Abstract
Green plants play a fundamental role in ecosystems, human health, and agriculture. As de novo genomes are being generated for all known eukaryotic species as advocated by the Earth BioGenome Project, increasing genomic information on green land plants is essential. However, setting standards for the generation and storage of the complex set of genomes that characterize the green lineage of life is a major challenge for plant scientists. Such standards will need to accommodate the immense variation in green plant genome size, transposable element content, and structural complexity while enabling research into the molecular and evolutionary processes that have resulted in this enormous genomic variation. Here we provide an overview and assessment of the current state of knowledge of green plant genomes. To date fewer than 300 complete chromosome-scale genome assemblies representing fewer than 900 species have been generated across the estimated 450,000 to 500,000 species in the green plant clade. These genomes range in size from 12 Mb to 27.6 Gb and are biased toward agricultural crops with large branches of the green tree of life untouched by genomic-scale sequencing. Locating suitable tissue samples of most species of plants, especially those taxa from extreme environments, remains one of the biggest hurdles to increasing our genomic inventory. Furthermore, the annotation of plant genomes is at present undergoing intensive improvement. It is our hope that this fresh overview will help in the development of genomic quality standards for a cohesive and meaningful synthesis of green plant genomes as we scale up for the future.
Keywords: annotation, reference genome, transcriptomes, Viridiplantae, whole-genome duplication (WGD)
The nearly half-million species of green plants (Viridiplantae) on the planet today (1, 2) are fundamental drivers of global ecosystems and are critical to human health and well being via their enormous contributions to agriculture, medicine, and natural ecological processes (3, 4). As the world is challenged as never before by habitat destruction, species loss, climate change, and dramatically modified community interactions, plant genomic information is critical to finding plant-based solutions to the existential challenges we face today. Yet, our knowledge of green plant genomes lags far behind that of some major clades on the tree of life, such as vertebrates, for which most genomics technologies have been tested and developed. Although around 13% (812 species) of the 6,480 eukaryotic species with whole-genome sequence information are green plants (International Nucleotide Sequence Database Collaboration [INSDC]) (5) (see below), fewer than 0.2% of all green plant species are represented by these 812 species.
The Earth BioGenome (EBP) (6–8) is a large, multiinstitutional consortium (currently 36 institutions in 16 countries) that aims to coordinate and unify the construction of de novo genomes across all eukaryotic life and has emphasized the importance of setting standards in methodology and genome quality in light of fast-advancing sequencing technologies. The EBP has facilitated conversations among genome initiatives focused on clades of animals (e.g., Vertebrate Genomes Project [VGP], 5000 Insect Genomes [i5K], Global Invertebrates Genome Alliance [GIGA]) resulting in a heightened recognition of the great heterogeneity in genome size and complexity that exists across these clades. However, the challenges of setting quality standards for genomic data across other diverse clades of life, including unicellular eukaryotes, fungi, other invertebrates, arthropods, and multiple lineages of plants (e.g., red algae, glaucophytes, and green plants), are immense.
The goals of generating genomes for all eukaryotic life on Earth are to revise and reinvigorate our understanding of biology, ecosystems, and evolution; to enable the conservation, protection, and regeneration of biodiversity; and to maximize returns on genomic information to society and human welfare (6, 8). Complete reference-quality genomes for all green plants will open up new scientific avenues, for example, to decipher the origin and phylogeny of plant life and the genomic basis of speciation, to determine how many species are currently on the planet, to unveil the genetic control of specialized plant traits, to track the complex functioning of ecosystems through species interactions, to preserve the genotypes of species before they disappear, to discover natural botanical compounds, which can be used to cure diseases of humans and other species, and to enhance the quality of our lives through improved crops (8).
The rapidly evolving palette of genomic and transcriptomic data available for green plants (9–12) (SI Appendix, Table S1) is advancing our knowledge of how genome size and complexity of Viridiplantae compare to other clades of life. Multispecies, genome-enabled analyses of chromatin structure are yielding new insights into the nature of plant gene regulatory elements and how they differ from regulatory elements in metazoan genomes (13). Comparative analyses aimed at effective translation of genotype to phenotype and meaningful interpretation of plant genome structure and evolution more generally will require a set of quality standards that can be applied across all green plants. Such standards will need to accommodate the immense variation in genome size, transposable elements (TEs), and other repetitive DNA content, ploidy, and related aspects of genome structure across the green tree of life (14), while enabling research into the molecular and evolutionary processes that have spawned this variation.
Here we provide an overview and assessment of the current state of knowledge of green plant genomes. We also address the challenges confronting the field of plant genomics in planning for genome sequencing programs and in setting quality standards. How do green plant genomes vary in size, structure, and complexity and differ from vertebrate, arthropod, and microbial genomes? How many green plant genomes have been sequenced, and how are these species with available genomes distributed across the plant clade? Which are the “dark clades” of green plant life that currently have little or no current genomic information? How complete are the available green plant genomes, and do they meet emerging standards for contiguity and quality? How do we establish a reasonable and practical set of standard goals for genome assembly and annotation sufficient to address specific questions about genome content, structure, and function? And how do we scale up in the future to focus finite resources and energies? Although current genome sequences for green plants are not always easily comparable because of extreme variations in assembly accuracy and structural annotations, the quality of green plant genomes are rapidly improving as advances in sequencing and genome assembly technologies are reducing costs and becoming more widely adopted.
Our goal is to provide a fresh and current overview of green plant genomes. Following a brief summary of what is known about size, complexity, and the extent of present-day data on genomes in green plants, we provide an evaluation of reference genomes for plants and what is required to characterize such genomes. An outline of the seemingly simple but actually daunting tasks of sampling green plants worldwide in compliance with relevant international policies and sequencing and annotating their diverse genomes is followed by our vision for moving the field of green plant genomes forward to meet the challenge of the EBP. We hope that the perspectives presented here will encourage global collaboration and participation, help set genomic quality standards across Viridiplantae, and develop strategies to guide current and future plant genome projects.
Welcome to the Real World: Green Plants
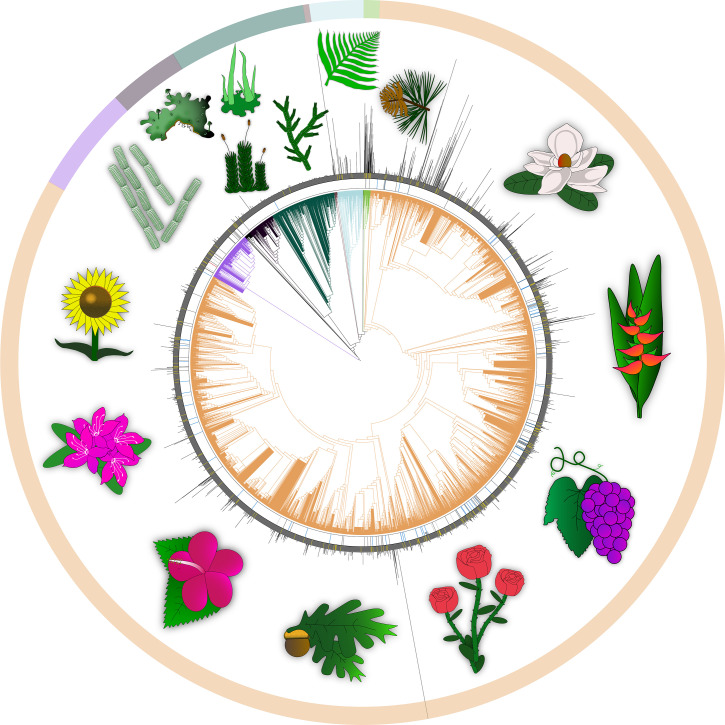
There is a huge disparity in genome size and complexity across the diverse clades of life. Many vertebrate genomes (especially mammals, birds, reptiles, and frogs) have relatively small, compact genomes ranging from 1 pg to 10 pg (i.e., ∼978 Mb to 9.8 Gb; animal genome size database; http://genomesize.com/statistics.php). However, across all animals there is an enormous range of variation (4,000-fold) from 0.02 pg (19.6 Mb) for a nematode (Pratylenchus coffeae), to 132.83 pg (129.9 Gb) for the lungfish (Protopterus aethiopicus) (http://genomesize.com/statistics.php). Among land plants (Fig. 1 and Dataset S1), genome size and composition vary similarly (largely within angiosperms) with a nearly 3,000-fold range in DNA content from a 1C value of 0.07 pg (65 Mb) for species of the corkscrew plant (Genlisea) to 152.2 pg (148.9 Gb) for the canopy plant (Paris japonica), one of the largest known genomes (15, 16) (https://cvalues.science.kew.org/). Mechanisms primarily responsible for the large variation in genome size observed in green plants include repeated episodes of whole-genome duplication (WGD; polyploidy), which is widespread in plants, especially ferns and flowering plants (11), and the dynamics of TE loss and gain. The number of protein-coding genes is fairly consistent in green plants: based on transcript-supported annotations the typical number is ∼40,000 (17), but with a range from 19,623 genes in duckweed (Spirodela polyehiza) to ∼50,000 in the tetraploid burclover (Medicago trunculata) and ∼75,000 in tetraploid cottons (Gossypium hirsutum and several other species) (18–21). Green plants, therefore, appear to have a higher number of genes than is observed in vertebrates, but with extensive variation and an increase in gene number due to high levels of recent as well as ancient polyploidy (11, 22, 23).
Fig. 1.
Light purple, green algae; black, liverworts; dark green, bryophytes; brown, hornworts; light blue, ferns and lycophytes; light green, gymnosperms; and orange, angiosperms. The inner circle shows the current state of genome sequencing with complete genome assemblies shown as red bars, chromosome-level assemblies as blue, scaffold assemblies as dark gray, and contig assemblies as light gray. The outer circle (filled in gray) shows taxa indicated with yellow bars for which transcriptome data are available. Genome data were surveyed from GenBank, European Molecular Biology Laboratory ( EMBL), and DNA Data Bank of Japan (DDBJ) in October 2020. Lines radiating out from the circle show genome sizes as C values (genomesize.com). The phylogenetic framework for all plants to genus level was extracted from the Open Tree of Life (opentreeoflife.org) in October 2020. Full supporting data are available in Dataset S1. Thanks to Keith Crandall and David Stern for assistance with this figure, with help from M.R.G.
Genome content also remains a source of complexity due to abundant pseudogenes, variable gene family expansions (some associated with chemical defense), and transposon activity (24). With regard to the latter, variation in the gain-and-loss dynamics of TEs is the greatest source of plant genome size variation, making plant genomes far more complex than vertebrate genomes (25). The relationship between plant genome size and TE content is generally linear within a given ploidy level (26) with TEs comprising as little as 3% in some genomes to nearly 85% in others; the larger the genome, the larger the percentage devoted to TE content.
Compared to the 72,478 species of vertebrates (27), the sheer size of the green plant clade, especially the angiosperms with ∼370,000 species (2), represents a huge challenge for genome sequencing as well as exciting opportunities for genome biologists. Over 70% of all species of flowering plants have not been sequenced for any DNA region, let alone a complete genome (28). Moreover, given their enormous habitat diversity and often remote, localized distributions, many plant species can only be sampled readily from herbarium specimens, limiting both the quantity and quality of DNA for gene and genome sequencing. Even more challenging is that green plants host highly diverse communities of microorganisms, including bacteria, fungi, protists, nematodes, and viruses (i.e., the plant microbiome), which occur in every accessible plant tissue—leaves, shoots, and roots (29). However, such microbiomes are also found in nonplants, increasing the difficulty of DNA extraction in almost all taxa. It is well known that numerous plant species contain large amounts of specialized metabolites, including polysaccharides, polyphenols, and other secondary metabolites that can interfere with DNA and RNA extraction and purification (30). These compounds can result in greatly reduced DNA/RNA yield and quality.
The State of the Art of Plant Genomes
The norm in the genomics research community is for DNA sequence data to be submitted to public archives, usually those maintained by the INSDC (5). These archives as of November 2020 contain 1,139 genomes from 812 species of Viridiplantae. Of these 812 species, 543 are angiosperms, 11 species are gymnosperms, 5 species are ferns and lycophytes, 8 species are bryophytes, and 249 species are green algae (Fig. 1 and Dataset S1). The genome assemblies range in size from 12 Mb (the genome of the green algal insect parasite Helicosporidium) to 27.6 Gb (the genome of the sugar pine, Pinus lambertiana), with a median of 517 Mb and a mean of 1.21 Gb. The taxonomic distribution is quite asymmetric and biased toward agricultural crops. For example, 135 genomes come from just one family, Poaceae, which contain the cereal grasses. Among these, 46 are accessions from the genus Oryza (rice). Brassicaceae (mustards) and Fabaceae (legumes) are also very well represented, with 96 and 55 complete genomes, respectively.
Many published plant genomes, although mostly complete at the sequence level, have highly fragmentary assemblies. Since 2016, the Vertebrate Genome Project (VGP) has been producing assemblies of diverse vertebrate genomes with a contig N50 of at least 1 Mb and a scaffold N50 of at least 10 Mb (two elements of the wider 3.4.2.QV40 phased metric). Of plant genomes, 302 meet the contig standard, 398 meet the scaffold standard, and only 232 meet both. Encouragingly, while only four plant genome assemblies meeting both standards were submitted prior to 2018, i.e., 0.6% of all plant genome assemblies submitted up to that date, 150 were submitted in 2020 alone, i.e., 36% of all assemblies submitted that year, indicating the increased power of recently developed sequencing and assembly methods. The largest archived assembly to date that meets both VGP standards is that of the 2.7-Gb genome of the opium poppy, Papaver somniferum (31). Of the 70 plant assemblies submitted so far in 2021, 8 have a contig N50 >10 Mb (including a new assembly of barley with a haploid genome size of 5.1 Gb and a contig N50 length of 69.6 Mb) (32). This is broadly in line with results now achieved for vertebrate genomes; for comparison, in 2020, the VGP published 13 assemblies with an N50 >10 Mb (https://hgdownload.soe.ucsc.edu/hubs/VGP/).
We emphasize that given the variation in size and structure of plant genomes, alternative quality standards may be more informative for plants. For example, a scaffold N50 of 10 Mb for a large genome with few chromosomes (some of which may be an order of magnitude larger than this size) may not be as informative as a standard that is scaled to the size of the genome. In any case, the quality and contiguity of assemblies for complex plant genomes is greatly improving with increasing accuracy of long-read sequences and evolving technologies for assembling contigs into chromosomal scaffolds for both haplotypes in diploid genomes. Undoubtedly, the accuracy of large, complex plant genome assemblies will improve yearly over the next decade or more. Rather than setting static minimal standards, we advocate best practices for optimizing assembly accuracy given continually improving state-of-the-art technologies.
What Do We Mean by a “Genome” in Plants? Why Are High-Quality Genomes Needed?
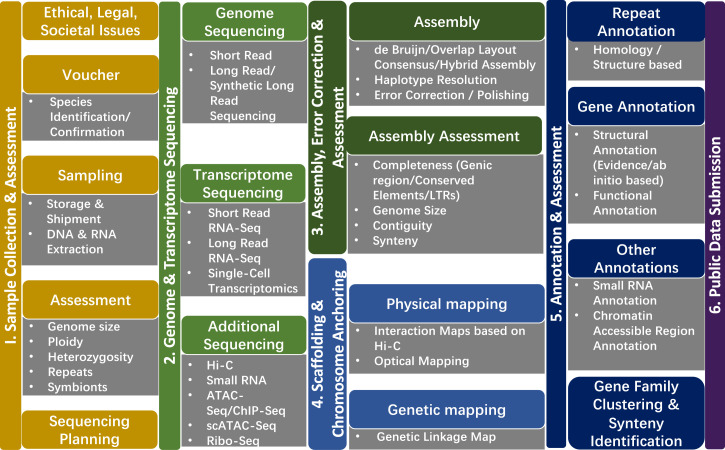
At its most complete, a reference genome conveys both the nucleotide sequence of all chromosomes and structural information (an annotation) that describes the arrangement of genes relative to each other, to noncoding sequences, to centromeres, and to chromosome ends (Fig. 2). However, most assembly and annotation efforts to date fall short of this comprehensive ideal and instead provide an estimate of gene space, i.e., the coding region of the genome. A “reference genome” is a standard against which other genomes can be compared and is typically derived from a single individual of a species, providing a basis for comparison with other individuals of the same species and with other species. However, what constitutes the “ideal reference genome” is not necessarily clear (33), and the concept of reference genome varies among research and user communities depending on factors ranging from genome size and complexity to resources and goals to be addressed with genomic data. Further phased versioning of genomes is required as the reference genome for a given species or genotype evolves from a compilation of gene space contigs to a chromosome assembly in which entire molecules are scaffolded (34) with chromosomal localization of genes. Moreover, the concept of reference genome is evolving; whereas it once referred to the genome of a single organism, the ideal reference genome today may actually be the pangenome (35).
Fig. 2.
A generalized plant genome workflow from sample collection through assembly and annotation to public data submission. The workflow follows from Left to Right. 1) Sample Collection and Assessment (yellow) to ethically and legally collect, identify, and voucher the reference specimen; obtain, store, ship, and extract DNA/RNA from the samples; as well as assess its biological qualities. 2) Genome and Transcriptome Sequencing (green) conducted using short- and long-read sequencing technologies. 3) Assembly, Error Correction, and Assessment (dark green) to determine sequence contiguity, completeness, and accuracy. 4) Scaffolding and Chromosome Anchoring (blue) to evaluate, elongate the scaffolds, and anchor the chromosomes. 5) Annotation and Assessment (dark blue) integrates transcriptomic resources and targeted sequencing to identify protein coding, repeat, and regulatory regions, and to provide biological context to the identified elements. 6) Public Data Submission (purple) to ensure open access of the sequence data and the derived assemblies and annotation.
Given the size and complexity of many plant genomes, the first step is typically to generate short-read data for preliminary genome assessment and gene space assembly. In fact, meeting the EBP’s goal of producing a genome sequence to represent each taxonomic family within 3 y (6) for many plant taxa, we may rely on short-read sequencing and assembly. This will yield draft assemblies sufficient for estimates of gene space and repeat content, but of limited utility for investigations of chromosomal organization. While it may be argued that these compilations of short-read assemblies should not be referred to as “genome sequences,” historical precedent has labeled them as such. In addition, such genomes, despite their shortcomings for evaluating genome structure, may contribute substantially to initial inferences of genome content and possibly even organization by revealing complexities that may result from gene family expansion or whole-genome duplication. Moreover, they may be informative regarding repeat evolution, provide markers for plant phylogeny, generate hypotheses for further investigation, and serve as important first steps toward more contiguous genome sequences. Nonetheless, we urge reserving the term genome for chromosome-level assemblies and explicitly referring to gene space or short-read assemblies as such.
The addition of long-read data can greatly improve the contiguity of assemblies based on short reads and enhance the value of short-read genome assemblies by improved inferences of gene and genome duplication, genome structure and rearrangements, and orthology (36). With the advent of PacBio HiFi sequencing (37), which produces highly accurate 10 to 30-kb circular consensus sequence (CCS) reads, ultralong Oxford Nanopore long-read protocols (38), Hi-C scaffolding (39), and optical mapping (e.g., BioNano) (40, 41) technologies, it is now possible to generate chromosomal and even haplotype assemblies of highly complex genomes. Innovations in sequencing technology, coupled with reductions in per-base sequencing costs, enable massive production of long-read data and are paving the way for chromosome-level assemblies for even the largest plant genomes. For example, a chromosome-level assembly for the model fern, Ceratopteris richardii (n = 39; 7.46 Gb of 9.25 Gb genome assembled), was recently completed as part of the Open Green Genomes project (https://phytozome-next.jgi.doe.gov/info/Crichardii_v2_1), and chromosomal assemblies of polyploid genomes are now possible (21, 42–44). Such high-quality genomes enable full-scale within- and between-genome synteny analyses that provide foundational views of genome structure, possible function, and evolution. Long-read sequencing of multiple genotypes within species is revealing structural variation contributing to trait variation (45). With recent advances in long-read sequencing and assembly technologies, such structural variation is evident in chromosomal haplotype assemblies for polyploid and highly heterozygous genomes (e.g., refs. 32, 44, 46). Until recently, such high-quality genomes were primarily produced for inbred crop and genetic model species, but they are increasingly emerging for plant species outside these criteria (e.g., Lindenbergia, Acorus, Joinvillea, and Pharus; https://phytozome-next.jgi.doe.gov/ogg/). As technologies advance, a plant reference genome will be a chromosome-level assembly that represents both the conserved and variable gene (and noncoding) regions of the genome across multiple individuals (pangenomes) and will serve multiple uses for diverse research communities, from plant breeding to genome evolution to function and adaptation.
Sampling the Dark Clades of the Green Tree of Life
Large branches of the green tree of life are not represented by any genomic DNA sequencing data (Fig. 1). The National Center for Biotechnology Information (NCBI) taxonomy contains just 155,935 species of green plants, and for many of these species only a limited number of genes has been sequenced. The 812 plant species with significant genome information are only a fraction of the over 412,000 species reported for Viridiplantae (∼8,000 green algal species, ∼20,000 species of bryophytes, ∼13,000 species of ferns and lycophytes, ∼1,000 species of gymnosperms, and ∼370,000 species of angiosperms) (2). Moreover, the true numbers of species within some of these taxonomic groupings for the most part remain unknown. Among the chlorophyte algae, 71 genomes are from just two classes, the Chlorophyceae and Trebouxiophyceae. Within land plants, the statistics above document the poor sampling of ferns and bryophytes (including mosses, hornworts, and liverworts). These taxa dominate some terrestrial ecosystems and play an important role in global carbon and nitrogen cycling (47). A chromosomal assembly for giant sequoia (Sequoiadendron giganteum) has recently been published (48), but most assemblies of large gymnosperm genomes are highly fragmented, and there is still room to improve contiguity of chromosomal scaffold assemblies. Of the 416 families of angiosperms (49), a full genome sequence exists for representatives of only 130, and 56 have just a single representative.
Locating suitable material of these missing taxa, obtaining fresh tissue, extracting high-molecular-weight DNA, and carrying out long-read sequencing to produce high-quality genome assemblies, scaffolds, and annotations, fall within the ambitions of the EBP, but will likely prove a lengthy and expensive task (Fig. 2). However, the majority of plant taxa have been taxonomically described, collected, and stored as dried specimens in herbaria worldwide. The ultimate goal of producing de novo reference genomes from across the plant tree of life will require well-preserved tissues collected specifically for use in genome sequencing projects in a way that minimizes degradation by rapid desiccation, freezing, and/or preservation.
It should be noted that although DNA degrades under herbarium storage, short-read sequencing is often still possible for phylogenomic analyses and characterization of genetic variation. Targeted capture and sequencing of specific genomic regions (50) and low-coverage shotgun (skim) sequencing (51) have been used to assemble and analyze nuclear genome segments and whole plastid genomes from herbarium specimens, including historical collections (52–55). Target capture and skim sequence data do not permit full nuclear genome assembly, but they can be extremely useful for phylogenetic reconstruction, allowing the exploration of evolutionary relationships and the testing of species hypotheses, in the absence of live material for inaccessible species (56).
To achieve the scale of genome sequencing highlighted here, a well-coordinated sampling strategy is encouraged that combines phylogenetic-based sampling with practicality that accounts for the diversity and complexity of the lineage. The Global Genome Initiative for Gardens (GGI-Gardens) (57) has proposed a sampling approach that aims to cover maximum phylogenetic distance by setting three targets, including the collection of at least one sample from 1) each family, 2) 50% of all genera, and 3) all species of vascular plants on Earth. Targets 1 and 2 should be complete for plants within the next 2 y. The gap analysis tool (https://globalgeno.me) developed by the GGI is used by GGI-Gardens to identify and prioritize genomic samples for collection and preservation (58). Collection priority must also account for the complexity of plant genomes. As de novo reference genomes are established for families and genera of green plants, care should be taken to select species with smaller and ideally diploid genomes. Taxa that are not well represented in living collections (i.e., nonvascular plants) will also require participation of specialists to provide tissue.
An invaluable source of fresh green plant material for genomic analysis lies in the vast diversity of living collections in botanic gardens around the world. Data from Botanic Gardens Conservation International (BGCI)’s Plant Search database (https://tools.bgci.org/plant_search.php) demonstrate that these living collections are home to at least 30% of the species, nearly 60% of the genera, and 75% of the families of vascular plants (59). The potential for botanic gardens to facilitate large-scale genomic research has been recognized for nearly a decade (60, 61), but until recently, large, coordinated collection efforts have not been feasible. The GGI-Gardens program was founded in 2015 to leverage living collections for the preservation of genome-quality tissue samples across the plant tree of life (62).
Since its inception, GGI-Gardens has collected tissues from more than 400 families and 4,500 genera of vascular plants for genomic work. To date, the partnership includes 38 botanical institutions around the world with 28 active collection programs. Each collection includes an herbarium voucher deposited in a recognized herbarium and at least one genomic tissue voucher of either silica dried and/or flash frozen tissue in liquid nitrogen (63), which are stored in biorepositories that are part of the Global Genome Biodiversity Network (GGBN) (64). All GGBN tissue vouchers and their associated metadata are made publicly available through the GGBN web portal (https://www.ggbn.org/ggbn_portal/). However, achieving comprehensive sequencing for all species will require even more ambitious efforts that focus on fieldwork, especially in the most biodiverse countries.
Participation in the global biodiversity genomics effort also requires an acute familiarity with international policies governing plant genetic resources, such as the International Treaty on Plant Genetic Resources for Food and Agriculture (ITPGRFA, https://fao.org/plant-treaty) (65) and the Nagoya Protocol, https://www.cbd.int/abs) (66), which stipulate policies for access and benefit sharing (ABS) of plant genetic resources (Fig. 2). Researchers can navigate these policies through help from the ABS Clearing House (https://absch.cbd.int), BGCI (67), and GGBN (68) to understand whether their genomics projects are compliant with international rules (see https://www.bgci.org/resources/bgci-tools-and-resources/abs-learning-package/).
The Rest of the Story: Annotation
The penultimate stage in completing a genome is structural and functional annotation, which provides both physical and biological context to the assembled sequence data (Fig. 2). A high-quality annotation is necessary to identify locations of coexpressed genes, the proximity of a variant to candidate genes, or regions associated with chromatin availability. Variation in the number of genes and their structure, provides a framework for examining morphological and physiological traits (35). Annotation artifacts may lead to incorrect inference of gene family evolution and function.
Genome annotation remains a time-consuming and computationally intensive process that combines numerous types of sequence analysis and heuristic prediction (69). Transcript evidence should sample from multiple tissue types. The process typically consists of, at minimum, 1) repeat masking of the genome, 2) using splice-aligning transcripts and proteins from the same/related species for evidence-based gene structure prediction, 3) using ab initio gene-finding algorithms to annotate possible gene structures, 4) combining the above data sources to create a set of possible gene structures, and 5) filtering the results through quality filters to find the most probable set of structures that represent full-length or near-full-length coding regions. Interest in resolving structures, such as long noncoding RNA (lnc-RNAs), an effort that requires additional analysis (70), is also increasing. Assessments of annotated plant genomes revealed that model systems, such as Arabidopsis and rice (Oryza), benefit from community-sourced improvements to existing references, while most plant genomes receive very little manual or computational intervention after public release (71, 72). Common errors include gene assignments to retroelements, conflicting models, frame inconsistencies, fragmented models, contaminant-sourced models, false positive pseudogenes, missed gene models, and structural errors (72, 73).
Even with substantial external evidence, annotation remains challenging and requires careful assessment (and reassessment) (74, 75). The genomics community generally favors the assembly of more genomes over revised genome annotations for existing references. This dichotomy has led to persistent errors that are propagated through public repositories that host and integrate the associated data (76). At the same time, highly curated and frequently updated genome annotations for model and crop plant species have major impacts on our understanding of gene structure, regulation, and function. Similarly, pangenome analyses are expanding our knowledge of gene function and genome dynamics. These deep, taxonomically focused (typically single species) resources will provide an ideal framework for gene annotation improvement in green plants (35).
For complex green plant genomes, current workflows for genome annotation remain a limitation. More efficient and accurate algorithms, alongside comprehensive transcriptomic resources, are needed. Annotation workflows should consider the likely signatures of WGD, fragmentation, and potential assembly error in the process. The repeat determination for highly repetitive plant genomes must integrate de novo, similarity, and structural identification methods for robust masking. In terms of external evidence for exonic support, use of long-read technologies, such as PacBio Iso-Seq or Oxford Nanopore, should be employed to generate full-length transcripts (77). Massive introns (≫100 kb) are not uncommon in large-genome plant species, and long-read transcript sequence data are enabling full-length annotation of these genes (48). In general, transcriptomic data should be sourced from the reference individual and be derived from a deep sampling of tissues and developmental stages. The continued reliance on short-read data, before and after de novo assembly, propagates errors into the process (78, 79). At the same time, it should be noted that long-read–derived assemblies (or transcripts) without adequate coverage can also introduce errors into the annotation process (80). Regardless of the source of the transcriptomic reads, annotation frameworks should extend their ab initio models to include features beyond primary sequence, such as RNA folding and functional motifs. For plant genomes with both multicopy genes and abundant pseudogenes, it may be necessary for workflows to additionally utilize ATAC-Seq (assays for transposase-accessible chromatin using sequencing for detection of open chromatin), ChIP-Seq (chromatin immunoprecipitation with parallel DNA seqeuncing for histone modifications), and Ribo-Seq (ribosome profiling) data for characterization of translation initiation sites (81, 82). ATAC-Seq and ChIP-Seq can also aid in identification and comparative analyses of cis-regulatory elements in sequenced plant genomes (13).
A Roadmap for Moving Forward with Green Plants
With whole-genome sequence data available for fewer than 0.2% of green plant species, we have a long way to go before even a majority of taxa, whether families, genera, or species, are characterized by reference genomes as envisioned by the EBP. Yet, we are confident that the final goal of the EBP “to sequence and annotate the genomes of all currently known eukaryotic species in 10 years” (6, 7) can be achieved for green plants. To move forward, researchers must consider efficient sampling schemes; genome quality standards sufficient to answer specific biological questions; optimal strategies for sequencing particular types of plant genomes; the incorporation of new sequencing, scaffolding, and annotation technologies; and selecting appropriate bioinformatics tools and workflows. Each of these efforts will need to be tackled separately, but all must be considered collectively for success.
Sampling schemes for green plants should continue to be based on a phylogenetic framework as advocated by the EBP (6–8) and employed in the One Thousand Plant Transcriptomes Initiative (1KP) (11, 83), the Open Green Genomes project (https://phytozome-next.jgi.doe.gov/ogg/), and the 10KP genomes project (12) (SI Appendix, Table S1). For the many clades of green plants that still lack a reference genome or are in need of additional genome sequencing efforts, it may be possible in some instances to focus on exemplars with small genome sizes. However, for some major clades of Viridiplantae in which all species have large genomes (e.g., gymnosperms and the flowering plant order Liliales with more than 1,750 species) the generation of chromosomal genome assemblies will require continued technological advances and substantial financial and human resources. At present, identification of phylogenetic gaps in available chromosomal genome assemblies is an important guide for prioritizing efforts to assemble and annotate large plant genomes.
As sequencing, assembly, and annotation costs are further reduced and long-read sequencing technologies improve, we are confident that it will be feasible to generate chromosomal assemblies for comparative genome analyses for taxa with large genomes. Until that time, no one set of recommendations can accurately define the quality for all plant genome assemblies, which in large part need to be assessed on the scientific questions for which they are being assembled. Many questions about plant phylogenetic relationships, taxonomic diversity, and ecological interactions can be examined with short-read assemblies, i.e., questions on gene diversity or gene evolution do not require a chromosome-level assembly. For questions focused on evolutionary change or conservation of gene order (synteny), or changes in genome content and structure following polyploidization, a chromosome-level assembly is essential.
Benchmarks for completeness and quality among green plant genome assemblies must assess both the assembly and annotation of particular genome categories. Genome size estimates, derived from k-mer analysis of short-read data, should be used to inform the initial strategy in relation to size, ploidy, repetitive content, and potential contaminants/symbionts (84). Recent informatic advancements have facilitated the assessment of k-mer frequencies in polyploids and highly heterozygous genomes with tools such as GenomeScope2 and Smudgeplot (85). Very accurate long-read data can also be used for this estimation (86). When polyploid signals remain challenging to resolve from short reads alone, additional assessments with flow cytometry and/or karyotyping methods have tremendous utility (16, 87). The quantity and composition of the repetitive content can also inform the optimal long-read versus short-read strategy. It should be noted that other challenges, such as bacterial symbionts in many bryophytes, low-yield high-molecular-weight DNA extractions from diminutive species, or exceptionally large genomes seen in many gymnosperms, will impact the selection of sequencing inputs.
Great technological and analytical progress in recent years has remedied many of the challenges posed in the assembly of the large and complex genomes that typify many green plants (21, 36, 41–44). Furthermore, helpful workflows have been provided to assist in rapid de novo genome assembly (88). For example, deep PacBio HiFi and/or Oxford Nanopore long-read data can be used to generate large multimegabase contigs, and Hi-C data can be employed to scaffold these contigs and generate pseudochromosomes as well as provide utility for polishing. Other data, not the least of which is a chromosome count, are also crucial for generating assessing chromosome-level assemblies. However, only a small percentage of green plants have even a single reported chromosome count. Genetic maps or a reference genome for a neighboring species, can also be of great utility, but are seldom available for most nonmodel species of Viridiplantae.
Regardless of type of sequence data or the availability of supporting data types (e.g., genetic maps or optical maps), care should be taken in the selection of the appropriate informatic tools (Fig. 2), including error correction, de novo assemblers, haplotype phasing, scaffolding, and genome annotation, as informatic approaches change as fast as, if not faster than, the sequencing technologies available, and the variation across tools on the end product is tremendous (89). Furthermore, after assembly and annotation, traditional metrics associated with contiguity remain useful, but are likely not sufficient for the assessment of complex genomes, especially those in darker clades of the green tree of life. Aspects of synteny, paralogs, gene families, and repeat structure can inform researchers of challenges that may not be reflected in scaffold totals and N50/L50 values (90–92). Gene space completeness as evaluated by single-copy benchmarks, including universal single-copy orthologues (BUSCO), core gene families (CoreGFs), or the online platform for plant comparative genomes PLAZA, should be considered in light of species composition of the source databases and methodologies (72). Percentage of the genome assembly and estimated genome size in chromosome scaffolds may also be more informative than N50/L50 statistics when comparing assemblies of plant genomes.
As technologies improve and as new applications for genome data arise, it is inevitable that the quality of the genome assembly for a species will be improved, and the genomic data will move from one level of standard to the next. We believe that the generation of genomic data for green plants will greatly accelerate over the next decade and beyond, and that more taxon-specific best practices for sequencing, assembly, and annotation should be encouraged and supported by the community of plant scientists. Such community-developed standards should be encouraged alongside long-range plans for tissue acquisition and improved standards of access and benefit sharing for genetic resources, broader collaboration, global participation, and open channels of communication. These efforts will provide critical cost and time savings, ensure that efforts are not wasted in acquiring and sequencing the same taxa, and reduce unnecessary competition for limited resources. Furthermore, it is encouraging that the coalition of institutions and projects now participating in the EBP are starting to identify and secure funding from local, national, and regional sources to cover current and future sample collection and sequencing expenses [e.g., Saudi Arabia’s support for sequencing the genomes of native date palm species, Chile’s project to sequence the genomes of plant species of the Atacama Desert, the EBP-Colombia Project (93), the Darwin Tree of Life Project in the United Kingdom (94), the Catalan Initiative for the Earth BioGenome Project, and the African BioGenome Project]. Such place-based and habitat-based activities will ensure that native and often difficult to obtain taxa will be included in global genome priorities (7).
Despite the many unique aspects of plant genomes, the model as envisioned by the EBP for genome sequencing and data sharing across eukaryotic life can efficiently be applied to plants. We recognize that acquiring this level of achievement will require considerable cost and effort, including significant investment in training at all steps of the genome sequencing–assembly–annotation workflow and at all stages of the academic pipeline. Global investment in workforce development will improve the pace and quality of plant genome sequencing and will yield scientists with skills to address pressing problems in agriculture, conservation, and the emerging bioeconomy. For both large projects and small laboratories, the generation of plant genomes is expected to accelerate across the green tree of life in the near future (86). Although the challenges are great for completing a genomic inventory of green plants, we are confident it will succeed in this era of rapidly expanding horizons and opportunities.
Supplementary Material
Acknowledgments
We thank Drs. Keith Crandall and David Stern at the George Washington University for their assistance with Fig. 1. We also greatly appreciate the efforts of the Earth BioGenome Project for encouraging, facilitating, and setting the standards for generating complete genomes for all life on the planet.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2115640118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Corlett R. T., Plant diversity in a changing world: Status, trends, and conservation needs. Plant Divers. 38, 10–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lughadha E. N., et al. , Counting counts: Revised estimates of numbers of accepted species of flowering plants, seed plants, vascular plants and land plants with a review of other recent estimates. Phytotaxa 272, 82–88 (2016). [Google Scholar]
- 3.Schery R. W., Plants for Man (Prentice-Hall, ed. 2, 1972). [Google Scholar]
- 4.Keddy P., Evolutionary ecology of plant–plant interactions: An empirical modelling approach. Ann. Bot. (Lond.) 99, 372–374 (2007). [Google Scholar]
- 5.Arita M., Karsch-Mizrachi I., Cochrane G., The international nucleotide sequence database collaboration. Nucleic Acids Res. 49 (D1), D121–D124 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewin H. A., et al. , Earth BioGenome Project: Sequencing life for the future of life. Proc. Natl. Acad. Sci. U.S.A. 115, 4325–4333 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewin H. A., et al. , The Earth BioGenome Project 2020: Starting the clock. Proc. Natl. Acad. Sci. U.S.A. 119, e2115635118 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blaxter M., et al. , Why sequence all eukaryotes? Proc. Natl. Acad. Sci. U.S.A. 119, e2115636118 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen J. M., Folk R. A., Soltis P. S., Soltis D. E., Guralnick R. P., Biodiversity synthesis across the green branches of the tree of life. Nat. Plants 5, 11–13 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Kersey P. J., Plant genome sequences: Past, present, future. Curr. Opin. Plant Biol. 48, 1–8 (2019). [DOI] [PubMed] [Google Scholar]
- 11.One Thousand Plant Transcriptomes Initiative, One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng S., et al. , 10KP: A phylodiverse genome sequencing plan. Gigascience 7, 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Z., et al. , The prevalence, evolution and chromatin signatures of plant regulatory elements. Nat. Plants 5, 1250–1259 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Exposito-Alonso M., Drost H.-G., Burbano H. A., Weigel D., The Earth BioGenome project: Opportunities and challenges for plant genomics and conservation. Plant J. 102, 222–229 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Pellicer J., Fay M. F., Leitch I. J., The largest eukaryotic genome of them all? Bot. J. Linn. Soc. 164, 10–15 (2010). [Google Scholar]
- 16.Pellicer J., Leitch I. J., The Plant DNA C-values database (release 7.1): An updated online repository of plant genome size data for comparative studies. New Phytol. 226, 301–305 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Sterck L., Rombauts S., Vandepoele K., Rouzé P., Van de Peer Y., How many genes are there in plants (... and why are they there)? Curr. Opin. Plant Biol. 10, 199–203 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Proost S., Pattyn P., Gerats T., Van de Peer Y., Journey through the past: 150 million years of plant genome evolution. Plant J. 66, 58–65 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Michael T. P., Jackson S., The first 50 plant genomes. Plant Genome 6, 10.3835/plantgenome2013.03.0001in (2013). [DOI] [Google Scholar]
- 20.Michael T. P., Plant genome size variation: Bloating and purging DNA. Brief. Funct. Genomics 13, 308–317 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Chen Z. J., et al. , Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat. Genet. 52, 525–533 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van de Peer Y., Mizrachi E., Marchal K., The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Soltis D. E., Visger C. J., Soltis P. S., The polyploidy revolution then…and now: Stebbins revisited. Am. J. Bot. 101, 1057–1078 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Claros M. G., et al. , Why assembling plant genome sequences is so challenging. Biology (Basel) 1, 439–459 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiao W.-B., Schneeberger K., The impact of third generation genomic technologies on plant genome assembly. Curr. Opin. Plant Biol. 36, 64–70 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Lee S.-I., Kim N.-S., Transposable elements and genome size variations in plants. Genomics Inform. 12, 87–97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Betts J., et al. , A framework for evaluating the impact of the IUCN Red List of threatened species. Conserv. Biol. 34, 632–643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Folk R. A., et al. , Challenges of comprehensive taxon sampling in comparative biology: Wrestling with rosids. Am. J. Bot. 105, 433–445 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Trivedi P., Leach J. E., Tringe S. G., Sa T., Singh B. K., Plant-microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Friar E. A., Isolation of DNA from plants with large amounts of secondary metabolites. Methods Enzymol. 395, 3–14 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Guo L., et al. , The opium poppy genome and morphinan production. Science 362, 343–347 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Mascher M., et al. , Long-read sequence assembly: A technical evaluation in barley. Plant Cell 33, 1888–1906 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ballouz S., Dobin A., Gillis J. A., Is it time to change the reference genome? Genome Biol. 20, 159 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchant D. B., et al. , The C-Fern (Ceratopteris richardii) genome: Insights into plant genome evolution with the first partial homosporous fern genome assembly. Sci. Rep. 9, 18181 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bayer P. E., Golicz A. A., Scheben A., Batley J., Edwards D., Plant pan-genomes are the new reference. Nat. Plants 6, 914–920 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Michael T. P., VanBuren R., Building near-complete plant genomes. Curr. Opin. Plant Biol. 54, 26–33 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Wenger A. M., et al. , Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 37, 1155–1162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dumschott K., Schmidt M. H.-W., Chawla H. S., Snowdon R., Usadel B., Oxford Nanopore sequencing: New opportunities for plant genomics? J. Exp. Bot. 71, 5313–5322 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieberman-Aiden E., et al. , Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belser C., et al. , Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps. Nat. Plants 4, 879–887 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Liu J., et al. , Gapless assembly of maize chromosomes using long-read technologies. Genome Biol. 21, 121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X., Zhang S., Zhao Q., Ming R., Tang H., Assembly of allele-aware, chromosomal-scale autopolyploid genomes based on Hi-C data. Nat. Plants 5, 833–845 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Bertioli D. J., et al. , The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 51, 877–884 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Lovell J. T., et al. , Genomic mechanisms of climate adaptation in polyploid bioenergy switchgrass. Nature 590, 438–444 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alonge M., et al. , Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell 182, 145–161.e23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lovell J. T., et al. , Four chromosome scale genomes and a pan-genome annotation to accelerate pecan tree breeding. Nat. Commun. 12, 4125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elbert W., et al. , Contribution of cryptogamic covers to the global cycles of carbon and nitrogen. Nat. Geosci. 5, 459–462 (2012). [Google Scholar]
- 48.Scott A. D., et al. , A reference genome sequence for giant sequoia. G3 (Bethesda) 10, 3907–3919 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chase M. W., et al. , An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 181, 1–20 (2016). [Google Scholar]
- 50.Johnson M. G., et al. , A universal probe set for targeted sequencing of 353 nuclear genes from any flowering plant designed using k-medoids clustering. Syst. Biol. 68, 594–606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng C.-X., et al. , Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods 14, 43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forrest L. L., et al. , The Limits of Hyb-Seq for herbarium specimens: Impact of preservation techniques. Front. Ecol. Evol. 7, 439 (2019). [Google Scholar]
- 53.Brewer G. E., et al. , Factors affecting targeted sequencing of 353 nuclear genes from herbarium specimens spanning the diversity of angiosperms. Front. Plant Sci. 10, 1102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kates H. R., Soltis P. S., Soltis D. E., Evolutionary and domestication history of Cucurbita (pumpkin and squash) species inferred from 44 nuclear loci. Mol. Phylogenet. Evol. 111, 98–109 (2017). [DOI] [PubMed] [Google Scholar]
- 55.R .Folk A., et al., High-throughput methods for efficiently building massive phylogenies from natural history collections. Appl. Plant Sci. 9, e11410 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shee Z. Q., Frodin D. G., Cámara-Leret R., Pokorny L., Reconstructing the complex evolutionary history of the Papuasian Schefflera radiation through herbariomics. Front Plant Sci 11, 258 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Linksy J., Gostel M. R., The Global Genome Initiative for Gardens: Conservation priorities at the interface of botanic gardens and biodiversity genomics. BGJournal. 18, 21–23 (2021). [Google Scholar]
- 58.Ali N. S., Trivedi C., Botanic gardens and climate change: A review of scientific activities at the Royal Botanic Gardens, Kew. Biodivers. Conserv. 20, 295–307 (2011). [Google Scholar]
- 59.Mounce R., Smith P., Brockington S., Ex situ conservation of plant diversity in the world’s botanic gardens. Nat. Plants 3, 795–802 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Dosmann M., Groover A., The importance of living botanical collections for plant biology and the “next generation” of evo-devo research. Front Plant Sci. 3, 137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westwood M., Cavender N., Meyer A., Smith P., Botanic garden solutions to the plant extinction crisis. Plants People Planet 3, 22–32 (2021). [Google Scholar]
- 62.Gostel M. R., Kelloff C., Wallick K., Funk V. A., A workflow to preserve genome-quality tissue samples from plants in botanical gardens and arboreta. Appl. Plant Sci. 4, 1600039 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Funk V. A., et al. , Guidelines for collecting vouchers and tissues intended for genomic work (Smithsonian Institution): Botany Best Practices. Biodivers. Data J. 5, e11625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seberg O., et al. , Global Genome Biodiversity Network: Saving a blueprint of the Tree of Life—A botanical perspective. Ann. Bot. 118, 393–399 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.FAO, International Treaty on Plant Genetic Resources for Food and Agriculture (Food and Agriculture Organization of the United Nations, 2002). [Google Scholar]
- 66.Buck M., Hamilton C., The Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from their Utilization to the Convention on Biological Diversity. Rev. Eur. Community Int. Environ. Law 20, 47–61 (2011). [Google Scholar]
- 67.Sharrock S., Davis K., Promoting the ABS-compliant use of plant resources in research and development. BGjournal 16, 18–21 (2019). [Google Scholar]
- 68.Droege G., et al. , The Global Genome Biodiversity Network (GGBN) Data Standard specification. Database (Oxford) 2016, baw125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Salzberg S. L., Next-generation genome annotation: We still struggle to get it right. Genome Biol. 20, 92 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harris Z. N., Kovacs L. G., Londo J. P., RNA-seq-based genome annotation and identification of long-noncoding RNAs in the grapevine cultivar ‘Riesling’. BMC Genomics 18, 937 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Veeckman E., Ruttink T., Vandepoele K., Are we there yet? Reliably estimating the completeness of plant genome sequences. Plant Cell 28, 1759–1768 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vaattovaara A., Leppälä J., Salojärvi J., Wrzaczek M., High-throughput sequencing data and the impact of plant gene annotation quality. J. Exp. Bot. 70, 1069–1076 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tello-Ruiz M. K., et al. , Gramene 2021: Harnessing the power of comparative genomics and pathways for plant research. Nucleic Acids Res. 49 (D1), D1452–D1463 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lovell J. T., et al. , The genomic landscape of molecular responses to natural drought stress in Panicum hallii. Nat. Commun. 9, 5213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Q., et al. , A chromosome-scale genome assembly of cucumber (Cucumis sativus L.). Gigascience 8, giz072 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klimke W., et al. , Solving the problem: Genome annotation standards before the data deluge. Stand. Genomic Sci. 5, 168–193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kovaka S., et al. , Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20, 278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trapnell C., et al. , Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoff K. J., Stanke M., Current methods for automated annotation of protein-coding genes. Curr. Opin. Insect Sci. 7, 8–14 (2015). [DOI] [PubMed] [Google Scholar]
- 80.Watson M., Warr A., Errors in long-read assemblies can critically affect protein prediction. Nat. Biotechnol. 37, 124–126 (2019). [DOI] [PubMed] [Google Scholar]
- 81.Zhang P., et al. , Genome-wide identification and differential analysis of translational initiation. Nat. Commun. 8, 1749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mendieta J. P., Marand A. P., Ricci W. A., Zhang X., Schmitz R. J., Leveraging histone modifications to improve genome annotations. G3 (Bethesda) 11, jkab263 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wickett N. J., et al. , Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. U.S.A. 111, E4859–E4868 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li F.-W., Harkess A., A guide to sequence your favorite plant genomes. Appl. Plant Sci. 6, e1030 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ranallo-Benavidez T. R., Jaron K. S., Schatz M. C., GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 11, 1432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hon T., et al. , Highly accurate long-read HiFi sequencing data for five complex genomes. Sci. Data 7, 399 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ferreira D. A. T., Praça-Fontes M. M., Vieira A. T., Nunes A. C. P., Clarindo W. R., Karyotype and nuclear DNA content variation in Passiflora L. Sci. Hortic. (Amsterdam) 272, 109532 (2020). [Google Scholar]
- 88.Driguez P., et al. , LeafGo: Leaf to Genome, a quick workflow to produce high-quality de novo plant genomes using long-read sequencing technology. Genome Biol. 22, 256 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang W., et al. , The draft nuclear genome assembly of Eucalyptus pauciflora: A pipeline for comparing de novo assemblies. Gigascience 9, giz160 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ou S., Chen J., Jiang N., Assessing genome assembly quality using the LTR Assembly Index (LAI). Nucleic Acids Res. 46, e126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Emms D. M., Kelly S., OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Haug-Baltzell A., Stephens S. A., Davey S., Scheidegger C. E., Lyons E., SynMap2 and SynMap3D: Web-based whole-genome synteny browsers. Bioinformatics 33, 2197–2198 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Huddart J. E. A., A. J. Crawford, A. L. Luna-Tapia, S. Restrepo, F. Di Palma, EBP-Colombia and the bioeconomy: Genomics in the service of biodiversity conservation and sustainable development. Proc. Natl. Acad. Sci. U.S.A. 119, e2115641119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Blaxter M., et al. , Sequence locally, think globally: The Darwin Tree of Life Project. Proc. Natl. Acad. Sci. U.S.A. 119, e2115642118 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.