Significance
Topoisomerases are crucial for genome maintenance and are targets for several chemotherapeutic agents. While anticancer drugs targeting topoisomerases can lead to secondary malignancies, there have been no descriptions of genetic defects in topoisomerases having roles in cancer development. Here we show that a somatic topoisomerase IIα mutation found in human tumors results in a mutator phenotype. We show that this mutation and the concomitant mutational signature, which we call ID_TOP2α, are associated with genomic rearrangements and with potentially oncogenic indel mutations in known driver genes. Our results shed new light on topoisomerase IIα function, on repair of trapped cleavage complexes, and on a likely oncogenic role for topoisomerases.
Keywords: topoisomerase II, duplications, cancer, indel mutational signature, yeast
Abstract
Topoisomerases nick and reseal DNA to relieve torsional stress associated with transcription and replication and to resolve structures such as knots and catenanes. Stabilization of the yeast Top2 cleavage intermediates is mutagenic in yeast, but whether this extends to higher eukaryotes is less clear. Chemotherapeutic topoisomerase poisons also elevate cleavage, resulting in mutagenesis. Here, we describe p.K743N mutations in human topoisomerase hTOP2α and link them to a previously undescribed mutator phenotype in cancer. Overexpression of the orthologous mutant protein in yeast generated a characteristic pattern of 2- to 4-base pair (bp) duplications resembling those in tumors with p.K743N. Using mutant strains and biochemical analysis, we determined the genetic requirements of this mutagenic process and showed that it results from trapping of the mutant yeast yTop2 cleavage complex. In addition to 2- to 4-bp duplications, hTOP2α p.K743N is also associated with deletions that are absent in yeast. We call the combined pattern of duplications and deletions ID_TOP2α. All seven tumors carrying the hTOP2α p.K743N mutation showed ID_TOP2α, while it was absent from all other tumors examined (n = 12,269). Each tumor with the ID_TOP2α signature had indels in several known cancer genes, which included frameshift mutations in tumor suppressors PTEN and TP53 and an activating insertion in BRAF. Sequence motifs found at ID_TOP2α mutations were present at 80% of indels in cancer-driver genes, suggesting that ID_TOP2α mutagenesis may contribute to tumorigenesis. The results reported here shed further light on the role of topoisomerase II in genome instability.
Topoisomerases are critical for managing the torsional stress associated with the DNA unwinding required for transcription and replication and for decatenating sister chromatids to allow their separation during mitosis. Type II topoisomerases resolve these topological problems by transiently nicking both DNA strands to create a double-strand break (DSB) through which an intact duplex can pass (1–3). During this reaction, the 5′ end of each nick is covalently linked to a topoisomerase monomer by a phosphotyrosyl bond (4). Humans have two genes encoding type II topoisomerases: hTOP2A on chromosome 17 and hTOP2B on chromosome 3 (5). The encoded proteins, hTOP2α and hTOP2β, have unique but overlapping functions. hTOP2α is expressed in proliferating cells and is essential for the viability of proliferating cells, while hTOP2β is also expressed in quiescent cells and plays an important role in regulating transcription (1, 6). Topoisomerases have been widely studied as chemotherapeutic targets, and several topoisomerase poisons, such as etoposide and doxorubicin, are commonly used in the clinic (5, 7). Clinically active topoisomerase-targeting agents cause elevated levels of Top2 covalent complexes that interfere with DNA metabolism, leading to the accumulation of DSBs that kill rapidly dividing cancer cells. In some cases, up-regulation of type II topoisomerases in cancer is a marker of poor prognosis (8, 9).
We recently described an allele of yeast topoisomerase II (yTop2) that is associated with elevated mutation rates (10). The product of this allele, the yTop2-F1025Y,R1128G protein (abbreviated here as yTop2-FY,RG) forms elevated levels of stabilized cleavage complexes, with the mutagenic repair of the resulting DSB specifically increasing 2- to 4-base pair (bp) duplications without elevating single-base substitutions (10). Here, we describe a mutant form of hTOP2α that is present in a small subset of tumors and is associated with a distinctive mutational signature comprising duplications similar to those reported in yeast as well as small deletions.
Results
Somatic hTOP2α p.K743N Mutations Associate with 2- to 4-bp Duplications.
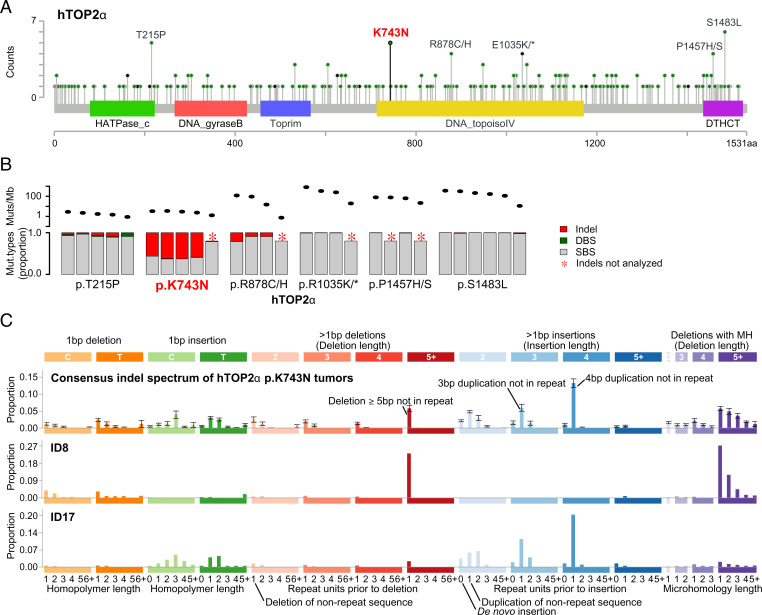
To determine whether defects in hTOP2α or hTOP2β contribute to carcinogenesis, we looked for recurrent mutations in 23,829 whole-exome and whole-genome sequenced tumors (11). hTOP2α showed several hotspots in various protein domains and hTOP2β showed a clear hotspot at p.R651 (Fig. 1A and SI Appendix, Fig. S1). Most recurrent hTOP2α mutations and hTOP2β p.R651 were in highly mutated (>10 mutations per megabase) tumors (Fig. 1B and Dataset S1). By contrast, hTOP2α p.K743N was observed in four whole-exome sequenced (WES) gastric cancers (GCs) and one whole-genome sequenced (WGS) cholangiocarcinoma (CCA_TH_19), none of which were highly mutated. Strikingly, all GCs carrying hTOP2α p.K743N showed elevated levels of small insertions and deletions (indels) (CCA_TH_19 had not been previously analyzed for indels).
Fig. 1.
hTOP2α hotspot mutations and associated mutagenesis. (A) Recurrent amino acid substitutions in hTOP2α (figure generated using MutationMapper) (43). Amino acid positions are marked on the x axis and domains are shown by colored rectangles with names below. (B) Total mutation counts and proportions of indel, DBS, and SBS mutations in hTOP2α hotspot carriers as reported by Alexandrov et al. (11). Ovals in the Top subpanel show the combined mutation load of indels, DBSs, and SBSs; bars in the Bottom panel show the proportions of indels (red), DBSs (green), and SBSs (gray). (C) Consensus indel mutation spectrum of tumors carrying the hTOP2α p.K743N substitution and the ID8 and ID17 indel mutational signatures (11). Each bar shows the mean proportion of indels in a given indel class; error bars show SEM. Abbreviations: DBS, doublet base substitution; MH, microhomology; Mut, mutation; SBS, single base substitution. Please see https://www.synapse.org/#!Synapse:syn11801742 for an explanation of the indel classes.
In recent years, much progress has been made on understanding mutational processes through the analysis of mutational signatures (11, 12). A mutational signature is a representation of the proportions of different types of mutations caused by a mutational process. For indels, the mutations are categorized into 83 types, reflecting the size of the indel and the surrounding sequence context (11). To date, 18 indel mutational signatures have been described, but the etiology for most remains unknown (13). To determine whether the indels in the hTOP2α p.K743N carriers match any previously described indel mutational signature, we plotted the indel mutation spectra for each of these tumors (SI Appendix, Fig. S2). The GC indel spectra were partly composed of a distinct pattern that was previously identified as indel mutational signature ID17, which is characterized by duplications of 2 to 4 bp in nonrepetitive sequences (11) (Fig. 1C). This preponderance of 2- to 4-bp duplications was previously observed in yeast carrying yTop2-FY,RG (10). Among 12,273 tumors with indels analyzed by Alexandrov et al. (11, 12), hTOP2α p.K743N and ID17 occurred only in these four GCs (P = 1.06 × 10−15, two-sided Fisher’s exact test). The Catalogue Of Somatic Mutations In Cancer (COSMIC) contained two additional tumors carrying hTOP2α p.K743N: one pancreatic adenocarcinoma and one prostate cancer, and WES was available for these (14, 15). Realignment and indel calling of these two tumors as well as CCA_TH_19 revealed ID17-like mutagenesis in all three (Fig. 1C and SI Appendix, Fig. S2).
yTop2-K720N Is DNA Damaging and Introduces Stalled Cleavage Complexes.
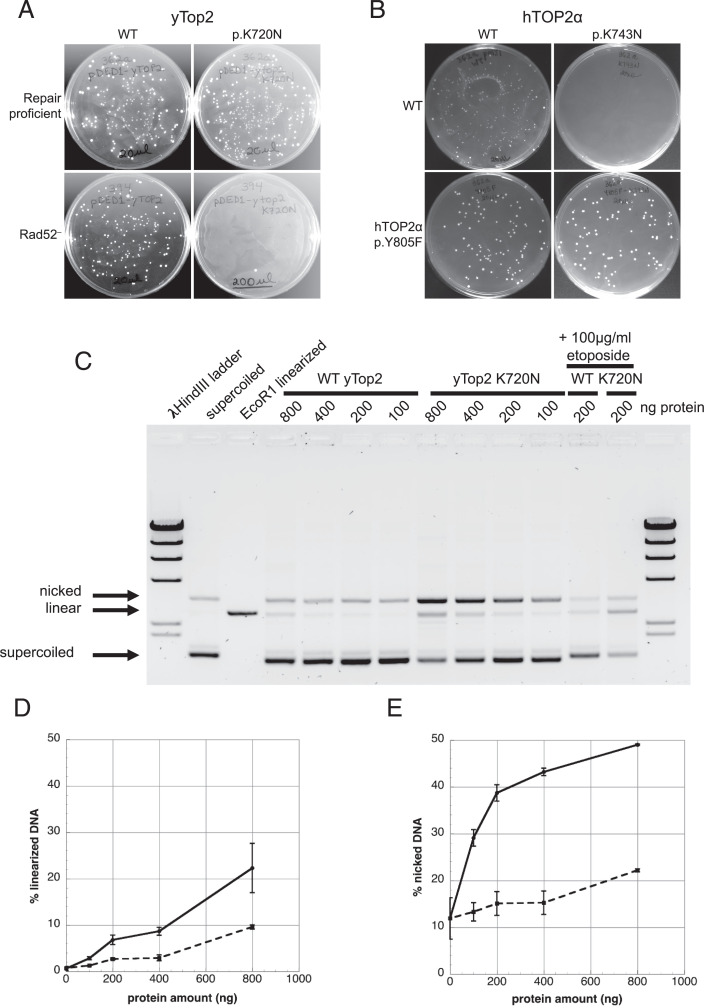
To investigate whether hTOP2α p.K743N causes the 2- to 4-bp duplications resembling ID17, we introduced the orthologous mutation (K720N) into the yTop2. We found that yTop2-K720N could be readily overexpressed in wild-type yeast but not in a strain lacking the homologous recombination repair pathway protein Rad52 (Fig. 2A). Strikingly, not even repair-proficient yeast tolerated overexpression of hTOP2α p.K743N, indicating that this protein likely introduces more DNA damage than yTop2-K720N (Fig. 2B). Overexpression of a cleavage-incompetent (p.Y805F) hTOP2α double mutant was tolerated, however, demonstrating that hTOP2α p.K743N toxicity is associated with its DNA cleavage activity (Fig. 2B).
Fig. 2.
yTop2-K720N effects on cell viability and DNA-damaging properties. (A) Viability after transformation of a plasmid overexpressing yTop2 or yTop2-K720N into wild-type (WT) and recombination-deficient (Rad52−) strains. (B) Viability following overexpression of hTOP2α or hTOP2α p.K743N in yeast. Introduction of p.Y805F into hTOP2α p.K743N rescued the toxicity. (C) yTop2-mediated cleavage of negatively supercoiled pUC18 in the presence of Mg2+ as a function of protein concentration. Samples with 200 ng WT yTop2 or yTop2-K720N proteins and 100 µg/mL etoposide are included as positive controls for DNA cleavage. (D) Quantitation of linear DNA and (E) nicked DNA generated by WT yTop2 (dashed lines) or yTop2-K720N (solid lines) protein; bars indicate SEM, n = 3.
The topoisomerase II homodimer relieves torsional stress in DNA by creating a transient DSB through which an intact duplex can pass. During this process each monomer covalently binds one of the DNA ends. We previously showed that yTop2-FY,RG lethality in the absence of Rad52 stems from trapped cleavage complexes, and we hypothesized that yTop2-K720N also results in trapped cleavage complexes (10). To investigate this, we overexpressed and purified yTop2-K720N. Wild-type yTop2 and yTop2-K720N had a similar ability to decatenate kinetoplast DNA, indicating comparable catalytic activity (SI Appendix, Fig. S3). We next assessed the ability of the purified yTop2-K720N to damage DNA by generating covalent complexes. Fig. 2C shows a standard cleavage assay with purified proteins and supercoiled pUC18 as a substrate. Wild-type Top2 showed low levels of covalent complexes that include either DSBs (giving rise to linearized plasmid DNA) or single-strand breaks (giving rise to nicked DNA). By contrast, plasmid DNA treated with yTop2-K720N protein gave rise to substantially higher levels of both linear and nicked DNA. Quantitation of the linear DNA in Fig. 2C shows that, at all protein concentrations examined, yTop2-K720N protein resulted in a two- to threefold increase in linearized plasmid DNA (Fig. 2D). A similar increase was seen when the level of nicked DNA was quantitated (Fig. 2E). Taken together, these results demonstrate a higher steady-state level of cleavage complexes in reactions with yTop2-K720N protein compared to wild-type yTop2. We also examined DNA cleavage when Ca2+ replaces Mg2+ as a divalent cation. Previous work showed that Ca2+ leads to elevated levels of covalent complexes compared to reactions in the presence of Mg2+ (16). In the presence of Ca2+, robust single- and double-strand cleavage was seen at low concentrations of yTop2-K720N protein, and higher concentrations of protein gave rise to elevated cleavage, compared to the wild-type Top2 protein (SI Appendix, Fig. S4). We conclude that the yTop2-K720N protein generates DNA damage through enzyme-mediated DNA cleavage, and that the hTOP2α p.K743N protein likely operates similarly.
yTop2-K720N Is Associated with ID17-Like Duplications.
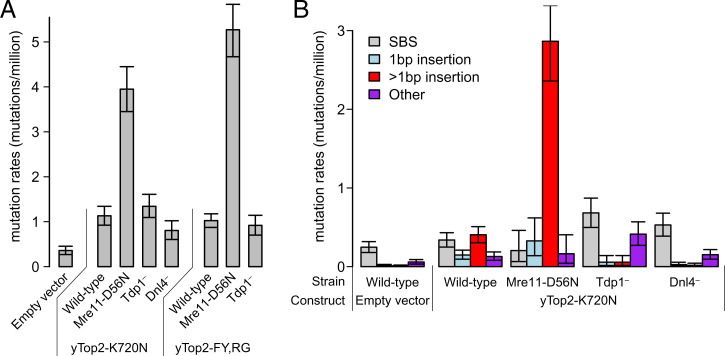
To examine mutagenesis associated with yTop-K720N, we employed a forward-mutation assay used previously to characterize the duplications associated with yTop2-FY,RG. This approach identifies inactivating mutations in the CAN1 locus that confer resistance to canavanine (10). The can1 mutation rate increased ∼4-fold in cells overexpressing yTop2-K720N compared to the control, and was similar to the elevated mutation rate associated with yTop2-FY,RG (Fig. 3A). Whereas yTop2-K720N had no effect on the rate of single-base substitutions, there was a 72-fold increase in insertions >1 bp (Fig. 3B). As seen in mutational signature ID17 and in the human hTOP2α p.K743N tumors, the most common duplication size was 4 bp (SI Appendix, Fig. S5), which is the distance between topoisomerase II-generated nicks that comprise the enzyme-induced DSBs (17). In the yeast data, both yTop2-FY,RG and yTop2-K720N were associated with fewer 3-bp duplications than in the human data. This is likely due to the experimental design, which selects for mutations that disrupt Can1 function; 3-bp insertions are in-frame events and are less likely to disrupt CAN1.
Fig. 3.
Mutations induced by yTop2-K720N. (A) Overall mutation rates at the yeast CAN1 locus with 95% confidence intervals. Data for empty vector and yTop2-FY,RG are from Stantial et al. (10). (B) Rates of various types of mutations at the same locus shown separately. Mre11-D56N, genetically inactivated nuclease subunit of the MRX complex; Tdp1, tyrosyl-DNA-phosphodiesterase I; Dnl4, DNA ligase required for NHEJ; SBS, single base substitution.
Insertions caused by yTop2-FY,RG depend on both removal of covalent yTop2 cleavage complexes from the DNA by tyrosyl-DNA phosphodiesterase I (Tdp1) and on subsequent DSB repair by nonhomologous end joining (NHEJ). To determine whether yTop2-K720N insertions have the same genetic requirements, we deleted TDP1 as well as DNL4, which encodes the ligase required for NHEJ. Loss of Dnl4 and Tdp1 both substantially reduced the overall rate of can1 mutants and almost completely eliminated insertions, confirming that the duplications depend on Tdp1 and NHEJ (Fig. 3 A and B). In addition, removal of yTop2-FY,RG cleavage complexes by the Mre11-Rad50-Sae2 complex initiates homologous recombination repair, which is predominantly error-free repair (10). As seen previously with the yTop2-FY,RG allele, cells with nuclease-dead Mre11 (Mre11-D56N) had an approximately fivefold higher duplication rate than yTop2-K720N cells with wild-type Mre11 (Fig. 3B).
All Indel Classes in p.K743N Tumors Show Features of hTOP2α-Associated Mutagenesis.
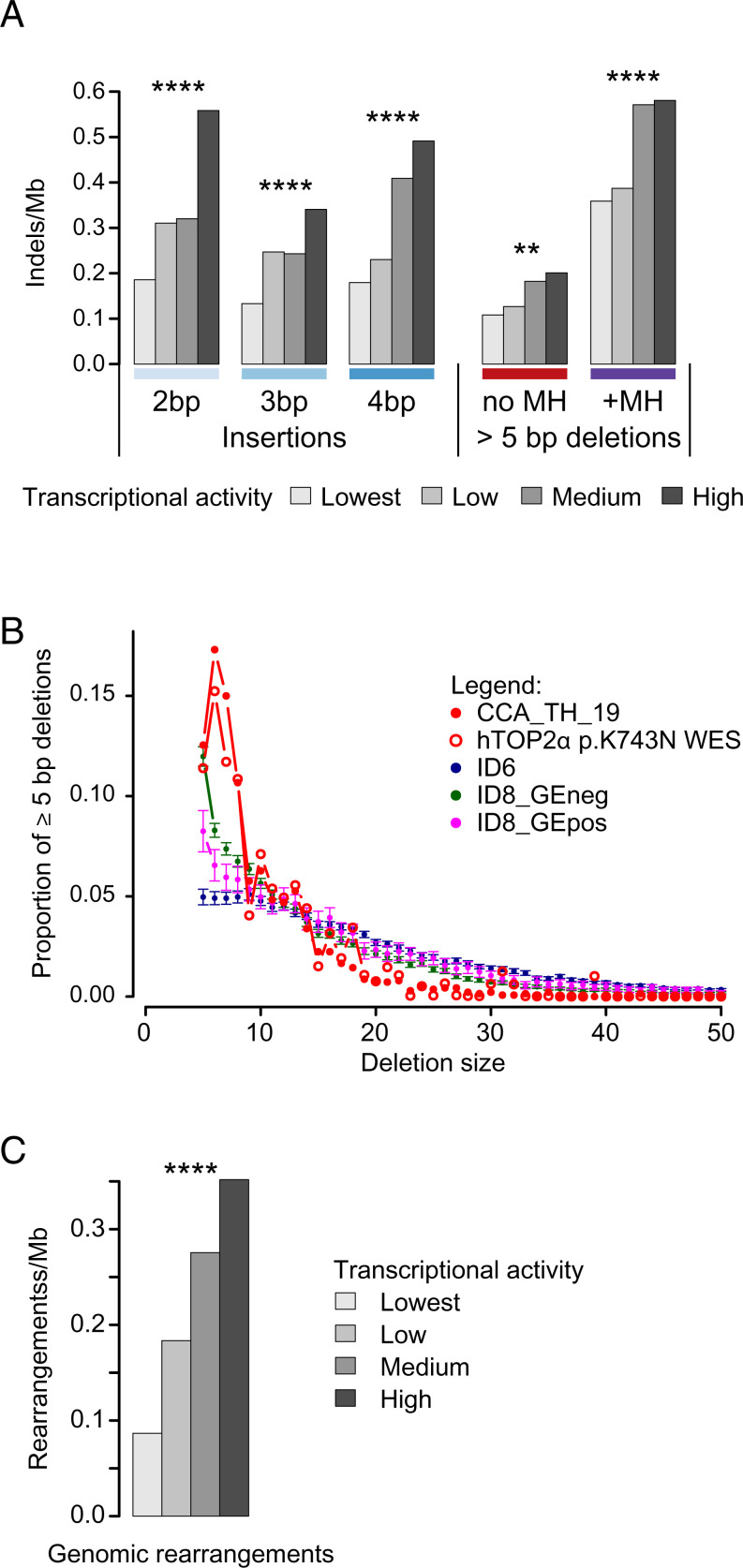
Having confirmed in the yeast model that hTOP2α p.K743N likely causes the ID17-like 2- to 4-bp duplications in p.K743N tumors, we next sought to confirm that other aspects of the mutagenesis observed in these tumors is consistent with the known biology of hTOP2α. Because hTOP2α cleavage is enriched in highly transcribed regions (18), we examined ID17 mutagenesis in relation to transcriptional activity. In-depth analysis in the only tumor with WGS, CCA_TH_19, showed increased mutational activity at highly transcribed regions (P = 6.13 × 10−53, one-sided Cochran–Armitage test, SI Appendix, Fig. S6). Notably, in addition to the ID17-like duplications of 2 to 4 bp, all other classes of indels were increased with transcriptional activity, which is consistent with known hTOP2α biology and suggests that many of them also stemmed from hTOP2α p.K743N mutagenesis (false discovery rate [FDR] <0.05 for all classes, one-sided Cochran–Armitage test, Fig. 4A and SI Appendix, Fig. S7A). As with CCA_TH_19, all WES samples also showed higher density of indel mutagenesis in more highly transcribed regions (P = 0.016, one-sided sign-test, SI Appendix, Fig. S7B).
Fig. 4.
Characterization of ID_TOP2α. (A) Positive correlation between transcriptional activity and 2- to 4-bp duplications and deletions ≥5 bp in CCA_TH_19. (B) Comparison of size distributions of deletions ≥5 bp in hTOP2α p.K743N carriers, in tumors with high counts of ID6 deletions, in tumors with high counts of ID8 deletions with increasing mutation density in highly transcribed genes (ID8_GEpos), and in tumors with high counts of ID8 deletions without this characteristic (ID8_GEneg). Please see SI Appendix for details. Compared to each of the other groups of tumors, hTOP2α p.K743N carriers were enriched for 6- to 8-bp deletions (P < 0.0001 for each of the pairwise comparisons, two-sided Wilcoxon rank-sum tests). For ID6, ID8_Geneg, and ID8_GEpos, means and 95% confidence intervals are plotted. For display purposes, all hTOP2α p.K743N WES data were combined; statistical comparisons between the groups were performed using the individual tumors. (C) Positive correlation between transcriptional activity and genomic rearrangements in CCA_TH_19. ** and **** indicate Benjamini–Hochberg false discovery rates of <0.01 and <0.0001, based on P values from two-sided Cochran–Armitage tests for trend. Mb, megabase; MH, microhomology.
hTOP2α p.K743N Is Also Associated with ID8-Like Deletions ≥5 bp.
We noted that the ID17-like pattern of duplications in the hTOP2α p.K743N tumors always co-occur with a pattern of deletions that resembles indel mutational signature ID8, which consists almost entirely of ≥5-bp deletions not occurring in repeats. Indeed, in three of the four indel spectra from hTOP2α p.K743N GCs, Alexandrov et al. assigned the ≥5-bp deletions to ID8 (11). Furthermore, the numbers of ID17-like mutations and ≥5-bp deletions were correlated in the p.K743N tumors. While this suggests that the signature of hTOP2α p.K743N might in fact be a combination of ID17-like mutations and deletions ≥5 bp, ID8 also occurs in the majority of tumors lacking hTOP2α p.K743N and in most cancer types. This observation led us to ask whether the ID8-like mutations in hTOP2α p.K743N tumors might in fact stem from the same unknown, but not hTOP2α-related, mutational processes that generate ID8. We took two approaches to investigating this question.
First, we examined association of mutation density with transcriptional activity. We compared the ID8-like deletions (deletions ≥5 bp not in repeats) in the hTOP2α p.K743N tumors to those in WGS tumors with indels analyzed by Alexandrov et al. (11) and the ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium (19). These deletions were positively correlated with transcriptional activity in only 124 tumors (Dataset S2); most of these tumors showed ID8 mutagenesis (82.6%). However, in the vast majority of tumors with ID8, transcriptional activity and ID8 density were uncorrelated.
Following up on this observation, because there is evidence of a general trend for enrichment of indels in genic regions (transcripts) versus intergenic regions across cancer types (20, 21), we dissected this enrichment by subcategories of indels in CCA_TH_19 and in non-hTOP2α p.K743N tumors (from ref. 19). We analyzed separately the following subcategories of indels: Duplications of 2 to 4 bp and deletions of ≥5 bp not in repeats with and without microhomology, that is, the deletions constituting the vast majority of ID8-like deletions (Fig. 1C). In CCA_TH_19 versus the other tumors, duplications of 2, 3, and 4 bp were more prevalent in genic regions (SI Appendix, Fig. S8). For deletions of ≥5 bp with microhomology, CCA_TH_19 had the second highest density of deletions in genic regions and clear enrichment for mutations in genic regions, but a few other tumors with high mutation counts also had enrichment for genic mutations, and many but not all tumors with lower mutation counts also had higher mutation densities in genic regions (SI Appendix, Fig. S9). For deletions ≥5 bp not in repeats and without microhomology, CCA_TH_19 had the second highest number of deletions and a clear genic enrichment for these deletions (SI Appendix, Fig. S9). Again however, most tumors with lower deletion counts had more genic than intergenic deletions, though some had more intergenic than genic deletions. In addition, eight tumors had genic enrichment similar or more extreme than CCA_TH_19. None of these eight had nonsilent mutations in TOP2α or β, and their mutational spectra were otherwise unremarkable (SI Appendix, Fig. S10).
Second, we considered whether there might be differences in the distributions of sizes of deletions not in repeats in hTOP2α p.K743N tumors versus other tumors. In particular, categorizing all deletions of length ≥5 bp as a single indel type might have obscured important differences in deletion-size distributions stemming from different mutational processes. To investigate this, we compared the sizes of deletions ≥5 bp in hTOP2α p.K743N tumors to those in other tumors with many deletions ≥5 bp. The latter were tumors with many deletions ascribed to ID6 or ID8 mutations. (ID6 consists mainly of deletions of ≥5 bp with microhomology; although it does not resemble the deletions in hTOP2α p.K743N tumors, it is included here as another signature dominated by deletions of ≥5 bp.) We divided the tumors into three groups for analysis: ID6 tumors, ID8 tumors with increased ID8 mutagenesis in highly transcribed regions, and ID8 tumors without increased mutagenesis in transcribed regions (ID6, ID8_Gepos, and ID8_GEneg, respectively, Fig. 4B and Dataset S2). Six- to 8-bp deletions were highly enriched in hTOP2α p.K743N tumors versus all other groups of tumors (P = 7.49 × 10−6, 1.94 × 10−5, and 1.97 × 10−5 for ID6, ID8_GEpos, and ID8_GEneg, respectively; two-sided Wilcoxon rank-sum tests). We also noted that hTOP2α p.K743N tumors had far more deletions of sizes 2, 3, and 4 bp not in repeats relative to deletions ≥5 bp compared to a sample of tumors dominated by ID8 mutagenesis (P < 0.0084 by one-sided Wilcoxon rank-sum test on the ratio counts of deletions of sizes 2, 3, and 4 to counts of deletions of ≥5 bp; SI Appendix, Table S1 and Figs. S11 and S12).
To summarize, in addition to the 2- to 4-bp duplications that were previously reported as ID17, hTOP2α p.K743N is associated with substantial numbers of deletions. These comprise 1) deletions of length ≥5 bp and not in repeats, which resemble ID8 deletions, except that deletions of lengths 6, 7, and 8 bp are relatively more abundant than in ID8-dominated tumors; and 2) deletions of 2, 3, and 4 bp that are also relatively much more abundant than in ID8-dominated tumors. The density of these deletions correlates with transcriptional activity, as do all the other major classes of indels in hTOP2α p.K743N tumors, which is consistent with the known biology of hTOP2α. Regarding the 2-, 3-, and 4-bp duplications, their unusual abundance and unusual genic enrichment and association with higher transcription (SI Appendix, Figs. S7 and S8) indicate that these are almost exclusively due to the hTOP2α p.K743N mutation. Regarding the deletions in hTOP2α p.K743N tumors, it is possible that some of these stem from the same mutational processes that generate ID8. However, the differences in size distribution of these deletions in hTOP2α p.K743N versus non-hTOP2α p.K743N tumors and, to some extent, the level of genic enrichment and association with transcriptional activity, suggest that many or most of these deletions stem from hTOP2α p.K743N. To gather further evidence of this, we next investigated the possibility of a sequence motif associated with hTOP2α p.K743N mutagenesis.
Sequence Motifs Associated with hTOP2α p.K734N Mutagenesis.
Because a consensus binding site of topoisomerase II has not been identified, we asked whether a consensus motif could be identified using our yeast and human mutagenesis data. The 4-bp duplications are a result of the 4-nt 5′ overhangs created by topoisomerase II cleavage. Following clean removal of yTop2 protein by Tdp1, the overhangs are completely filled and the blunt ends are ligated by NHEJ. Shorter duplications are proposed to result from the partial hybridization of the overhangs prior to gap filling and ligation. Consequently, only the 4-bp duplications indicate the precise cleavage sites (17). The 4-bp duplicated sequences in the yTop2-FY,RG and yTop2-K720N strains were different, with ACCT being most commonly duplicated in the yTop2-K720N strain and ATAA being most commonly duplicated in the yTop2-FY,RG strain (SI Appendix, Fig. S13 and Dataset S4). Interestingly, duplications in the human data showed different preferences, with AGCT being the most commonly duplicated tetranucleotide. Although we previously suggested that the AT richness of duplications in the yTop2-FY,RG strain might facilitate strand separation (10), the current data suggest there is at most only weak sequence specificity for the 4 bp between the topoisomerase-generated nicks.
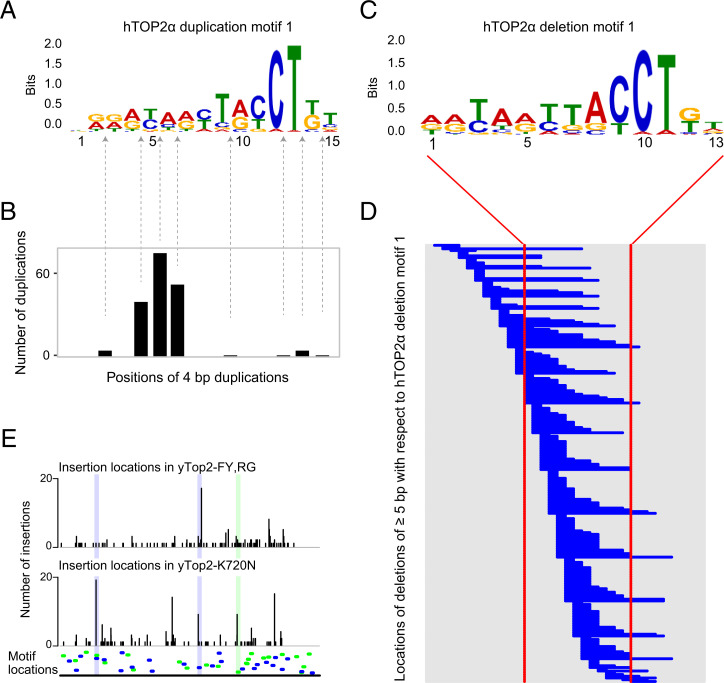
We used the MEME analysis software to search for sequence motifs associated with indels in hTOP2α p.K743N-mutated tumors (22). In CCA_TH_19, we searched independently for motifs near duplications of length 4, 3, and 2 bp and of lengths 4 and 3 bp combined, and also near deletions of length 2, 3, 4, or ≥5 bp not in repeats. In the combined data from all hTOP2α p.K743N tumors with WES, we searched for motifs near duplications of 3 and 4 bp combined and also near deletions of length 2, 3, 4, or ≥5 bp not in repeats. MEME extracted two motifs across all searches in the CCA_TH_19 data, one of which was also detected near both duplications and deletions in the combined whole-exome data (Fig. 5A and SI Appendix, Figs. S14 and S15). The most commonly detected motif had a strong preference for [TC]T[AG]CCT> on the right. The second discovered motif had a strong preference for TTCA on the right.
Fig. 5.
Similar motifs associated with duplications and deletions in hTOP2α p.K743N tumors. (A) Duplication motif 1, extracted from sequences surrounding 4-bp duplications in CCA_TH_19 using MEME (22). See SI Appendix, Figs. S14 and S15 for similar motifs extracted from other duplication sizes and from hTOP2α p.K743N tumors with WES. (B) Positions of duplications in relation to the motif in A as detected by MAST (23) with E values <10. (C) Deletion motif 1 extracted from sequences overlapping deletions of length 2, 3, 4, or ≥2 bp not in repeats in CCA_TH_19. (D) Positions of deletions relative to the motif in C. Each horizonal blue line indicates the position and extent of a deletion. Shown are deletions in or near sequences in which MAST detected the deletion with E value <10. (E) Overlap between the motif in A and insertion mutations in yeast can1 mutation data. Bottom shows the locations of hTOP2α sequence motifs as detected by FIMO using the motif in A as the query, ordered by P value (most significant at the Bottom). Motifs are in blue (detected on the reference genome strand) or green (detected on the opposite strand). The panels above depict the locations of insertions >1 bp in the yTop2-FY,RG and yTop2-K720N data. Several hotspots are highlighted.
While tumors without the hTOP2α p.K743N mutation have few duplications of 2, 3, and 4 bp, the pattern of deletions in hTOP2α p.K743N-mutated tumors somewhat resembles mutational signature ID8. We therefore searched for motifs near deletions of 2, 3, 4 or ≥5 bp, not in repeats, in three tumors with spectra dominated by ID8 (SI Appendix, Fig. S12). No motif resembling either of the two hTOP2α p.K743N deletion motifs (SI Appendix, Fig. S15) was detected (SI Appendix, Fig. S16). We also used MAST (23) to search for the two hTOP2α p.K743N deletion motifs near deletions in CCA_TH_19 and the ID8-dominated control tumors. Compared to the control tumors, CCA_TH_19 was highly enriched for both motifs (minimum odds ratio across all controls and both motifs = 6.7, maximum P value <10−13 by Fisher’s exact one-sided test, SI Appendix, Table S2). These findings provide evidence in addition to the difference in size distribution (Fig. 4B and SI Appendix, Fig. S11) and high mutation counts, that many of the ID8-like mutations in the hTOP2α p.K743N tumors stem from the hTOP2α p.K743N mutation, rather than from the unknown but much more common mutational process or processes responsible for ID8. Thus, we consider the indel mutation pattern in the hTOP2α p.K743N tumors to be an indel mutational signature that combines ID17 and features of ID8 but with a different distribution of deletion sizes, which we call ID_TOP2α (Fig. 1C and Dataset S3).
Although hTOP2α is a homodimer and the constituent monomers would be expected to have a similar consensus recognition site on complementary strands flanking the cleavage site, the discovered motifs are not palindromic, and the sequence context preferences are much stronger to one side of the motif. The Discussion presents a possible biochemical explanation for this observation.
To investigate the locations of the discovered motifs in relation to insertions and deletions, we used MAST (23) to detect sequences matching the motifs extracted from the 4-bp duplications and from the deletions of length 2, 3, 4, or ≥5 bp, not in repeats (Fig. 5 A and B, SI Appendix, Fig. S17). For duplication motif 1, the locations of duplications of 3 and 4 bp had pronounced modes 2 bp to the left of [TC]T[AG]CCT>, and the 2-bp duplications had a pronounced mode 1 bp to the left of this sequence. With respect to deletion motif 1, the sites of deletions were distributed more broadly near the motifs (Fig. 5C and D and SI Appendix, Fig. S17). Please refer to SI Appendix, Fig. S17 for the sites of duplications and deletions with respect to duplication and deletion motifs 2.
We also examined whether the insertions (including duplications) observed in yeast occur near the 4 bp-insertion motif (Fig. 5A). FIMO (24) detected 48 motif sites in the CAN1 locus (Fig. 5E and Dataset S5, tab “FIMO CAN1”). There was no enrichment of yTop2-FY,RG insertions at motif sites, but there was a 1.68-fold enrichment of yTop2-K720N insertions (P < 6 × 10−11, one-sided binomial test; see also Dataset S6).
hTOP2α p.K743N Is Associated with Genomic Rearrangements.
Stabilized hTOP2α and yTop2 cleavage complexes induce genomic rearrangements (25–27), which led us to investigate whether hTOP2α p.K743N is similarly associated with genomic rearrangements. Because genomic rearrangement breakpoints are strongly depleted in exons, and WGS was available only for CCA_TH_19, we compared the rearrangements previously reported for this tumor with those in the other cholangiocarcinomas in the same cohort (28). CCA_TH_19 had the most rearrangements, consisting mainly of deletions (41.5%) and interchromosomal translocations (27.6%) (SI Appendix, Fig. S18). Like the indels, rearrangements were enriched in highly transcribed regions (P = 2.56 × 10−12, one-sided Cochran–Armitage test; SI Appendix, Fig. S19). Enrichment in more highly transcribed regions was also observed for the following subclasses of genomic rearrangements: large deletions, interchromosomal translocations, intrachromosomal translocations and large insertions (FDRs 1.93 × 10−2, 4.00 × 10−9, 1.33 × 10−2, and 1.24 × 10−3 respectively; one-sided Cochran–Armitage tests). Compared to other cholangiocarcinomas, the rearrangement breakpoints in CCA_TH_19 were more strongly associated with higher transcription (SI Appendix, Fig. 20).
A MEME search for motifs at the sites of rearrangements in CCA_TH_19 yielded a motif that resembles motif 2 as extracted from hTOP2α p.K743N-mutated tumors (SI Appendix, Figs. S14A and S21 and Dataset S5). We then used MAST to map the hTOP2α p.K743N deletion motifs (SI Appendix, Fig. S15A) to the regions near genomic rearrangement breakpoints, considering each breakpoint separately (Dataset S5). There was a strong enrichment for both motifs at the breakpoints in CCA_TH_19 compared to other cholangiocarcinomas from the same study (28): For deletion motif 1, the odds ratio for enrichment was 115; for deletion motif 2, it was 48 (P < 10−81 and P < 10−48, respectively, by Fisher’s two-sided tests on the data in SI Appendix, Table S3; see also SI Appendix, Fig. S17 E and F). These results support a role for hTOP2α p.K743N in formation of the genomic rearrangements in CCA_TH_19.
Cancer Driver Mutations Fit ID_TOP2α Mutagenesis.
There was a total of 45 indels in COSMIC cancer-driver genes in hTOP2α p.K743N tumors, and each tumor had several such indels (Dataset S7). None of these indels occurred in any of ∼2,700 genomes reported in ref. 19, and at least one of the hTOP2α p.K743N motifs mapped to sequences near 36 of these indels. (We used 4-bp duplication motifs 1 and 2 as queries against duplication sites in the cancer genes and deletion motifs 1 and 2 as queries against deletion sites; SI Appendix, Figs. S14A and S15A). The analogous counts for the tumors in ref. 19 are 2,812 indels in total, of which one or more motifs mapped to sequences near 61 indels (Dataset S8). The odds ratio for enrichment in hTOP2α p.K743N tumors was 177 (P < 10−47 by Fisher’s two-sided exact test). Indeed, six of the seven hTOP2α p.K743N-mutated tumors had frameshift mutations in the key tumor suppressor genes PTEN and TP53. There was also a 15 bp, in-frame deletion in BRAF, which has been reported to be oncogenic (29). At least one of the hTOP2α p.K743N motifs mapped near each of these indels. These specific examples and the overall enrichment for the hTOP2α p.K743N motifs across all 45 indels in known cancer-driver genes in the hTOP2α p.K743N tumors support the hypothesis that ID_TOP2α mutagenesis contributed to tumorigenesis in these tumors.
Discussion
Topoisomerases are critical for genome maintenance. Here, we have identified a somatic alteration in hTOP2α (p.K743N) in human tumors that causes a remarkably specific pattern of indel mutagenesis that we name ID_TOP2α. We showed that the orthologous mutation in yeast Top2 (yTop2-K720N) leads to enzyme-mediated DNA damage, both in cells and with the purified enzyme in vitro. Overexpression of the orthologous alteration in yeast recapitulated the 2- to 4-bp duplications observed in the tumors. Duplications of 3 bp were depleted in the yeast data when compared to the human tumors, as these in-frame deletions are less likely to disrupt function of the reporter gene used. Also of note, the deletions in ID_TOP2α were nearly absent in the yeast mutation spectra, while these constituted 23% of indels in ID_TOP2α (Fig. 1C). We postulate this difference is due to the absence of theta-mediated end joining (TMEJ) in yeast (30). TMEJ is the major alternative to classical NHEJ in humans and uses ≥2 bp of homology to facilitate DSB repair. Although the precise molecular mechanisms remain to be elucidated, TMEJ is known to cause small deletions (31, 32). Therefore, we postulate that in human tumors, the deletions in ID_TOP2α are the result of TMEJ.
We have only observed the ID_TOP2α pattern of indel mutagenesis in the presence of hTOP2α p.K743N, but expect that other somatic hTOP2α alterations will be associated with the ID_TOP2α-like mutator phenotype as more human cancers are sequenced. This is supported by the nearly identical pattern of 2- to 4-bp duplications associated with the yeast yTop2-FY,RG and yTop2-K720N proteins, which affect different functional domains of yTop2: yTop2-K720N is located within the DNA cleavage domain, while F1025 and R1128 are in the C-terminal dimer interface. Indeed, we previously described another, synthetically generated, DNA-damaging hTOP2α allele that is located in yet another domain (33). Finally, it should be noted that overexpression of wild-type yTop2 also induces 2- to 4-bp duplications, albeit less strongly than the mutant enzymes (10).
It is known that topoisomerase II creates nicks on complementary strands that are 4 bp apart, and we found that the sequence preference for the 4 bp between the nicks is variable between different yeast variants and human p.K743N (SI Appendix, Fig. S13). Nevertheless, there is strong evidence that the DNA sequence influences the mutagenic outcome associated with yTop2-K720N. For this allele, none of the sites of recurrent 4-bp duplications also showed 2-or 3-bp duplications, nor were 4-bp duplications common at sites of recurrent 2-bp duplications (Dataset S6).
Using the sites of indels in CCA_TH_19 ID_TOP2α, we identified hTOP2α p.K743N mutagenesis motifs that likely overlap or flank hTOP2α cleavage sites (Fig. 5 A and C and SI Appendix, Figs. S14 and S15). Strikingly, although hTOP2α functions as a homodimer, the sequence context preference at the 3′ side of the discovered motif was far stronger. This could be due to the dominant role of the first monomer in determining a nick site. DNA binding of topoisomerase II monomer half sites has limited sequence preference. Once the first monomer nicks, the kinetics of cleavage of the second strand is ∼10-fold faster than first-strand nicking (34–37). In this model, binding of the first monomer and the associated nick occur in a sequence-specific manner, after which the second nick follows quickly, with far less sequence specificity. There are likely also other factors that affect the selectivity of mutational induction. These might include chromatin structure, the stability of the cleavage complexes at particular sites, interactions between the trapped protein and other DNA metabolic processes such as replication and transcription, or factors related to the removal of TOP2α that had been trapped on DNA.
The yTop2-K720N–associated mutations mostly fall within hTOP2α p.K743N duplication motif 1 (Fig. 5E). However, the mutations associated with yTop2-FY,RG are not statistically enriched for this motif and mostly do not overlap it. Furthermore, even for the yeast ortholog of hTOP2α p.K743N, one of the main hotspots for 4-bp duplications was not located at a predicted hTOP2α p.K743N binding motif. We consider several explanations for this. First, the yeast CAN1 locus is only ∼1,800 bp. This provides very limited sequence complexity as a substrate for possible mutations and potentially enriches for mutations not optimally fitting topoisomerase II cleavage sites. Second, although topoisomerase II is highly evolutionarily conserved, and yTop2 and hTOP2α are very similar in terms of amino acid sequence, there could be differences of sequence context preference between the yeast and human proteins. Third, the topoisomerase II mutants examined here might have a sequence specificity different from that of the wild-type protein. In light of these considerations, we consider the discovered motifs specific to hTOP2α p.K743N. Additional studies will be required to determine whether the motifs described herein are generally applicable to all eukaryotic topoisomerase II enzymes.
In conclusion, we identified an indel mutator phenotype that is caused by the hTOP2α p.K743N protein. This phenotype generates a characteristic pattern of indels that we name ID_TOP2α, which consists of de novo 2- to 4-bp duplications together with deletions of 2, 3, 4, or ≥5 bp not in repeats that somewhat resemble ID8, but that have a different size distribution, including more deletions of 2, 3, and 4 bp and, for deletions ≥5 bp, more deletions of sizes 6 to 8 bp. In hTOP2α p.K743N tumors, two groups of sequence motifs are highly enriched near these indels and at genome rearrangement breakpoints. There are also indels that match ID_TOP2α and that are matched by hTOP2α p.K743N motifs in key cancer drivers such as BRAF, PTEN, and TP53, suggesting that hTOP2α p.K743N may have contributed to tumorigenesis. Increased sensitivity of yeast to topoisomerase poisons in the presence of yTop2-FY,RG (10) suggests that ID_TOP2α could be a biomarker for increased tumor vulnerability to topoisomerase II inhibitors. Further studies with mammalian cells expressing mutant topoisomerases will be needed to assess this possibility.
Methods
Data Sources.
Published mutation spectra from 23,829 tumors were used (https://www.synapse.org/#!Synapse:syn11726601/) (11). Variant calls for 2,780 WGS samples from the ICGC/TCGA (International Cancer Genome Consortium/The Cancer Genome Atlas) Pan-Cancer Analysis of Whole Genomes Consortium and gene expression data for a subset of these were obtained from the ICGC data portal (https://dcc.icgc.org/releases/current/Projects/) (19). Sequencing reads from samples 04-112 and 10T were kindly provided by Peter S. Nelson (Fred Hutchinson Cancer Research Center and University of Washington, Seattle, Washington) and Fergus J. Couch (Mayo Clinic, Rochester, Minnesota) (14, 15). Sequencing reads from CCA_TH_19, for which indels were not previously analyzed, were downloaded from the European Genome-phenome Archive (EGAS00001001653). Genomic rearrangements for 70 whole-genome sequenced cholangiocarcinomas were obtained from the SI Appendix of the associated publication (28). The COSMIC Cancer Gene Census was used for identification of known cancer driver genes (38).
Reanalysis of Short-Read Sequencing Data.
Read alignment, variant calling, and filtering were performed as described previously (39), except that reads were aligned to GRCh38.p7. For analysis of genomic rearrangement occurrence as a function of transcriptional activity, the transcriptional activity at the location of the genomic rearrangement breakpoint with the highest transcriptional activity was taken.
Mutational Signature Analysis.
We used the classification for indel mutational signatures as proposed previously (11); for details see https://www.synapse.org/#!Synapse:syn11801742. Mutational signatures were plotted using ICAMSv2.1.2.9000 (https://github.com/steverozen/ICAMS).
Correlation between Transcriptional Activity and Mutagenesis in WGS Data.
For each tumor type, genes were assigned to one of four gene expression bins. For every sample, variants were grouped by expression bins by linking the genomic position of the variant to the genes assigned to expression bins. Mutation density was reported as events per megabase to compensate for differences in size of the total gene expression bins. A Cochran–Armitage test for trend was performed to determine statistical significance. As not all ICGC projects have RNA-sequencing data available, for some ICGC cohorts we used RNA-sequencing data from similar cohorts. For details, see Dataset S2.
Definition of ID6-High and ID8-High Tumors in WGS Data.
For comparison of the size distributions of deletions ≥5 bp in ID6-high and ID8-high tumors, we classified tumors using the existing indel signature assignments (11). ID6-high tumors were defined as follows. Let m be the median number of ID6 mutations among tumors with > 0 ID6 mutations. Then ID6-high tumors were those with > m ID6 mutations. ID8-high tumors were selected analogously, but excluding any tumors that had ID6 mutagenesis. ID8 tumors were further divided into samples that did (ID8_GEpos) or did not (ID8_GEneg) show significantly higher mutation density in loci with high transcription versus loci with low transcription.
Motif Discovery and Detection.
Motifs were detected with the MEME web server (https://meme-suite.org/meme/tools/meme, version 5.4.1) generally using default parameters, except sometimes increasing the number of motifs to return (Fig. 5 A and B and SI Appendix, Figs. S14–S17, and S21). For deletions, sequences were submitted spanning 15 bp 5′ from the deletion start to 15 bp 3′ from the deletion end. For insertions and genomic rearrangement breakpoints, sequences spanning 15 bp to either side of the insertion or breakpoint were submitted (22). Motifs of interest were directly exported from MEME to MAST version 5.4.1 (23) or FIMO version 5.4.1 (24) to search for the presence of the motifs in other sequences (Dataset S5, Fig. 5, and SI Appendix, Fig. S17).
Yeast Methods.
Haploid strains used for mutation analyses were RAD5 derivatives of W303 [ade2-1 his3-11,15 ura3-1 leu2-3,112 trp1-1 can1-100 rad5-G535R] (40). Strain/plasmid construction details and relevant growth conditions are described in SI Appendix. Fluctuation analysis using liquid cultures was used to determine rates of canavanine resistance (Can-R); independent Can-R mutants for mutation-type analysis were isolated on solid medium. Mutation rates were calculated using webSalvador (https://websalvador.eeeeeric.com/). Mutation-type rates were calculated as previously described (10). Additional details are in SI Appendix.
Wild-type and mutant Top2 proteins were overexpressed in yeast as N-terminally His-tagged proteins using the plasmid pTRB378 and were purified by nickel affinity chromatography as previously described (41). DNA decatenation and cleavage assays were performed as previously described (10, 42). Quantitation of the drug-independent Top2 cleavage activity was performed using Bio-Rad Image Lab version 6.0.1 software.
Statistics.
Statistical analyses were performed in R v.3.6.3; multiple testing correction was done according to the Benjamini–Hochberg method using the p.adjust function. Cochran–Armitage tests were performed using the DescTools package.
Supplementary Material
Acknowledgments
We thank Peter S. Nelson and Fergus J. Couch for kindly providing sequencing data. This work was supported by a Khoo Postdoctoral Fellowship Award (KPFA) (Duke–NUS KPFA/2018/0027) (to A.B.), a Singapore National Medical Research Council award MOH-000032/MOH-CIRG18may-0004 (to S.G.R.), and NIH grant GM118077 (to S.J.-R.).
Footnotes
Reviewers: N.O., Vanderbilt University; and E.R., Harvard Medical School.
Competing interest statement: S.G.R., A.B., and one of the reviewers of this manuscript, Professor E.R., participated in the Pan-Cancer Analysis of Whole Genomes (PCAWG) Consortium, which comprised over 700 researchers from around the world. S.G.R., A.B., and Professor E.R. did not directly work or publish together.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114024119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
Previously published data were used for this work (10).
References
- 1.Vos S. M., Tretter E. M., Schmidt B. H., Berger J. M., All tangled up: How cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 12, 827–841 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pendleton M., Lindsey R. H. Jr., Felix C. A., Grimwade D., Osheroff N., Topoisomerase II and leukemia. Ann. N. Y. Acad. Sci. 1310, 98–110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J. C., Moving one DNA double helix through another by a type II DNA topoisomerase: The story of a simple molecular machine. Q. Rev. Biophys. 31, 107–144 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Corbett K. D., Berger J. M., Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 33, 95–118 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Pommier Y., Leo E., Zhang H., Marchand C., DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17, 421–433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pommier Y., Sun Y., Huang S. N., Nitiss J. L., Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 17, 703–721 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang X., et al. , A comprehensive review of topoisomerase inhibitors as anticancer agents in the past decade. Eur. J. Med. Chem. 171, 129–168 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Heestand G. M., Schwaederle M., Gatalica Z., Arguello D., Kurzrock R., Topoisomerase expression and amplification in solid tumours: Analysis of 24,262 patients. Eur. J. Cancer 83, 80–87 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren L., Liu J., Gou K., Xing C., Copy number variation and high expression of DNA topoisomerase II alpha predict worse prognosis of cancer: A meta-analysis. J. Cancer 9, 2082–2092 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stantial N., et al. , Trapped topoisomerase II initiates formation of de novo duplications via the nonhomologous end-joining pathway in yeast. Proc. Natl. Acad. Sci. U.S.A. 117, 26876–26884 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexandrov L. B., et al. , PCAWG Mutational Signatures Working Group; PCAWG Consortium, The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexandrov L. B., et al. , Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain, Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbes S. A., et al. , COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 45 (D1), D777–D783 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar A., et al. , Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat. Med. 22, 369–378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy S. J., et al. , Genetic alterations associated with progression from pancreatic intraepithelial neoplasia to invasive pancreatic tumor. Gastroenterology 145, 1098–1109.e1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osheroff N., Zechiedrich E. L., Calcium-promoted DNA cleavage by eukaryotic topoisomerase II: Trapping the covalent enzyme-DNA complex in an active form. Biochemistry 26, 4303–4309 (1987). [DOI] [PubMed] [Google Scholar]
- 17.Sander M., Hsieh T., Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J. Biol. Chem. 258, 8421–8428 (1983). [PubMed] [Google Scholar]
- 18.Yu X., et al. , Genome-wide TOP2A DNA cleavage is biased toward translocated and highly transcribed loci. Genome Res. 27, 1238–1249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium, Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rheinbay E., et al. , PCAWG Drivers and Functional Interpretation Working Group; PCAWG Structural Variation Working Group; PCAWG Consortium, Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 578, 102–111 (2020).32025015 [Google Scholar]
- 21.Imielinski M., Guo G., Meyerson M., Insertions and deletions target lineage-defining genes in human cancers. Cell 168, 460–472.e414 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey T. L., Elkan C., Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 (1994). [PubMed] [Google Scholar]
- 23.Bailey T. L., Gribskov M., Combining evidence using p-values: Application to sequence homology searches. Bioinformatics 14, 48–54 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Grant C. E., Bailey T. L., Noble W. S., FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gómez-Herreros F., et al. , TDP2 suppresses chromosomal translocations induced by DNA topoisomerase II during gene transcription. Nat. Commun. 8, 233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sciascia N., et al. , Suppressing proteasome mediated processing of topoisomerase II DNA-protein complexes preserves genome integrity. eLife 9, e53447 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felix C. A., Kolaris C. P., Osheroff N., Topoisomerase II and the etiology of chromosomal translocations. DNA Repair (Amst.) 5, 1093–1108 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Jusakul A., et al. , Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov. 7, 1116–1135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foster S. A., et al. , Activation mechanism of oncogenic deletion mutations in BRAF, EGFR, and HER2. Cancer Cell 29, 477–493 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Wyatt D. W., et al. , Essential roles for polymerase θ-mediated end joining in the repair of chromosome breaks. Mol. Cell 63, 662–673 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carvajal-Garcia J., et al. , Mechanistic basis for microhomology identification and genome scarring by polymerase theta. Proc. Natl. Acad. Sci. U.S.A. 117, 8476–8485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schimmel J., van Schendel R., den Dunnen J. T., Tijsterman M., Templated insertions: A smoking gun for polymerase theta-mediated end joining. Trends Genet. 35, 632–644 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Walker J. V., et al. , A mutation in human topoisomerase II alpha whose expression is lethal in DNA repair-deficient yeast cells. J. Biol. Chem. 279, 25947–25954 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Lee S., et al. , DNA cleavage and opening reactions of human topoisomerase IIα are regulated via Mg2+-mediated dynamic bending of gate-DNA. Proc. Natl. Acad. Sci. U.S.A. 109, 2925–2930 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mueller-Planitz F., Herschlag D., DNA topoisomerase II selects DNA cleavage sites based on reactivity rather than binding affinity. Nucleic Acids Res. 35, 3764–3773 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deweese J. E., Burgin A. B., Osheroff N., Using 3′-bridging phosphorothiolates to isolate the forward DNA cleavage reaction of human topoisomerase IIalpha. Biochemistry 47, 4129–4140 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deweese J. E., Osheroff N., The DNA cleavage reaction of topoisomerase II: Wolf in sheep’s clothing. Nucleic Acids Res. 37, 738–748 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sondka Z., et al. , The COSMIC cancer gene census: Describing genetic dysfunction across all human cancers. Nat. Rev. Cancer 18, 696–705 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boot A., et al. , Characterization of colibactin-associated mutational signature in an Asian oral squamous cell carcinoma and in other mucosal tumor types. Genome Res. 30, 803–813 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas B. J., Rothstein R., Elevated recombination rates in transcriptionally active DNA. Cell 56, 619–630 (1989). [DOI] [PubMed] [Google Scholar]
- 41.Blower T. R., et al. , A complex suite of loci and elements in eukaryotic type II topoisomerases determine selective sensitivity to distinct poisoning agents. Nucleic Acids Res. 47, 8163–8179 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nitiss J. L., Soans E., Rogojina A., Seth A., Mishina M., Topoisomerase assays. Curr. Protoc. Pharmacol. 57, 3.3.1–3.3.27 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao J., et al. , Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.
Previously published data were used for this work (10).