Significance
Reactive compounds cause cellular damage that is suspected to contribute to aging and neurodegenerative diseases. Oxidative stress and environmental factors likely contribute to this. Here we report that an enzyme mutated in Parkinson’s disease can prevent damage of metabolites and proteins caused by a metabolite from the central pathway of sugar metabolism. Inactivation of this enzyme in model systems, ranging from flies to human cells, leads to the accumulation of a wide range of damaged metabolites and proteins. Thus, this enzyme represents a highly conserved strategy to prevent damage in cells that metabolize sugars. Overall, we discovered a fundamental link between carbohydrate metabolism and a type of cellular damage that might contribute to the development of Parkinson’s disease.
Keywords: glycolysis, protein damage, posttranslational modification, Parkinson's disease, metabolite damage
Abstract
Cells are continuously exposed to potentially dangerous compounds. Progressive accumulation of damage is suspected to contribute to neurodegenerative diseases and aging, but the molecular identity of the damage remains largely unknown. Here we report that PARK7, an enzyme mutated in hereditary Parkinson’s disease, prevents damage of proteins and metabolites caused by a metabolite of glycolysis. We found that the glycolytic metabolite 1,3-bisphosphoglycerate (1,3-BPG) spontaneously forms a novel reactive intermediate that avidly reacts with amino groups. PARK7 acts by destroying this intermediate, thereby preventing the formation of proteins and metabolites with glycerate and phosphoglycerate modifications on amino groups. As a consequence, inactivation of PARK7 (or its orthologs) in human cell lines, mouse brain, and Drosophila melanogaster leads to the accumulation of these damaged compounds, most of which have not been described before. Our work demonstrates that PARK7 function represents a highly conserved strategy to prevent damage in cells that metabolize carbohydrates. This represents a fundamental link between metabolism and a type of cellular damage that might contribute to the development of Parkinson’s disease.
Parkinson’s disease is the second most common neurodegenerative disease (1). It is characterized by progressive debilitating motor symptoms (i.e., spontaneous shaking, stiffness, slow movements, and postural instability) (1). Currently, there is no causal therapy, and this is in part due to our insufficient knowledge about the pathogenesis of this disease. Loss of a vulnerable neuron population (i.e., dopaminergic neurons in the substantia nigra) plays a key role in this disease, and the formation of insoluble aggregates of the protein α-synuclein (Lewy bodies) seems to contribute to disease progression.
Most cases of Parkinson’s disease occur at old age, a major risk factor for neurodegenerative diseases in general (2). Cells are continuously exposed to reactive compounds that can damage cellular components. The progressive accumulation of damage likely contributes to aging and age-associated diseases (3, 4). Why some individuals develop Parkinson’s disease and most others don’t, is unknown. Evidence from rare hereditary cases indicates that the failure to eliminate damaged mitochondria can play an important role in the development of Parkinson’s disease (5–7). It is also increasingly clear that the neurons affected in Parkinson’s disease are particularly sensitive to oxidative stress and toxins (8–10).
Reactive oxygen species are clearly a major cause of cellular damage (11). Yet, physiological metabolic activity produces many other dangerous compounds (4) that are often overlooked. We are only starting to understand the pathways that preempt or repair this type of damage (12, 13). In the present paper we reveal that the enzyme PARK7 prevents damage caused by a metabolite from glycolysis, the key metabolic pathway for carbohydrates in most cells and organisms.
Inactivating mutations in PARK7 (also called DJ1) lead to autosomal recessive Parkinson’s disease (14). Patients present with early-onset disease that slowly progresses and is accompanied by the formation of Lewy bodies (15, 16). PARK7 knockout mice develop hypokinesia and defects in the same neuronal population affected by nonhereditary Parkinson’s disease (17–19). Surprisingly, PARK7 also plays a role in cancer (20, 21) and a wide range of cellular functions (reviewed in refs. 22 and 23) that include the protection from reactive oxygen species (24–30), the maintenance of mitochondrial function (31, 32), and chaperone activity (33, 34). Yet, it remained enigmatic which enzymatic activity of PARK7 was required for these functions.
PARK7 is part of an evolutionary conserved enzyme family that relies on a key cysteine residue for its activity (35, 36). In PARK7, this cysteine residue is easily oxidized and has been reported to serve as a scavenger for reactive oxygen species (24–30). Yet, it remained unclear how the oxidized protein would be reduced again to maintain its protective function. Recently, evidence has been presented that PARK7 (and bacterial homologs) can remove covalent adducts produced by a reaction between the metabolite methylglyoxal and amino groups on proteins, metabolites, and nucleic acids (37–40). These studies seemed to represent a potential breakthrough, since they suggested that Parkinson’s disease might be caused by the failure to maintain proteins and DNA “clean” and functional. More recently, these findings have been disputed (41, 42), and convincing evidence has been presented that this apparent deglycating activity is not due to the active removal of these adducts, but rather due to the degradation of free methylglyoxal, which is in rapid equilibrium with these modifications (43, 44). Consistent with this, we did not observe an increase of methylglyoxal-modified proteins or nucleotides in several PARK7 knockout cell lines (SI Appendix, Fig. S1).
We therefore hypothesized that the real enzymatic function of PARK7 that underlies its pleiotropic cellular effects might still be unknown. Using a combination of biochemical, genetic, and analytical approaches in human cell lines, mouse brain, and fruit flies, we discovered that PARK7 prevents damage of metabolites and proteins that is caused by the glycolytic intermediate 1,3-bisphosphoglycerate (1,3-BPG). Remarkably, some patients with genetic inactivation of the enzyme that utilizes 1,3-BPG also develop early-onset Parkinson’s disease (45), indicating that damage caused by 1,3-BPG may play a pathogenetic role in patients without PARK7 mutations as well.
Results
PARK7 Prevents Accumulation of a New Class of Metabolites.
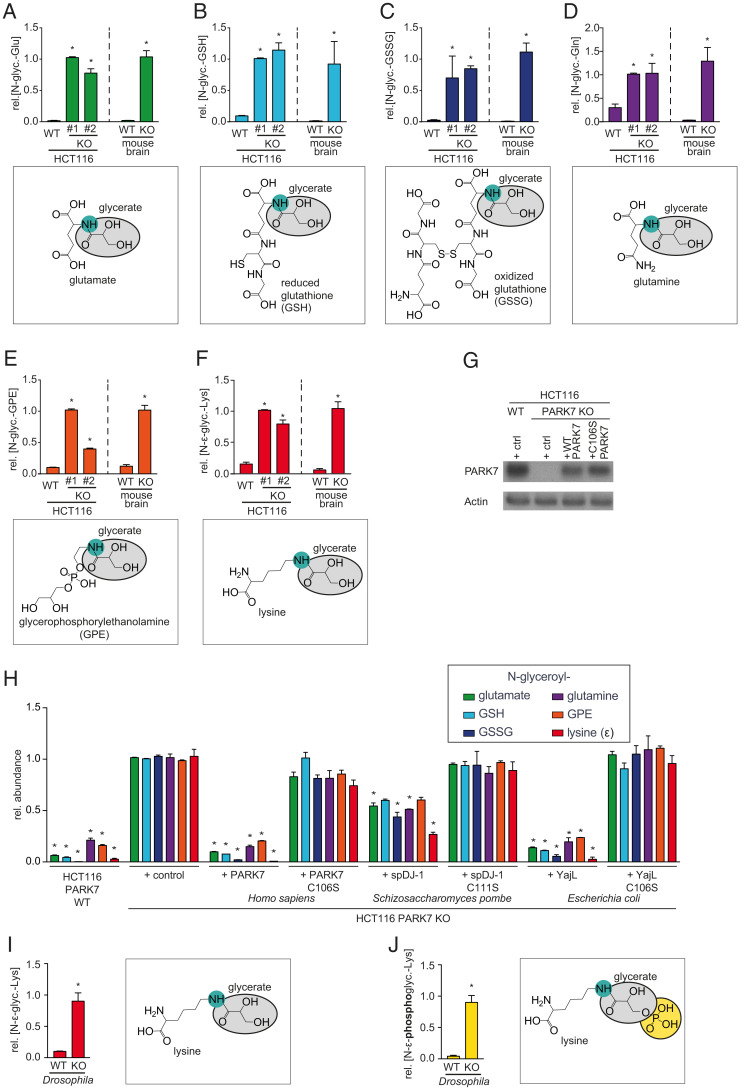
The molecular function of PARK7 should be detectable in a variety of experimental settings, given that PARK7 has been found to be the most stably expressed protein across a wide variety of tissues and organisms (46). Hence, we analyzed extracts from cancer cell lines, mouse brain, and Drosophila melanogaster with a highly sensitive liquid chromatography-mass spectrometry (LC-MS) approach. We observed that the concentrations of several unknown metabolites were increased 3- to 50-fold in PARK7 knockout cell lines and mouse brain (Fig. 1 A–F and SI Appendix, Fig. S2), whereas these compounds were close to the limit of detection in wild-type samples. We identified these metabolites as glycerate-adducts of the physiological metabolites glutamate (Glu), reduced (GSH), and oxidized (GSSG) glutathione, glutamine (Gln), glycerophosphorylethanolamine (GPE), and lysine (Lys, ε-amino group) based on the observed m/z, fragmentation pattern and elution time in comparison to synthetic standards (Dataset S1). In all cases, glycerate groups were attached to free amino groups (N-glyceroyl). To ensure that these changes were due to the loss of the enzymatic activity of PARK7, we re-expressed PARK7 or its homologs in knockout cell lines (Fig. 1G). This revealed that wild-type PARK7 can prevent accumulation of these modified metabolites, whereas PARK7 with a mutation in the key catalytic cysteine residue (C106S) did not have any effect (Fig. 1H). Of note, expression of the PARK7 homolog from Escherichia coli (yajL) in PARK7 knockout cells prevented the accumulation of all these modified metabolites, and expression of the Schizosaccharomyces pombe homolog spDJ-1 significantly reduced the abundance of four of six modified metabolites (Fig. 1H). In both cases, a mutation in the key catalytic cysteine abolished this effect. These results demonstrate that this function of PARK7 is highly conserved during evolution.
Fig. 1.
PARK7 and its orthologs prevent accumulation of glycerate- and phosphoglycerate-modified metabolites. (A–F) N-glyceroyl-glutamate (A), N-glyceroyl-GSH (B), N-glyceroyl-GSSG (C), N-glyceroyl-glutamine (D), N-glyceroyl-glycerophosphoethanolamine (GPE) (E), and N-ε-glyceroyl-lysine (F) were quantified by LC-MS in HCT116 wild-type and two PARK7 knockout clones, as well as wild type and Park7 knockout mouse brain. (G and H) Western blot analysis (G) and quantification of the indicated metabolites (H) in wild-type or PARK7 knockout HCT116 cells, where wild-type or catalytic mutant PARK7, yajL, or spDJ1 were re-expressed. “Control vector” denotes cells transduced with lentiviruses with an empty expression cassette. (I and J) Quantification of the indicated metabolites in DJ1β knockout and wild type flies. Values are means ± SEM (n ≥ 3) and were normalized within each metabolite. *P < 0.05 in comparison to the wild-type condition (A–F, I, and J) and the knockout control with the control vector (G and H).
Interestingly, D. melanogaster DJ1β knockouts showed not only an increase in glycerate-modified lysine (Fig. 1I), but also in phosphoglycerate-modified lysine (Fig. 1J).
A Glycolytic Metabolite Is Responsible for the Formation of Glycerate-Modified Metabolites.
Glycerate-modified metabolites have not been described before, but Moellering and Cravatt (47) have previously reported that proteins can be modified with phosphoglycerate when the glycolytic metabolite 1,3-BPG spontaneously reacts with amino groups in lysine side-chains. Furthermore, they observed that cells can dephosphorylate phosphoglycerate-modified peptides by unknown phosphatases. We reasoned that if amino groups in proteins can react with 1,3-BPG, then amino groups in metabolites might also undergo the same reaction. In a second step, the resulting phosphoglycerate adducts could be dephosphorylated by phosphatases to produce glycerate adducts (Fig. 2A).
Fig. 2.
A glycolytic metabolite is responsible for the formation of glycerate-modified metabolites. (A) Schematic representation of our working model for glycerate and phosphoglycerate modifications of metabolites and proteins. (B) Experimental setup for the continuous production of 1,3-BPG by PGK, ATP and 3-phosphoglycerate (3-PG) in the presence of metabolites with amino groups in vitro. (C and D) 1,3-BPG levels (C) and phosphoglycerate-modified metabolites (D) were quantified by LC-MS in a reaction where metabolites were exposed to continuously produced 1,3-BPG. (E and H) Western blot analysis (E and G) and metabolite quantification by LC-MS (F and H) in PARK7 knockout (E and F) or wild-type (G and H) cells expressing PGK1 or GAPDH shRNAs or nonsilencing control shRNAs. For the in vitro experiment, values are means ± SEM of three independent experiments. For each metabolite, values were normalized to the median of the 1.5-h timepoint for the 1,3-BPG production reaction. Asterisks denote statistical significance (*P < 0.05) compared to this condition. For the cellular experiment, values are means ± SEM of three independent experiments performed in triplicates. For each metabolite and within each experiment, values were normalized to the median of the values observed in knockout cells expressing the control shRNA. Asterisks indicate *P < 0.05 in comparison to the shRNA control condition.
We first tested this hypothesis in vitro. To this end, we produced 1,3-BPG with phosphoglycerate kinase in the presence of the six metabolites that were modified in PARK7 knockout models (Fig. 2 B–D). Under these conditions, we observed a continuous increase in phosphoglycerate-modified metabolites (Fig. 2D), consistent with 1,3-BPG being the source for these modifications.
Next, we investigated whether 1,3-BPG was required for the formation of glycerate-modified metabolites in cells. To do this, we modulated cellular 1,3-BPG levels. On the one hand, we knocked down the enzyme that produces 1,3-BPG in glycolysis, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and, on the other hand, we knocked down the enzyme that consumes it, phosphoglycerate kinase (PGK) (Fig. 2 A and E). In PARK7 knockout cells, we observed an increase in glycerate-modified metabolites when PGK was knocked down, whereas levels were decreased when GAPDH was knocked down (Fig. 2F). Similar tendencies were also observed when we knocked down these enzymes in wild-type cells, though the low abundance made quantifications difficult (Fig. 2 G and H). Overall, these observations are consistent with glycerate-modifications forming first as phosphoglycerate modifications, followed by subsequent dephosphorylation by yet unknown phosphatases. This model would also explain why DJ1β-deficient D. melanogaster show increases of both N-glyceroyl-lysine and N-phosphoglyceroyl-lysine (Fig. 1 I and J).
PARK7 Deficiency Leads to the Accumulation of Proteins Damaged by Phosphoglycerate and Glycerate Modifications.
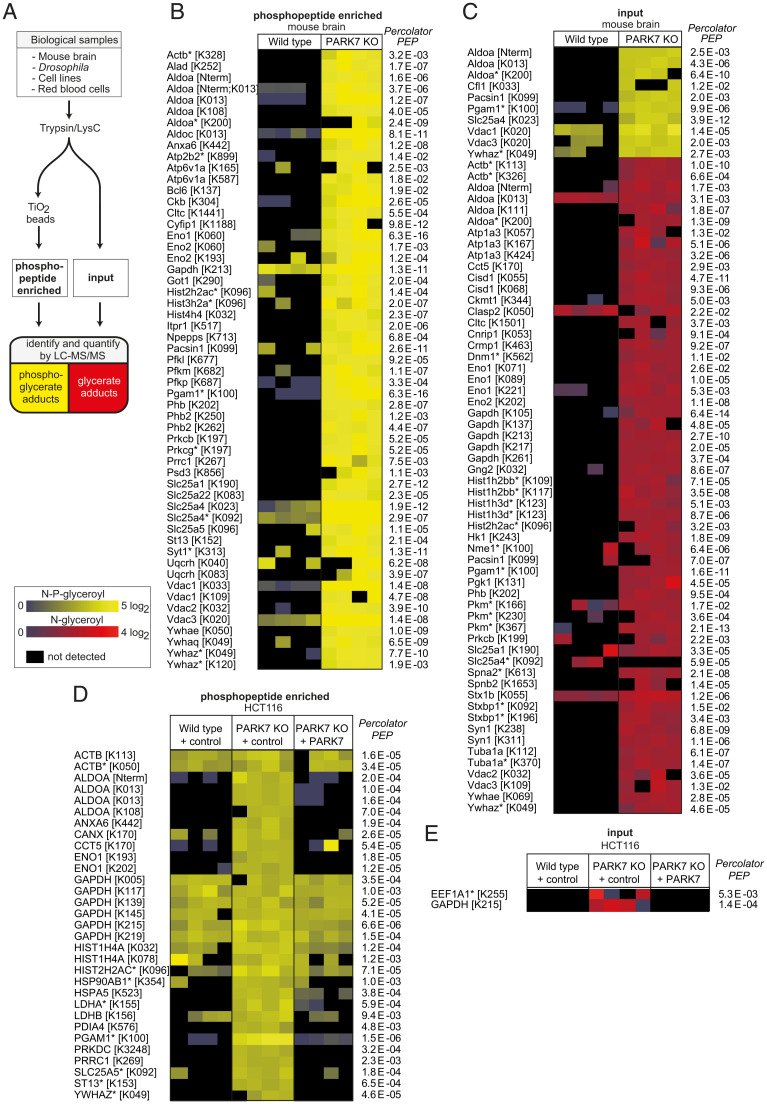
Free lysine with phosphoglycerate or glycerate modifications was increased in PARK7/DJ1β-deficient samples. We therefore wondered whether proteins with these modifications on lysine residues would also accumulate. To test this, we analyzed proteolytic digests from mouse brain using nano–LC-MS. The identification of phosphoglycerate-modified peptides was greatly facilitated by using common phosphopeptide-enrichment protocols (Fig. 3A). Thus, we found 62 phosphoglycerate-modified peptides from 51 proteins (see Fig. 3B and SI Appendix, Fig. S3A and Datasets S2, S3, and S5 for comparison with standards and MS3), which were exclusively detectable or of much higher abundance in PARK7 knockout samples.
Fig. 3.
Glycerate- and phosphoglycerate-modified proteins accumulate in PARK7 deficiency. (A) schematic representation of the experimental setup. (B–E) LC-MS analysis of proteolytically digested mouse brain (B and C) or cancer cell line (D and E) samples with (phosphopeptide enriched in B and D) or without (input in C and E) phosphopeptide enrichment. N-phosphoglyceroyl-lysine modifications are presented in yellow and N-glyceroyl-lysine modifications are presented in red. PARK7 wild type or knockout HCT116 cells were transduced with a lentiviral vector driving expression of wild type PARK7 or an empty expression cassette (control). Peptides levels were normalized to the abundance of the corresponding proteins and normalized to the median of the abundances across samples. Asterisks indicate peptides that could be derived from more than one protein isoform. Raw data are available online (Dataset S2 Proteomic data, and ProteomeXchange (62) under the accession number PXD029410.
Phosphoglycerate-modified metabolites were almost completely converted into glycerate-modified metabolites in vivo. Glycerate-modified proteins have never been described before, but we reasoned that many phosphoglycerate-modified proteins might also be converted to glycerate-modified proteins, which may accumulate in PARK7 knockout samples. We therefore analyzed proteolytic digests of mouse brain samples without prior phosphopeptide enrichment. This revealed 76 glycerate-modified peptides from 46 proteins, and 12 phosphoglycerate-modified peptides from 10 proteins (Fig. 3C and SI Appendix, Fig. S3B and Datasets S2, S4, and S5 for comparison with standards and MS3). Again, almost all modified peptides were found at much higher levels in PARK7 knockout samples.
Next, we investigated whether the accumulation of modified proteins could be rescued by re-expression of PARK7 in knockout cells. We found 30 phosphoglycerate- and 2 glycerate-modified peptides from 21 proteins in PARK7-deficient HCT116 cells. Most modified peptides were much more abundant than in wild-type cells, and re-expression of PARK7 in these cell lines brought their abundance down to wild-type levels (Fig. 3 D and E), proving that the observed changes were caused by the loss of PARK7.
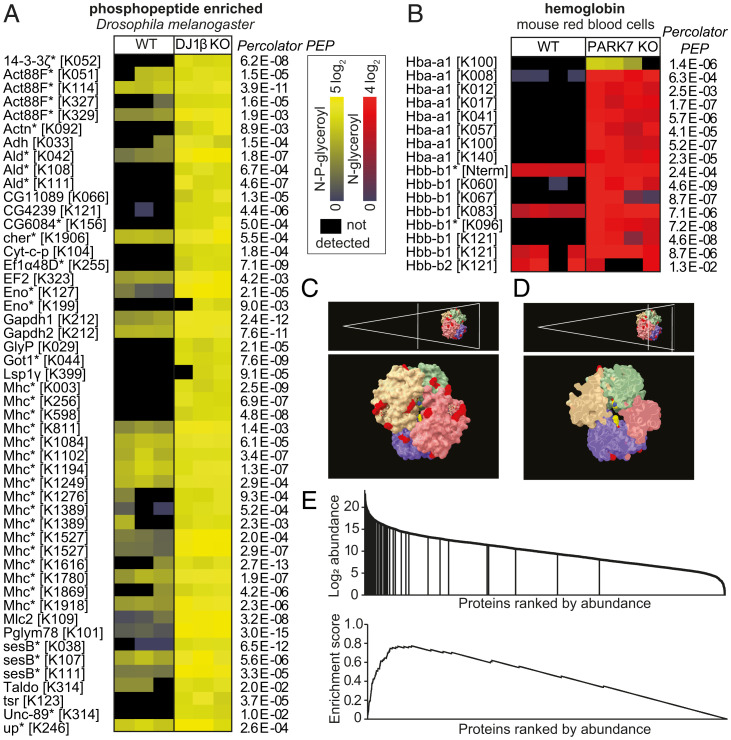
Samples from DJ1β-deficient D. melanogaster contained 51 phosphoglycerate-modified peptides from 26 different proteins, which were either absent or much less abundant in samples from wild-type flies (Fig. 4A). Curiously, we were unable to detect any glycerate-modified proteins in Drosophila DJ1β knockout samples. This indicated that the phosphatases acting on phosphoglycerate modifications in mouse brain and HCT116 cells might be absent in Drosophila.
Fig. 4.
Accessibility determines whether glycerate- or phosphoglycerate modifications occur. (A and B) LC-MS analysis of proteolytically digested wild type and DJ1β knockout D. melanogaster samples after phosphopeptide enrichment (A), or red blood cell lysates without prior enrichment (B). Data are presented as in Fig. 3. (C and D) Localization of residues modified with glycerate (red) and phosphoglycerate (yellow) on the hemoglobin tetramer (PDB ID code 3HRW). Note that the only phosphoglycerate-modified residue, K100 of the hemoglobin α1 chain, is found on the inside of the tetramer, as indicated by the cross section of hemoglobin (D). (E) Enrichment of phosphoglycerate- and glycerate-modified peptides among highly abundant proteins in mouse brain (P < 10−6). The enrichment score was calculated as described previously (68).
At this point, it was unclear why some lysines were modified with phosphoglycerate and others with glycerate. To better understand this, we decided to analyze a single protein in depth. We chose to focus on hemoglobin, which represents more than 95% of the protein content of red blood cells. We found glycerate-modifications on 12 lysine residues and the N terminus of hemoglobin at much higher levels in PARK7 knockout mice than in wild-type controls (Fig. 4B). All of these modifications affected surface-exposed lysine residues (Fig. 4C), consistent with the idea that not only a reactive metabolite but also a phosphatase needs to have access to the site of modification. In contrast, only a single phosphoglycerate modification could be quantified. Remarkably, it affected a residue located on the inside of the hemoglobin tetramer, suggesting that access for phosphatases might be limited, thereby preventing the conversion of phosphoglycerate into glycerate (Fig. 4D). This indicated that the accessibility determines which lysine residues retain phosphoglycerate modifications and which ones become dephosphorylated. Remarkably, we found evidence that more than half of the surface-exposed lysine residues of the hemoglobin tetramer can be modified with glycerate.
A nonenzymatic modification of amino groups would be expected to occur in an indiscriminate manner, depending on the accessibility and the protonation state of the amino group. Consistent with this, we not only observed modified lysine residues, but also modified N termini of proteins (Figs. 3 B–D and 4B). Modifications were mainly found in highly abundant proteins (Fig. 4E), which are expected to be more easily detected. This includes the glycolytic enzymes that had previously been found by Moellering and Cravatt (47) to be phosphoglycerate-modified. Several affected proteins are known to interact with phosphorylated organic acids (e.g., enolase, phosphoglycerate mutase, phosphoglycerate kinase, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycolate phosphatase, the mitochondrial adenine nucleotide translocator SLC25A4, and hemoglobin) and might have an intrinsic affinity for 1,3-BPG. However, this is not a requirement since more than half of all accessible lysine residues in hemoglobin were found to be modified, suggesting that most proteins can be modified and potentially damaged by glycerate and phosphoglycerate modifications. Thus, many modified proteins of lower abundance undoubtedly remain to be discovered.
PARK7 Prevents Damage Caused by a Glycolytic Metabolite without Destroying It.
The striking increase in glycerate- and phosphoglycerate-modified proteins and metabolites upon PARK7 loss-of-function indicated that PARK7 either removes these modifications (hypothesis 1 and 2 in Fig. 5A) or prevents their formation (hypothesis 3 in Fig. 5A). To distinguish between these models, we quantified newly synthesized glycerate-modified metabolites in cells by following the incorporation of 13C atoms after treatment with U-13C-glucose. Given that the glycerate-modification is expected to incorporate three carbons from glucose, we assumed that newly formed glycerate-modified metabolites would be detected as m+3 forms, whereas the preexisting fraction would present as m+0 (Fig. 5A). We performed these experiments in an HCT116 PARK7 knockout clone where we acutely induced the expression of either wild-type or catalytic mutant C106S PARK7 from a doxycycline-inducible promoter (Fig. 5B). If PARK7 prevents the formation of newly synthesized glycerate-adducts, we expected that it would only reduce the abundance of the m+3 fraction, whereas both the m+0 and the m+3 fraction would be affected if PARK7 removed the glycerate modification. Following induction of PARK7 expression, the m+3 fraction was more than twofold lower, whereas no change was observed when mutant PARK7 was expressed (Fig. 5 C–H). This demonstrates that PARK7 does not actively remove glycerate adducts. Furthermore, recombinant PARK7 did not have any activity on phosphoglycerate adducts in vitro (Fig. 5I). Taken together, PARK7 prevents the formation of phosphoglycerate and glycerate modifications of metabolites, but does not remove them.
Fig. 5.
PARK7 prevents formation of the glycerate and phosphoglycerate modifications but does not remove them. (A) Rationale for the 13C tracing experiment to determine whether PARK7 prevents formation of adducts or removes them. Six hours after induction of PARK7 expression in PARK7 knockout (KO) cell lines, cells were incubated with U-13C- glucose for 6 h. (B) Western blot of acute doxycycline-induced re-expression of PARK7 in PARK7 KO HCT116 cells. (C–H) De novo synthesized (black, m+3) and residual old (white, m+0) N-glyceroyl-glutamate (C and D), N-glyceroyl-GSH (E and F), and phosphoglycerate (G and H) were assessed by following 13C incorporation upon treatment with U-13C-glucose after acute re-expression of wild-type or mutant PARK7 in PARK7 knockout cell lines. Results are expressed in arbitrary units (C, E, and G) or as the fraction of each metabolite in the m+3 form (D, F, and H). (I) Phosphoglycerate-modified metabolites were incubated overnight ± PARK7 and quantified by LC-MS. Values are means ± SD of one representative experiment out of three (C–H) or means ± SEM of three independent experiments (I) (n = 3) and were normalized for each metabolite to the median of the PARK7-treated condition (I). Asterisks denote statistical significance (*P < 0.05) compared to the noninduced condition with wild-type vector (C–H) or the buffer control (I). AU, arbitrary units.
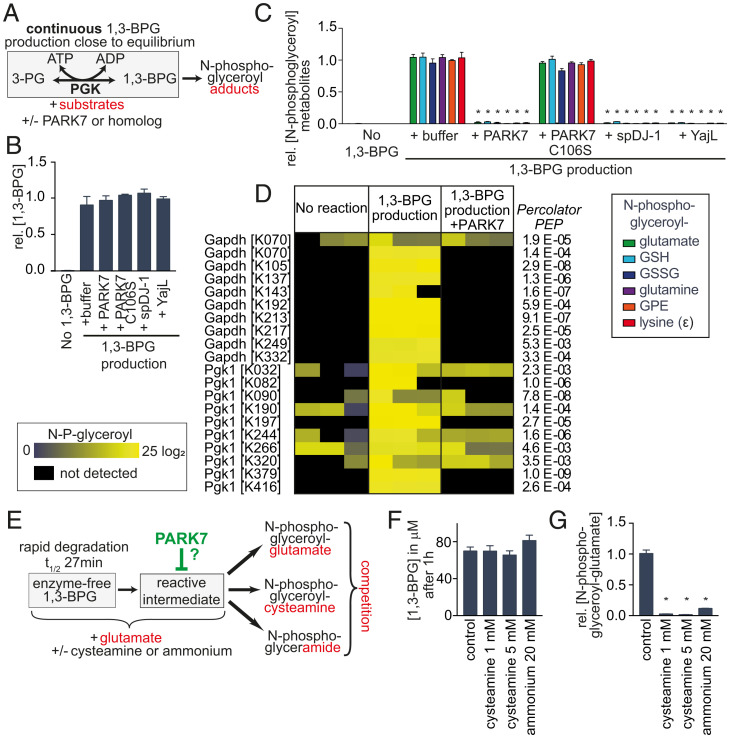
This was puzzling since our experiments (Fig. 2) and the work of Moellering and Cravatt (47) had indicated that 1,3-BPG causes the formation of phosphoglycerate adducts. How could PARK7 prevent formation of these adducts without destroying this important metabolite and affecting glycolysis? To resolve this question, we recapitulated the formation of adducts in vitro (Fig. 6 A–C). We produced 1,3-BPG in the presence of metabolites with free amino groups (Fig. 6A). This led to the formation of phosphoglycerate-modified metabolites, similar to what we had previously observed (Fig. 2 B–D). In contrast, formation of phosphoglycerate-modified metabolites was almost completely prevented when this reaction was performed in the presence of PARK7 or its distant homologs spDJ-1 (S. pombe) or yajL (E. coli) (Fig. 6C). Strikingly, 1,3-BPG levels remained unchanged (Fig. 6B). Of note, PARK7 carrying a mutation in the key catalytic cysteine residue, C106S, did not affect adduct formation nor 1,3-BPG levels (Fig. 6 B and C). Similar results were also observed when we incubated metabolites with enzyme-free 1,3-BPG (SI Appendix, Fig. S4). Again, addition of recombinant PARK7 did not change concentrations of 1,3-BPG (SI Appendix, Fig. S4B), but almost completely prevented the formation of phosphoglycerate-modified metabolites. These findings clearly demonstrated that PARK7 prevents 1,3-BPG-dependent modifications without affecting 1,3-BPG levels.
Fig. 6.
PARK7 prevents damage caused by a glycolytic metabolite without destroying this metabolite. (A) Experimental setup for the continuous production of 1,3-BPG by PGK, ATP, and 3-phosphoglycerate (3-PG) in the presence or absence of recombinant PARK7 or putative orthologs. (B and C) 1,3-BPG levels (B) and phosphoglycerate-modified metabolites (C) were quantified by LC-MS after 4-h incubation of metabolites with continuous 1,3-BPG production ± PARK7, its catalytic mutant C106S or its E. coli (yajL) or S. pombe (spDJ-1) homologs. (D) N-phosphoglyceroyl modifications of proteins were quantified by LC-MS after an overnight reaction where 1,3-BPG was produced by PGK, ATP, and 3P-glycerate. The protein GAPDH was included to allow parallel experiments, where 1,3-BPG was produced from glyceraldehyde 3-phosphate (SI Appendix, Fig. S5). For details see Dataset S2. (E–G) Experimental setup to assess the competition between cysteamine or ammonium and glutamate for a reaction with 1,3-BPG or a putative reactive degradation product (E), spectrophotometric quantification of 1,3-BPG concentrations after 1 h (F), and quantification of N-glyceroyl-glutamate after 2 h by LC-MS (G). Values are means ± SEM of three independent experiments (B, C, F, and G) (n = 3), and were normalized to the “1,3-BPG + buffer” condition within each experiment separately for each metabolite. Asterisks denote statistical significance (*P < 0.05) compared to the “1,3-BPG + buffer” condition (C and G).
To test whether PARK7 could also prevent phosphoglycerate modifications on proteins, we set up reactions that produced 1,3-BPG via PGK, but where GAPDH was also present in the reaction mixture (similar to Fig. 6A). The analysis of protein modifications revealed numerous phosphoglycerate-modifications on both GAPDH and PGK, which were strongly reduced in abundance in the presence of PARK7 (Fig. 6D). Of note, comparable results were also obtained when 1,3-BPG was produced via GAPDH (SI Appendix, Fig. S5)
Taken together, these observations unequivocally demonstrate that PARK7 prevents the formation of 1,3-BPG–dependent modifications without affecting 1,3-BPG levels. Thus, PARK7 must destroy a reactive intermediate that spontaneously forms from 1,3-BPG and avidly reacts with amino groups. To demonstrate this, we set up a competition experiment between glutamate and the nucleophile cysteamine (Fig. 6E). If glutamate reacted directly with 1,3-BPG, then formation of phosphoglycerate-modified glutamate should only be decreased by the addition of cysteamine if this addition depletes 1,3-BPG levels. In contrast, if cysteamine and glutamate compete in a reaction with a reactive degradation product of 1,3-BPG, then the addition of cysteamine should reduce the formation of phosphoglycerate-modified glutamate without affecting 1,3-BPG levels. As shown in Fig. 6F, the addition of 1 mM cysteamine did not affect 1,3-BPG levels. In contrast, phosphoglycerate-modified glutamate production was reduced by more than 90% (Fig. 6G). This demonstrates that 1,3-BPG is not directly responsible for the formation of phosphoglycerate adducts (Fig. 7A). We also noted that even weaker nucleophiles (such as ammonium ions) can prevent the formation of adducts and react with the reactive intermediate (Fig. 6G), making direct identification by MS difficult.
Fig. 7.
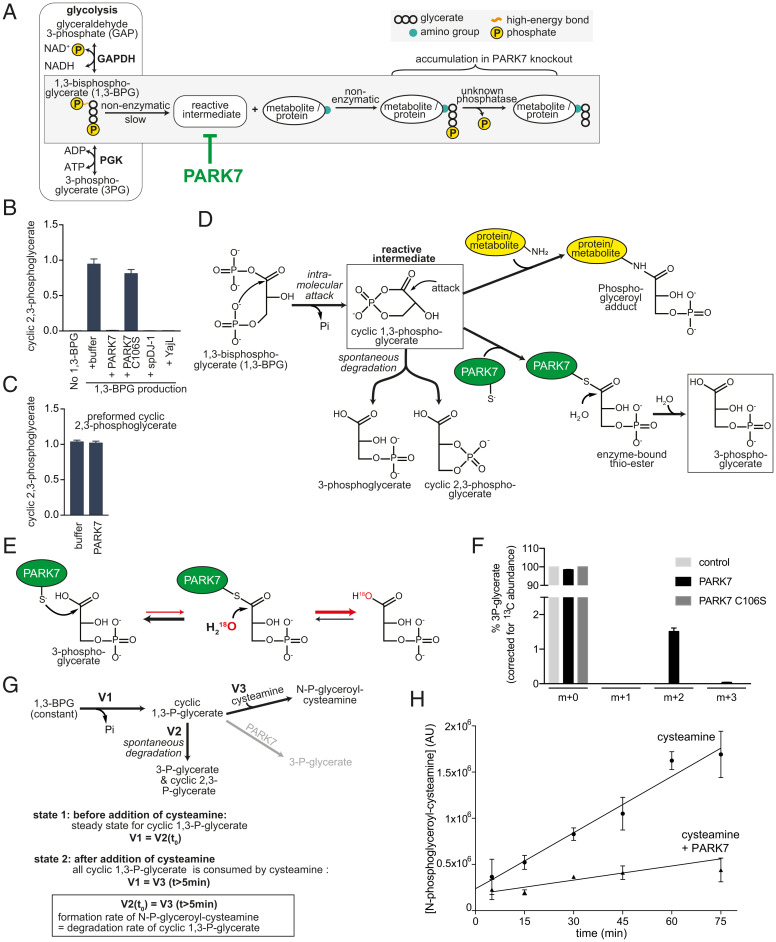
Tentative identification and characterization of the reactive compound as cyclic-1,3-phosphoglycerate. (A) Schematic representation of the formation of N-glyceroyl- and N-phosphoglyceroyl-modifications in proteins and metabolites. (B) Quantification of an ion with an m/z of 166.9751 by LC-MS in reactions, where 1,3-BPG was continuously produced in the presence or absence of PARK7 and related proteins (same setup and samples as Fig. 6 A–C). The metabolite was identified as cyclic-2,3-phosphoglycerate (SI Appendix, Fig. S6). (C) Cyclic-2,3-phosphoglycerate was quantified after preformed cyclic-2,3-phosphoglycerate was incubated in presence of PARK7 enzyme or its storage buffer. (D) Working model of cyclic-1,3-phosphoglycerate as the reactive intermediate, its formation, its fates and its degradation by PARK7. (E) Exchange reaction with H218O expected to occur if PARK7 forms 3-phosphoglycerate via a thioester intermediate. While the equilibrium of the thioester hydrolysis is far on the side of 3-phosphoglycerate, incorporation of 18O into 3-phosphoglycerate in the presence of PARK7 would indicate that a thioester has been formed transiently. (F) 3-phosphoglycerate was incubated with wild-type PARK7, C106S mutant PARK7 or storage buffer in the presence of 60% H218O. The isotopic distribution of 3-P-glycerate was analyzed by LC-MS and is presented here after correction for the natural isotope abundance of 13C. The presence of the m+2 indicates the incorporation of one 18O atom. (G) Experimental system to determine the half-life of the reactive intermediate. The experiment is based on the observation that cysteamine almost completely depletes the reactive intermediate, since it prevents modification of other nucleophiles (see Fig. 6 E–G). In a reaction where 1,3-BPG levels are kept stable, levels of cyclic-1,3-phosphoglycerate are expected to reach a steady state (state 1, formation rate = degradation rate). When we added cysteamine, this led to a rapid appearance of N-phosphoglyceroyl-cysteamine to levels that are expected to be similar to the preexisting cyclic-1,3-phosphoglycerate levels. Subsequently, N-phosphoglyceroyl-cysteamine levels increased at a constant rate that is expected to be identical to the formation rate of cyclic-1,3-phosphoglycerate. (H) Quantification of N-phosphoglyceroyl-cysteamine after addition of cysteamine or cysteamine and PARK7 to a reaction where 1,3-BPG levels were maintained constant by continuous production via phosphoglycerate kinase. Values are means ± SEM of three independent experiments.
Tentative Identification of the Reactive Metabolite as Cyclic-1,3-Phosphoglycerate Formed by an Intramolecular Attack from 1,3-BPG.
To glean insights into the identity of the elusive reactive metabolite, we searched for products of 1,3-BPG that were absent in in vitro reactions containing PARK7. This revealed a metabolite with an m/z of 166.9751, which was absent when we produced 1,3-BPG via phosphoglycerate kinase in the presence of PARK7 or its orthologs (Fig. 7B, presenting the same samples as in Fig. 6C). Based on its coelution with a synthetic standard, its resistance to alkaline phosphatase, the loss of a carboxylic acid function in MS2, and based on its hydrolysis into both 2-phosphoglycerate and 3-phosphoglycerate, this metabolite was identified as cyclic-2,3-phosphoglycerate (SI Appendix, Fig. S6). Interestingly, while PARK7 prevented the formation of cyclic-2,3-phosphoglycerate, it was unable to degrade it (Fig. 7C). This indicated that cyclic-2,3-phosphoglycerate is not the elusive PARK7 substrate but rather formed downstream of this substrate.
To form cyclic-2,3-phosphoglycerate, either the hydroxyl group on carbon 2 or the phosphate group on carbon 3 need to be activated, for example by attaching a good leaving group. Coming from 1,3-BPG, the most parsimonious explanation is the formation of cyclic-1,3-phosphoglycerate, which can form by an intramolecular attack of 1,3-BPG (Fig. 7D). A nucleophilic attack by amino groups would explain the formation of phosphoglycerate-adducts. Likewise, we hypothesized that a nucleophilic attack on carbon 1 by the catalytic cysteine residue of PARK7 would lead to the formation of a thioester, which would then hydrolyze to form 3-P-glycerate (Fig. 7D). In such a model, PARK7 should be able to convert 3-P-glycerate to the enzyme-linked thioester, even if the equilibrium of the hydrolysis is expected to be far on the side of 3P-glycerate. To test this, we incubated 3-P-glycerate in the presence or absence of PARK7 in a reaction where 60% of the water molecules contained 18O (Fig. 7E). If PARK7 could form a thioester with 3P-glycerate, we would expect that hydrolysis of the thioester back to 3-phosphoglycerate would incorporate 18O into the molecule. When we incubated 3-P-glycerate in the presence of PARK7, we observed a twofold increase in 3-phosphoglycerate with a two-unit mass increase. After correction for natural isotope distribution of 13C, this corresponds to 18O incorporation in 1.5% of 3-P-glycerate molecules, whereas none was incorporated in presence of C106S mutant PARK7 or without enzyme (Fig. 7F). These observations are consistent with a nucleophilic attack of PARK7 on carbon 1 of cyclic-1,3-phosphoglycerate, which is eventually converted to 3P-glycerate.
As indicated above, the reactive intermediate was very sensitive to the presence of nucleophiles, which likely precluded its detection by LC-MS. Nevertheless, we wondered whether the kinetics of adduct formation might allow us to determine its half-life. We reasoned that the concentration of the reactive intermediate should reach a steady-state in a reaction with constant 1,3-BPG levels (Fig. 7G, state 1). Under these conditions, the rate of the formation and the rate of the degradation of the reactive metabolite should be identical. While we could not determine the concentrations directly, we were able to determine adduct formation when we added cysteamine at concentrations that almost completely depleted this metabolite (Figs. 6G and 7G, state 2). This led to the rapid formation of N-phosphoglyceroyl-cysteamine within <5 min, by reacting with the preexisting reactive metabolite (Fig. 7H). Subsequently, N-phosphoglyceroyl-cysteamine increased much slower but at a constant rate, which should correspond to the rate of the formation of the reactive intermediate (Fig. 7G, state 2). Based on this rate, the reactive intermediate present in steady state before addition of cysteamine would have needed 12 min to be formed (SD = 3.8 min). Given that formation rate and degradation rate of the reactive metabolite in steady state should be identical, we estimate that the half-life of the reactive intermediate is 8.4 min. Of note, addition of PARK7 at the same time as cysteamine led to the formation of N-phosphoglyceroyl-cysteamine at 4× lower rate. This indicates that 75% of the reactive metabolite is being destroyed by PARK7 even in the presence of a 50-fold excess of the nucleophile cysteamine.
Discussion
PARK7 Solves a Universal Problem of Glycolysis.
Degradation of glucose in glycolysis produces two molecules of ATP. This can only be achieved via the formation of 1,3-BPG. Thus, damage caused by this metabolite has to be dealt with in all cells that perform glycolysis, explaining the functional conservation of PARK7 during evolution. Our data shows that PARK7 achieves the seemingly impossible task of preventing the damage caused by 1,3-BPG without affecting levels of this key glycolytic metabolite. This reveals a so far unknown chemical property of 1,3-BPG: it does not directly modify amino groups efficiently. Rather, this modification proceeds mostly via a reactive intermediate, which can be eliminated by PARK7 before it reacts with amino groups of metabolites and proteins. In fact, the necessity for PARK7 to compete with amino groups might explain why PARK7 is highly abundant and why its concentration is kept the same across different cell types and organisms (46). PARK7 destroys a reactive metabolite that was not previously known to exist. While we were unable to detect this metabolite by mass spectrometry, indirect evidence indicates that it is cyclic-1,3-phosphoglycerate, which can form by an intramolecular attack of 1,3-BPG.
The discovery that PARK7 destroys a short-lived reactive metabolite might stimulate the search for other enzymes that serve to inactivate other short-lived reactive metabolites that might escape detection by mass spectrometry. Like PARK7, such enzymes might play a role in preventing cellular damage and the development of degenerative diseases.
A Solution to the Long-Standing Enigma of PARK7 Function?
More than 2,000 papers have been published on PARK7. It is therefore legitimate to ask whether the PARK7 function reported here is the core function or whether it is simply one more function for a multifunctional protein. Indeed, many molecular functions have been reported in vitro, but the expected changes in knockout or knockdown models were modest or absent. In contrast, we found very strong changes of glycerate or phosphoglycerate modified metabolites and proteins in PARK7/DJ1 knockout cell lines, mice, and flies, which support our in vitro data. Furthermore, we find that even a bacterial ortholog can perform the same function, indicating that the function reported here drove the evolutionary conservation of this protein and is responsible for the diverse cellular functions of PARK7.
Some confusion exists in the literature about the role of PARK7 in the detoxification of the glycolytic side-product methylglyoxal and the removal of the damage that is caused by methylglyoxal adducts on proteins, nucleic acids, and metabolites (reviewed in ref. 48). We and others have found that PARK7 can act on methylglyoxal (43, 44), and strong overexpression of PARK7 can prevent formation of methylglyoxal-modified proteins (40). However, this activity does not seem to be required in cells that maintain a functional glutathione-dependent glyoxalase function (SI Appendix, Fig. S1) (40, 42, 44, 49, 50). Furthermore, while several groups have demonstrated that incubation of reversibly modified proteins, nucleotides, and metabolites with PARK7 can indeed reduce these modifications (37–39, 51, 52), it has recently been demonstrated that this apparent deglycase activity of PARK7 results from the degradation of free methylglyoxal that is in fast equilibrium with hemithioacetals and hemiaminals (44). Consistent with this, we and others did not observe any increase in methylglyoxal-modified metabolites and proteins when PARK7 was inactivated alone in mice, cell lines and flies (SI Appendix, Fig. S1) (42, 44).
Widespread, but Rare, Damage of Metabolites and Proteins Provides an Explanation for the Pleiotropic Effects of PARK7.
We found glycerate or phosphoglycerate modifications on more than 80 different proteins in PARK7-deficient mouse brain, which were almost absent in wild-type controls. Modifications were mainly detected on very abundant proteins (Fig. 4E). Yet, the in-depth characterization of modifications on hemoglobin demonstrated that more than half of the accessible lysine residues can be modified (Fig. 4 C and D). Thus, with increasing sensitivity of MS approaches and better enrichment protocols, many other modified proteins and modification sites will be discovered. Even though only a small fraction of each protein is modified with glycerate or phosphoglycerate, the large variety of modified proteins provides an explanation for the pleiotropic effects of PARK7, given that some of these modifications lead to changes in protein function (47). For several reasons, it is difficult to pin-point which modified protein is responsible for which cellular phenotype in PARK7 deficient organisms. First, we cannot prevent these modifications on individual proteins. Second, it is likely that modifications of several proteins synergize to provoke a cellular phenotype. Last but not least, in the context of Parkinson’s disease, rare substoichiometric protein modifications might serve as seeds for protein aggregates that contribute to the progression by driving the formation of Lewy bodies. Even in PARK7-deficient patients, the disease takes decades to develop, suggesting that protein aggregation remains inefficient, perhaps because most damaged proteins are degraded. Thus, it will be daunting to find an experimental setup that faithfully recapitulates a process that takes decades in vivo. Of note, an additional layer of complexity is added by the accumulation of glycerate-modified metabolites. Among these, N-glyceroyl-glutathione might interfere with glutathione-dependent redox homeostasis and detoxification, thereby contributing to the vulnerability of sensitive neuron populations such as dopaminergic neurons (8, 9).
The Phosphatases That Convert N-Phosphoglyceroyl to N-Glyceroyl Adducts Remain Elusive.
While we present unequivocal evidence that PARK7 prevents the formation of phosphoglycerate- and glycerate-modified proteins and metabolites, we do not know how N-phosphoglyceroyl adducts are converted into N-glyceroyl adducts (Fig. 7A). Yet, in mammalian PARK7 knockout models, all phosphoglycerate-modified metabolites and many phosphoglycerate-modified proteins are dephosphorylated. Glycerate adducts of metabolites and proteins have not been reported before, but Moellering and Cravatt (47) had reported that phosphoglycerate-modified peptides could be converted to glycerate-modified peptides by unknown phosphatases. Future studies will be needed to reveal the molecular identity and the role of these phosphatases in cells.
Possible Connection to Other Hereditary Forms of Parkinson’s Disease.
PARK7 deficiency leads to oxidative stress (24–28), disturbs mitochondrial metabolism (24, 53), and can increase glycolytic flux (54). This phenotype partially overlaps with the phenotype that is observed upon inactivation of other genes mutated in early-onset Parkinson’s disease (53). Several of these play a role in mitophagy (e.g., PINK1 and Parkin) (5–7), a mitochondrial quality-control mechanism. When they are inactivated, this can lead to mitochondrial dysfunction, increased reactive oxygen species production, and increased glycolytic flux (8, 9, 55). Oxidative stress and mitochondrial dysfunction have received most of the attention as a cause for neuronal cell death. However, an increase in neuronal glycolytic flux can also affect the viability of neurons. For example, a forced activation at the level of phosphofructokinase reduces viability of neurons, whereas an inhibition at this step protects neurons from detrimental effects of ischemia-reperfusion injury and excitotoxicity (56–58). These observations have been explained by reciprocal changes of the flux in the pentose phosphate pathway, which via the production of NADPH normally helps cells to resist against oxidative damage (56). Nevertheless, activation of phosphofructokinase may also increase 1,3-BPG levels and thereby the formation of phosphoglycerate and glycerate adducts. Thus, it is conceivable that modified proteins and metabolites may contribute to the phenotype observed in other models of hereditary Parkinson’s disease and neuronal toxicity. This might explain why PARK7 can rescue part of the phenotype of PINK1 deficiency (53).
New Roads for Preventive or Therapeutic Interventions?
While patients with genetic PARK7 deficiency are rare, the catalytic cysteine 106 of PARK7 is easily inactivated by oxidative stress (59). This suggests that PARK7 might be intermittently inactivated even in individuals without mutations in this protein. As a consequence, modified metabolites might form, and specific modified proteins might accumulate over the course of the lifetime of an organism. To what extent this contributes to the pathogenesis of nonhereditary Parkinson’s disease will need to be investigated.
The formation of phosphoglycerate and glycerate adducts is expected to depend on the cellular concentration of 1,3-BPG. Consistent with this, we observed that knockdown of PGK (i.e., the enzyme that consumes 1,3-BPG) leads to increased levels of glycerate-modified metabolites even when PARK7 is fully functional (Fig. 2 G and H). Therefore, accumulation of modified metabolites or proteins might play a role in the development of early-onset Parkinsonism in a subset of patients with PGK1 deficiency (45).
In reverse, a reduction of cellular 1,3-BPG levels might slow down disease progression by reducing the formation of phosphoglycerate and glycerate adducts. This therapeutic goal may be achieved by stimulating glycolytic enzymes downstream of 1,3-BPG or by inhibiting upstream enzymes. At the same time, these interventions need to ensure that overall energy metabolism and the delicate metabolic interplay between different cell types in the brain are maintained (60). It has recently been found that PGK activators can improve the outcome of experimental Parkinson’s models (61). It is tempting to speculate that these effects were partially caused by a reduction in phosphoglycerate and glycerate adducts. In any case, our work suggests that dietary or pharmacologic interventions to reduce cellular 1,3-BPG levels should be explored as future therapeutic or preventive approaches.
Methods
A detailed description of the methods is contained in SI Appendix.
Resource Availability.
Further information and requests for resources and reagents should be directed to G.T.B. Proteomics datasets have been submitted to ProteomeXchange (62) under the accession no. PXD029410.
Plasmids.
Vectors for CRISPR/Cas9-mediated gene-targeting were generated based on the vector pX459 as described in Sanjana et al. (63). Plasmids for the lentiviral expression of shRNAs were produced in a system related to the miR-E system (64). Constitutive and inducible expression in mammalian cells was achieved using lentiviral constructs driven by a CMV promoter (65) or a doxycycline-inducible promoter from the pTRIPZ plasmid (Openbiosystem), respectively.
Cell Culture.
We generated knockout clones by transient transfection with CRISPR/Cas9 plasmids using Lipofectamine 2000, followed by 48-h puromycin selection. Recombinant lentiviruses were produced by transient transfection of HEK293T cells with lentiviral vectors and packaging plasmids psPAX2 and pMD2.G using calcium phosphate coprecipitation (66). The virus-containing supernatant was recovered to infect target cell lines, followed by a 3-d puromycin selection.
Animals.
Drosophila DJ1β[Δ93] and tissues of the mouse line B6.Cg-Park7tm1Shn/J were snap-frozen and underwent metabolomic and proteomic analysis. Mouse experiments were performed following the European and Belgian legislation for care and use of laboratory animals, and approved by the University’s Animal Welfare Committee.
Production of Recombinant Proteins and In Vitro Assays.
Recombinant proteins were produced in E. coli and purified via a hexahistidine tag. The activity of these proteins (in combination with commercially available enzymes) was assessed by following product formation with LC-MS. Concentrations of 1,3-BPG were determined using a coupled enzymatic assay.
LC-MS Analysis of Metabolites.
Metabolites were analyzed using an Agilent 6550 ion funnel mass spectrometer in negative mode, in comparison to synthetic standards. Their abundance was quantified using the MS1 [M-H]− peak area.
Nano-LC-MS Analysis of Peptides.
After reduction, alkylation and proteolytic digestion, peptides were analyzed by an Orbitrap Fusion Lumos tribrid mass spectrometer, and (phospho)glycerate-modified peptides were identified based on the HCD MS2 spectra using Proteome Discoverer 2.4 (Thermo). Peptides were quantified using MS1 precursor intensity. We confirmed the identity of several peptides by MS3 and by comparison to synthetic standards.
Western Blotting.
Cell lysates were prepared in RIPA buffer and Western blots were performed as previously described (67). Primary antibodies were anti-PARK7 (Santa-Cruz, clone D4, 1 : 1000), anti-PGK1 (Proteintech 17811–1-AP, 1:4,000), anti-GAPDH (Proteintech 60004–1-Ig, 1:20,000), anti–β-actin (Sigma, clone AC-15, 1:5,000), antiphospho-p53 (Cell Signaling #9284; 1:1,000), anti–P-H2AX (Millipore; 1:1,000), anti-GLO1 (1:1,000) and antimethylglyoxal (recognizing the MG-H1 modification of arginines, Cell Biolabs STA-011, 1:5,000) diluted in Tris-buffered saline (150 mM NaCl, 20 mM Tris-Cl pH 7.4, 0.05% Tween-20) containing 2% bovine serum albumin (Sigma).
Supplementary Material
Acknowledgments
We thank Feng Zhang (Massachusetts Institute of Technology, Cambridge, MA), Eric Fearon (University of Michigan, Ann Arbor, MI) and Didier Trono (University of Geneva, Switzerland) for material; Jean-François Collet for comments on the manuscript; and J. P. Bolanos (University of Salamanca) for initial help with the mouse model. This project has received funding from WELBIO grants (to E.V.S. and G.T.B.), the European Research Council under the European Union’s Horizon 2020 research and innovation programme (Grant agreement 771704, to G.T.B.), the Fondation Contre le Cancer (G.T.B.), a FRIA fellowship (to I.P.H.), a TELEVIE fellowship (to M.B.), an Incentive grant of the FNRS (to G.T.B), Deutsche Forschungsgemeinschaft SFB1118 Grant (to A.T.), as well as support from the Université Catholique de Louvain and from the Fonds Maurange.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2111338119/-/DCSupplemental.
Data Availability
Mass spectrometry proteomics data have been deposited at ProteomeXchange, www.proteomexchange.org/ (accession no. PXD029410) via the PRIDE partner repository (62). Processed mass spectrometry data are presented in Datasets S2–S5. All other study data are included in the main text and supporting information.
References
- 1.Poewe W., et al. , Parkinson disease. Nat. Rev. Dis. Primers 3, 17013 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Hou Y., et al. , Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 15, 565–581 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Gladyshev V. N., Aging: Progressive decline in fitness due to the rising deleteriome adjusted by genetic, environmental, and stochastic processes. Aging Cell 15, 594–602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Golubev A., Hanson A. D., Gladyshev V. N., Non-enzymatic molecular damage as a prototypic driver of aging. J. Biol. Chem. 292, 6029–6038 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McWilliams T. G., Muqit M. M., PINK1 and Parkin: Emerging themes in mitochondrial homeostasis. Curr. Opin. Cell Biol. 45, 83–91 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Mouton-Liger F., Jacoupy M., Corvol J. C., Corti O., PINK1/Parkin-dependent mitochondrial surveillance: From pleiotropy to Parkinson’s disease. Front. Mol. Neurosci. 10, 120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pickrell A. M., Youle R. J., The roles of PINK1, Parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 85, 257–273 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burbulla L. F., et al. , Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson’s disease. Science 357, 1255–1261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dias V., Junn E., Mouradian M. M., The role of oxidative stress in Parkinson’s disease. J. Parkinsons Dis. 3, 461–491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markey S. P., Johannessen J. N., Chiueh C. C., Burns R. S., Herkenham M. A., Intraneuronal generation of a pyridinium metabolite may cause drug-induced parkinsonism. Nature 311, 464–467 (1984). [DOI] [PubMed] [Google Scholar]
- 11.Sies H., Berndt C., Jones D. P., Oxidative stress. Annu. Rev. Biochem. 86, 715–748 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Linster C. L., Van Schaftingen E., Hanson A. D., Metabolite damage and its repair or pre-emption. Nat. Chem. Biol. 9, 72–80 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Bommer G. T., Van Schaftingen E., Veiga-da-Cunha M., Metabolite repair enzymes control metabolic damage in glycolysis. Trends Biochem. Sci. 45, 228–243 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Bonifati V., et al. , Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 299, 256–259 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Abbas M. M., et al. , Early onset Parkinson’s disease due to DJ1 mutations: An Indian study. Parkinsonism Relat. Disord. 32, 20–24 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Taipa R., et al. , DJ-1 linked parkinsonism (PARK7) is associated with Lewy body pathology. Brain 139, 1680–1687 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Goldberg M. S., et al. , Nigrostriatal dopaminergic deficits and hypokinesia caused by inactivation of the familial parkinsonism-linked gene DJ-1. Neuron 45, 489–496 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Rousseaux M. W., et al. , Progressive dopaminergic cell loss with unilateral-to-bilateral progression in a genetic model of Parkinson disease. Proc. Natl. Acad. Sci. U.S.A. 109, 15918–15923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim R. H., et al. , Hypersensitivity of DJ-1-deficient mice to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrindine (MPTP) and oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 102, 5215–5220 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim R. H., et al. , DJ-1, a novel regulator of the tumor suppressor PTEN. Cancer Cell 7, 263–273 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Vasseur S., et al. , DJ-1/PARK7 is an important mediator of hypoxia-induced cellular responses. Proc. Natl. Acad. Sci. U.S.A. 106, 1111–1116 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biosa A., et al. , Recent findings on the physiological function of DJ-1: Beyond Parkinson’s disease. Neurobiol. Dis. 108, 65–72 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Mencke P., et al. , The role of DJ-1 in cellular metabolism and pathophysiological implications for Parkinson’s disease. Cells 10, 347 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi S. Y., et al. , DJ-1 links muscle ROS production with metabolic reprogramming and systemic energy homeostasis in mice. Nat. Commun. 6, 7415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Billia F., et al. , Parkinson-susceptibility gene DJ-1/PARK7 protects the murine heart from oxidative damage in vivo. Proc. Natl. Acad. Sci. U.S.A. 110, 6085–6090 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guzman J. N., et al. , Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature 468, 696–700 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canet-Avilés R. M., et al. , The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. U.S.A. 101, 9103–9108 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clements C. M., McNally R. S., Conti B. J., Mak T. W., Ting J. P., DJ-1, a cancer- and Parkinson’s disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc. Natl. Acad. Sci. U.S.A. 103, 15091–15096 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taira T., et al. , DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 5, 213–218 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinumi T., Kimata J., Taira T., Ariga H., Niki E., Cysteine-106 of DJ-1 is the most sensitive cysteine residue to hydrogen peroxide-mediated oxidation in vivo in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 317, 722–728 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Thomas K. J., et al. , DJ-1 acts in parallel to the PINK1/Parkin pathway to control mitochondrial function and autophagy. Hum. Mol. Genet. 20, 40–50 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Irrcher I., et al. , Loss of the Parkinson’s disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum. Mol. Genet. 19, 3734–3746 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Zhou W., Zhu M., Wilson M. A., Petsko G. A., Fink A. L., The oxidation state of DJ-1 regulates its chaperone activity toward alpha-synuclein. J. Mol. Biol. 356, 1036–1048 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Shendelman S., Jonason A., Martinat C., Leete T., Abeliovich A., DJ-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation. PLoS Biol. 2, e362 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei Y., Ringe D., Wilson M. A., Ondrechen M. J., Identification of functional subclasses in the DJ-1 superfamily proteins. PLOS Comput. Biol. 3, e10 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Wilson M. A., Collins J. L., Hod Y., Ringe D., Petsko G. A., The 1.1-A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson’s disease. Proc. Natl. Acad. Sci. U.S.A. 100, 9256–9261 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuda N., et al. , Parkinson’s disease-related DJ-1 functions in thiol quality control against aldehyde attack in vitro. Sci. Rep. 7, 12816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richarme G., et al. , Guanine glycation repair by DJ-1/Park7 and its bacterial homologs. Science 357, 208–211 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Richarme G., et al. , Parkinsonism-associated protein DJ-1/Park7 is a major protein deglycase that repairs methylglyoxal- and glyoxal-glycated cysteine, arginine, and lysine residues. J. Biol. Chem. 290, 1885–1897 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng Q., et al. , Reversible histone glycation is associated with disease-related changes in chromatin architecture. Nat. Commun. 10, 1289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfaff D. H., Fleming T., Nawroth P., Teleman A. A., Reply to Richarme: Evidence against a role of DJ-1 in methylglyoxal detoxification. J. Biol. Chem. 292, 12784–12785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfaff D. H., Fleming T., Nawroth P., Teleman A. A., Evidence against a role for the parkinsonism-associated protein DJ-1 in methylglyoxal detoxification. J. Biol. Chem. 292, 685–690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J. Y., et al. , Human DJ-1 and its homologs are novel glyoxalases. Hum. Mol. Genet. 21, 3215–3225 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Andreeva A., et al. , The apparent deglycase activity of DJ-1 results from the conversion of free methylglyoxal present in fast equilibrium with hemithioacetals and hemiaminals. J. Biol. Chem. 294, 18863–18872 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morales-Briceño H., et al. , Parkinsonism in PGK1 deficiency implicates the glycolytic pathway in nigrostriatal dysfunction. Parkinsonism Relat. Disord. 64, 319–323 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Wiśniewski J. R., Mann M., A proteomics approach to the protein normalization problem: Selection of unvarying proteins for MS-based proteomics and western blotting. J. Proteome Res. 15, 2321–2326 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Moellering R. E., Cravatt B. F., Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science 341, 549–553 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jun Y. W., Kool E. T., Small substrate or large? Debate over the mechanism of glycation adduct repair by DJ-1. Cell Chem. Biol. 27, 1117–1123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galligan J. J., et al. , Methylglyoxal-derived posttranslational arginine modifications are abundant histone marks. Proc. Natl. Acad. Sci. U.S.A. 115, 9228–9233 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chaudhuri J., et al. , A Caenorhabditis elegans model elucidates a conserved role for TRPA1-Nrf signaling in reactive α-dicarbonyl detoxification. Curr. Biol. 26, 3014–3025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richarme G., Dairou J., Parkinsonism-associated protein DJ-1 is a bona fide deglycase. Biochem. Biophys. Res. Commun. 483, 387–391 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Richarme G., Marguet E., Forterre P., Ishino S., Ishino Y., DJ-1 family Maillard deglycases prevent acrylamide formation. Biochem. Biophys. Res. Commun. 478, 1111–1116 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Hao L. Y., Giasson B. I., Bonini N. M., DJ-1 is critical for mitochondrial function and rescues PINK1 loss of function. Proc. Natl. Acad. Sci. U.S.A. 107, 9747–9752 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Requejo-Aguilar R., et al. , DJ1 represses glycolysis and cell proliferation by transcriptionally up-regulating Pink1. Biochem. J. 467, 303–310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Requejo-Aguilar R., et al. , PINK1 deficiency sustains cell proliferation by reprogramming glucose metabolism through HIF1. Nat. Commun. 5, 4514 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Herrero-Mendez A., et al. , The bioenergetic and antioxidant status of neurons is controlled by continuous degradation of a key glycolytic enzyme by APC/C-Cdh1. Nat. Cell Biol. 11, 747–752 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Burmistrova O., et al. , Targeting PFKFB3 alleviates cerebral ischemia-reperfusion injury in mice. Sci. Rep. 9, 11670 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodriguez-Rodriguez P., Fernandez E., Almeida A., Bolaños J. P., Excitotoxic stimulus stabilizes PFKFB3 causing pentose-phosphate pathway to glycolysis switch and neurodegeneration. Cell Death Differ. 19, 1582–1589 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meulener M. C., Xu K., Thomson L., Ischiropoulos H., Bonini N. M., Mutational analysis of DJ-1 in Drosophila implicates functional inactivation by oxidative damage and aging. Proc. Natl. Acad. Sci. U.S.A. 103, 12517–12522 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonvento G., Bolaños J. P., Astrocyte-neuron metabolic cooperation shapes brain activity. Cell Metab. 33, 1546–1564 (2021). [DOI] [PubMed] [Google Scholar]
- 61.Cai R., et al. , Enhancing glycolysis attenuates Parkinson’s disease progression in models and clinical databases. J. Clin. Invest. 129, 4539–4549 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perez-Riverol Y., et al. , The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47 (D1), D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanjana N. E., Shalem O., Zhang F., Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fellmann C., et al. , An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 5, 1704–1713 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Ury B., Potelle S., Caligiore F., Whorton M. R., Bommer G. T., The promiscuous binding pocket of SLC35A1 ensures redundant transport of CDP-ribitol to the Golgi. J. Biol. Chem. 296, 100789 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jordan M., Wurm F., Transfection of adherent and suspended cells by calcium phosphate. Methods 33, 136–143 (2004). [DOI] [PubMed] [Google Scholar]
- 67.Collard F., et al. , A conserved phosphatase destroys toxic glycolytic side products in mammals and yeast. Nat. Chem. Biol. 12, 601–607 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry proteomics data have been deposited at ProteomeXchange, www.proteomexchange.org/ (accession no. PXD029410) via the PRIDE partner repository (62). Processed mass spectrometry data are presented in Datasets S2–S5. All other study data are included in the main text and supporting information.