. (I) Serum concentrations over time of test antibodies in BALB/c mice (n = 5 mice per group). Animals were infected with the Newman variants Δclfa-spa+ or Δclfa-spaKKAA or PBS (mock) treated 12 h after the administration of test antibodies. (J) ICs of test antibodies Tefi/TefiR and cognate antigen ClfA-A were injected in BALB/c mice (n = 5 mice per group) along with PBS, SpA, or SpAKKAA, and the remaining fraction of test antibodies was measured over time. Data are presented as mean ± SEM and representative of at least two independent experiments (A, B, and H). Significant differences were identified by two-way ANOVA with Bonferroni posttests (**P < 0.01; *P < 0.05).
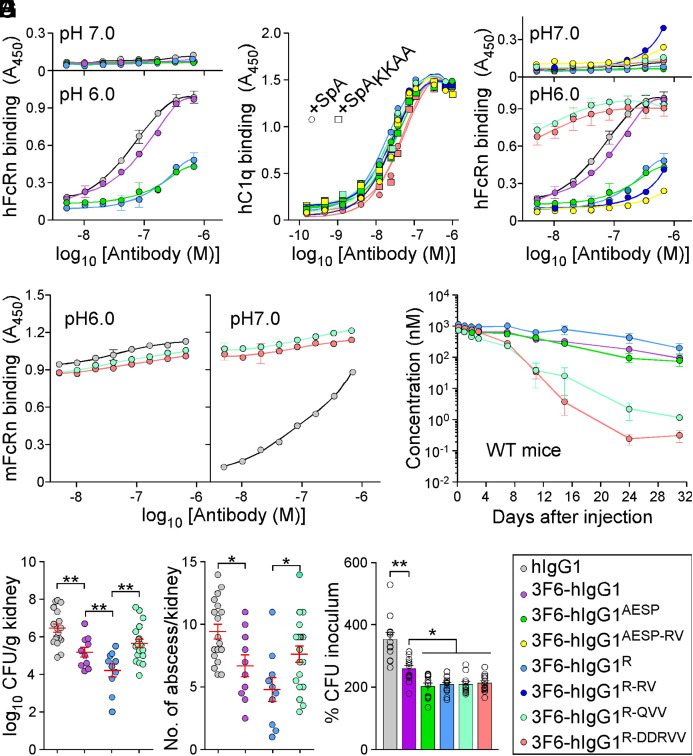
Engineering 3F6 Antibodies for Human Use.
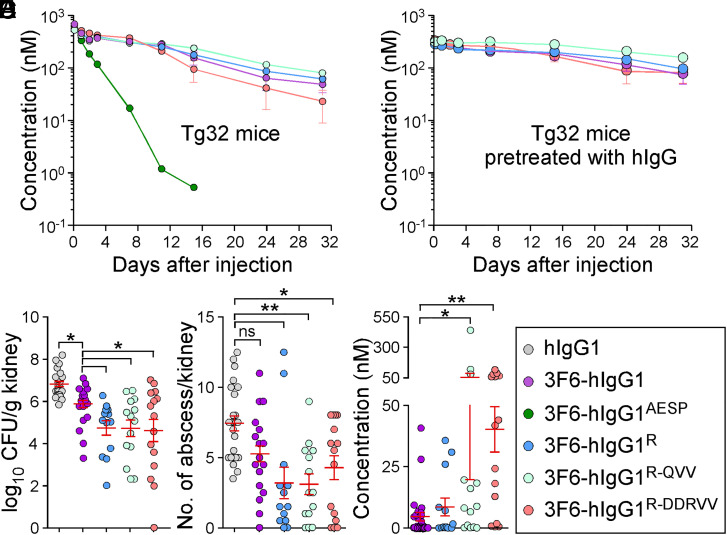
Ig-Fc mutations that decrease SpA-binding may also alter interactions with FcRn and the trade-off for escaping protein A-mediated immune evasion may be reduced antibody half-life. To investigate this possibility, interactions between purified antibodies and FcRn were first measured by ELISA. 3F6-hIgG1 and 3F6-hIgG1R bound mFcRn similarly at pH 6.0 and 7.0, while 3F6-hIgG1AESP displayed lower binding at both pH (SI Appendix, Fig. S5A). As compared to hIgG1or 3F6-hIgG1, 3F6-hIgG1R and 3F6-hIgG1AESP displayed a weaker association with hFcRn at pH6.0 (Fig. 5A and SI Appendix, Table S1). KD values for hFcRn at pH 6.0 were also obtained using SPR and measured as 538 nM for 3F6-hIgG1, 1,189 nM for 3F6-hIgG1R, and 9,825 nM for 3F6-hIgG1AESP, suggesting that 3F6-hIgG1AESP may have a shorter serum half-life (SI Appendix, Fig. S5B). An earlier structure-guided design suggested that substitutions at residues 256, 286, 307, 311, and 378 may ameliorate antibody–FcRn interactions (38). Combinations RV (T307R/A378V), QVV (T307Q/Q311V/A378V), and DDRVV (T256D/N286D/T307R/Q311V/A378V) were introduced in four test antibodies 3F6-hIgG1AESP-RV, 3F6-hIgG1R-RV, 3F6-hIgG1R-QVV and 3F6-hIgG1R-DDRVV (SI Appendix, Fig. S5C). The substitutions did not interfere with hC1q/SpA-binding (Fig. 5B) and, except for R-DDRVV, did not alter antibody binding to Newman or spaKKAA (SI Appendix, Fig. S5 D–F). When subjected to size exclusion chromatography, all 3F6 variants eluted as monomers (SI Appendix, Fig. S5G). QVV and DDRVV substitutions markedly increased 3F6-hIgG1R binding to hFcRn at pH 6.0 but not at pH 7.0 (Fig. 5C). 3F6-hIgG1R-QVV and 3F6-hIgG1R-DDRVV interacted strongly with mFcRn at pH 7.0 (Fig. 5D), which correlated with their rapid turnover (Fig. 5E and SI Appendix, Table S3) and increased immunogenicity in BALB/c mice (SI Appendix, Fig. S5H). The reduced half-life of 3F6-hIgG1R-QVV in mice abrogated any gain of protection against MRSA observed with 3F6-hIgG1R (Fig. 5 F and G). Nonetheless, when OPK activity was tested in whole human blood, 3F6-hIgG1R-QVV and 3F6-hIgG1R-DDRVV performed better than 3F6-hIgG1 and similarly to 3F6-hIgG1AESP and 3F6-hIgG1R (Fig. 5H). Next, humanized FcRn Tg32 mice (39) were used to measure the half-life and therapeutic activity of test antibodies. 3F6-hIgG1AESP that failed to bind hFcRn in vitro was rapidly turned over in Tg32 animals, while R435 variants behaved as the parent 3F6 antibody (Fig. 6A). Accordingly, Tg32 mice did not develop strong antibody responses against test antibodies (SI Appendix, Fig. S6S2). Since mIgG interacts poorly with hFcRn, Tg32 mice were also pretreated with hIgG to better mimic human conditions (40). All engineered antibodies retained high serum concentrations with the slowest turnover observed for 3F6-hIgG1R-QVV (Fig. 6B and SI Appendix, Table S3) and were further tested for efficacy (Fig. 6 C and D). In Tg32 mice unlike in BALB/c animals, 3F6-hIgG1 protected partially by reducing bacterial loads (but not abscess lesions) as compared to hIgG1 (Fig. 6 C and D). Whereas in BALB/c animals, 3F6-hIgG1R-QVV provided less protection than 3F6-hIgG1R against MRSA challenge (Fig. 5 F and G); in Tg32 mice, this difference was no longer observed. In fact, both 3F6-hIgG1R-QVV and 3F6-hIgG1R-DDRVV performed as well as 3F6-hIgG1R, with a small but statistical improvement over 3F6-hIgG1 (Fig. 6 C and D). Therapeutic superiority correlated with enhanced serum concentration of antibodies (Fig. 6E).
Fig. 5.
Amino acid substitutions for improved interactions with hFcRn. (A–D) ELISA-binding curves (n = 3 assays) of hFcRn (at pH 6.0 and 7.0) (A and C), hC1q (B), or mFcRn (pH 6.0 and 7.0) (D) to test antibody variants including control hIgG1 and 3F6-hIgG1. ELISA plates were coated with serially diluted test antibodies (color code in I). (B) ELISA-binding curves of purified hC1q to test antibodies were assessed in the presence of SpA or SpAKKAA (n = 3 assays). Binding curves were mostly superimposable. (E) Serum concentrations over time of 3F6-hIgG1 and variants (AESP, R, R-QVV, and R-DDRVV) following injection in BALB/c mice (n = 4 mice per group). (F and G) Enumeration of bacterial loads (F) and kidney surface abscesses (G) as described in Fig. 3 in animals pretreated with antibody hIgG1, 3F6-hIgG1, 3F6-hIgG1R, or 3F6-hIgG1R-QVV (n = 10 to 20 mice). Data are presented as mean ± SEM and analyzed with one-way ANOVA with Tukey’s multiple-comparison test (**P < 0.01; *P < 0.05). (H) OPK activity of test antibodies (color code in I) toward S. aureus MW2 in anticoagulated freshly drawn human blood (n = 12 donors). Data were plotted as described in Fig. 3 and significance calculated with one-way ANOVA and Tukey’s multiple-comparison test (*P < 0.05; **P < 0.01). (I) Color code for test antibodies used in this figure. Data are presented as mean ± SEM and representative of two independent experiments (A–D).
Fig. 6.
Antibody efficacy in the humanized FcRn (Tg32) mice. (A and B) Serum concentrations over time of 3F6-hIgG1R and variants R-QVV and R-DDRVV following injection (5 mg/kg) in Tg32 mice without (A) or with (B) pretreatment with pooled hIgG at 100 mg/kg (n = 5 mice per group). (C and D) Enumeration of bacterial loads (C) and kidney surface abscesses (D) as described in Fig. 3 in Tg32 animals pretreated with antibody hIgG1, 3F6-hIgG1, 3F6-hIgG1R, 3F6-hIgG1R-QVV, or 3F6-hIgG1R-DDRVV challenged for 15 d with S. aureus MW2 (n = 13 to 17 mice). (E) Serum concentration of test antibodies that remained after 15 d infection with S. aureus MW2. Sera were from animals shown in C. (F) Color code for test antibodies used in this figure. Data are presented as mean ± SEM and analyzed with one-way ANOVA and Tukey’s multiple-comparison test in C–E (**P < 0.01; *P < 0.05).
Discussion
S. aureus is responsible for both healthcare- and community-associated infections (41, 42). Infections are treated with antibiotics. However, the treatment of S. aureus infections can be complicated by disease severity, high recurrence, a dwindling antibiotic arsenal against methicillin-resistant strains, and treatment length (43). Some infections such as skin and soft-tissue collections also require incision and drainage. Failure to drain abscesses delays resolution and favors the development of antibiotic resistance (44). For all these reasons, it seems reasonable to expand efforts toward the development of antibodies that could aid in the treatment of S. aureus diseases. Several envelope determinants have been considered as targets of opsonophagocytic antibodies but failed in clinical trials (43). None of these approaches accounted for the ability of SpA to bind the Fc region of IgG, the effector region of antibodies that interacts with FcγRs, C1q, and FcRn. The possibility that SpA may affect all effector functions of antibodies and the implication for the therapeutic activity of antibodies has not been addressed.
Here, we report that SpA binding to IgG-Fc in vitro obstructs further association with C1q and FcRn but not with recombinant FcγRs. The Fc region of antibodies with CDRs directed against SpA (3F6) or ClfA (Tefi) were engineered to prevent SpA and Sbi interference with C1q recruitment. Antibody variants with AESP and R substitutions that disrupted SpA/Sbi interference were found to improve the therapeutic and OPK activities of 3F6 and Tefi antibodies in a mouse model of MRSA bloodstream infection and in human whole blood, respectively. Further, blocking CR3 prevented S. aureus killing in whole blood, suggesting that 3F6 variants enhanced iC3b opsonization to promote phagocytic uptake. We propose that the OPK activity of 3F6 candidate antibodies correlates with enhanced activation of C1q at the bacterial cell surface. We cannot rule out the involvement of other effector or complement activities.
Our experiments confirmed that SpA and FcRn compete for Fc binding in a manner that correlated with binding affinity to antibodies. SpA interacts with hIgG1 and mIgG2 with high affinity but fails to bind the Fc fragment of most other immunoglobulins including hIgG3, IgM, IgE, IgA, and mIgG1 that do not interact with FcRn and display shorter half-lives in vivo (3, 40, 45, 46). By competing for FcRn-binding, SpA effectively reduced the half-life of hIgG1 and mIgG2. Depletion was very effective and observed following injection of SpA in animals as well as during infection with S. aureus. We reported earlier that SpA is key to the colonization success of S. aureus in a mouse model (21, 47), implying that colonization could also accelerate the turnover of hIgG1. This evolutionary adaptation represents a challenge for the engineering of therapeutic antibodies. Breaking apart Fc–SpA interactions restores C1q recruitment at the cost of reducing the half-life of antibody. Unsurprisingly, the 3F6-hIgG1R and 3F6-hIgG1AESP variants displayed a weaker association with hFcRn at pH6.0 as compared to3F6-hIgG1. To circumvent this problem, amino acid substitutions shown to enhance hFcRn-binding (38) were introduced in the sequences of 3F6 variants. As the substitutions resulted in increased binding to mFcRn at 7.0 (indicative of short half-life), transgenic Tg32 animals with an mFcRn α-chain replaced with the hFcRn α-chain (39) were used for testing. Predictably, 3F6-hIgG1AESP was rapidly turned over in Tg32 mice; surprisingly, 3F6-hIgG1R displayed a similar half-life as 3F6-hIgG1. Variant 3F6-hIgG1R-QVV was even more stable than 3F6-hIgG1; the variant afforded significant protection against bacterial burdens and abscess lesions in animal-infected tissues. We conclude that R-QVV substitutions may be best suited for the development of antibodies against S. aureus in humans.
All FcγRs in their soluble form were found to interact with bacteria via nonspecific SpA–Fc complexes. It is unclear whether such interactions also occur between bacteria and immune cells and what the outcome of such interactions may be. Possibly, S. aureus exploits such interactions to its advantage. SpA released from bacteria and bound to Fc or to legitimate IgG–antigen complexes may also bind FcγRs for uptake; once in the endocytic compartment, SpA will obstruct further FcRn binding. In antigen-presenting cells, FcRn interactions with large IgG ICs has been shown to mediate the secretion of inflammatory cytokines and enhance intracellular routing for further antigen processing (48). Thus, SpA-mediated FcRn avoidance may contribute multiple immune evasion strategies, including reduced half-life of hIgG1 antibodies and reduced immune responses.
In conclusion, it may be possible to develop therapeutic antibodies against surface determinants of S. aureus. Such determinants should be abundant, conserved, preferentially key to the pathogenic program of S. aureus, and the antibodies should be designed to escape neutralization by SpA and other proteins, such as Sbi that have evolved to bind Ig-Fc (10). SpA represents an ideal target for such antibodies.
Materials and Methods
Ethics Statement.
The University of Chicago’s Institutional Review Board reviewed, approved, and supervised the protocol used for all experiments utilizing blood from human volunteers, and informed-consent forms were obtained from all participants. Animal research was performed in accordance with institutional guidelines following experimental protocol review, approval, and supervision by the Institutional Animal Care and Use Committee at The University of Chicago. Experiments with S. aureus were performed in biosafety level 2 (BSL2)/animal BSL2 containment upon review by The University of Chicago Institutional Biosafety Committee.
Bacterial Strains, Mammalian Cell Lines, and Growth Media.
S. aureus strains Newman and its variants (spaKK, spaAA, spaKKAA, Δspa, ΔclfA-spa+, and ΔclfA-spaKKAA), USA300 LAC and its variant Δspa, and MW2 were grown in tryptic soy broth or agar at 37 °C. Escherichia coli strain DH5α and BL21 (DE3) were grown in Luria broth or agar with 100 μg/mL ampicillin at 37 °C. Suspension serum-free adapted FreeStyleTM 293-F (human embryonic kidney [HEK]-293F) cells were cultured in FreeStyleTM 293 Expression Medium (Life Technologies) and maintained in a 5% CO2 humidified incubator at 37 °C.
Construction, Expression, and Purification of 3F6-hIgG1 and Tefi Variants.
Recombinant 3F6-hIgG1 cloned into the expression vector pVITRO1-102.1F10-IgG1/λ (Addgene no. 50366) was as described previously (29) and served as a template for further mutagenesis. The heavy- and light-chain genes of Tefi were synthesized from published sequences (22, 49) by Integrated DNA Technologies, Inc (IDT) and swapped into the pVITRO1 plasmid encoding 3F6-hIgG1. Primers 5′-AACCGCTACACGCAGAAGAGCCTCTC-3′ and 5′-GTAGCGGTTGTGCAGAGCCTCATGCAT-3′ were used to generate the TefiR and 3F6-hIgG1R variants. For other variants, partial Fc genes with specific mutation(s) were synthesized by IDT and swapped into the vector encoding Tefi or 3F6-hIgG1 using the polymerase incomplete primer extension (PIPE) method (21). PIPE products were transfected into HEK-293F cells using polyethylenimine and transfectants selected with hygromycin B (400 μg/mL) before expansion using TripleFlask Cell Culture Flasks (Thermo Fisher Scientific). Antibodies were affinity purified from supernatants of expanded cultures on protein G resin (GenScript) and dialyzed against PBS as described (29). Dialyzed antibodies were separated by size exclusion high performance liquid chromatography using a Tricorn Superose 6 Increase 10/300 GL column. Briefly, up to 200 μL of antibody suspension at concentrations ranging between 2 and 3 mg/mL was injected in PBS pH 7.2 over the column. PBS at pH 7.2 was used as the eluent and run at 0.4 mL/min.
Enzyme-Linked Immunosorbent Assays.
Binding measurements were performed in microtiter plates (Nunc Maxisorp) coated with either 10 nM test antibodies, 30 nM recombinant proteins (SpAKKAA, SpA, Sbi, or SbiKKAA), or 1 × 106 colony-forming units (CFU) of S. aureus bacteria, in 0.1 M carbonate buffer (pH 9.5) at 4 °C overnight or at 37 °C for 2 h. Wells were blocked by 2% (wt/vol) bovine serum albumin (BSA) before incubation with serial concentrations of test antibodies or His-tagged proteins. Binding capacity was quantified following incubation with HRP-conjugated goat anti-hIgG (Promega, W403B, 1:15,000) or with the HisProbe-HRP conjugate (Thermo Fisher Scientific, 15165). For competition experiments with C1q, microtiter plates were coated with serial dilutions of purified test antibodies overnight. After blocking, recombinant SpA or SpAKKAA was added along with 20 µg/mL human or mouse C1q (CompTech, A099 and M099), and C1q binding was detected with HRP-conjugated sheep anti-C1q (Bio-Rad, 2221–5004P). The molar ratio of test antibody over recombinant SpA/SpAKKAA was 5:1. To measure the binding of hC1q and FcγRs to bacteria, microtiter plates were first coated with bacteria at 37 °C for 2 h and blocked with BSA before adding 10 μg/mL test antibody and either 20 µg/mL hC1q, 10% NHS (CompTech), or 2 μg/mL His-tagged hFcγRs (SinoBiological). Bound hC1q binding was detected with HRP-conjugated sheep anti-C1q (Bio-Rad, 2221–5004P) and bound FcγR with the HisProbe-HRP conjugate (Thermo Fisher Scientific, 15165). For competition experiments with hFcRn or mFcRn, microtiter plates were coated with serial dilutions of purified test antibodies overnight. After blocking, 30 nM recombinant SpA or SpAKKAA was added to each well at either pH 6.0 or pH 7.0 along with 2 μg/mL biotinylated FcRn (Immunitrack). Bound FcRn was detected with HRP-conjugated streptavidin (New England Biolabs). All plates were developed using Opt enzyme immunoassay (EIA) reagent (BD Biosciences). Recombinant SpA/Sbi proteins and their variants were purified from E. coli as described (19). Values of dissociation constants reported in this study are listed in SI Appendix, Tables S1 and S2.
SPR Analysis.
SPR experiments were performed with a Biacore 8K instrument by the Biophysics Core of the University of Illinois Chicago. Ligands were bound to sensor chip nitrilotriacetic acid. Analytes were provided in PBS containing 0.05% Tween-20 and 1% BSA at indicated pH. The sensor-chip surfaces were activated with 2 mM nickel sulfate and regenerated with 300 mM ethylenediamine tetraacetic acid, respectively. In total, 500 nM of test articles (SpA, SpAKKAA, and hFcRn) were immobilized at a flow rate of 25 µL/min. To measure interactions with SpA or SpAKKAA ligands, analytes (3F6-hIgG1 and its variants) were used at a concentration range of 10 to 0.045 nM in a threefold serial dilution. To measure interactions with hFcRn, analytes were used at a concentration range of 2.5 to 0.02 μM with a threefold serial dilution. The association and dissociation rates were measured at a continuous flow rate of 25 µL/min and analyzed using the single-cycle kinetics. Dissociation constants were determined from two independent experiments.
C3a Release Assay and C4d Measurement.
In total, 1 × 108 CFU of live bacteria were suspended in 900 μL GVB++ buffer (CompTech, B100) with 100 μL NHS or C1q-depleted human serum (CompTech, A300) and incubated at 37 °C for 10, 30, or 60 min. Complement activation was quenched by adding 1 mL gelatin veronal sodium buffer with EDTA (CompTech, B105), and the samples were spun at 14,000 × g for 5 min. Amounts of C3a in supernatants were determined using the MicroVue C3a Plus EIA kit (Quidel, A031) according to the manufacturer’s instructions. To measure C4d deposition in the envelope of staphylococci, reaction mixtures were removed after 0 and 10 min, spun at 14,000 × g for 5 min, and bacteria in the pellets were treated with lysostaphin in 0.5 M Tris buffer (pH 7.0) for 30 min at 37 °C. C4d levels in cell lysates were determined using the MicroVue C4d EIA kit (Quidel, A009) according to the manufacturer’s instructions.
Measuring Serum Concentrations of Antibodies in Mice.
To determine the effect of SpA on mIgG, BALB/c mice (6 to 7 wk of age) obtained from Jackson Laboratory were injected into the peritoneal cavity with 100 μg recombinant, endotoxin-free, vaccine-grade SpA or SpAKKAA or into the periorbital venous plexus with 1 × 107 CFU of live bacteria. After 0, 4, and 6 h and 1, 2, 3, 5, 8, 9, 11, and 15 d, periorbital venous blood was obtained, and serum samples analyzed by ELISA. Briefly, microtiter plates were coated overnight with 10,000× dilution of serum sample. After blocking, wells were incubated with goat anti-mIgG1, -mIgG2a, -mIgG2b, or mIgG3 (Novus Biologicals). Serum mIgG concentrations were calculated using a standard curve of purified mIgG1, mIgG2a, mIgG2b, and mIgG3 (Biolegend) at a range of 1 to 1,500 ng/mL Microtiter plates coated with recombinant SpAKK (100 ng/well) were used to measure the VH3-clonal mIgG in serum samples. To investigate the impact of infection on hIgG, 100 μg of purified hIgG1 or hIgG3 (Sigma-Aldrich) were first injected into the peritoneal cavity of μMT mice (6 to 7 wk of age). A total 16 h later, animals were infected with 1 × 107 CFU of live bacteria or mock (PBS) into the periorbital venous plexus. Serum samples were collected at 4 h and 3 and 11 d after antibody injection, and serum concentrations of hIgG were calculated using a standard curve of purified hIgG1 and hIgG3 at a range of 1 to 50 ng/mL. To examine the effect of soluble SpA/SpAKKAA on test therapeutic antibodies, BALB/c mice (6 to 7 wk of age) were injected into the peritoneal cavity with 100 μg test antibody plus 4 μg recombinant, endotoxin-free, vaccine-grade SpA/SpAKKAA (molar ratio 5:1). Control mice were injected with 100 μg test antibody plus PBS. After 4 h and 1, 2, 3, 7, 11, and 15 d, serum samples were collected, and serum concentrations of test antibody were detected in microtiter plates coated with matched antigens (SpAKKAA or ClfA-A). To examine the impact of infection on therapeutic antibodies, 100 μg of test antibodies Tefi/TefiR were first injected into the peritoneal cavity of BALB/c mice (6 to 7 wk of age). After 16 h, animals were challenged intravenously with mock (PBS) or 1 × 107 CFU of ΔclfA-spa+ or ΔclfA-spaKKAA. Serum concentrations of test antibody were measured following serum sampling at 4 h and 1, 2, 3, 7, 11, and 15 d post infection using microtiter plates coated with ClfA-A (19). Tefi/TefiR concentrations in samples were calculated using a standard curve of purified Tefi/TefiR diluted in the mouse serum at a range of 0.1 to 150 ng/mL. To measure the persistence of 3F6-hIgG1 and its variants in Tg32 mice (B6.Cg-Fcgrttm1Dcr Tg(FCGRT)32Dcr/DcrJ), animals (n = 5, 6 to 7 wk of age) were either pretreated or not with hIgG (100 mg/kg body weight) before 100 μg test antibody was delivered by intraperitoneal injection. After 4 h and 1, 2, 3, 7, 11, 15, 24, and 31 d, periorbital venous blood was obtained, and test antibody in serum samples was captured by SpAKKAA-coated plates. Serum antibody concentrations were calculated using a standard curve of each test antibody diluted in mouse serum at a range of 1 to 500 ng/mL Pharmacokinetic (PK) parameters were determined for each group of mice with a noncompartmental analysis model using PKSolver (50).
Therapeutic Evaluation of Test Antibodies in Animals.
Animals (6 to 8 wk of age) were treated with 100 μg of indicated test antibody 12 to 18 h before challenge with S. aureus USA400 (MW2). Bacterial cultures were grown to an absorbance at 600 nm of 0.42, and bacteria were washed in PBS once and adjusted to a suspension of 6.5 × 107 CFU/mL. In total, 100 μL of this suspension was injected into the periorbital venous plexus of animals anesthetized with ketamine/xylazine (100 and 20 mg/kg). On day 15, mice were killed by carbon dioxide inhalation and necropsied to remove kidneys. Surface abscesses visible on intact kidneys were enumerated. The second kidney was weighed, homogenized, serially diluted, and plated on agar to count bacterial burden in tissues (CFU per gram of tissue).
Staphylococcal Survival in Human Blood.
Test or control antibody (2.5 μg) was added to 0.5 mL of freshly drawn human blood anticoagulated with 5 µg/mL desirudin or heparin. Where indicated, 5 μg/mL of anti-CR3 or anti-CR4 (BioLegend; Catalog no. 301302 or Catalog no. 301616) was added to the blood for 10 min prior to infection with bacteria. At time 0, a 50-μl bacterial suspension in PBS (5 × 106 CFU) was added to the blood, and tubes were incubated for 0 or 60 min at 37 °C. 0.5 mL streptokinase lysis buffer (PBS containing 0.5% saponin, 100 U streptokinase, 50 µg trypsin, 1 µg DNase, and 5 µg RNase) was added to each sample for 10 min at 37 °C prior to plating on agar for CFU enumeration. Assays were performed in duplicate and repeated for reproducibility.
Statistical Analyses.
All statistical analyses were performed using GraphPad Prism version 7.04 (GraphPad Software, Inc., La Jolla). Statistically significant differences were calculated by using statistical methods, as indicated. P values of <0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Blake Sanders for careful reading of the manuscript and Tatyana Golovkina, Hwan Keun Kim, Vilasack Thammavongsa, Lai-Xi Wang, and members of our laboratory for discussion. This project was supported by funds from the National Institute of Allergy and Infectious Diseases, NIH under award AI148543.
Footnotes
Competing interest statement: The authors are inventors of patents describing therapeutic antibodies against Staphylococcus aureus. D.M. declares a competing financial interest as a founder of ImmunArtes LLC, a University of Chicago startup company that aims to develop vaccines and therapies against S. aureus infections.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114478119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Vidarsson G., Dekkers G., Rispens T., IgG subclasses and allotypes: From structure to effector functions. Front. Immunol. 5, 520 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee C.-H., et al. , IgG Fc domains that bind C1q but not effector Fcγ receptors delineate the importance of complement-mediated effector functions. Nat. Immunol. 18, 889–898 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stapleton N. M., et al. , Competition for FcRn-mediated transport gives rise to short half-life of human IgG3 and offers therapeutic potential. Nat. Commun. 2, 599 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes-Jones N. C., Gardner B., Reaction between the isolated globular sub-units of the complement component C1q and IgG-complexes. Mol. Immunol. 16, 697–701 (1979). [DOI] [PubMed] [Google Scholar]
- 5.Duncan A. R., Winter G., The binding site for C1q on IgG. Nature 332, 738–740 (1988). [DOI] [PubMed] [Google Scholar]
- 6.Diebolder C. A., et al. , Complement is activated by IgG hexamers assembled at the cell surface. Science 343, 1260–1263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ricklin D., Reis E. S., Mastellos D. C., Gros P., Lambris J. D., Complement component C3 - The “Swiss Army Knife” of innate immunity and host defense. Immunol. Rev. 274, 33–58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berends E. T., et al. , Distinct localization of the complement C5b-9 complex on Gram-positive bacteria. Cell. Microbiol. 15, 1955–1968 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Bjanes E., Nizet V., More than a pore: Nonlytic antimicrobial functions of complement and bacterial strategies for evasion. Microbiol. Mol. Biol. Rev. 85, e00177-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thammavongsa V., Kim H. K., Missiakas D., Schneewind O., Staphylococcal manipulation of host immune responses. Nat. Rev. Microbiol. 13, 529–543 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deisenhofer J., Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochemistry 20, 2361–2370 (1981). [PubMed] [Google Scholar]
- 12.Graille M., et al. , Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: Structural basis for recognition of B-cell receptors and superantigen activity. Proc. Natl. Acad. Sci. U.S.A. 97, 5399–5404 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falugi F., Kim H. K., Missiakas D. M., Schneewind O., Role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. MBio 4, e00575–e13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wines B. D., Powell M. S., Parren P. W., Barnes N., Hogarth P. M., The IgG Fc contains distinct Fc receptor (FcR) binding sites: The leukocyte receptors Fc γ RI and Fc γ RIIa bind to a region in the Fc distinct from that recognized by neonatal FcR and protein A. J. Immunol. 164, 5313–5318 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Becker S., Frankel M. B., Schneewind O., Missiakas D., Release of protein A from the cell wall of Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 111, 1574–1579 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forsgren A., Svedjelund A., Wigzell H., Lymphocyte stimulation by protein A of Staphylococcus aureus. Eur. J. Immunol. 6, 207–213 (1976). [DOI] [PubMed] [Google Scholar]
- 17.Pauli N. T., et al. , Staphylococcus aureus infection induces protein A-mediated immune evasion in humans. J. Exp. Med. 211, 2331–2339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H. K., Falugi F., Missiakas D. M., Schneewind O., Peptidoglycan-linked protein A promotes T cell-dependent antibody expansion during Staphylococcus aureus infection. Proc. Natl. Acad. Sci. U.S.A. 113, 5718–5723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H. K., Cheng A. G., Kim H.-Y., Missiakas D. M., Schneewind O., Nontoxigenic protein A vaccine for methicillin-resistant Staphylococcus aureus infections in mice. J. Exp. Med. 207, 1863–1870 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H. K., Falugi F., Thomer L., Missiakas D. M., Schneewind O., Protein A suppresses immune responses during Staphylococcus aureus bloodstream infection in guinea pigs. MBio 6, e02369-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X., Sun Y., Missiakas D., Schneewind O., Staphylococcus aureus decolonization of mice with monoclonal antibody neutralizing protein A. J. Infect. Dis. 219, 884–888 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganesh V. K., et al. , Lessons from the crystal structure of the S. aureus surface protein clumping factor A in complex with tefibazumab, an inhibiting monoclonal antibody. EBioMedicine 13, 328–338 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim H. K., et al. , Protein A-specific monoclonal antibodies and prevention of Staphylococcus aureus disease in mice. Infect. Immun. 80, 3460–3470 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Loghem E., Frangione B., Recht B., Franklin E. C., Staphylococcal protein A and human IgG subclasses and allotypes. Scand. J. Immunol. 15, 275–278 (1982). [DOI] [PubMed] [Google Scholar]
- 25.Sauer-Eriksson A. E., Kleywegt G. J., Uhlén M., Jones T. A., Crystal structure of the C2 fragment of streptococcal protein G in complex with the Fc domain of human IgG. Structure 3, 265–278 (1995). [DOI] [PubMed] [Google Scholar]
- 26.Cruz A. R., et al. , Staphylococcal protein A inhibits complement activation by interfering with IgG hexamer formation. Proc. Natl. Acad. Sci. U.S.A. 118, e2016772118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai H.-S., et al. , The Fc domain of immunoglobulin is sufficient to bridge NK cells with virally infected cells. Immunity 47, 159–170.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng A. G., et al. , Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 23, 3393–3404 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X., et al. , Glycosylation-dependent opsonophagocytic activity of staphylococcal protein A antibodies. Proc. Natl. Acad. Sci. U.S.A. 117, 22992–23000 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomer L., et al. , Antibodies against a secreted product of Staphylococcus aureus trigger phagocytic killing. J. Exp. Med. 213, 293–301 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pokhrel S., et al. , Complement receptor 3 contributes to the sexual dimorphism in neutrophil killing of Staphylococcus aureus. J. Immunol. 205, 1593–1600 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukácsi S., Nagy-Baló Z., Erdei A., Sándor N., Bajtay Z., The role of CR3 (CD11b/CD18) and CR4 (CD11c/CD18) in complement-mediated phagocytosis and podosome formation by human phagocytes. Immunol. Lett. 189, 64–72 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Gordon D. L., Rice J. L., McDonald P. J., Regulation of human neutrophil type 3 complement receptor (iC3b receptor) expression during phagocytosis of Staphylococcus aureus and Escherichia coli. Immunology 67, 460–465 (1989). [PMC free article] [PubMed] [Google Scholar]
- 34.Burmeister W. P., Huber A. H., Bjorkman P. J., Crystal structure of the complex of rat neonatal Fc receptor with Fc. Nature 372, 379–383 (1994). [DOI] [PubMed] [Google Scholar]
- 35.Kitamura D., Roes J., Kühn R., Rajewsky K., A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature 350, 423–426 (1991). [DOI] [PubMed] [Google Scholar]
- 36.Prokesová L., et al. , Cleavage of human immunoglobulins by serine proteinase from Staphylococcus aureus. Immunol. Lett. 31, 259–265 (1992). [DOI] [PubMed] [Google Scholar]
- 37.Pietrocola G., Nobile G., Rindi S., Speziale P., Staphylococcus aureus manipulates innate immunity through own and host-expressed proteases. Front. Cell. Infect. Microbiol. 7, 166 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Booth B. J., et al. , “Extending human IgG half-life using structure-guided design” in MAbs (Taylor & Francis, 2018), pp. 1098–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petkova S. B., et al. , Enhanced half-life of genetically engineered human IgG1 antibodies in a humanized FcRn mouse model: Potential application in humorally mediated autoimmune disease. Int. Immunol. 18, 1759–1769 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Ober R. J., Radu C. G., Ghetie V., Ward E. S., Differences in promiscuity for antibody-FcRn interactions across species: Implications for therapeutic antibodies. Int. Immunol. 13, 1551–1559 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Kimmig A., et al. , Management of Staphylococcus aureus bloodstream infections. Front. Med. (Lausanne) 7, 616524 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tong S. Y., Davis J. S., Eichenberger E., Holland T. L., Fowler V. G. J. Jr., Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sause W. E., Buckley P. T., Strohl W. R., Lynch A. S., Torres V. J., Antibody-based biologics and their promise to combat Staphylococcus aureus infections. Trends Pharmacol. Sci. 37, 231–241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skov R., et al. , Update on the prevention and control of community-acquired meticillin-resistant Staphylococcus aureus (CA-MRSA). Int. J. Antimicrob. Agents 39, 193–200 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Sasso E. H., Silverman G. J., Mannik M., Human IgM molecules that bind staphylococcal protein A contain VHIII H chains. J. Immunol. 142, 2778–2783 (1989). [PubMed] [Google Scholar]
- 46.Ljungberg U. K., et al. , The interaction between different domains of staphylococcal protein A and human polyclonal IgG, IgA, IgM and F(ab’)2: Separation of affinity from specificity. Mol. Immunol. 30, 1279–1285 (1993). [DOI] [PubMed] [Google Scholar]
- 47.Sun Y., et al. , Staphylococcal protein A contributes to persistent colonization of mice with Staphylococcus aureus. J. Bacteriol. 200, e00735-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiao S.-W., et al. , Dependence of antibody-mediated presentation of antigen on FcRn. Proc. Natl. Acad. Sci. U.S.A. 105, 9337–9342 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.J. J. Weems, Jr, et al. , Phase II, randomized, double-blind, multicenter study comparing the safety and pharmacokinetics of tefibazumab to placebo for treatment of Staphylococcus aureus bacteremia. Antimicrob. Agents Chemother. 50, 2751–2755 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y., Huo M., Zhou J., Xie S., PKSolver: An add-in program for pharmacokinetic and pharmacodynamic data analysis in Microsoft Excel. Comput. Methods Programs Biomed. 99, 306–314 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.