Abstract
We have isolated a gram-positive, weakly acid-alcohol-fast, irregular rod-shaped bacterium from cultures of blood from a 5-year-old girl with acute myelogenous leukemia. This isolate was compared with 14 other strains including reference strains of Tsukamurella species by a polyphasic approach based on physiological and biochemical properties, whole-cell short-chain fatty acid and mycolic acid analyses, DNA-DNA hybridization, and sequencing of the 16S rRNA gene. This isolate represents a new taxon within the genus Tsukamurella for which we propose the name Tsukamurella strandjordae sp. nov. Our study also revealed that Tsukamurella paurometabola ATCC 25938 represents a misnamed Tsukamurella inchonensis isolate and confirms that Tsukamurella wratislaviensis belongs to the genus Rhodococcus.
Tsukamurellae are members of the mycolic acid-containing aerobic actinomycetes. The genus was created in 1988 to accommodate a group of chemically unique organisms characterized by a series of very long chain (68 to 76 carbons) highly unsaturated (two to six double bonds) mycolic acids, in addition to possessing meso-diaminopimelic acid and arabinogalactan, common to the genus Corynebacterium (6). The type species, Tsukamurella paurometabola, described by Steinhaus as Corynebacterium paurometabolum in 1941, was originally isolated from the mycetomes and ovaries of bed bugs (27). The first human isolate of Tsukamurella was reported in 1971 as Gordona aurantiaca (33). Four additional species were proposed in the 1990s. Tsukamurella wratislaviensis was isolated from soil (10). Strains of Tsukamurella inchonensis, Tsukamurella pulmonis, and Tsukamurella tyrosinosolvens have been isolated only from human specimens, and all were associated with clinical disease (34–36). We have encountered an unusual isolate in multiple cultures of blood from a 5-year-old girl with acute myelogenous leukemia who presented with sepsis. On the basis of physiological and biochemical characteristics, analysis of cell components, DNA-DNA hybridizations, and the 16S rRNA gene sequence, we propose that this strain represents a new taxon within the genus Tsukamurella to which we assign the name Tsukamurella strandjordae sp. nov. Our study compared this isolate to reference and clinical strains of the other Tsukamurella species and found that T. paurometabola ATCC 25938 is a misnamed T. inchonensis isolate.
MATERIAL AND METHODS
Strains.
The study included type strains of all proposed Tsukamurella species. T. paurometabola ATCC 8368 and T. wratislaviensis ATCC 51786 were purchased from the American Type Culture Collection (ATCC). A. F. Yassin generously provided the type strains T. pulmonis ATCC 700081 and T. inchonesis ATCC 700082. Type strain T. tyrosinosolvens DSMZ 44234 was purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ; Braunschweig, Germany). Other purchased reference strains included strains T. paurometabola ATCC 25938 and T. tyrosinosolvens DSMZ 44316. The strain of T. strandjordae, the subject of this study, was isolated from a 5-year-old girl with acute myelogenous leukemia who presented with sepsis. T. strandjordae was the sole bacterial isolate in four consecutive blood samples for culture drawn over 1-week period. All broth cultures became positive within 24 h (BACTEC 9240 with Peds PLUS medium; Becton Dickinson, Sparks, Md.). Isolates from all four cultures were analyzed. A detailed clinical history has been published elsewhere (4). In addition, we analyzed seven clinical isolates of other Tsukamurella species, including three T. pulmonis, two T. inchonensis, and two T. tyrosinosolvens isolates that were referred to our laboratory for identification between 1993 and 2000. Species-level identification of the clinical isolates was achieved by the polyphasic approach outlined below.
Media and morphology.
Routine mycobacterial identification tests were inoculated with growth from Middlebrook 7H11 agar (Remel, Lenexa, Kans.). Temperature response studies were done on Lowenstein-Jensen slants (Remel) and read after 1 week. The urease test was performed by the Murphy Hawkins disk method (BBL, Cockeysville, Md.). The lysozyme test was performed with lysozyme broth and included a glycerol broth control (Remel). For analyses by gas-liquid chromatography (GLC) with the Microbial Identification System (MIS; MIDI, Newark, Del.) by the mycobacteriological method and for high-performance liquid chromatography (HPLC) analysis, cells were grown on Middlebrook 7H11 agar (Remel). For analysis by the MIDI clinical method, all nonmycobacterial biochemical tests, and evaluation of gross colonial morphology, cells were grown on Trypticase soy agar supplemented with 5% sheep blood (TSA; Remel). For inoculation into API 50 CH strips (see below), cells were grown on nutrient agar (Remel). To assess colonial micromorphology, cells were grown on tap water agar. Air-dried smears, obtained from growth after 2 days in brain heart infusion broth, were stained with Gram and modified Kinyoun stains.
Biochemical identification tests.
Mycobacterial identification tests and tests for hydrolysis of xanthine, hypoxanthine, and tyrosine were performed by standard procedures (3, 18). The results of tests with xanthine and hypoxanthine were read weekly for 3 weeks, and the results of tests with tyrosine were read weekly for 4 weeks. Testing with the API Coryne system (version 2.0), the API 20C AUX system, the API 50 CH system, and the APIZYM system (bio Mérieux Vitek, Hazelwood, Mo.) and the RapID CB Plus system (Remel) was done according to the manufacturers' instructions. Reactions with the API Coryne system were read after incubation for 24 h for enzymes and for 2, 4, and 7 days for sugars. The API 20C AUX strips were read at 2 and 6 days, and the results were reported at 6 days. The APIZYM and RapID CB Plus systems were incubated for 4 h. For the API 50 CH strips, inocula were suspended in API 50 CHB broth (bioMérieux) and tests were read daily, with the final reading taken at 5 days. Incubation beyond 5 days gave weak reactions in many wells that were interpreted as nonspecific. Testing with commercial strips was done twice, and the results were read by two observers to evaluate reproducibility. Only assimilation results were considered for the API 50 CH system because oxidation and fermentation reactions were weak, difficult to interpret, and nonreproducible on repeated testing. Sugar assimilations in the API 20C AUX and API 50 CH systems were read as positive when heavy growth in the cupules obscured the underlying stripes.
Susceptibility testing.
E-tests (AB BIODISK, Solna, Sweden) were done according to the manufacturer's instructions. Inocula were prepared by touching five colonies with an applicator stick and suspending the cells in 3 ml of Mueller-Hinton broth supplemented with Tween 80 to a final concentration of 0.7%. Cell suspensions were vortexed, and large clumps were allowed to settle before dilution to match a 0.5 McFarland turbidity standard. Six E-test strips were applied to each inoculated 150-mm plate of Mueller-Hinton agar. The plates were incubated in an aerobic atmosphere at 35°C, and the E-test MICs were read daily for 3 days.
GLC.
Whole-cell short-chain fatty acid analyses were performed by GLC with the MIS essentially as described by Leonard et al. (16). The manufacturer's protocol was followed for cell growth, harvesting, saponification, methylation, extraction, and chromatography with a Hewlett-Packard 5890 Series II gas chromatograph with an electronic pulse control and sample controller. Data were integrated and analyzed with a Hewlett-Packard Vectra computer with MIDI Sherlock Library Generation Software (version 1.06) to compare the experimental chromatographic profiles with both the Clinical library (version 4.0) and the Mycobacterial library (version 3.8) of MIDI.
HPLC.
The whole-cell mycolic acid extraction procedure and the internal standards were as described by Springer et al. (25). The Beckman HPLC system consisted of a 507 autosampler, a 126 pump, a detector set at 260 nm, and a cartridge-style ultrasphere column (4.5 mm by 7.5 cm with 3-μm particles) with a guard column and a Dell Dimension (XPS P100 C) computer system loaded with Beckman System Gold Nouveau software. Samples were analyzed at 35°C by the standard method recommended by the HPLC User's Group. Relative retention times were calculated by assigning “0” to the injection time and “1” to the peak of the high-molecular-weight internal standard. These standards were used to convert the absolute retention times of peaks to relative retention times. To evaluate pattern reproducibility within species, HPLC analyses were performed at least twice for all isolates.
DNA-DNA hybridizations.
DNA-DNA hybridizations were performed by dot blot analysis. Chromosomal DNA was prepared from cells that were incubated with shaking at 37°C for 48 h in 200 ml of brain heart infusion broth (Difco Laboratories, Detroit, Mich). DNA extraction was done by the protocol described by Thierry et al. (28). Chemiluminescent probe labeling, hybridization, and signal detection with the NE Blot Phototope kit (New England Biolabs, Inc., Beverly, Mass.) and the PolarPlex Chemiluminescent Blotting kit (New England Biolabs, Inc.) were performed as recommended by the manufacturer, with hybridization and washing temperatures set at 68°C. The hybridization signal intensity was evaluated visually and graded semiquantitatively on a scale of 0 to 4, with 0 indicating no signal detected and 4 indicating a strong signal from homologous DNA-DNA hybridization derived from a single isolate.
16S rRNA gene sequencing.
The 16S rRNA genes of T. paurometabola type strain ATCC 8368, T. paurometabola ATCC 25938, T. inchonensis type strain ATCC 700082, and T. strandjordae were sequenced in their entirety. The 16S rRNA gene PCR amplification was performed with primers 8FPL and DG74 (positions 8 to 27 and 1522 to 1540 [Escherichia coli numbering], respectively) (12, 22) on a thermocycler (model 9700, Perkin-Elmer Applied Biosystems, Foster City, Calif.) as described previously (14). After column purification of the PCR products (Microcon 100; Amicon, Beverly, Mass.), primers 8FPL, 516F, 516R, 806F, 806R, 1175F, 1175R (8, 22), and DG74 were used for cycle sequencing with the Reaction Ready Big Dye terminator kit (Perkin-Elmer Applied Biosystems) on a Perkin-Elmer Applied Biosystems model 9700 thermocycler. Sequencing products were column purified (CentriSep, Adelphia, N.J.) and analyzed on an API PRISM 377 (Perkin-Elmer Applied Biosystems). For all other strains, after full gene amplification, we performed partial sequencing of the first 500 bp with primers 8FPL and 516R. Sequences were assembled and edited by using Sequencher software (version 3.1; Gene Codes Corp., Ann Arbor, Mich.).
Phylogenetic analysis.
The 16S rRNA gene sequence of T. strandjordae and partial sequences of clinical isolates were searched against the GenBank database by using the gapped BLAST program (1). The first 15 matches of GenBank sequences to the T. strandjordae sequence all corresponded to Tsukamurella species entries. Among these, the following type and reference strain sequences were retrieved from GenBank: T. paurometabola type strain ATCC 8368 (accession nos. X80628 and Z46751), T. paurometabola ATCC 25938 (accession no. Z36933), T. paurometabola strain M334 (accession no. Z37151), T. inchonensis type strain ATCC 700082 (accession no. X85955), T. pulmonis type strain ATCC 700081 (accession no. X92981), T. pulmonis strain KUL50 (accession no. AF001011), T. tyrosinosolvens type strain DSMZ 44234 (accession no. Y12246), T. tyrosinosolvens strains D-1411, D-1437 and D-1498 (accession nos. Y12248, Y12247, and Y12245, respectively), T. wratislaviensis (accession no. Z37138), and Tsukamurella spumae (accession no. Z37150, an invalidly named species). Sequences were also retrieved from GenBank for representative reference strains of closely related mycolic acid-containing genera including Rhodococcus equi (accession no. X80614), Rhodococcus rhodocrous (accession no. X79288), Nocardia asteroides (accession no. X84850), Nocardia otitidis-N. caviarum (accession no. X80611), Mycobacterium chelonae (accession no. M29559), Dietzia maris (accession no. X81920), Gordonia terrae (accession no. AF154833), Skermania piniformis (accession no. Z35435), Williamsia murale (accession no. Y17384), and Corynebacterium diphtheriae (accession no. X82059). These sequences, along with the complete sequences generated in the present study for T. strandjordae, T. paurometabola type strain ATCC 8368, T. paurometabola ATCC 25938, and T. inchonensis type strain ATCC 700082, were aligned by using the CLUSTAL_X program (29). The multiple-sequence-alignment output was trimmed and manually edited with the JalView program (M. Clamp, European Bioinformatics Institute [http://circinus.ebi.ac.uk:6543/jalview/]) and Microsoft Word (Microsoft, Redmond, Wash.). Phylogenetic analyses were performed with the PHYLIP program (J. Felsenstein, Department of Genetics, University of Washington [http://evolution.genetics.washington.edu/phylip.html]) with the neighbor-joining, maximum-likelihood and maximum-parsimony algorithms by using the default parameters with the NEIGHBOR, DNAML, and DNAPARS, programs, respectively. Bootstrap analysis (7) with 1,000 replications for each algorithm was performed with the SEQBOOT program in the PHYLIP software package. Alignment of the Tsukamurella 16S rRNA gene sequences derived from reference strains and submitted by different investigators with those of related members of the order Actinomycetales allowed us to discern species-specific signature sequences, even though infrageneric sequence similarities among Tsukamurella species, with the exception of T. wratislaviensis, exceeded 99% (data not shown). These signature sequences were used for species-level identification of the clinical isolates.
Nucleotide sequence accession numbers.
The complete 16S rRNA gene sequences of T. strandjordae, T. paurometabola ATCC 8368, T. paurometabola ATCC 25938, and T. inchonensis ATCC 700082 have been deposited in GenBank under the following accession nos.: AF283283, AF283280, AF283282, and AF283281 respectively.
RESULTS
Microscopic and colonial morphologies.
With the exception of T. paurometabola and T. wratislaviensis, colonies of Tsukamurella species shared common features. Colonies of T. strandjordae, T. inchonensis, T. pulmonis, and T. tyrosinosolvens, visible after 48 h of incubation on Middlebrook 7H11 agar or TSA, were up to 5 mm in diameter, dry, rough, velvety, and flat and slightly raised in the central one-third to one-half of the colony with fringed edges. Colonies of T. pulmonis and T. tyrosinosolvens were nonpigmented gray-tan and dark tan or buff, respectively. In contrast, colonies of T. strandjordae and T. inchonensis including T. paurometabola ATCC 25938 (identified as T. inchonensis [see below]) were indistinguishable and were pigmented yellow to orange. Colonies of the T. paurometabola type strain had a distinctive fried-egg appearance. These colonies were 2 to 5 mm in diameter, smooth creamy with entire edges, gray, and flat with a tan raised center. Colonies of T. wratislaviensis were up to 3 mm in diameter, gray, rough, and uniformly raised. The colonial morphology on tap water agar visualized under ×20 magnification for T. strandjordae, T. inchonensis, T. pulmonis, and T. tyrosinosolvens showed a biphasic pattern: complex branching forms with a frost-like appearance of substrate hyphae resembling rapidly growing mycobacteria and coccobacillary forms arranged in a classic diphtheroid fashion without production of aerial hyphae. In contrast, T. paurometabola and T. wratislaviensis grew only at the surface of the agar as coccobacilli in diphtheroid arrangements. The Gram staining morphologies for T. strandjordae, T. inchonensis, T. pulmonis, and T. tyrosinosolvens were identical. Cells consisted of long, rod-shaped forms resembling nontuberculous mycobacteria that were gram positive, and by staining with the modified Kinyoun stain, the cells were irregularly positive throughout the smear. Gram staining was generally nonhomogeneous and sometimes beaded. Cells of T. paurometabola type strain ATCC 8368 were diphtheroid, Gram stain variable, and modified Kinyoun stain positive. Cells of T. wratislaviensis were gram-positive coccobacilli and were occasionally positive by staining with the modified Kinyoun stain.
Physiological and biochemical tests.
Standard biochemical tests for acid-fast bacilli showed that all isolates except T. wratislaviensis were resistant to lysozyme, positive for semiquantitative catalase and 68°C heat-stable catalase, Tween 80 hydrolysis at 5 days, urease, pyrazinamidase, Fe uptake, and tolerance to 5% NaCl at 1 week and negative for nitrate reduction after 2 weeks and arylsulfatase after 3 and 14 days. All except T. wratislaviensis and two isolates of T. tyrosinosolvens grew on MacConkey agar without crystal violet after 11 days, but growth after 5 days was inconsistent among all species. None of the isolates grew anaerobically. The results obtained with the API Coryne (version 2.0), RapID CB Plus, and API 20C AUX systems are summarized in Table 1. The profile numbers obtained at 2 days with the API Coryne system corresponded either to Corynebacterium aquaticum (profile no. 2550004) or R. equi (profile no. 2150004) or gave no identification (profile no. 2551004). In the RapID CB Plus system, except for profile no. 0623531, which had no match in the database, all other profiles corresponded to R. equi. T. wratislaviensis gave profile no. 0207531, which also corresponds to R. equi. T. strandjordae displayed a unique profile in the API 20C AUX system and the API Coryne system after 4 days of incubation (Table 1).
TABLE 1.
Biochemical profiles of Tsukamurella species with three commercial systems
| Identification system | Biochemical profile (no. of isolates) for the following Tsukamurella species (no. of isolates):
|
||||
|---|---|---|---|---|---|
| T. strandjordae (1) | T. paurometabola (1) | T. inchonensis (4)d | T. pulmonis (4) | T. tyrosinosolvens (4) | |
| API Corynea | |||||
| 2 days | 2550004 | 2550004 | 2150004 (2), 2550004 (1), 2551004 (1) | 2150004 | 2150004 |
| 4 days | 2553004 | 2550104 | 2150004 (2), 2550004 (1), 2551004 (1) | 2152004 (1) | 2152004 (2) |
| 7 days | 2553004 | 2550104 | 2153004 (1) | 2152004 (1) | 2152004 (2) |
| RapID CB Plusb | 0627511 | 0647511 | 0607511 (3), 0623531 (1) | 0627511 | 0607511 (2), 0627511 (1), 0627531 (1) |
| API 20C AUXc | 6063160 | 6040160 | 6167171 (2), 2127171 (1), 2067171 (1) | 6062160 (2), 6040120 (1), 6042160 (1) | 6167171 (1), 6167771 (1), 2147171 (1) |
The Rapid Coryne strip contained the following reactive ingredients in the ordered groups of three as follows: nitrate reduction, pyrazinamidase, and pyrrolidonyl arylamidase; alkaline phosphatase, β-glucuronidase, and β-galactosidase, α-glucosidase, N-acety-β-glucosaminidase, and esculin; urease, gelatin hydrolysis, and fermentation control; glucose, ribose, and xylose; mannitol, maltose, and lactose; and sucrose and glycogen (catalase was the 21st test). Reaction changes observed after 2 days were all weakly positive.
RapID CB panel tests, ordered in groups of three except for the last two tests, are as follows: glucose, sucrose, and ribose; maltose, p-nitrophenyl-α-d-glucoside, and p-nitrophenyl-β-d-glucoside; p-nitrophenyl-N-acetyl-β-d-glucosaminide, p-nitrophenyl-glycoside, and o-nitrophenyl-β-d-galactoside; p-nitrophenylphosphate, fatty acid ester, and proline-β-naphthylamide; tryptophane-β-naphthylamide, pyrrolidine-β-naphthylamine, and leucyl-glycine-β-naphthylamide; leucine-β-naphthylamide, urea, and potassium nitrate; and catalase and yellow pigment. Reactions were recorded after 4 h.
API 20 C AUX biochemicals, ordered in groups of three except for the first two and the last two tests, are as follows: glucose and glycerol; 2-keto-d-gluconate, l-arabinose, and d-xylose; adonitol, xylitol, and galactose; inositol, sorbitol, and α-methyl-d-glucoside; N-acetyl-d-glucosamine, cellobiose, and lactose; maltose, sucrose, and trehalose; and melezitose and raffinose. The first cupule is a growth control and is not scored. Reactions were recorded after 6 days.
Includes T. paurometabola ATCC 25938.
The sugar assimilation results obtained with the API 20C AUX and API 50 CH systems were reproducible on repeated testing and when the tests were evaluated by two observers. Among the commercial systems, the API 50 CH system was the most helpful in distinguishing T. paurometabola and T. pulmonis from the other Tsukamurella species. Testing with the API 50 CH system in combination with standard biochemicals for mycobacteria, temperature responses, and degradation agars allowed us to identify all tsukamurellae to the species level. All sugars included in the API 20C AUX system are part of the API 50 CH system, which contains 30 additional tests. However, both systems were used because the sugar utilization results obtained with both the API Coryne and the RapID CB Plus systems were uniformly negative. Some discrepancies were observed between the API 20C AUX and the API 50 CH systems (Table 2). However, within each system, sugar assimilation results were generally reproducible on repeated testing. All 14 tested isolates utilized glucose, galactose, saccharose, trehalose, d-fructose, d-turanose, gluconate, and N-acetyl-d-glucosamine as a sole carbon source; and all failed to utilize l-arabinose, d-xylose, adonitol, cellobiose, lactose, raffinose, erythritol, l-xylose, d-fucose, l-sorbose, rhamnose, dulcitol, melibiose, starch, glycogen, β-gentiobiose, d-lyxose, d-tagatose, l-arabitol, amygdaline, 5-ketogluconate, β-methylxyloside, and α-methyl-d-mannoside as a sole carbon source. Differential results for Tsukamurella species with the API 20C AUX and the API 50CH systems and degradation agars and their temperature responses are presented in Table 2. All strains uniformly hydrolyzed esculin in the API Coryne, RapID CB Plus, and API 50 CH systems. The leucine-β-naphthylamide test was positive in both the RapID CB Plus system and the API ZYM system for all tsukamurellae including T. wratislaviensis. It is noteworthy that the urease test with the urea disk was positive for all isolates, although it was negative in the API Coryne and RapID CB Plus systems. The reactions in the API ZYM system were identical for all Tsukamurella strains except T. wratislaviensis. As recommended by the manufacturer, reactions were graded on a scale of 0 to 5, with reactions graded 0 to 2 considered negative. They were positive for 2-naphthylphosphate, 2-naphthylcaprylate, l-leucyl-2-naphthylamide, naphthol-AS-BI-phosphate, 2-naphthyl-α-d-glucopyranoside, and bromo-2-naphthyl-β-d-glucopyranoside. T. wratislaviensis could be distinguished from other Tsukamurella species by being negative for 2-naphthylphosphate, 2-naphthylcaprylate, and bromo-2-naphthyl-β-d-glucopyranoside and positive for l-valyl-2-naphthylamide and l-cystyl-2-naphthylamide.
TABLE 2.
Differential properties and sugar assimilation results with the API 20C AUX and API 50 CH systemsa
| Property or substrateb | Result for the following species (no. of isolates):
|
||||
|---|---|---|---|---|---|
| T. strandjordae (1) | T. paurometabola (1) | T. inchonensis (4) | T. pulmonis (4) | T. tyrosinosolvens (4) | |
| Growth at 28°Cc | + | + | + | −/+ | +/− |
| Growth at 35°Cc | + | + | + | + | + |
| Growth at 42°C | − | − | + | − | − |
| Growth at 45°C | − | − | − | − | − |
| Xanthined | − | − | − | − | − |
| Hypoxanthined | − | − | + | + | + |
| Tyrosined | − | − | +/− | − | + |
| Glycerol (20C) | + | + | +/− | + | + |
| Glycerol (50 CH) | − | + | +/− | +/− | −/+ |
| 2-Ketogluconate (20C) | − | − | +/− | − | + |
| 2-Ketogluconate (50 CH) | + | − | + | − | + |
| Xylitol (20C) | + | − | + | +/− | + |
| Xylitol (50 CH) | + | − | +/− | +/− | +/− |
| Inositol (20C) | + | − | + | − | + |
| Inositol (50 CH) | + | − | + | − | + |
| α-Methyl-d-glucoside (20C) | − | − | + | − | + |
| α-Methyl-d-glucoside (50 CH) | + | − | + | − | − |
| Maltosee (20C) | − | − | + | − | + |
| Maltosee (50 CH) | + | − | + | − | + |
| Melezitose (20C) | − | − | + | − | + |
| Melezitose (50 CH) | + | − | + | − | + |
| d-Arabinose (50 CH) | − | − | +/− | +/− | +/− |
| Ribosee (50 CH) | − | − | +/− | − | +/− |
| d-Mannose (50 CH) | + | − | + | + | +/− |
| Mannitole (50 CH) | + | − | + | + | + |
| Arbutine (50 CH) | + | − | −/+ | − | +/− |
| Salicin (50 CH) | + | − | +/− | − | +/− |
| Inulin (50 CH) | + | − | + | − | + |
| l-Fucose (50 CH) | + | − | +/− | + | + |
| d-Arabitol (50 CH) | + | − | + | + | + |
+ and −, assimilation results were positive and negative, respectively, for all isolates; +/−, two or more strains were positive; −/+, one strain was positive. T. wratislaviensis was not tested with either the API 50 CH or the API 20C AUX system.
20C, the API 20C AUX system; 50 CH, the API 50 CH system.
T. wratilaviensis grew at 28°C but failed to grow at 35°C.
In degradation agars, T. wratislaviensis hydrolyzed tyrosine but failed to hydrolyze xanthine and hypoxanthine.
The substrates were not metabolized in either the API Coryne or the RapID CB Plus. system.
Lipid analyses.
The results of short-chain fatty acid methyl ester (FAME) analyses by GLC chromatography are shown in Table 3. T. paurometabola and T. wratislaviensis are the only Tsukamurella species included in the current Clinical (version 4.0) and Environmental (Trypticase soy broth agar; version 4.0) libraries of the MIDI database, and analyses of FAMEs with chain lengths of 20 carbons or less have not previously been reported for other Tsukamurella species. Similar to other aerobic actinomycetes, the FAME contents of all tsukamurellae consist of a combination of straight-chain saturated and monounsaturated fatty acids and, with the exception of a single clinical isolate of T. pulmonis, tuberculostearic acid (C18:0 10 methyl). The proportions of some FAMEs exhibited wide variation within species and variations for a single isolate between the clinical and mycobacteriological methods. In addition, the fatty acid profiles overlapped among various Tsukamurella species and with those of other aerobic actinomycetes and mycobacteria in the MIDI Clinical and Mycobacterial libraries, respectively. However, by the clinical method, T. pulmonis could be distinguished from other tsukamurellae by a higher proportion of 20:1ω9C fatty acids (>4.5%), whereas all other species contained <2% 20:1ω9C fatty acids. Interestingly, even though T. wratislaviensis is included in the Clinical library of the MIDI database, it was misidentified as Nocardia brasiliensis by the MIS with a high degree of probability (similarity index, 0.664).
TABLE 3.
Short-chain fatty acid content of 15 strains of Tsukamurella analyzed with the MIS
| Fatty acid | Mean ± SD fatty acid content (%)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
T. strandjordae
|
T. paurometabola
|
T. inchonensis
|
T. pulmonis
|
T. tyrosinosolvens
|
T. wratislaviensis
|
|||||||
| Clinical method | Mycobacteriological method | Clinical method | Mycobacteriological method | Clinical method | Mycobacteriological method | Clinical method | Mycobacteriological method | Clinical method | Mycobacteriological method | Clinical method | Mycobacteriological method | |
| C12:0 | —f | — | — | — | — | — | — | — | — | 0.37 ± 0.02 | — | — |
| C14:0 | 4.94 | 5.36 | 3.81 | 5.15 | 4.83 ± 0.82 | 3.48 ± 0.93 | 4.05 ± 0.69 | 3.28 ± 0.57 | 3.93 ± 0.45 | 5.95 ± 1.34 | 3.57 | 3.52 |
| C15:0 | 1.00 | 0.36 | — | — | 0.68 ± 0.21 | 0.5 ± 0.25 | 0.75 ± 0.14 | 0.27 ± 0.01 | 0.81 ± 0.24 | — | 3.92 | 2.52 |
| C16:1ω9 | — | 0.86 | — | 1.68 | 0.62 ± 0.98 | 1.35 ± 0.55 | 1.48 ± 0.42 | 2.42 ± 0.43 | — | 1.01 ± 0.28 | — | 0.60 |
| C16:1ω6 | — | 2.73 | — | 10.59 | — | 4.62 ± 1.69 | — | 4.91 ± 1.75 | — | 3.15 ± 1.26 | — | 8.66 |
| C16:0 | 40.36 | 40.69 | 36.89 | 36.72 | 38.88 ± 2.67 | 37.74 ± 2.6 | 33.73 ± 3.54 | 31.08 ± 2.68 | 42.89 ± 1.52 | 39.32 ± 2.44 | 35.45 | 30.06 |
| C17:1ω8 | — | — | — | — | — | — | — | — | — | — | 3.90 | 4.69 |
| C17:0 | 1.78 | 0.40 | 1.57 | — | 1.04 ± 0.29 | 0.65 ± 0.42 | 1.44 ± 0.37 | 0.32 ± 0.01 | 1.36 ± 0.57 | — | 2.68 | 3.80 |
| C17:0 10 methyl | — | — | — | — | — | — | — | — | — | — | 2.01 | — |
| C18:1ω9 | 23.70 | 17.72 | 26.43 | 13.39 | 24.79 ± 2.58 | 17.94 ± 6.76 | 30.86 ± 6.82 | 30.23 ± 12.15 | 29.59 ± 3.83 | 20.11 ± 6.23 | 21.47 | 22.28 |
| C18:0 | 2.18 | 11.13 | 3.30 | 6.66 | 2.78 ± 1.31 | 9.41 ± 4.67 | 4.95 ± 1.54 | 8.21 ± 2.24 | 2.72 ± 0.77 | 8.67 ± 3.5 | 2.44 | 2.34 |
| TBSAa | 16.58 | 18.51 | 16.49 | 25.81 | 16.7 ± 4.37 | 24.28 ± 7.83 | 10.07 ± 8.34 | 17.28 ± 12.72 | 13.21 ± 1.52 | 20.63 ± 3.74 | 12.73 | 12.02 |
| C20:1ω9 | 1.65 | — | 1.69 | — | 1.59 ± 0.29 | — | 5.03 ± 0.34 | — | 1.18 ± 0.24 | — | — | — |
| C20:0 | — | 1.87 | — | — | — | 1.26 ± 0.99 | 0.39 ± 0.5 | 2.15 ± 0.99 | — | 1.23 ± 0.65 | — | 0.58 |
| Sum 1b | — | 0.37 | — | — | — | 0.52 ± 0.23 | — | — | — | — | — | 1.00 |
| Sum 3c | 7.81 | — | 9.20 | — | 7.07 ± 2.46 | — | 6.33 ± 2.85 | — | — | — | 11.83 | 7.28 |
| Sum 5d | — | — | 0.62 | — | 0.68 ± 0.31 | — | 0.74 ± 0.42 | — | 0.84 ± 0.46 | — | — | — |
| Sum 6e | — | — | — | — | 0.97 ± 0.58 | — | 0.75 ± 0.3 | — | — | — | — | — |
TBSA, tuberculostearic acid (C18:0 10 methyl).
Sum 1, summed feature 1 (C16:0 8 methyl or C16:0 10 methyl; these fatty acids elute at the same time and therefore cannot be differentiated).
Sum 3, summed feature 3 (C16:1ω7 or C15 iso 2OH).
Sum 5, summed feature 5 (C18:2ω6,9 or C18:0 Ante).
Sum 6, summed feature 6 (C19:1ω9 or C19:1ω1).
—, fatty acid not detected.
HPLC analysis of mycolic acids yielded similar chromatographic profiles for all Tsukamurella species with the exception of T. wratislaviensis. These profiles consisted of a characteristic single cluster of poorly resolved peaks that were not sufficiently distinctive for species identification. The overall relative retention times of these peaks ranged from 6.25 to 7.50 min, but the last peak to emerge was often hard to define. The chromatographic profile of T. wratislaviensis was unique compared to those for the other tsukamurellae and exhibited two clusters of peaks that eluted much earlier: the early peaks were obscured by the solvent front and the last peak emerged at 5 min. These data suggest that T. wratislaviensis possesses shorter-chain mycolic acids than other Tsukamurella species.
Antimicrobial susceptibility.
By using the National Committee for Clinical Laboratory Standards (NCCLS) interpretive guidelines for the family Enterobacteriaceae and Staphylococcus species, T. paurometabola could be distinguished by its high level of resistance to imipenem (MIC, >32 μg/ml), whereas all other isolates were susceptible to imipenem (MICs, <1 μg/ml for all except one clinical isolate of T. inchonensis, for which the imipenem MIC was 4 μg/ml). T. wratislaviensis was susceptible to cefoxitin (MIC, 8 μg/ml), whereas the other isolates were highly resistant to cefoxitin-(MICs, >256 μg/ml). T. strandjordae and two clinical isolates of T. pulmonis were intermediately susceptible to gentamicin, but all other strains were fully susceptible to gentamicin. The susceptibility profile of T. strandjordae with the other antimicrobials tested otherwise resembled those of strains of T. pulmonis, T. inchonensis, and T. tyrosinosolvens (see description of new taxon below). A more detailed description of the antimicrobial susceptibility of the Tsukamurellae will be reported elsewhere (M. A. Schwartz et al., submitted for publication).
DNA-DNA hybridizations, 16S rRNA gene sequencing, and phylogenetic analyses.
DNA-DNA hybridizations and 16S rRNA gene sequencing confirmed that T. strandjordae is a novel bacterium and allowed us to assign all clinical isolates to their respective species. Hybridization experiments, performed twice with T. strandjordae, showed strong hybridization signals (4+) with homologous DNA, weak signals (1+ or 2+) with DNAs from isolates of T. inchonensis, T. paurometabola, T. pulmonis, and T. tyrosinosolvens, and no signal with DNA from T. wratislaviensis (data not shown). T. paurometabola ATCC 25938 displayed a strong hybridization signal (4+) with DNA from T. inchonensis type strain ATCC 70082 and homologous DNA, weak hybridization signals (1+ or 2+) with DNAs from T. strandjordae, T. pulmonis, and T. tyrosinosolvens, and no signal with DNAs from T. wratislaviensis and the type strain T. paurometabola ATCC 8368. Except for homologous DNA, T. wratislaviensis exhibited no hybridization with the DNA of any other isolate. Similarly, isolates of T. pulmonis, T. inchonensis, and T. tyrosinosolvens displayed signals of 3+ or 4+ within species and with homologous DNA and signals of 0, 1+, or, less commonly, 2+ with DNAs from isolates of other species.
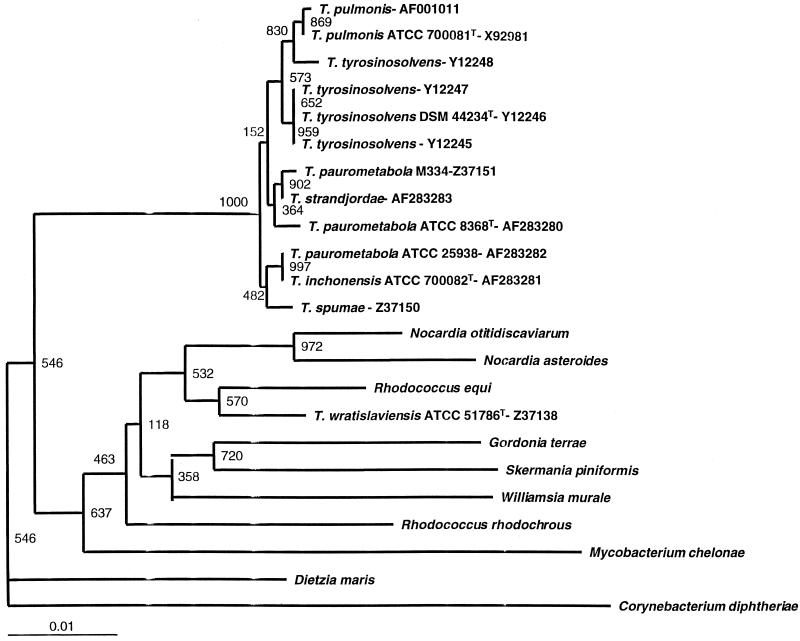
Our hybridization results were supported by 16S rRNA gene sequencing. Strain pairs that displayed strong DNA-DNA hybridization signals (3+ or 4+) had identical 16S rRNA gene signature sequences in pairwise sequence comparisons. Likewise, the 16S rRNA gene sequence of T. wratislaviensis differed by more than 4% from the rRNA gene sequences of all other Tsukamurella species tested, as predicted from DNA-DNA hybridization data. Except at a single position, the sequence of T. strandjordae was identical to that found in GenBank for a strain designated T. paurometabola M334; however, its sequence differed from that of T. paurometabola ATCC 8368 at 9 positions, including 2 insertions or deletions, and from that of T. paurometabola ATCC 25938 at 10 positions. Phylogenetic analyses with three treeing algorithms showed that T. strandjordae and T. paurometabola strain M334 formed a distinct branch within the Tsukamurella clade with strong bootstrap support (Fig. 1) and thus merit assignment to a novel species. Pairwise sequence comparisons with other Tsukamurella species showed that T. strandjordae differed from T. inchonensis type strain ATCC 700082 at 10 positions, from T. pulmonis type strain ATCC 700081 at 6 positions, from T. tyrosinosolvens type strain DSMz 44234 at 4 positions, from T. spumae at 8 positions including 2 insertions or deletions, and from T. wratislaviensis at 61 positions. We also found that the complete 16S rRNA gene sequence of T. paurometabola ATCC 25938 is 100% identical to that of T. inchonensis type strain ATCC 700082. These data, together with DNA-DNA hybridization and phenotypic data, provide strong evidence that T. paurometabola ATCC 25938 is a misnamed T. inchonensis strain.
FIG. 1.
Neighbor-joining dendrogram based on 1,311 consecutive positions of the 16S rRNA gene with 1,000 bootstrap replications showing the phylogenetic relationships of T. strandjordae and T. paurometabola (T. inchonensis) ATCC 25938 with other tsukamurellae and select related corynebacterineae of the order Actinomycetales. C. diphtheriae was chosen as the outgroup. Numbers at each node correspond to the bootstrap values. GenBank nucleotide sequence accession numbers are also included for the tsukamurellae. The sequences in GenBank for T. paurometabola ATCC 8368 (accession no. Z46751 and X80628), T. paurometabola ATCC 25938 (accession no. Z36933), and T. inchonensis ATCC 70082 (accession no. X85955) were identical to those generated in this study and were therefore not included in the dendrogram.
DISCUSSION
The taxonomic history of the genus Tsukamurella has been reviewed extensively by Yassin et al. (34). The type species T. paurometabola, isolated from the ovaries of bed bugs, was initially described by Steinhaus as Corynebacterium paurometabolum. In a comprehensive numerical phenetic study by Jones (13), the type strain of C. paurometabolum ATCC 8368 was found to be quite distinct from Corynebacterium species. The first human isolate of Tsukamurella, named Gordona aurantiaca, was identified in the sputum of a patient with tuberculosis, but its causal role in disease was questioned (33). In 1985, G. aurantiaca was renamed Rhodococcus aurantiacus by Tsukamura and assigned a new type strain, ATCC 25938 (31). Chemical and numerical taxonomic studies of C. paurometabolum and G. aurantiaca (5, 9) revealed that they had similar cellular compositions. Thus, in 1988, on the basis of these data, the >99% similarity of the 16S rRNA gene sequences, and the phylogenetic positions of strain ATCC 8368 and strain 25938 among other members of the order Actinomycetales, Collins and colleagues (6) concluded that they be assigned to a new genus, Tsukamurella, and that they belong to a single species, Tsukamurella paurometabolum. They designated ATCC 8368 as the type strain. Yassin later correctly renamed T. paurometabolum T. paurometabola (34). Since the proposal of the genus in 1988, clinical infections associated with tsukamurellae were mostly attributed to T. paurometabola; however, the published descriptions of these clinical isolates do not resemble the description of T. paurometabola type strain ATCC 8368. Auerbach et al. (2) first noted heterogeneity within the genus Tsukamurella in 1992. They reported a common-source outbreak of pseudoinfection in 10 patients caused by T. paurometabola resulting from laboratory contamination. Similar to our observations, they found that the new type strain, ATCC 8368, differed from the outbreak isolates and from the original type strain, ATCC 25938, in terms of colonial morphology, biochemistry, antimicrobial susceptibility, and ribotyping. McNabb et al. (17) also found that strains ATCC 8368 and ATCC 25938 differed in their cellular fatty acid contents, suggesting the possibility of separate species. From 1995 to 1997, Yassin et al. (34–36) proposed three additional species, T. pulmonis, T. inchonensis and T. tyrosinosolvens, all of which were isolated from human samples.
By comparing type and reference strains of Tsukamurella to clinical isolates, we were able to identify another Tsukamurella species closely related to but distinct from T. pulmonis, T. inchonensis, T. paurometabola, and T. tyrosinosolvens. Our patient's isolate, T. strandjordae, was the sole bacterium grown repeatedly from cultures of blood from a 5-year-old girl with acute myelogenous leukemia and thus adds to the growing list of aerobic actinomycetes causing opportunistic infections. This organism is characterized by a unique 16S rRNA gene sequence and does not hybridize strongly with any other Tsukamurella species. The 16S rRNA sequence of our unique isolate, T. strandjordae, differs at only one position from that listed in GenBank for another strain, M334, identified as T. paurometabola. Therefore, we believe that strain M334 most likely belongs to our newly proposed species, T. strandjordae. We could not obtain strain M334 to compare it with our isolate. However, strain M334 was included in a polyphasic taxonomic study of Gordonia and Tsukamurella (10), in which it exhibited low-level DNA-DNA hybridization (<45%) with other Tsukamurella strains including strain N663, synonymous with T. paurometabola ATCC 25938. However, the current type strain, T. paurometabola ATCC 8368, was not included in that study. Considering a DNA homology threshold of 70% for species definition (26), these data suggest that strain M334 is closely related to other Tsukamurella strains tested in that study but is a separate species. Our study also showed that T. paurometabola strain ATCC 25938, formerly G. aurantiaca, belongs to T. inchonensis on the basis of the 100% identity of its 16S rRNA gene sequence to the 16S rRNA gene sequence of T. inchonensis type strain ATCC 70082, the high level of DNA-DNA hybridization between the two strains, and the concordant phenotypic data for the two strains.
The pathogenic role of tsukamurellae has been debated. Since the initial report by Tsukamura and Kawakami (32) of a lung infection that mimicked tuberculosis caused by G. aurantiaca in 1982, several cases of human infection have been described primarily in immunocompromised patients or patients with predisposing conditions but occasionally in immunocompetent hosts as well (11, 15, 19–21, 23, 24, 30; R. F. Haft, C. L. Shapiro, N. M. Gantz, J. C. Christenson, and R. J. Wallace, Jr., Letter, Clin. Infect. Dis. 15:883, 1992; R. S. Jones, T. Fekete, A. L. Truant, and V. Satishchandran, Letter, Clin. Infect. Dis. 18:830–832, 1994; K. K. Lai, Letter, Clin. Infect. Dis. 17:285–287, 1993; D. Rey, D. De Briel, R. Heller, P. Fraisse, M. Partisani, M. Leiva-Mena, and J. M. Lang, Letter, AIDS 9:1379, 1995). Some reports indicate that these organisms were isolated concomitantly with other bacteria and that they were considered environmental contaminants. However, cases of catheter-related infection, peritonitis, fatal meningitis, and bone infection in which tsukamurellae were the sole isolates suggest that these organisms are potential opportunistic pathogens. In some of these patients, Tsukamurella species were repeatedly isolated over a several-month period and could not be eradicated, despite polyantimicrobial therapy (30, 32).
On the basis of its sugar utilization profile and short-chain fatty acid content, T. strandjordae could be easily distinguished from T. paurometabola and T. pulmonis, but distinction from T. inchonensis and T. tyrosinosolvens was based on a few tests, namely, temperature responses and hydrolysis of hypoxanthine and tyrosine. With few discrepancies, our study of 15 strains of Tsukamurella generally showed good agreement between our phenotypic data and those reported by Yassin et al. for T. paurometabola, T. inchonensis, T. pulmonis, and T. tyrosinosolvens (34–36). Testing for sugar utilization with the API 50 CH system showed excellent correlation with published data based on tests with conventional biochemicals. However, with degradation agars, we found inconsistencies among different studies and with our results (2, 24, 34–36). Moreover, in our study we found discrepancies between different commercial systems. For example, it is noteworthy that sugar utilization tests and the urease tests were negative throughout or occasionally weakly positive in the API Coryne and RapID CB Plus systems. The test for β-galactosidase reportedly one of the distinguishing features between Tsukamurella and Rhodococcus species (32), in the API Coryne system was negative for >50% of the strains tested. Tsukamurella strains which lack β-galactosidase activity have been reported by others (21, 30). The discrepancies that were encountered could be attributed to the lack of metabolic substrates in basal media that would favor expression of certain characteristics in some organisms, phenotypic variability within species, or variation in experimental conditions.
Analysis of short-chain fatty acids was helpful only for the separation of T. pulmonis and T. wratislavensis from other Tsukamurella species, but distinction from other aerobic actinomycetes and Mycobacterium species was inconsistent. We did not expect our fatty acid profiles to be comparable to those of McNabb et al. (17) since we used different media (TSA and Middlebrook 7H11 agar versus Trypticase soy agar), different culture incubation periods (48 versus 96 h), and a different temperature (28 versus 35°C), as recommended by the manufacturer. These parameters are well recognized to have a critical impact when these profiles are used for bacterial identification. Mycolic acid analyses by HPLC are useful in distinguishing clinical isolates of tsukamurellae from related mycolic acid-containing bacteria but do not distinguish the different species.
Another interesting observation in our study was that T. wratislaviensis is distant from other Tsukamurella species both in phenotype and by its 16S rRNA sequence. T. wratislaviensis was originally found in soil and to our knowledge has never been isolated from a human source. We found that T. wratislaviensis contains mycolic acids of a shorter chain length compared to those for other Tsukamurella species. Our phylogenetic data revealed that T. wratislaviensis forms a phyletic line distinct from the Tsukamurella clade (100% boostrap support) and clusters with Rhodococcus and Nocardia species. A BLAST search of the sequences in GenBank showed that T. wratislaviensis is most closely related to Rhodococcus opacus with 99.7% 16S rRNA gene sequence (GenBank accession no. X80631) similarity, strongly suggesting that T. wratislaviensis belongs to the genus Rhodococcus (36).
Description of Tsukamurella strandjordae sp. nov.
Tsukamurella strandjordae sp. nov. (strandjordae, in honor of Paul Strandjord, founder and chair of the Department of Laboratory Medicine, University of Washington, from 1969 to 1994).
Cells are strictly aerobic, long, gram-positive rods and are slightly acid-fast with the modified Kinyoun stain. Abundant growth is observed after 48 h on Trypticase soy agar, Middlebrook 7H11 agar, nutrient agar, and MacConkey agar without crystal violet. Colonies are rough and pigmented tan to yellow, and range in size from 2 to 5 mm in diameter. The organism grows at 28 and 35°C but not at 42°C. On tap water agar it produces short, branching substrate hyphae and diphtheroid forms but no aerial hyphae. It does not hydrolyze xanthine, hypoxanthine, or tyrosine. It is positive for semi quantitative catalase and 68°C heat-stable catalase, Tween 80 hydrolysis at 5 days, urease, pyrazinamidase, 5% NaCl tolerance, and iron uptake. It does not hydrolyze nitrate at 2 weeks and is negative for arylsulfatase at 3 and 14 days. The numerical profile with the API Coryne system after 2 days is 2550004, that with the RapID CB Plus system after 4 h is 0627511, and that with the API 20C AUX system after 6 days is 6063160. In the API 50 CH system it can utilize galactose, d-glucose, d-fructose, d-mannose, maltose, saccharose, trehalose, melezitose, d-turanose, l-fucose, gluconate, inositol, mannitol, sorbitol, d-arabitol, l-arabitol, α-methyl-d-glucoside, N-acetylglucosamine, arbutine, salicine, and 2-ketogluconate as a sole carbon source and hydrolyzed esculin. It does not utilize erythritol, d-arabinose, l-arabinose, ribose, d-xylose, l-xylose, adonitol, β-methylxyloside, l-sorbose, rhamnose, dulcitol, α-methyl-d-mannoside, amygdaline, cellobiose, lactose, melibiose, inuline, d-raffinose, starch, glycogen, xylitol, β-gentiobiose, d-lyxose, d-fucose, l-fucose, gluconate, or 5-ketogluconate as a sole carbon source. In the API ZYM system it is positive only for 2-naphthylphosphate, 2-naphthylcaprylate, l-leucyl-2-naphthylamide, 2-naphthyl-α-d-glucopyranoside, and bromo-2-naphthyl-β-d-glucopyranoside. It contains short, straight-chain saturated and monounsaturated fatty acids and tuberculostearic acid and long-chain mycolic acids, forming a single cluster of peaks on HPLC. It can be distinguished from T. paurometabola and T. pulmonis by assimilation of inositol, d-mannose, arbutine, salicin, and inulin and can be distinguished from T. inchonensis and T. tyrosinosolvens by the absence of growth at 42°C and a lack of assimilation of hypoxanthine and tyrosine. By the E-test it is susceptible to imipenem, amikacin, clarithromycin, ciprofloxacin, and trimethoprim-sulfamethoxazole. It has intermediate susceptibility to gentamicin and is resistant to cefoxitin, cefotaxime, tobramycin, erythromycin, and azithromycin (Schwartz et al., submitted). It is characterized by a unique 16S rRNA gene sequence (GenBank accession no. AF283283). The type strain has been deposited at the ATCC as strain ATCC BAA-173.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Auerbach S B, McNeil M M, Brown J M, Lasker B A, Jarvis W R. Outbreak of pseudoinfection with Tsukamurella paurometabolum traced to laboratory contamination: efficacy of joint epidemiological and laboratory investigation. Clin Infect Dis. 1992;14:1015–1022. doi: 10.1093/clinids/14.5.1015. [DOI] [PubMed] [Google Scholar]
- 3.Brown J M, McNeil M M, Desmond E P. Nocardia, Rhodococcus, Gordona, Actinomadura, Streptomyces, and other actinomycetes of medical importance. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: American Society for Microbiology; 1999. pp. 370–398. [Google Scholar]
- 4.Clausen C, Wallis C K. Bacteremia caused by Tsukamurella species. Clin Microbiol Newsl. 1994;16:6–8. [Google Scholar]
- 5.Collins M D, Jones D. Lipid composition of Corynebacterium paurometabolum. FEMS Microbiol Lett. 1982;13:13–16. [Google Scholar]
- 6.Collins M D, Smida J, Dorsch M, Stackebrandt E. Tsukamurella gen. nov. harboring Corynebacterium paurometabolum and Rhodococcus aurantiacus. Int J Syst Bacteriol. 1988;38:385–391. [Google Scholar]
- 7.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 8.Fredricks D N, Relman D A. Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate. J Clin Microbiol. 1998;36:2810–2816. doi: 10.1128/jcm.36.10.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodfellow M, Orlean P A B, Collins M D, Alshamaony L, Minnikin D E. Chemical and numerical taxonomy of strains received as Gordona aurantiaca. J Gen Microbiol. 1978;109:57–68. [Google Scholar]
- 10.Goodfellow M, Zakrzewska-Czerwinska J, Thomas E G, Mordarski M, Ward A C, James A L. Polyphasic taxonomic study of the genera Gordona and Tsukamurella including the description of Tsukamurella wratislaviensis sp. nov. Zentbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig. 1991;275:162–178. doi: 10.1016/s0934-8840(11)80063-0. [DOI] [PubMed] [Google Scholar]
- 11.Granel F, Lozniewski A, Barbaud A, Lion C, Dailloux M, Weber M, Schmutz J L. Cutaneous infection caused by Tsukamurella paurometabolum. Clin Infect Dis. 1996;23:839–840. doi: 10.1093/clinids/23.4.839. [DOI] [PubMed] [Google Scholar]
- 12.Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones D. Numerical taxonomy of coryneform and related bacteria. J Gen Microbiol. 1975;87:52–96. doi: 10.1099/00221287-87-1-52. [DOI] [PubMed] [Google Scholar]
- 14.Kattar M M, Chavez J F, Limaye A P, Rassoulian-Barrett S L, Yarfitz S L, Carlson L C, Houze Y, Swanzy S, Wood B L, Cookson B T. Application of 16S rRNA gene sequencing to identify Bordetella hinzii as the causative agent of fatal septicemia. J Clin Microbiol. 2000;38:789–794. doi: 10.1128/jcm.38.2.789-794.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larkin J A, Lit L, Sinnott J, Wills T, Szentivanyi A. Infection of a knee prosthesis with Tsukamurella species. South Med J. 1999;92:831–832. doi: 10.1097/00007611-199908000-00019. [DOI] [PubMed] [Google Scholar]
- 16.Leonard R B, Nowowiejski D J, Warren J J, Finn D J, Coyle M B. Molecular evidence of person-to-person transmission of a pigmented strain of Corynebacterium striatum in intensive care units. J Clin Microbiol. 1994;32:164–169. doi: 10.1128/jcm.32.1.164-169.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNabb A, Shuttleworth R, Behme R, Colby W D. Fatty acid characterization of rapidly growing pathogenic aerobic actinomycetes as a means of identification. J Clin Microbiol. 1997;35:1361–1368. doi: 10.1128/jcm.35.6.1361-1368.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metchock B G, Nolte F S, Wallace R J., Jr . Mycobacterium. In: Murray P R, Barron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: American Society for Microbiology; 1999. pp. 399–437. [Google Scholar]
- 19.Min Y, Asano M, Kohanawa M, Minagawa T. Movement disorders in encephalitis induced by Rhodococcus aurantiacus infection relieved by the administration of l-dopa and anti-T-cell antibodies. Immunology. 1999;96:10–15. doi: 10.1046/j.1365-2567.1999.00659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osoagbaka O U. Evidence for the pathogenic role of Rhodococcus species in pulmonary diseases. J Appl Bacteriol. 1989;66:497–506. doi: 10.1111/j.1365-2672.1989.tb04570.x. [DOI] [PubMed] [Google Scholar]
- 21.Prinz G, Ban E, Fekete S, Szabo Z. Meningitis caused by Gordona aurantiaca (Rhodococcus aurantiacus) J Clin Microbiol. 1985;22:472–474. doi: 10.1128/jcm.22.3.472-474.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Relman D A. Universal bacterial 16S rRNA amplification and sequencing. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C.: American Society for Microbiology; 1993. pp. 489–495. [Google Scholar]
- 23.Rey D, Fraisse P, Riegel P, Piemont Y, Lang J M. Tsukamurella infections. Review of the literature apropos of a case. Pathol Biol (Paris) 1997;45:60–65. . (In French.) [PubMed] [Google Scholar]
- 24.Shapiro C L, Haft R F, Gantz N M, Doern G V, Christenson J C, O'Brien R, Overall J C, Brown B A, Wallace R J., Jr Tsukamurella paurometabolum: a novel pathogen causing catheter-related bacteremia in patients with cancer. Clin Infect Dis. 1992;14:200–203. doi: 10.1093/clinids/14.1.200. [DOI] [PubMed] [Google Scholar]
- 25.Springer B, Wu W K, Bodmer T, Haase G, Pfyffer G E, Kroppenstedt R M, Schroder K H, Emler S, Kilburn J O, Kirschner P, Telenti A, Coyle M B, Bottger E C. Isolation and characterization of a unique group of slowly growing mycobacteria: description of Mycobacterium lentiflavum sp. nov. J Clin Microbiol. 1996;34:1100–1107. doi: 10.1128/jcm.34.5.1100-1107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 27.Steinhaus E. A study of the bacteria associated with thirty species of insects. J Bacteriol. 1941;42:757–790. doi: 10.1128/jb.42.6.757-790.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thierry D, Vincent V, Clement F, Guesdon J L. Isolation of specific DNA fragments of Mycobacterium avium and their possible use in diagnosis. J Clin Microbiol. 1993;31:1048–1054. doi: 10.1128/jcm.31.5.1048-1054.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsukamura M, Hikosaka K, Nishimura K, Hara S. Severe progressive subcutaneous abscesses and necrotizing tenosynovitis caused by Rhodococcus aurantiacus. J Clin Microbiol. 1988;26:201–205. doi: 10.1128/jcm.26.2.201-205.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsukamura M, Ikuya Y. Rhodococcus sputi sp. nov., nom. rev., and Rhodococcus aurantiacus sp. nov., nom. rev. Int J Syst Bacteriol. 1985;35:364–368. [Google Scholar]
- 32.Tsukamura M, Kawakami K. Lung infection caused by Gordona aurantiaca (Rhodococcus aurantiacus) J Clin Microbiol. 1982;16:604–607. doi: 10.1128/jcm.16.4.604-607.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsukamura M, Mizuno S. A new species Gordona aurantiaca occurring in sputa of patients with pulmonary disease. Kekkaku. 1971;46:93–98. . (In Japanese.) [PubMed] [Google Scholar]
- 34.Yassin A F, Rainey F A, Brzezinka H, Burghardt J, Lee H J, Schaal K P. Tsukamurella inchonensis sp. nov. Int J Syst Bacteriol. 1995;45:522–527. doi: 10.1099/00207713-45-3-522. [DOI] [PubMed] [Google Scholar]
- 35.Yassin A F, Rainey F A, Brzezinka H, Burghardt J, Rifai M, Seifert P, Feldmann K, Schaal K P. Tsukamurella pulmonis sp. nov. Int J Syst Bacteriol. 1996;46:429–436. doi: 10.1099/00207713-46-2-429. [DOI] [PubMed] [Google Scholar]
- 36.Yassin A F, Rainey F A, Burghardt J, Brzezinka H, Schmitt S, Seifert P, Zimmermann O, Mauch H, Gierth D, Lux I, Schaal K P. Tsukamurella tyrosinosolvens sp. nov. Int J Syst Bacteriol. 1997;47:607–614. doi: 10.1099/00207713-47-3-607. [DOI] [PubMed] [Google Scholar]