Significance
The high genetic diversity of HIV-1 continues to be a major barrier to the development of therapeutics for prevention and treatment. Here, we describe the design of an antibody platform that allows assembly of a highly avid, multispecific molecule that targets, simultaneously, the most conserved epitopes on the HIV-1 envelope glycoprotein. The combined multivalency and multispecificity translates into extraordinary neutralization potency and pan-neutralization of HIV-1 strains, surpassing that of the most potent anti-HIV broadly neutralizing antibody cocktails.
Keywords: HIV-1, antibody, neutralization, protein engineering
Abstract
Deep mining of B cell repertoires of HIV-1–infected individuals has resulted in the isolation of dozens of HIV-1 broadly neutralizing antibodies (bNAbs). Yet, it remains uncertain whether any such bNAbs alone are sufficiently broad and potent to deploy therapeutically. Here, we engineered HIV-1 bNAbs for their combination on a single multispecific and avid molecule via direct genetic fusion of their Fab fragments to the human apoferritin light chain. The resulting molecule demonstrated a remarkable median IC50 value of 0.0009 µg/mL and 100% neutralization coverage of a broad HIV-1 pseudovirus panel (118 isolates) at a 4 µg/mL cutoff—a 32-fold enhancement in viral neutralization potency compared to a mixture of the corresponding HIV-1 bNAbs. Importantly, Fc incorporation on the molecule and engineering to modulate Fc receptor binding resulted in IgG-like bioavailability in vivo. This robust plug-and-play antibody design is relevant against indications where multispecificity and avidity are leveraged simultaneously to mediate optimal biological activity.
Despite decades of research, no effective vaccine or cure exists for HIV type I (HIV-1). However, the fact that a small proportion of HIV-1–infected individuals develop antibodies with exceptional neutralization potency across circulating HIV-1 isolates highlights the potential for antibody-mediated control of HIV-1. Since the first generation of broadly neutralizing antibodies (bNAbs) 2F5 (1), 4E10 (2, 3), 2G12 (4), and b12 (5, 6) were isolated, the number of bNAbs has dramatically increased due to implementation of new technologies such as Env-specific single B cell sorting (7–9), antibody cloning and high-throughput neutralization assays (10–13), and, more recently, proteomic deconvolution (14). Several HIV-1 bNAbs have now been described that primarily target six conserved sites on the trimeric HIV envelope glycoprotein (Env), including the V1/V2 loops at the trimer apex, V3 loop glycans, the CD4 binding site (CD4bs), the gp120-g41 interface, the Env silent face, and the membrane-proximal external region (MPER) (7, 9, 11–20).
Interest in bNAbs as therapeutic agents in the fight against HIV-1 arises from the potent antiviral activity observed in challenge studies in macaques (21–25) and humanized mice (26–29), and from the reduced viremia achieved in infected humans following infusion of bNAbs (30–34). In addition, antibodies possess key advantages in comparison to oral antiretroviral therapy: they have longer circulating half-lives, and can form immune complexes that enhance host immunity to the virus. These observations have led to the clinical evaluation of antibody-based therapies to confer protection against HIV-1 acquisition through passive administration of bNAbs (35), and efforts to control and/or clear HIV-1 in infected individuals (31–33).
The recent Antibody Mediated Prevention (AMP) trials explored the ability of bNAb VRC01 to confer passive immunity against HIV-1. In these studies, antibody breadth and potency inferred from TZM‐bl neutralization assays were proposed as effective predictors of antibody efficacy in humans. Specifically, inhibition of 80% of viral infection with a concentration below 1 µg/mL (IC80 < 1 µg/mL) was established as the potency threshold that a biotherapeutic needs to achieve in order to confer protection against a specific HIV-1 strain (35). VRC01 only met that threshold against 30% of HIV-1 strains in the trials and, hence, failed to confer broad protection, highlighting a critical need for more potent and broadly acting molecules. While such breadth of coverage could be achieved by administration of multiple bNAbs, despite recent IgG engineering efforts (36–40), potency may still limit the therapeutic efficacy of antibody mixtures.
Here, we overcome the immense sequence diversity of HIV-1 with extraordinary neutralization potency by engineering the human apoferritin subunit to drive multimerization of three different HIV-1 bNAbs on a single molecule. The resulting multispecific, multiaffinity antibody (Multabody) was able to achieve pan-neutralization (100% virus coverage) with a median IC50 value of 0.0009 µg/mL (0.4 pM). The Multabody design described herein represents a robust and powerful plug-and-play platform to multimerize antibodies in order to enhance their neutralization of HIV-1 across the broadest range of isolates.
Results
Potency of HIV-1 bNAbs Can Be Enhanced with Avidity.
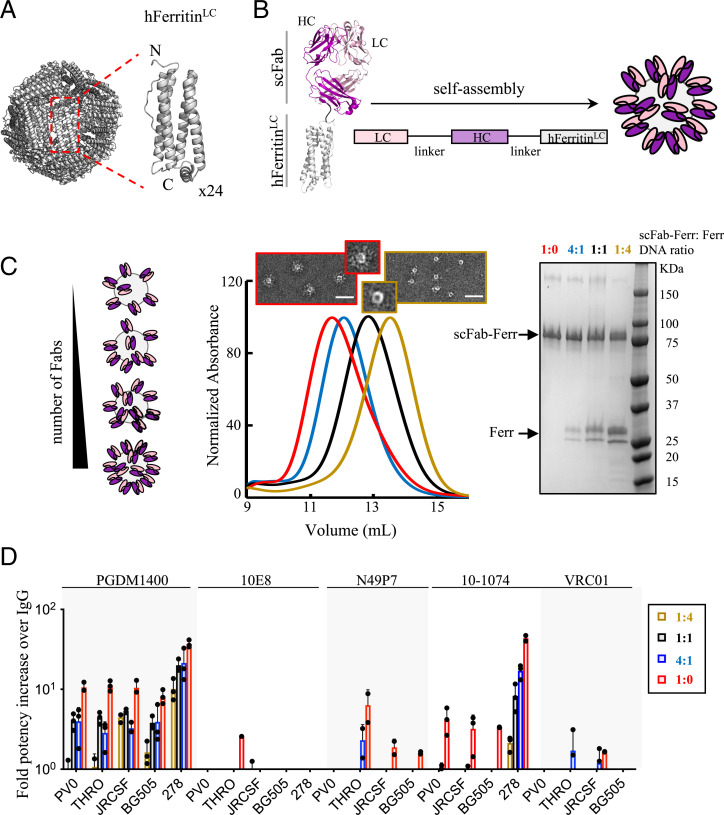
Apoferritin is a spherical nanocage of ∼6-nm hydrodynamic radius formed by the self-oligomerization of 24 identical subunits (Fig. 1A). To investigate the impact of multivalency on neutralization potency, we used the self-assembly properties of the light chain of human apoferritin to multimerize fragments of antigen binding (Fabs) derived from the most potent and broad HIV-1 bNAbs, which target different HIV-1 Env epitopes. Apoferritin subunits were genetically fused to single-chain Fabs (scFabs). The scFabs were generated using flexible linkers between the light and heavy chains to ensure correct Fab heterodimerization. Apoferritin self-assembly drove multimerization of the scFab and displayed the antibody fragments at the nanocage periphery (Fig. 1B). Different densities of multimerized Fabs were achieved by cotransfection of scFab-human apoferritin–encoding plasmids together with different ratios of nongenetically modified human apoferritin (Fig. 1C and SI Appendix, Fig. S1). The ability of the scFab-apoferritin fusions to block HIV-1 infection was compared to the corresponding IgGs using a small HIV-1 pseudovirus (PsV) panel (Fig. 1D). Strikingly, PGDM1400, one of the most potent anti-HIV bNAbs described to date, showed 10- to 40-fold higher neutralization potency when multimerized via the light chain of apoferritin compared to its conventional IgG format. The bNAb 10-1074 also showed a considerable improvement in neutralization potency (4- to 40-fold), whereas bNAbs 10E8, N49P7, and VRC01 showed no effect or more modest enhancements.
Fig. 1.
HIV-1 bNAb multimerization increases neutralization potency. Schematic of the self-assembly of (A) apoferritin (24 subunits) and (B) scFab-apoferritin fusions. Fab light chain (LC) and heavy chain (HC) are shown in light and dark pink, respectively, and are connected to the N terminus of the light chain of human apoferritin (gray) through a glycine-glycine-serine–like flexible linker (black). (C) Schematic representation of different Fab densities displayed on human apoferritin. Cotransfection of scFab-human apoferritin–encoding plasmids together with unconjugated apoferritin at ratios of 1:4 (dark yellow), 1:1 (black), 4:1 (blue), and 1:0 (red) resulted in molecules with different scFab valency, as confirmed by elution volumes in size exclusion chromatography (SEC) and less unconjugated apoferritin in sodium dodecyl sulfate polyacrylamide gel electrophoresis. Negative-stain electron micrographs of the samples with the lowest and highest scFab valency are shown. (Scale bar, 50 nm.) (D) Avidity effect on neutralization of five bNAbs against a five-PsV panel (PVO.04, JRCSF, BG505 T332N, THRO4156.18, and t278-50). Fold potency increase was calculated as the parental IgG IC50 (micrograms per milliliter) divided by the Fab-apoferritin fusion IC50 (micrograms per milliliter). Fold potency increase analyses were omitted in the following cases: N49P7-t278-50, VRC01-T278-50, and 10-1074-THRO4156.18 due to neutralization resistance. Bars (± SD) represent the mean value from n = 3 biologically independent samples.
Multabodies Potently and Broadly Neutralize HIV-1.
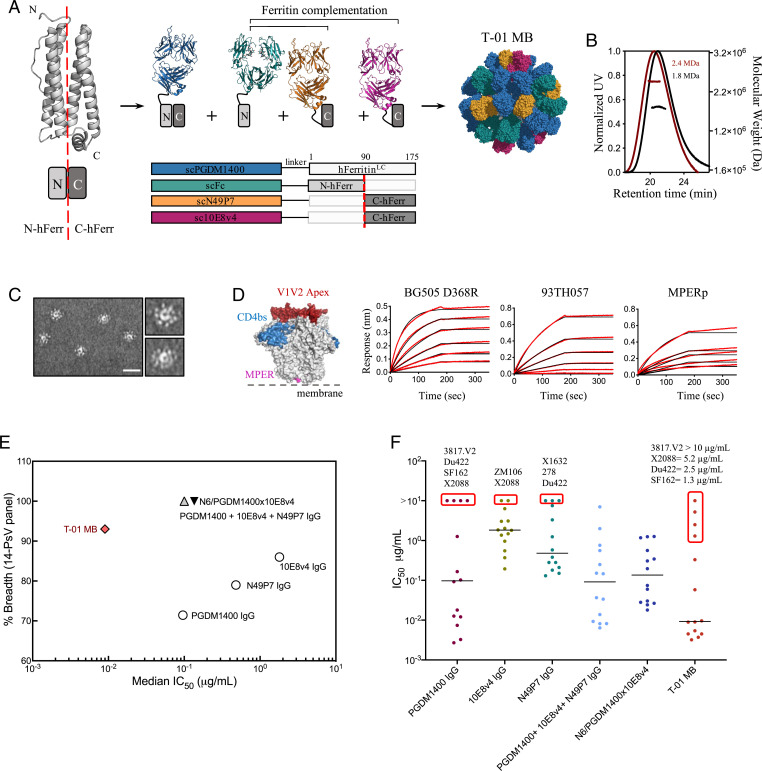
In view of these results, we sought to increase the coverage of PGDM1400 using our previously described Multabody platform based on an apoferritin split design (41). The strategy consists of the separation of the four-helix apoferritin subunit into two halves (N-ferritin and C-ferritin) and their N-terminal fusion to scFabs of different specificities (Fig. 2A). This approach allows inclusion of a higher number of Fabs on the surface of the nanocage, resulting in a final molecule with higher avidity. In addition, the design allows the efficient combination of three different antibody specificities as well as a fragment crystallizable (Fc) to endow the molecule with IgG-like properties, such as ease of purification leveraging protein A affinity (SI Appendix, Fig. S2). Specifically, we combined scFab PGDM1400 with scFabs of the near–pan-neutralizing antibodies 10E8v4 [a modified 10E8 with improved solubility (42)] and N49P7, and the single-chain construct of the Fc (scFc) of human IgG1 isotype (Fig. 2A). To explore whether a Multabody could also be designed that cross-targets the HIV-1 Env and its primary receptor, CD4, we replaced N49P7 with Ibalizumab (iMab), a CD4-directed postattachment inhibitor that has been shown to effectively inhibit HIV-1 entry (43, 44) (SI Appendix, Fig. S3A). The resulting trispecific Multabodies, termed T-01 MB and T-02 MB, formed highly decorated and homogeneous particles of around 2.4 MDa (Fig. 2 B and C and SI Appendix, Fig. S3 B and C) with thermostability similar to the corresponding IgGs (SI Appendix, Fig. S4). Epitope engagement by the trispecific Multabodies was assessed in binding kinetics experiments using epitope-specific molecules: BG505 SOSIP.664 D368R (PGDM1400), 93TH057 gp120/CD4 (N49P7/iMab), and an MPER peptide (10E8v4) (SI Appendix, Fig. S5). Binding to the three epitope-specific antigens with high apparent binding affinities and no detectable dissociation confirms the presence of the three antibody specificities in the Multabodies (Fig. 2D and SI Appendix, Fig. S3D).
Fig. 2.
Design, assembly, and neutralization profile of HIV-1 Multabodies against a 14-PsV panel. (A) Schematic of the human apoferritin split design that drives heterodimerization of scFab-human apoferritin subunits. (B) Size exclusion chromatography in-line with multiangle light scattering of 24-mer PGDM1400 scFab-apoferritin particles (black) and T-01 MB (dark red). The molar mass of each elution peak (lines under ultraviolet [UV] absorbance) is shown in megadaltons. (C) Negative-stain electron micrographs of T-01 MB. (Scale bar, 50 nm.) (D) Concentration-response curves for binding of T-01 MB to multiple epitopes. PGDM1400, N49P7, and 10E8 binding sites are colored in red, blue, and pink, respectively, on the surface representation of the HIV-1 Env trimer (gray). Red lines represent raw data; black lines represent global fits. (E) Breadth (cutoff IC50 set at 10 µg/mL) and median IC50 values (micrograms per milliliter) of T-01 MB (red diamond), parental bNAbs (white circles), IgG combination (gray triangle), and the N6/PGDM1400x10E8v4 trispecific antibody (black triangle). The 14-PsV panel was selected based on susceptibility and resistance to the parental IgGs. (F) Individual IC50 values (micrograms per milliliter) to each PsV variant. The solid line denotes the median neutralization IC50 of all 14 viral strains. Those pseudoviruses that show the highest neutralization resistance are highlighted by a red box. IC50 values in D and E were calculated from three biological replicates.
Neutralization potency and breadth of the Multabodies was first assessed against a panel of 14 PsVs in a standardized in vitro TZM-bl neutralization assay (45). The 14-PsV panel was designed to include low-sensitivity PsVs, with at least one PsV resistant to each bNAb being evaluated (cutoff IC50 set at 10 µg/mL). The IC50 value and breadth of the Multabodies were compared to 1) each individual IgG, 2) an IgG mixture that contains the same relative amount of each IgG present in the Multabody and 3) the N6/PGDM1400x10E8v4 trispecific antibody (46). T-01 MB and T-02 MB displayed 93% and 100% breadth (cutoff IC50 set at 10 µg/mL) against this panel, with median IC50 values of 0.009 μg/mL (3.9 pM) and 0.008 μg/mL (3.5 pM), respectively (Fig. 2E and SI Appendix, Fig. S3E and Table S1). As such, there was a decrease of approximately one and two orders of magnitude in the median IC50 values when calculated in micrograms per milliliter and nanomoles for the Multabodies compared to the IC50 values of the IgG mixtures and the trispecific antibody, respectively. Inspection of individual IC50 values revealed that PsVs that are resistant to PGDM1400 IgG neutralization were also less sensitive to the Multabodies (Fig. 2F and SI Appendix, Fig. S3E and Table S1). These data suggested that the neutralization property of the Multabodies is heavily dependent on one out of the three antibody specificities within the particle, in this case, PGDM1400.
Engineering the Apoferritin Scaffold.
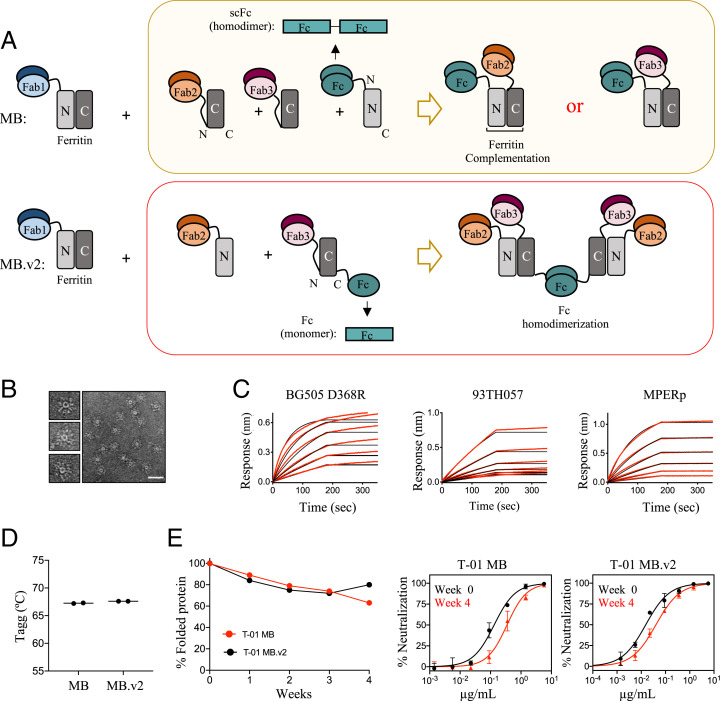
To further improve the neutralization properties of the Multabody, we introduced some modifications to its design and made a second-generation version (MB.v2). In the original MB, the scFc is located at the N terminus of the N-ferritin half, and only one Fab, either Fab2 or Fab3, is incorporated in the Multabody per each functional Fc homodimer (Fig. 3 A, Top). In comparison, the optimized MB.v2 contains a higher number of Fabs per Fc homodimer. To attain this, a monomeric Fc fragment (i.e., one Fc chain) and a scFab are positioned at the C terminus and the N terminus, respectively, of the C-ferritin half (Fig. 3 A, Bottom and SI Appendix, Fig. S6A). As a result, dimerization of a functional Fc homodimer drives assembly of the MB.v2 particle together with split ferritin complementation and ferritin subunit oligomerization (Fig. 3A and SI Appendix, Fig. S6B). Importantly, homodimerization to form one functional Fc ensures assembly of four Fabs different from PGDM1400 (i.e., two Fab2 and two Fab3), thus favoring a more balanced avidity for each of the three Fabs in the fully assembled MB.v2.
Fig. 3.
Engineering and biophysical characterization of Multabody v2. (A) The second-generation Multabody design displays two distinct features in comparison to the original Multabody design: 1) The Fc (green) is fused to the C terminus of the second half of apoferritin in the split ferritin design, and 2) the scFc domain (green), fused to the C terminus of an apoferritin half protomer, is reverted to a monomeric Fc chain. Dimerization of each Fc in MB.v2 drives assembly of four Fabs (two Fab2 and two Fab3, Bottom) while only one Fab is assembled per Fc into the previous MB version (Top). (B) Negative-stain electron micrographs of T-01 MB.v2. (Scale bar, 50 nm.) (C) Concentration-response curves for binding of T-01 MB.v2 to multiple epitopes. Red lines represent raw data; black lines represent global fits. (D) Comparison of Tagg and (E) long-term stability under temperature stress conditions (10 mg/mL; 40 °C) of the two different Multabody versions. PsV neutralization (mean values ± SD for two technical replicates) comparison at week 0 vs. week 4 is shown.
The optimized Multabody design was tested in the T-01 background (PGDM1400, N49P7, 10E8v4) that targets three epitopes on HIV-1 Env. The resulting Multabody (T-01 MB.v2) assembled into well-formed spherical particles with no significant differences in morphology compared to the previously characterized T-01 MB (Fig. 3B). Antigen binding to BG505 SOSIP.664 D368R, 93TH057 gp120, and MPER peptide confirmed correct folding of the three Fab specificities in T-01 MB.v2 (Fig. 3C). In addition, the new Multabody version preserves the same high thermal stability reported for T-01 MB, with an aggregation temperature (Tagg) of 67 °C (Fig. 3D). Multabodies were concentrated to 10 mg/mL and subjected to an accelerated stability test by incubating them at 40 °C for 4 wk. Assessment of the amount of soluble protein over time revealed that the Multabodies were highly stable under these conditions, with over 70% of the sample remaining soluble for 30 d. Stability was further confirmed by only a modest loss in neutralization potency observed for the Multabodies at week 4 in comparison to their potency at week 0 (Fig. 3E).
Pharmacokinetics of Multabodies Is Similar to Corresponding IgGs.
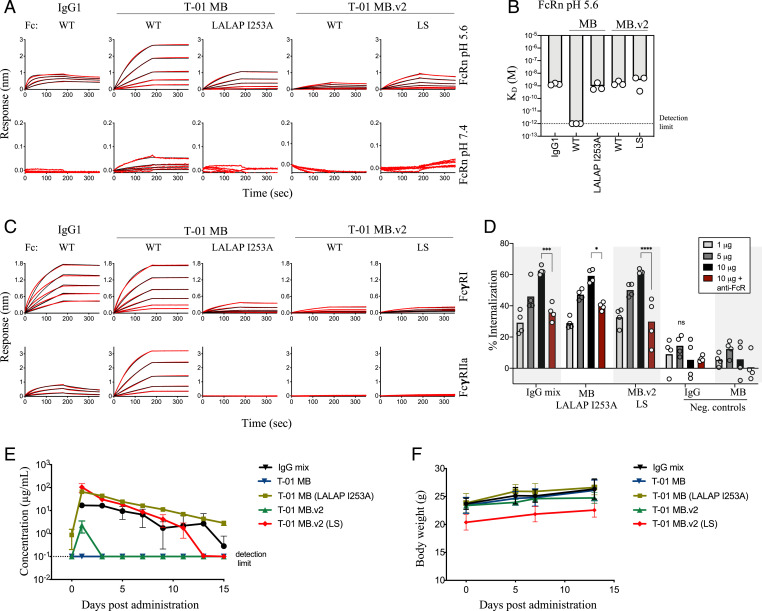
The antibody Fc domain has the capacity to interact with a variety of receptors, including Fc gamma receptors (FcγRs) and the neonatal Fc receptor (FcRn), conferring effector functions and in vivo half-life, respectively. However, Fc avidity can negatively impact the circulation time of molecules with multiple Fc fragments (41, 47). Indeed, T-01 MB showed strong binding to Fc receptors, including to human FcRn at physiological pH (Fig. 4 A and B), and high- and low-affinity FcγRs (Fig. 4C). Hence, we introduced the unique combination of LALAP (L234A, L235A, and P329G) and I253A mutations in the Fc of T-01 MB to decrease binding to FcγR and FcRn, respectively, and achieve binding comparable to that observed for an IgG1 molecule (Fig. 4 A–C). T-01 MB.v2 showed a more similar binding profile to IgG1, with comparable binding to human FcRn at acidic pH and no binding at physiological pH, even in the case of the half-life extension mutations LS (M428L/N434S) (Fig. 4 A and B). Binding of T-01 MB.v2 to FcγRs yielded a low binding profile similar to that obtained with the LALAP FcR-silencing mutations in T-01 MB (Fig. 4C). The different binding patterns observed for the two MB versions are likely due to the different arrangements of the Fc fragments within the molecules (Fig. 3A and SI Appendix, Fig. S6A). Despite low FcγRI binding, phagocytosis experiments using antigen-coated beads showed that both Multabody formats induced Fc-dependent internalization in THP-1 cells at levels similar to those achieved with the corresponding IgG mixture (Fig. 4D).
Fig. 4.
Fine tuning of Fc on Multabody for IgG-like characteristics. (A) Concentration-response curves for pH-dependent binding to human FcRn by T-01 MB and T-01 MB.v2. (B) Comparison of the FcRn apparent binding affinities (KD) at acidic pH between MBs and an IgG1; n = 3 biologically independent samples are shown. Apparent KD lower than 10−12 M (dotted black line) is beyond the instrument detection limit. (C) Concentration-response curves to the high-affinity human FcγRI (Top) and low-affinity human FcγRIIa (Bottom). (D) Dose-dependent phagocytosis determined as a percentage increase in internalization of 93TH057 gp120-coated fluorescent microspheres compared to a no-antibody control. Anti-human FcR binding inhibitor antibody was added to block Fc-mediated internalization (dark red). Data were analyzed by two‐way ANOVA with Tukey’s multiple comparisons test. Each group was compared to the IgG negative control. *P < 0.05, ***P < 0.001, and ****P < 0.0001. IgG and MB samples with no affinity for the antigen-coated beads were added as control samples; n = 4 biologically independent samples. ns, not significant. (E) Serum levels after subcutaneous administration of 5 mg/kg of Multabodies or parental IgG mixture in female NCG immunodeficient mice. (F) Body weight upon administration of 5 mg/kg of molecules in NCG mice. Mean values ± SD for n = 3 mice are shown in E and F.
Next, we examined the in vivo bioavailability of both Multabody formats with and without engineered Fc. A single dose of 5 mg/kg was administered subcutaneously in NOD/Shi-scid/IL-2Rγnull (NCG) immunodeficient mice, and the amount of each molecule in the sera was measured every 2 d for 15 consecutive days. As expected from the in vitro characterization, only Fc-engineered Multabodies that had an IgG-like binding profile showed days of in vivo exposure with a similar rate of decay as the parental IgG mixture (Fig. 4E). Multabody administration was well tolerated, with no decrease in body weight (Fig. 4F) or visible adverse effects.
Extraordinary Potency and Pan-Neutralization Breadth Achieved by MB.v2.
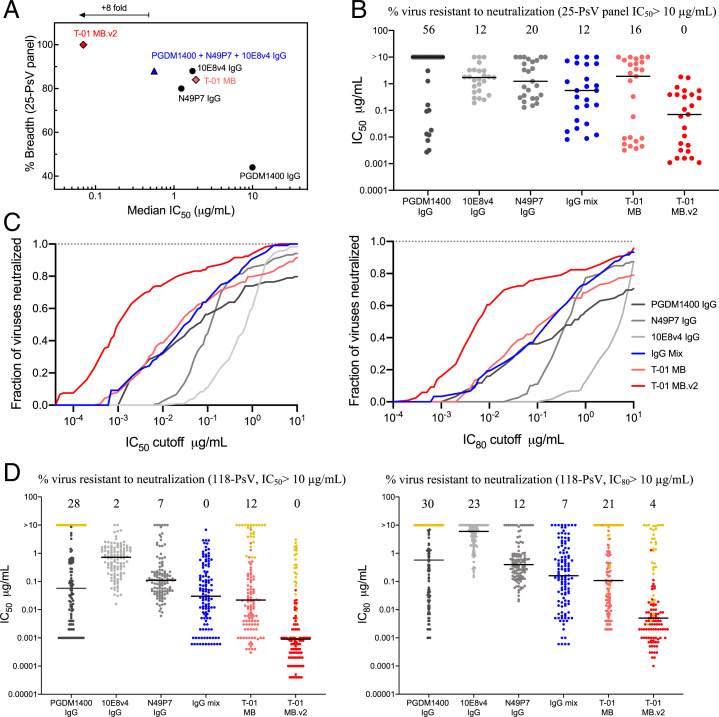
We assessed the neutralization profile of T-01 MB.v2 against a PsV panel generated through addition of 11 HIV-1 strains highly resistant to PGDM1400 to our previous panel. The resulting 25-PsV panel contains 56% of PsV variants resistant (cutoff IC50 set at 10 µg/mL) to PGDM1400 IgG neutralization (Fig. 5 A and B). As expected, breadth and potency of the T-01 MB was greatly affected in the presence of PGDM1400-resistant PsVs (Fig. 5 A and B and SI Appendix, Table S1). However, as engineered, the neutralization profile of antibodies N49P7 and 10E8v4 were more dominant in T-01 MB.v2, allowing this optimized Multabody to achieve pan-neutralization while preserving the enhanced neutralization potency previously observed for this type of molecule (Fig. 5 A and B and SI Appendix, Table S1). When tested against an extended multiclade panel of 118 PsVs, T-01 MB.v2 matched the pan-neutralization breadth of the corresponding IgG mixture (100% virus coverage, cutoff IC50 set at 10 µg/mL), yet displayed a remarkable neutralization potency (Fig. 5 C and D and SI Appendix, Fig. S7A and Table S2). Specifically, the IgG mixture and T-01 MB were only able to neutralize 9% and 8%, respectively, of the PsVs with an IC50 value of 0.001 µg/mL, while, in the case of T-01 MB.v2, 50% of the PsVs were still neutralized with an IC50 value of only 0.001 µg/mL (Fig. 5C). Remarkably, Multabodies achieved a median IC50 value of only 0.0009 µg/mL (0.4 pM) and hence achieved pan-neutralization 32- and 490-fold more potently in mass and molarity, respectively, compared to the IgG mixture (Fig. 5D and SI Appendix, Table S2). In addition, the IC80 of T-01 MB.v2 confirmed its superior neutralization propensity over both the individual IgGs and the IgG mixture, neutralizing 96% of all viral strains tested with a median IC80 value of 0.005 µg/mL (2.2 pM) (Fig. 5 C and D and SI Appendix, Fig. S7A and Table S2). Importantly, Multabodies also blocked infection of primary peripheral blood mononuclear cells (PBMCs) with the replication-competent CXCR4-tropic HIV-1 IIIB strain (SI Appendix, Fig. S7B), showing enhanced potency over the matched IgG mix, and without any impact on cell viability (SI Appendix, Fig. S7C).
Fig. 5.
Broad and potent neutralization by Multabody v2 against extended HIV-1 PsV panels. (A) Breadth and median IC50 values (micrograms per milliliter) of T-01 Multabody versions (different shades of red diamond), individual IgGs (black circles), and IgG mixture (blue triangle) against a 25-PsV panel with 56% of PsV variants resistant to PGDM1400 neutralization. (B) Individual IC50 values (micrograms per milliliter) for each PsV variant. IC50 values in A and B were calculated from three biological replicates. (C) Potency (IC50)-breadth (Left) and potency (IC80)-breadth (Right) curve comparison of T-01 Multabody versions, as well as parental IgGs and an IgG mixture against an extended multiclade panel of 118 HIV-1 PsV variants. (D) Individual IC50 (Left) and IC80 (Right) values for each PsV variant in C. Yellow dots correspond to IC50 values from PsVs that are highly resistant to PGDM1400 neutralization. The solid line in B and D denotes the median IC50 neutralization titer of all viral strains in each panel.
Discussion
The recent AMP clinical trials have highlighted the anticipated importance of both potency and breadth of bNAbs to be viable therapeutics capable of protecting against HIV-1 infection. Leveraging the principle of antibody avidity used in our previously described Multabody technology to improve the potency of antibodies against severe acute respiratory syndrome coronavirus 2 (41), here we have engineered a second-generation Multabody platform to deliver exceptional neutralization breadth and potency against the vast sequence diversity of HIV-1.
The most distinctive feature of the optimized Multabody design in comparison to the first-generation Multabody format (41) is the relative number of Fabs that self-associate per functional Fc domain. In contrast to the 1:1 Fc: 10E8v4/N49P7 ratio imposed by the design of the previously described Multabody platform, in the design of T-01 MB.v2, two N49P7 Fabs and two 10E8v4 Fabs are incorporated into the MB per homodimeric Fc. The higher number of these two Fabs in the optimized Multabody favors their avidity and, consequently, their greater contribution to the neutralization signature of the particle. This is in contrast to the T-01 MB, which primarily relies on the neutralization properties of PGDM1400. The more balanced contribution of each of the antibodies is reflected in the better functional properties of T-01 MB.v2, which displayed cross-clade neutralization coverage of 100% at a median IC50 value of 0.0009 µg/mL. In addition, viral infection by 83% of the 118 PsVs tested was blocked by T-01 MB.v2, with an IC80 value below 1 µg/mL, which has been recently proposed as the potency threshold required to confer in vivo protection in humans (35). However, it remains unclear whether that predictor of protection should be considered in mass or molarity. Indeed, despite a similar hydrodynamic radius and geometrical size for the Multabody compared to an IgM (41), the Multabody is ∼10× heavier in molar mass compared to an IgG. Therefore, if molarity is the relevant in vivo measure associated with protection, then T-01 MB.v2 exhibits an extraordinarily low median IC50 value of 0.4 pM. Correspondingly, a potency of IC80 below 6.7 nM (1 µg/mL molar equivalent for an IgG) was achieved in 96% of the 118 HIV-1 PsV strains. These remarkable neutralization properties surpass those obtained with previously described bispecific and trispecific antibodies (46, 48–50). In these antibody formats, the limited avidity precludes combination of both high avidity and multispecificity, and, consequently, potency and breadth are restricted to that of the parental mAbs.
There is a growing trend in the field of biotherapeutics toward the development of molecules with high valency. Strategies range from the generation of dodeca-valency IgM-like molecules (51, 52) upon addition of the mu tailpiece of IgM to the constant region of IgG to the design of alternative antibody formats. Among them are fusions of Fabs in a linear head-to-tail manner (53), appended IgGs (54–56), or diabody combination in tandem (Tamdabs) (57) or fused to the CH3 of an IgG (di-diabody) (58). In addition, the use of multimerization scaffolds, such as p53 (59), leucine zipper helixes (60), streptavidin (61), barnase-barstar modules (62), viral-like nanoparticles (63), and, more recently, de novo antibody cage-forming proteins (64), has been employed to overcome the limitation of IgG bivalency and improve the bioactivity of antibodies. Although attractive, these approaches face different challenges for their successful development as therapeutic agents. Multimeric antibody formats that rely on variable fragments of antibodies are often associated with low stability and, consequently, a high propensity to aggregate (65). Furthermore, dissociation of noncovalent fusions dictated by the affinity constant of the complex can limit the in vivo long-term stability of the molecule. In sharp contrast, Multabodies build on full IgG components (Fab and Fc) that are fused to the thermostable, functionally silent human apoferritin light-chain scaffold, and thus are highly stable IgG-like molecules even under thermal stress. A mouse surrogate Multabody previously administered subcutaneously in immunocompetent C57BL/6 mice showed undetectable levels of antidrug antibodies, similarly to its parent IgG, providing proof of principle for the potentially low intrinsic immunogenicity of the Multabody platform (41). Future studies in higher organisms will help determine the immunogenicity of Multabodies encoded by human-derived sequences, which we propose might be dictated predominantly by the properties of the underlying antibody sequences.
Bioavailability of large biologics is an additional challenge associated with engineered approaches to increase avidity (63). Multabodies have been engineered to include Fc domains and hence enable FcRn-mediated recycling of the molecule. As a consequence of Fc avidity, binding of T-01 MB to FcRn at pH 6.0 was improved by orders of magnitude; however, the simultaneous affinity improvement at pH 7.4 limits the application of this strategy toward enhancing half-life, and several mutations were needed to decrease Fc binding to FcRn and FcγR in order to not surpass the binding affinity observed for IgGs. Such enhanced Fc receptor avidity was not observed in the case of T-01 MB.v2, where fusion of the Fc chains at the C terminus of apoferritin leads to the formation of particles with inverted and more distantly located Fc domains and, consequently, reduced Fc avidity. Similar to a previous study with a mouse surrogate Multabody (41), Fc avidity modulation strategies successfully resulted in Multabody molecules with a rate of decay over time that is remarkably similar to that of the parental IgG mixture. In addition to the favorable pharmacokinetic profile, Multabodies in both formats that possess residual binding to FcγR induced Fc-mediated phagocytosis in vitro to levels similar to the parental IgG mixture, at least in a THP-1 system that coexpresses both FcγRI and FcγRIIa (66). Future experiments will be needed to fully characterize the capacity of the Multabodies to trigger immune effector functions and their in vivo implications.
From the limited number of antibody specificities we characterized in this study, we observed that antibodies targeting epitopes located at the apex of the HIV-1 Env trimer such as PGDM1400 seem to experience the greatest benefit to neutralization potency when formulated as Multabodies. This increase in potency was less apparent as the epitope was located closer to the viral membrane, as in the case of 10E8v4. The dependence on epitope location for potency enhancement could be further impacted by the low surface spike density (67), the arrangement of those sparse Env trimers on the HIV-1 surface (68), or the accessibility of certain epitopes that may be more or less sterically occluded. In light of this, it will be interesting to explore how the potency of antibodies against viruses with higher surface densities and closely spaced spikes can be enhanced by the Multabody platform, and further determine the impact of epitope location on potency enhancement mediated by avidity.
Replacement of N49P7 with iMab—a CD4-directed postattachment inhibitor—resulted in a functional Multabody with potent neutralization activity, demonstrating that cross-targeting of viral epitopes and cellular receptors can be attained with this type of particle. These data raise the interesting possibility that the Multabody technology could also be implemented in other fields that promote the binding of receptors across separate entities, such as cell–cell interactions in immunotherapy. Overall, our protein engineering study demonstrates the versatility of human apoferritin as a modular nanocage to build antibody avidity and multispecificity jointly toward enhanced functionality.
Materials and Methods
Expression and Purification of Fab-Only Apoferritin Multimers.
Genes encoding the light chain of human apoferritin and the scFab-human apoferritin fusions were synthesized and cloned by GeneArt (Life Technologies) into the pHLsec expression vector; 200 mL of HEK 293F cells (Thermo Fisher Scientific) were seeded at a density of 0.8 × 106 cells/mL in Freestyle expression media and incubated with 125 rpm oscillation at 37 °C, 8% CO2, and 70% humidity in a Multitron Pro shaker (Infors HT). Within 24 h of seeding, cells were transiently transfected using 50 μg of filtered DNA preincubated for 10 min at room temperature (RT) with the transfection reagent FectoPRO (Polyplus Transfections) at a 1:1 ratio. Plasmids encoding scFab-human apoferritin and human apoferritin were mixed at a ratio of 1:4, 1:1, 4:1, and 1:0. After 6 d to 7 d, cell suspensions were harvested by centrifugation at 5,000 × g for 15 min, and the supernatants were filtered through a 0.22-μm Steritop filter (EMD Millipore). The particles were purified by affinity chromatography to the Fab and eluted after a wash. Fractions containing protein were pooled, concentrated, and loaded onto a Superose 6 10/300 GL size exclusion column (GE Healthcare) in 20 mM sodium phosphate, pH 8.0, 150 mM NaCl.
Design, Expression, and Purification of Multabodies.
Genes encoding scFab and scFc fragments linked to half ferritin were generated by deletion of residues 1 to 90 (C-Ferritin) and 91 to 175 (N-Ferritin) of the light chain of human apoferritin. Furthermore, protein L binding specificity for iMab-C-Ferritin was disrupted by site-directed mutagenesis of alanine 12 of the antibody light chain to a proline residue (69). Transient transfection of T-01 MB in HEK 293F cells was obtained by mixing 66 μg of plasmids PGDM1400 scFab-human apoferritin:scFc-N-Ferritin:N49P7 scFab-C-Ferritin:10E8v4 scFab-C-Ferritin in a 4:2:1:1 ratio. In the case of the T-02 MB, plasmid N49P7 scFab-C-Ferritin was substituted by iMab scFab-C-Ferritin. In the case of the T-01 MB.v2, 63 μg of plasmids PGDM1400 scFab-human apoferritin:N49P7 scFab-N-Ferritin:10E8v4 scFab-C-Ferritin-Fc in a 3:1:1 ratio were used. The DNA mixture was filtered and incubated at RT with 63 μL of FectoPRO before adding it to the cell culture. Based on the necessary heterooligomerization to drive self-assembly, purification of Multabodies with the four components was achieved by a two-step affinity purification: protein A HP column (GE Healthcare) with 20 mM Tris pH 8.0, 3 M MgCl2 and 10% glycerol elution buffer (Fc binding), and protein L (GE Healthcare) (PGDM1400 binding, as 10E8 and N49P7 do not bind to protein L, and iMab-protein L binding was disrupted by the A12P mutation). A buffer exchange step was performed between the two affinity chromatography steps using a PD-10 desalting column (GE Healthcare). Fractions containing the protein were concentrated and further purified by gel filtration on a Superose 6 10/300 GL column (GE Healthcare) in 20 mM sodium phosphate pH 8.0, 150 mM NaCl.
Negative-Stain Electron Microscopy.
Three microliters of Multabody at a concentration of ∼0.02 mg/mL was added to a carbon-coated copper grid for 30 s and stained with 3 μL of 2% uranyl formate. Staining excess was immediately removed from the grid using Whatman No. 1 filter paper, and an additional 3 μL of 2% uranyl formate was added for 20 s. Grids were imaged using a field emission FEI Tecnai F20 electron microscope operating at 200 kV and equipped with an Orius charge-coupled device camera (Gatan Inc).
Biolayer Interferometry.
Binding kinetics measurements were conducted using an Octet RED96 BLI system (Pall ForteBio) in phosphate-buffered saline (PBS) pH 7.4, 0.01% bovine serum albumin (BSA) and 0.002% Tween. A unique His-tagged ligand for each of the Multabody components and Fc receptors was selected and loaded onto nickel nitrilotriacetic acid (Ni-NTA) biosensors to reach a signal response of 0.8 nm. Association rates were measured by transferring the loaded biosensors to wells containing serial dilutions of the Multabodies (10, 5, 2.5, 1.25, 0.65, and 0.32 nM) or IgGs (500, 250, 125, 62.5, 31.2, and 15.6 nM). Dissociation rates were measured by dipping the biosensors into buffer-containing wells. The duration of each of these two steps was 180 s. To achieve selective binding to PGDM1400, a D368R mutation in the CD4bs of the BG505 SOSIP.664 trimer was introduced, and, consequently, binding of N49P7 to this antigen was disrupted. Similarly, the gp120 subunit 93TH057, soluble CD4, and hFcRn in complex with β2-microglobulin were produced as the ligands for N49P7, iMab, and Fc, respectively. Binding to 10E8v4 was tested using a His-tagged MPER peptide (HHHHHHNEQELLELDKWASLWNWFNITNWLWYIKKKK, purchased from GenScript). Recombinantly expressed hFcγRI and hFcγRIIa were used to measure binding affinities of the IgGs and Multabodies. Ni-NTA purification followed by size exclusion chromatography in 20 mM phosphate, pH 8.0, 150 mM NaCl buffer was used for purification of BG505 SOSIP.664 D368R, CD4, 93TH057, hFcRn, hFcγRI, and hFcγRIIa.
Size Exclusion Chromatography in Line with Multiangle Light Scattering.
A MiniDAWN TREOS and an Optilab T-rEX refractometer (Wyatt) were used in-line with an Agilent Technologies 1260 infinity II HPLC. Fifty micrograms of 24-mer PGDM1400 scFab-ferritin fusion, T-01 MB, and T-02 MB were loaded onto a Superose 6 10/300 (GE Healthcare) column in 20 mM sodium phosphate, pH 8.0, 150 mM NaCl. Data collection and analysis were performed using the ASTRA software (Wyatt).
Stability Measurements.
The melting temperature (Tm) and Tagg of Multabodies, parental IgGs, the 12-mer homo-oligomeric Fabs and Fc, and the N6/PGDM1400x10E8v4 trispecific antibody were determined using a UNit system (Unchained Labs). Tm was obtained by measuring the barycentric mean fluorescence, while Tagg was determined as the temperature at which 50% increase in the static light scattering at a 266-nm wavelength relative to baseline was observed. Samples were concentrated to 1.0 mg/mL and subjected to a thermal ramp from 25 °C to 95 °C with 1 °C increments. The average and the SE of three independent measurements were calculated using UNit analysis software.
Stability was further analyzed under accelerated stress conditions. Multabodies diluted in 20 mM sodium phosphate pH 8.0, 150 mM NaCl, were concentrated to 10 mg/mL and incubated at 40 °C for 4 wk. Each week, the percentage of properly folded protein was calculated based on the soluble content from SEC. A sample before (week 0) and after (week 4) the incubation period was assessed in a PsV neutralization assay to compare the functional activity of the molecules.
Virus Production and TZM-bl Neutralization Assays.
A panel of 14 HIV-1 pseudotyped viruses was generated by cotransfection of 293T cells with the HIV-1 subtype B backbone NL4-3.Luc.R−E plasmid (AIDS Research and Reference Reagent Program [ARRRP]) and the plasmid encoding the full-length Env clone, as previously described (45). HIV isolates X2088.c09, ZM106.9, and 3817.v2.c59 were kindly provided by the Collaboration for AIDS Vaccine Discovery (CAVD), and pCNE8, 1632_S2_B10, THRO4156.18, 278-50, ZM197M.PB7, SF162, t257-31, Du422.1, and BG505 were from NIH ARRRP. Mutation T332N in the BG505 Env expression vector was introduced by site-directed mutagenesis using the KOD-Plus mutagenesis kit (Toyobo). The extended 25 HIV-1 pseudotyped panel was generated by adding HIV-1 isolates p1054.TC4.1499, 6535, ZM214M.PL15, AC10.29, p16845, P6244_13.B5.4576, pM246F_C1G, TRJO4551, QH0692, and pCAAN5342 obtained from NIH ARRRP. Neutralization was determined in a single-cycle neutralization assay using the standard TZM-bl neutralization assay (45). Briefly, IgGs and Multabodies were incubated with a 10 to 15% tissue culture infectious dose of PsV for 1 h at 37 °C prior to a 44- to 72-h incubation with TZM-bl cells. Virus neutralization was monitored by adding Britelite plus reagent (PerkinElmer) to the cells and measuring luminescence in relative light units using a Synergy Neo2 Multi-Mode Assay Microplate Reader (BioTek Instruments). HIV-1 Env PsVs in the extended multiclade panel of 118 PsVs were generated by transfection in 293T cells of Env expression plasmids with full-length, Env-defective HIV genome SG3dEnv. HIV-1 PsVs were incubated with Multabodies (primary concentration of 10 µg/mL and titrated sixfold seven times) for 1 h at 37 °C before TZM-bl cells were added. Luciferase expression was quantified 48 h after infection upon cell lysis and the addition of luciferin substrate (Promega). For the neutralization assays done with parental IgGs, historical data at the Center for Virology and Vaccine Research, Harvard Medical School was used (primary concentration of 50 µg/mL and titrated fivefold seven times). A cutoff limit of 10 μg/mL was used to determine antibody breadth.
Antibody-Dependent Phagocytosis.
Five microliters of red fluorescent Neutravidin microspheres (Invitrogen, F8775) were washed twice with PBS + 0.1% BSA and incubated with 10 μg of biotinylated 93TH057 gp120 antigen. Biotinylation was performed using the EZ-link Sulfo-NHS biotinylation kit (Thermo Scientific, 2143) following the manufacturer’s instructions. The final volume was brought to 200 μL with PBS/0.1% BSA and incubated overnight with rotation at 4 °C. Beads were washed twice before use to remove unbound protein, and resuspended in 200 μL per 5 μL of unlabeled bead volume.
Immune complexes were formed by incubating 93TH057 gp120-coated fluorescent beads (10 μL per sample) with 10 μL of 1, 5, and 10 μg of Multabody or antibody preparations for 2 h at 37 °C. THP-1 cells (ATCC TIB-202) were maintained at fewer than 5 × 105 cells/mL in RMPI + 10% fetal bovine serum (FBS; Wisent), and added to immune complexes at a concentration of 5 × 104 cells per well (in 200 μL) before incubation for 16 h at 37 °C, 5% CO2. After incubation, cells were pelleted and washed with PBS before staining with Live Dead Fixable Violet stain (Invitrogen, L34995) according to the manufacturer’s protocol. Cells were washed with PBS and fixed with 2% paraformaldehyde for 20 min at RT. Fixed cells were pelleted and washed once with fluorescence-activated cell sorter (FACS) buffer [PBS + 10% FBS, 0.5 mM (ethylenedinitrilo)tetraacetic acid] and analyzed on an LSRII Flow Cytometer (BD Biosciences). Data were analyzed in FlowJo (BD Biosciences), and phagocytosis was quantified as a percentage increase in phagocytosis compared to 93TH057 gp120-coated beads in the absence of antibody. Anti-human FcR binding inhibitor antibody (Invitrogen, 14-9161-73) was added to the indicated samples at the recommended concentration as an additional control.
PBMC Infection.
PBMCs were obtained from three healthy blood donors, with all donors providing written informed consent. The study was approved by the University of Toronto's Research Ethics Board (protocol #00037384). Blood was collected in heparinized vacutainers (BD Biosciences), and PBMCs were subsequently isolated using density centrifugation with Lymphoprep (StemCell Technologies, Cat#07861). PBMCs were activated with phytohemagglutinin (PHA; Gibco) in the presence of recombinant human IL-2 (50 U/mL) in complete Roswell Park Memorial Institute (RPMI) medium (Wisent), containing 10% FBS (Wisent), streptomycin at 100 µg/mL, and penicillin at 100 U/mL for 72 h prior to HIV-1 infection. After 3 d of activation, HIV-1 infection of cells was performed by addition of the CXCR4-tropic laboratory isolate IIIB (150 pg of p24 Gag antigen per well) to triplicate cultures of activated PBMCs in round-bottom 96-well plates seeded with 2 × 105 cells per well in RPMI + 10% FBS + 25 U/mL of IL-2. Before overlaying the cells with virus, Multabodies (T-01 MB and T-01 MB.v2) or IgG mixture were preincubated with virus for 1 h at RT. Infected cells were cultured in the presence/absence of Multabody or antibody controls at doses ranging from 0.01 μg/mL to 10 μg/mL, as indicated. The levels of HIV-1 replication were assessed by measuring the extracellular release of p24 Gag protein in cell-free culture supernatants tested at day 7 postinfection using a high-sensitivity AlphaLISA p24 detection kit (PerkinElmer) on a BioTek Synergy Plate Reader, according to the manufacturer’s protocols.
Cell Viability and Flow Cytometry.
On day 7 of infection, cells were fixed in 2% paraformaldehyde (PFA) and harvested for viability testing via absolute counting by flow cytometry performed using a BD LSRFortessa (Becton Dickinson). Cell viability was determined by comparison of the total live-gated cell counts in Multabody- or antibody-treated wells to the number of cells recovered from untreated control wells. Cell viability data were analyzed using FACSDiva.
Pharmacokinetic Studies.
In vivo studies were performed using three 6-wk-old female NCG (NCG strain code 572, Charles River Laboratories) immunodeficient mice per group. Mice were hosted by groups of four or six. Each mouse was uniquely identified. Animals were housed in a ventilated cage (type II [16 × 19 × 35 cm, floor area = 500 cm2]) under the following controlled conditions: 22 °C, 55% humidity, and 12:12-h light dark cycle 7 AM:7 PM. This study was reviewed and approved by the local committee in charge of the ethics of animal experimentation (CELEAG). T-01 MB composed of the scFab of antibodies PGDM1400, N49P7, and 10E8v4 and scFc fragments of IgG1 Fc containing 1) no mutations and 2) the effector function silencing mutations L234A, L235A, and P329G (LALAP) and the I253A mutation were used in the study. In addition, T-01 MB.v2 composed of the same antibody specificities with 1) no Fc mutations and 2) with the half-life extension mutations (M428L/N434S) in the IgG1 Fc were included. Mice received a single subcutaneous injection of 5 mg/kg of Multabodies or the control samples (an IgG mixture matching the Fab specificity of the Multabody) in 200 μL of PBS (pH 7.5). Blood samples were collected at multiple time points, and serum samples were assessed for levels of circulating antibodies by enzyme-linked immunosorbent assay. Briefly, 96-well Pierce Nickel-Coated Plates (Thermo Fisher) were coated with 50 μL at 0.5 μg/mL of each of the His6x-tagged antigens recognized by the MB—BG5050 D368R SOSIP.664 trimer, gp120 subunit 93TH057, and MPER peptide—to determine circulating sample concentrations using reagent-specific standard curves for IgGs and Multabodies. HRP-Protein A (Invitrogen) was used as a secondary molecule, and the chemiluminescence signal was quantified using the Epoch 2 Microplate spectrophotometer with the software BioTek Gen5 3.03.
Supplementary Material
Acknowledgments
The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases: TZM-bl cells (ARP-8129; contributed by Dr. John C. Kappes and Dr. Xiaoyun Wu); anti–HIV-1 gp160 monoclonal antibody (N6/PGDM1400x10E8v4) (ARP-13390; contributed by Drs. Ling Xu and Gary Nabel); HIV-1 NL4-3 ΔEnv Vpr luciferase reporter vector (pNL4-3.Luc.R-E-) (ARP-3418; contributed by Dr. Nathaniel Landau and Aaron Diamond); plasmids pcDNA3.1 D/V5-His TOPO-expressing HIV-1 Env/Rev (ARP-11017, ARP-11018, ARP-11024, and ARP-11022; contributed by Drs. David Montefiori, Feng Gao, and Ming Li); plasmid pcDNA3.1(+)-expressing HIV-1 Env/Rev (ARP-11037; contributed by Drs. B. H. Hahn and D. L. Kothe); plasmid pcDNA3.1 D/V5-His TOPO-expressing HIV-1 Env/Rev (ARP-11308; contributed by Drs. D. Montefiori, F. Gao, C. Williamson, and S. Abdool Karim); plasmid pcDNA3.1 V5-His TOPO-expressing HIV-1 Env/Rev (ARP-11309; contributed by Drs. B. H. Hahn, Y. Li, and J. F. Salazar-Gonzalez); HIV-1 BG505 Env expression vector (BG505.W6M.ENV.C2) (ARP-11518; contributed by Dr. Julie Overbaugh); HIV-1 Env expression vector (CRF02_AG clone 257) (ARP-11599; contributed by Drs. D. Ellenberger, B. Li, M. Callahan, and S. Butera); plasmid pcDNA3.1 V5-His TOPO-expressing HIV-1 CNE8 Env (ARP-12653; contributed by Drs. Linqi Zhang, Hong Shang, David Montefiori, Tsinghua University (Beijing, China), China Medical University (Beijing, China), and Duke University (Durham, NC); HIV-1 SF162 gp160 expression vector (ARP-10463; contributed by Drs. Leonidas Stamatatos and Cecilia Cheng-Mayer); plasmid pcDNA3.1 V5-His TOPO-expressing HIV-1 Env/Rev (ARP-11034; contributed by Drs. B. H. Hahn, X. Wei, and G. M. Shaw); plasmid pcDNA3.1/V5-His TOPO-expressing HIV Env/Rev (ARP-11038; contributed by Drs. B. H. Hahn and D. L. Kothe); plasmid pcDNA3.1 V5-His TOPO-expressing HIV-1 Env/Rev (ARP-11310; contributed by Drs. B. H. Hahn, Y. Li, and J. F. Salazar-Gonzalez); HIV-1 Env expression vector (p16845 env) (ARP-11503; contributed by Drs. R. Paranjape, S. Kulkarni, and D. Montefiori); HIV-1 1054 Env expression vector (p1054.TC4.1499) (ARP-11561) and 6244 Env expression vector (p6244_13.B5.4576) (ARP-11566; contributed by Drs. Beatrice H. Hahn, Brandon F. Keele, and George M. Shaw); HIV-1 ZM246F Env expression vector (pZM246F_C1G) (ARP-11830; contributed by Dr. Beatrice Hahn); HIV-1 Env expression vector (CRF02_AG clone 278) (ARP-11605; contributed by Drs. Michael Thomson, Ana Revilla, Elena Delgado, David Montefiori, Sonia Pérez Castro, Centro Nacional de Microbiologia, Instituto de Salud Carlos III (Majadahonda, Madrid, Spain), Complejo Hospitalario Santa María Madre (Orense, Spain), Duke University (Durham, NC), and the CAVD; and NL4-3 Env expression vector (pDOLHIVenv) (from Dr. Eric Freed and Dr. Rex Risser). The following reagents were kindly provided by CAVD: X2988, ZM106.9, and 3817. We thank S. Tabruyn and F. Arbogast for their assistance with in vivo studies. We thank the SickKids-University Health Network Flow Cytometry Facility. This work was supported by Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant 6280100058 (J.-P.J.) and by Operating Grant PJ4-169662 from the Canadian Institutes of Health Research (CIHR; B.T. and J.-P.J.). This research was also supported by the European Union’s Horizon 2020 research and innovation program under Marie Sklodowska-Curie Grant 790012 (E.R.), a Hospital for Sick Children Restracomp Postdoctoral Fellowship (C.B.A.), an NSERC postgraduate doctoral scholarship (T.Z.), a predoctoral fellowship from the Basque Government (PRE_2019_2_0046) (S.I.), the Canadian Institute for Advanced Research (CIFAR) Azrieli Global Scholar program (J.-P.J.), the Ontario Early Researcher Awards program (J.-P.J.), and the Canada Research Chairs program (B.T. and J.-P.J.). This work was supported, in part, by NSERC Discovery Grant RGPIN-2019-06442 and CIHR Project Grant–Priority Announcement PJH-175379 to C.G., and a CIHR Canada Graduate Scholarship (CGS-M) to J.B. Further support was obtained from the Spanish Ministry of Science, Innovation and Universities (MCIU) with the support of the Spanish Research Agency/The European Regional Development Fund (AEI/FEDER) (RTI2018-095624-B-C21) (J.L.N.) and the Basque Government (IT1196-19) (J.L.N.). Biophysical data were collected at the Structural & Biophysical Core facility supported by the Canada Foundation for Innovation and Ontario Research Fund.
Footnotes
Competing interest statement: The Hospital for Sick Children has applied for patents concerning the Multabody platform technology that are related to this work. B.T. and J.-P.J. are founders of Radiant Biotherapeutics and are members of its Scientific Advisory Board.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112887119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix. Requests for material can be directed to J.-P.J. (jean-philippe.julien@sickkids.ca). All materials and reagents will be available upon establishing a material transfer agreement.
References
- 1.Conley A. J., et al. , Neutralization of divergent human immunodeficiency virus type 1 variants and primary isolates by IAM-41-2F5, an anti-gp41 human monoclonal antibody. Proc. Natl. Acad. Sci. U.S.A. 91, 3348–3352 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stiegler G., et al. , A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 17, 1757–1765 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Zwick M. B., et al. , Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75, 10892–10905 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchacher A., et al. , Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retroviruses 10, 359–369 (1994). [DOI] [PubMed] [Google Scholar]
- 5.Barbas C. F. III, et al. , Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc. Natl. Acad. Sci. U.S.A. 89, 9339–9343 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton D. R., et al. , Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266, 1024–1027 (1994). [DOI] [PubMed] [Google Scholar]
- 7.Wu X., et al. , Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheid J. F., et al. , Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458, 636–640 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Sok D., et al. , Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc. Natl. Acad. Sci. U.S.A. 111, 17624–17629 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker L. M., et al. , Protocol G Principal Investigators, Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doria-Rose N. A., et al. , NISC Comparative Sequencing Program, Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509, 55–62 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J., et al. , Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker L. M., et al. , Protocol G Principal Investigators, Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sajadi M. M., et al. , Identification of near-pan-neutralizing antibodies against HIV-1 by deconvolution of plasma humoral responses. Cell 173, 1783–1795.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheid J. F., et al. , Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoofs T., et al. , Broad and potent neutralizing antibodies recognize the silent face of the HIV envelope. Immunity 50, 1513–1529.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J., et al. , Identification of a CD4-binding-site antibody to HIV that evolved near-pan neutralization breadth. Immunity 45, 1108–1121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pejchal R., et al. , A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334, 1097–1103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blattner C., et al. , Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity 40, 669–680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mouquet H., et al. , Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl. Acad. Sci. U.S.A. 109, E3268–E3277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baba T. W., et al. , Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6, 200–206 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Hessell A. J., et al. , Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat. Med. 15, 951–954 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hessell A. J., et al. , Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 5, e1000433 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofmann-Lehmann R., et al. , Postnatal pre- and postexposure passive immunization strategies: Protection of neonatal macaques against oral simian-human immunodeficiency virus challenge. J. Med. Primatol. 31, 109–119 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Hofmann-Lehmann R., et al. , Postnatal passive immunization of neonatal macaques with a triple combination of human monoclonal antibodies against oral simian-human immunodeficiency virus challenge. J. Virol. 75, 7470–7480 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Velden Y. U., et al. , Short communication: Protective efficacy of broadly neutralizing antibody PGDM1400 against HIV-1 challenge in humanized mice. AIDS Res. Hum. Retroviruses 34, 790–793 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Klein F., et al. , HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 492, 118–122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deruaz M., et al. , Protection of humanized mice from repeated intravaginal HIV challenge by passive immunization: A model for studying the efficacy of neutralizing antibodies in vivo. J. Infect. Dis. 214, 612–616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horwitz J. A., et al. , HIV-1 suppression and durable control by combining single broadly neutralizing antibodies and antiretroviral drugs in humanized mice. Proc. Natl. Acad. Sci. U.S.A. 110, 16538–16543 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehandru S., et al. , Adjunctive passive immunotherapy in human immunodeficiency virus type 1-infected individuals treated with antiviral therapy during acute and early infection. J. Virol. 81, 11016–11031 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caskey M., et al. , Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522, 487–491 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caskey M., et al. , Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat. Med. 23, 185–191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lynch R. M., et al. , VRC 601 Study Team, Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci. Transl. Med. 7, 319ra206 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Ledgerwood J. E., et al. , VRC 602 Study Team, Safety, pharmacokinetics and neutralization of the broadly neutralizing HIV-1 human monoclonal antibody VRC01 in healthy adults. Clin. Exp. Immunol. 182, 289–301 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corey L., et al. , HVTN 704/HPTN 085 and HVTN 703/HPTN 081 Study Teams, Two randomized trials of neutralizing antibodies to prevent HIV-1 acquisition. N. Engl. J. Med. 384, 1003–1014 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rudicell R. S., et al. , NISC Comparative Sequencing Program, Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J. Virol. 88, 12669–12682 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon Y. D., et al. , Surface-matrix screening identifies semi-specific interactions that improve potency of a near pan-reactive HIV-1-neutralizing antibody. Cell Rep. 22, 1798–1809 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rujas E., et al. , Functional optimization of broadly neutralizing HIV-1 antibody 10E8 by promotion of membrane interactions. J. Virol. 92, e02249–e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rujas E., et al. , Affinity for the interface underpins potency of antibodies operating in membrane environments. Cell Rep. 32, 108037 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diskin R., et al. , Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 334, 1289–1293 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rujas E., et al. , Multivalency transforms SARS-CoV-2 antibodies into ultrapotent neutralizers. Nat. Commun. 12, 3661 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon Y. D., et al. , Optimization of the solubility of HIV-1-neutralizing antibody 10E8 through somatic variation and structure-based design. J. Virol. 90, 5899–5914 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacobson J. M., et al. , Safety, pharmacokinetics, and antiretroviral activity of multiple doses of ibalizumab (formerly TNX-355), an anti-CD4 monoclonal antibody, in human immunodeficiency virus type 1-infected adults. Antimicrob. Agents Chemother. 53, 450–457 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuritzkes D. R., et al. , Antiretroviral activity of the anti-CD4 monoclonal antibody TNX-355 in patients infected with HIV type 1. J. Infect. Dis. 189, 286–291 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Montefiori D. C., Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol. Biol. 485, 395–405 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Xu L., et al. , Trispecific broadly neutralizing HIV antibodies mediate potent SHIV protection in macaques. Science 358, 85–90 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qureshi O. S., et al. , Multivalent Fcγ-receptor engagement by a hexameric Fc-fusion protein triggers Fcγ-receptor internalisation and modulation of Fcγ-receptor functions. Sci. Rep. 7, 17049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asokan M., et al. , Bispecific antibodies targeting different epitopes on the HIV-1 envelope exhibit broad and potent neutralization. J. Virol. 89, 12501–12512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan S. N., et al. , Targeting the HIV-1 spike and coreceptor with bi- and trispecific antibodies for single-component broad inhibition of entry. J. Virol. 92, e00384-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steinhardt J. J., et al. , Rational design of a trispecific antibody targeting the HIV-1 Env with elevated anti-viral activity. Nat. Commun. 9, 877 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith R. I., Coloma M. J., Morrison S. L., Addition of a mu-tailpiece to IgG results in polymeric antibodies with enhanced effector functions including complement-mediated cytolysis by IgG4. J. Immunol. 154, 2226–2236 (1995). [PubMed] [Google Scholar]
- 52.Olafsen T., Rasmussen I. B., Norderhaug L., Bruland Ø. S., Sandlie I., IgM secretory tailpiece drives multimerisation of bivalent scFv fragments in eukaryotic cells. Immunotechnology 4, 141–153 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Miller K., et al. , Design, construction, and in vitro analyses of multivalent antibodies. J. Immunol. 170, 4854–4861 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Wu C., et al. , Simultaneous targeting of multiple disease mediators by a dual-variable-domain immunoglobulin. Nat. Biotechnol. 25, 1290–1297 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Klein C., Schaefer W., Regula J. T., The use of CrossMAb technology for the generation of bi- and multispecific antibodies. MAbs 8, 1010–1020 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steinmetz A., et al. , CODV-Ig, a universal bispecific tetravalent and multifunctional immunoglobulin format for medical applications. MAbs 8, 867–878 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kipriyanov S. M., et al. , Bispecific tandem diabody for tumor therapy with improved antigen binding and pharmacokinetics. J. Mol. Biol. 293, 41–56 (1999). [DOI] [PubMed] [Google Scholar]
- 58.Lu D., et al. , Di-diabody: A novel tetravalent bispecific antibody molecule by design. J. Immunol. Methods 279, 219–232 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Kubetzko S., Balic E., Waibel R., Zangemeister-Wittke U., Plückthun A., PEGylation and multimerization of the anti-p185HER-2 single chain Fv fragment 4D5: Effects on tumor targeting. J. Biol. Chem. 281, 35186–35201 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Pack P., Müller K., Zahn R., Plückthun A., Tetravalent miniantibodies with high avidity assembling in Escherichia coli. J. Mol. Biol. 246, 28–34 (1995). [DOI] [PubMed] [Google Scholar]
- 61.Kipriyanov S. M., et al. , Affinity enhancement of a recombinant antibody: Formation of complexes with multiple valency by a single-chain Fv fragment-core streptavidin fusion. Protein Eng. 9, 203–211 (1996). [DOI] [PubMed] [Google Scholar]
- 62.Deyev S. M., Waibel R., Lebedenko E. N., Schubiger A. P., Plückthun A., Design of multivalent complexes using the barnase*barstar module. Nat. Biotechnol. 21, 1486–1492 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Hoffmann M. A. G., et al. , Nanoparticles presenting clusters of CD4 expose a universal vulnerability of HIV-1 by mimicking target cells. Proc. Natl. Acad. Sci. U.S.A. 117, 18719–18728 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Divine R., et al. , Designed proteins assemble antibodies into modular nanocages. Science 372, eabd99947 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wörn A., Plückthun A., Different equilibrium stability behavior of ScFv fragments: Identification, classification, and improvement by protein engineering. Biochemistry 38, 8739–8750 (1999). [DOI] [PubMed] [Google Scholar]
- 66.Fleit H. B., Kobasiuk C. D., The human monocyte-like cell line THP-1 expresses Fc gamma RI and Fc gamma RII. J. Leukoc. Biol. 49, 556–565 (1991). [DOI] [PubMed] [Google Scholar]
- 67.Zhu P., et al. , Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc. Natl. Acad. Sci. U.S.A. 100, 15812–15817 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chojnacki J., et al. , Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science 338, 524–528 (2012). [DOI] [PubMed] [Google Scholar]
- 69.Graille M., et al. , Complex between Peptostreptococcus magnus protein L and a human antibody reveals structural convergence in the interaction modes of Fab binding proteins. Structure 9, 679–687 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix. Requests for material can be directed to J.-P.J. (jean-philippe.julien@sickkids.ca). All materials and reagents will be available upon establishing a material transfer agreement.