Abstract
Chronic wasting disease (CWD) is a deleterious brain proteinopathy caused by a pathogenic form of prion protein (PrPSc), which is converted from a benign form of prion protein (PrPC) encoded by the prion protein gene (PRNP). In elk, the M132L single nucleotide polymorphism (SNP) of the PRNP gene likely plays a pivotal role in susceptibility to CWD. However, the association of the M132L SNP with susceptibility to CWD has not been evaluated in Korean elk to date. To estimate the association of the M132L SNP with susceptibility to CWD in Korean elk, we investigated the genotype and allele frequencies of the M132L SNP by amplicon sequencing and performed association analysis between CWD-positive and CWD-negative elk. In addition, we performed a meta-analysis to evaluate the association between the M132L SNP and susceptibility to CWD in quantitatively synthesized elk populations. Furthermore, we estimated the effect of the M132L SNP on elk PrP using in silico programs, including PolyPhen-2, PROVEAN, AMYCO and Swiss-PdbViewer. We did not identify a significant association between the M132L SNP of PRNP and susceptibility to CWD in Korean elk. The meta-analysis also did not identify a strong association between the M132L SNP of PRNP and susceptibility to CWD in quantitatively synthesized elk populations. Furthermore, we did not observe significant changes in structure, amyloid propensity or electrostatic potential based on the M132L SNP in elk PrP. To the best of our knowledge, this was the first report of an association analysis and meta-analysis in Korean elk and quantitatively synthesized elk populations, respectively.
Keywords: Elk (Cervus canadensis), prion, CWD, PRNP, M132L, polymorphism, SNP, meta-analysis
Introduction
Prion diseases are infectious brain proteinopathies caused by a pathogenic form of prion protein (PrPSc) converted from an endogenous form of prion protein (PrPC) and are classified into several types in a wide range of mammalian hosts: Creutzfeldt–Jakob disease (CJD), fatal familial insomnia (FFI) and Gerstmann-Sträussler-Scheinker syndrome (GSS) in humans; scrapie in sheep and goats; bovine spongiform encephalopathy (BSE) in cattle; and chronic wasting disease (CWD) in the Cervidae family (1–7). After CWD was first reported in mule deer in the USA in the 1967, CWD has been reported globally, including in North America, South Korea and Scandinavia, in variable cervid species, such as white-tailed deer, elk, moose, red deer and reindeer (8–11). To date, CWD is a fatal and incurable disorder, and its exact origin has not been elucidated to date.
Prion protein (PrP), which is encoded by the prion protein gene (PRNP), is the template of and PRNP gene variations are the primary factor in determining susceptibility to CWD (8, 12). In elk, the M132L single nucleotide polymorphism (SNP) of the PRNP gene, which is equivalent to codon 129 in the human PRNP gene, plays a pivotal role in susceptibility to CWD. A previous case-control study reported that the M132L SNP of the PRNP gene is associated with vulnerability to CWD in elk in the USA (13). The M132 allele was more frequently observed in CWD-positive elk, and the L132 allele was not observed in CWD-positive elk. In addition, CWD-inoculated elk PrP transgenic mice with the L132 allele showed resistance to CWD compared to elk PrP transgenic mice with M132 allele (14). Furthermore, experimentally CWD-infected elk carrying the 132LL genotype showed extended survival times compared to elk carrying the 132MM and 132ML genotypes (15). However, another case-control study did not report the association between the M132L SNP and susceptibility to CWD in the USA (16). In addition, an in vitro conversion assay using real-time quaking-induced conversion reactions (RT-QuIC) showed that regardless of the genotype of PRNP codon 132, the conversion efficiency of CWD was high when the genotype of this codon was matched (15). Thus, the effect of the PRNP M132L SNP on the risk of CWD remains controversial.
In Korea, >12,000 elk are bred to obtain meat and antlers as food and medicine, respectively (https://www.mafra.go.kr). Korean elk were originally imported from North America, and CWD in Korean elk has been reported sporadically (17). Given that CWD is a highly infectious prion disease and can be transmitted by peripheral body fluids, including urine, tears and saliva, it is very important to breed elk that are genetically resistant to CWD for preemptive control of CWD (18). However, the M132L SNP, one of the important genetic factors of CWD, has not been investigated in Korean elk to date.
To evaluate the association of the PRNP M132L SNP with susceptibility to CWD in Korean elk, we investigated the genotype and allele frequencies of this SNP by amplicon sequencing and performed association analysis between CWD-positive and CWD-negative elk. In addition, we performed an integrative evaluation of the association between the PRNP M132L SNP and susceptibility to CWD in quantitatively synthesized elk populations by meta-analysis. Furthermore, we estimated the effect of the M132L SNP on elk PrP using in silico programs, including PolyPhen-2, PROVEAN, AMYCO, and Swiss-PdbViewer (19–21).
Materials and Methods
Ethics Statements
All experimental procedures were approved according to the recommendations of the Institutional Animal Care and Use Committee of Jeonbuk National University (IACUC Number: JBNU-2019-0076). All experiments were performed in accordance with the Korea Experimental Animal Protection Act.
Subjects
A total of 253 brain samples were collected from animal farms in the Republic of Korea where CWD has occurred in elk. CWD tests were performed on all brain samples by the Animal and Plant Quarantine Agency (APQA) in the Republic of Korea using the HerdChek BSE-Scrapie Antigen Kit (IDEXX, USA) and western blot analysis. In 253 elk, 49 elk were infected with CWD.
Genomic DNA Extraction
Genomic DNA was eluted from 20 mg brain tissue using a QIAamp DNA Mini Kit (Qiagen, USA) following the manufacturer's instructions.
Genetic Analysis of the M132L SNP of the Elk PRNP Gene
The elk PRNP gene (accession number: FJ590751.1) was amplified from the genomic DNA using sense and antisense gene-specific primers. The sequences of the primers were as follows: PRNP-F (ATGGTGAAAAGCCACATAGGC) and PRNP-R (ACACTTGCCCCTCTTTGGTA). Polymerase chain reaction (PCR) was performed using GoTaq® DNA Polymerase (Promega, Fitchburg, Wisconsin, USA). The PCR mixture contained 20 pmol of each primer, 5 μl of 10× Taq DNA polymerase buffer, 1 μl of 10 mM dNTP mixture and 2.5 units of Taq DNA polymerase. The PCR conditions for the PRNP-F and PRNP-R primers were 95°C for 2 min for denaturation; 35 cycles of 94°C for 45 s, 59°C for 45 s, and 72°C for 1 min 30 s; 1 cycle of 72°C for 10 min for extension. PCR was performed using an S-1000 Thermal Cycler (Bio–Rad, Hercules, California, USA). The PCR products were purified using a PCR Purification Kit (Thermo Fisher Scientific, Bridgewater, New Jersey, USA) and sequenced using an ABI 3730 automatic sequencer (ABI, Foster City, California, USA). Sequencing results were read by Finch TV software (Geospiza Inc., Seattle, USA), and M132L SNP genotyping of the elk PRNP gene was performed.
Literature Search
A literature search was performed using the PubMed database to identify studies regarding the M132L SNP of the PRNP gene from CWD-affected elk. The following search terms were used: “elk,” “PRNP,” “prion,” “CWD,” or “chronic wasting disease” combined with “SNP,” “polymorphism,” or “susceptibility” (the last search update was performed on July 30, 2021). We also supplemented our search results by checking the reference lists of all the relevant studies, including original articles and reviews. Eligible studies met the following inclusion criteria: (1) regarding the association between the PRNP M132L SNP and CWD; (2) case-control study; (3) genetic information on the M132L SNP of CWD-affected elk; (4) full text; and (5) published in English. The exclusion criteria were as follows: (1) case reports and (2) insufficient genotype data.
Meta-Analysis
The strength of the integrated association between the M132L SNP of the PRNP gene and susceptibility to CWD was estimated by meta-analysis. The pooled odds ratios with 95% confidence intervals were estimated using additive (M vs. L), recessive (MM vs. ML+LL), dominant (MM+ML vs. LL), and overdominant (ML vs. MM+LL) genetic models and homozygote (MM vs. LL) and heterozygote (MM vs. ML and ML vs. LL) comparisons. Heterogeneity was calculated based on the P-value and I2-value. Fixed effect models were selected to calculate the pooled odds ratios according to the value of the I2-test. Publication bias was calculated using Egger's weighted regression methods. Meta-analysis was performed using the meta package of the R program (https://www.r-project.org/).
In silico Analysis of the Impact of the M132L SNP on Elk PrP
PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/index.shtml) and PROVEAN (http://provean.jcvi.org/seq_submit.php) were used to predict the impact of the M132L SNP. PolyPhen-2 was predicted based on several characteristics of the target protein, including the sequence, structural and phylogenetic information characterizing the variation. PolyPhen-2 yields the following estimation results, including benign, possibly damaging, or probably damaging, based on scores ranging from 0.0 to 1.0. The PROVEAN scores were calculated based on clustering basic local alignment search tool (BLAST) hits according to the homologs collected from a database (the NCBI nr database). PROVEAN scores were classified as follows: below −2.5 indicates “deleterious,” and above−2.5 indicates “neutral.” AMYCO analysis was performed to estimate the effect of the M132L SNP on the aggregation propensity of elk PrP based on the combined score of the pWALTZ and PAPA algorithms. An AMYCO score <0.45 indicates low amyloid properties.
3D Structure and Electrostatic Potential Analyses
The nuclear magnetic resonance (NMR) structure of elk PrP was obtained from the RCSB Protein Data Bank (PDB ID: 1XYW). The electrostatic potential according to the M132L SNP was also analyzed using the Swiss-PdbViewer 4.1 program (https://spdbv.vital-it.ch/).
Statistical Analysis
Statistical analyses were performed using SAS version 9.4 (SAS Institute Inc., USA). The differences in genotype and allele frequencies of the PRNP gene between CWD-negative and CWD-positive animals were compared using the χ2-test and Fisher's exact test. The Hardy-Weinberg equilibrium (HWE) test was performed using Haploview version 4.2 (Broad Institute, Cambridge, MA, USA).
Results
No Association Between the M132L SNP of the Elk PRNP Gene and Susceptibility to CWD in the Republic of Korea
To investigate the association between the M132L SNP of the elk PRNP gene and susceptibility to CWD, we performed amplicon sequencing and genotyping in 253 elk, including 204 CWD-negative elk and 49 CWD-positive elk. Detailed information on genotype and allele frequencies is described in Table 1. Notably, the genotype and allele frequencies of the M132L SNP were not significantly different between CWD-negative and CWD-positive elk in the Republic of Korea (Table 1).
Table 1.
Comparison of genotype and allele frequencies of the M132L single nucleotide polymorphism (SNP) in the prion protein gene (PRNP) between chronic wasting disease (CWD)-affected elks and matched healthy elks.
| Authors | Year | Country | Total, n | Genotype frequencies, n | P-valuea | Allele frequencies, n | P-valueb | HWE | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MM | ML | LL | M | L | ||||||||
| O'Rourke et al. (13) | 1999 | USA | CTL | 344 | 230 | 102 | 12 | 0.0312 | 562 | 126 | 0.0084 | 0.8677 |
| CWD | 43 | 37 | 6 | 0 | 80 | 6 | 0.6229 | |||||
| Perucchini et al. (16) | 2008 | USA | CTL | 248 | 162 | 80 | 6 | 0.6927 | 404 | 92 | 0.4294 | 0.2872 |
| CWD | 94 | 66 | 26 | 2 | 158 | 30 | 0.7621 | |||||
| In this study | 2021 | South Korea | CTL | 204 | 144 | 49 | 11 | 0.9326 | 337 | 71 | 0.8000 | 0.0188 |
| CWD | 49 | 35 | 12 | 2 | 82 | 16 | 0.4681 | |||||
The number of genotype frequencies in each column indicates the number of genotypes. The number reported in each “Allele frequencies” column indicates the number of alleles. The number “Total, n” indicates the number of animals. Bold text indicates statistically significant results (P < 0.05). CTL, matched healthy elks; CWD, CWD-affected elk; HWE, Hardy-Weinberg equilibrium; P-valuea: Based on the comparison of genotype frequencies between healthy controls and CWD-affected animals; P-valueb: Based on the comparison of allele frequencies between healthy controls and CWD-affected animals.
Evaluation of the Association Between the M132L SNP of the Elk PRNP Gene and Susceptibility to CWD by Meta-Analysis
We searched 210 research articles following the search terms “PRNP,” “prion,” “CWD,” or “chronic wasting disease” combined with “SNP” or “polymorphism” or “susceptibility” (the last search update was performed on July 30, 2021) in PubMed. After excluding duplicate and irrelevant articles, a total of 2 eligible studies were extracted from the PubMed database based on our inclusion and exclusion criteria.
To identify an association between the M132L SNP of the elk PRNP gene and susceptibility to CWD in each previous study, we performed an association analysis in 2 previous studies performed in the USA (Table 1). Notably, the genotype and allele frequencies of the M132L SNP of the elk PRNP gene exhibited a strong association (P < 0.05) with susceptibility to CWD in a study by O'Rourke et al. (13) (Table 1). However, the genotype and allele frequencies of the M132L SNP of the elk PRNP gene did not show a strong association with vulnerability to CWD in a study by Perucchini et al. (16).
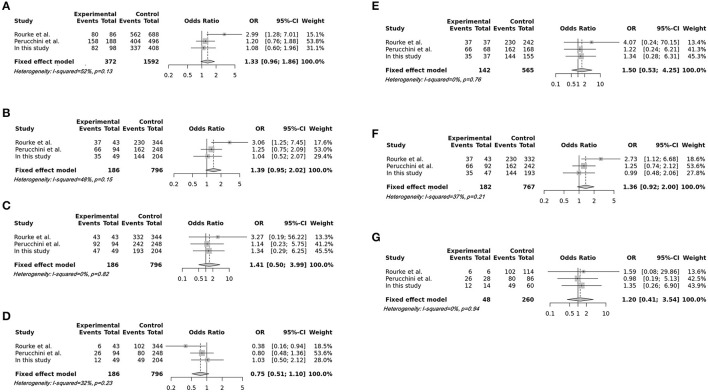
In total, 796 CWD-negative elk and 186 CWD-positive elk were included in the meta-analysis. The integrated association was evaluated by pooled odds ratios with 95% confident intervals using additive, recessive, dominant, and overdominant genetic models and homozygote and heterozygote comparisons. Heterogeneity was not observed (I2 <50%). Then, we used a fixed-effect model for the meta-analysis (Table 2). Notably, all meta-analyses using several genetic models and homozygote and heterozygote comparisons showed no association between the risk of CWD and the M132L SNP of the elk PRNP gene (Figure 1, Table 2). To examine potential publication bias, Egger's tests were performed, and publication bias was not observed in this meta-analysis (P > 0.1).
Table 2.
Meta-analysis of the association between the M132L SNP of the PRNP gene and susceptibility to CWD.
| Genetic model | Association test | Heterogeneity | Publication bias | ||||
|---|---|---|---|---|---|---|---|
| Odds ratio | 95% confidence interval | P-value | Model | P-value | I 2 | Egger's test P-value | |
| Additive model (M vs. L) | 1.3325 | [0.9570; 1.8553] | 0.6246 | Fixed | 0.13 | 0.52 | 0.3852 |
| Recessive model (MM vs. ML+LL) | 1.3873 | [0.9546; 2.0161] | 0.6029 | Fixed | 0.15 | 0.48 | 0.4978 |
| Dominant model (MM+ML vs. LL) | 1.4120 | [0.4998; 3.9891] | 1 | Fixed | 0.82 | 0.00 | 0.1956 |
| Overdominant model (ML vs. MM+LL) | 0.7506 | [0.5113; 1.1019] | 1 | Fixed | 0.23 | 0.32 | 0.6099 |
| MM vs. LL | 1.4962 | [0.5266; 4.2505] | 1 | Fixed | 0.76 | 0.00 | 0.1181 |
| MM vs. ML | 1.3585 | [0.9238; 1.9978] | 0.8363 | Fixed | 0.21 | 0.37 | 0.5972 |
| ML vs. LL | 1.2004 | [0.4067; 3.5433] | 1 | Fixed | 0.94 | 0.00 | 0.6146 |
Figure 1.
Forest plots of the association between the M132L single nucleotide polymorphism (SNP) of the elk prion protein gene (PRNP) and susceptibility to chronic wasting disease (CWD). (A) Forest plots of the association between the M132L SNP of the elk PRNP gene and susceptibility to CWD in the additive model (M vs. L). (B) Forest plots of the association between the M132L SNP of the elk PRNP gene and susceptibility to CWD in the recessive model (MM vs. ML+LL). (C) Forest plots of the association between the M132L SNP of the elk PRNP gene and susceptibility to CWD in the dominant model (MM+ML vs. LL). (D) Forest plots of the association between the M132L SNP of the elk PRNP gene and susceptibility to CWD in the overdominant model (ML vs. MM+LL). (E) Forest plots of the association between the M132L SNP of the elk PRNP gene and susceptibility to CWD in the homozygote comparison (MM vs. LL). (F) Forest plots of the association between the M132L SNP of the elk PRNP gene and susceptibility to CWD in the heterozygote comparison (MM vs. ML). (G) Forest plots of the association between the M132L SNP of the elk PRNP gene and susceptibility to CWD in the heterozygote comparison (ML vs. LL).
In silico Evaluation of the Effect of the M132L SNP on Elk PrP
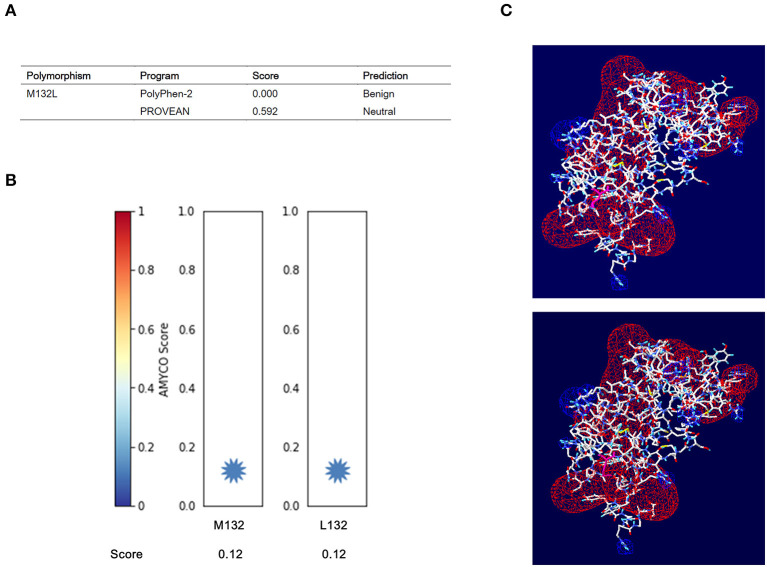
We estimated the impact of the M132L SNP of the PRNP gene on elk PrP using PolyPhen-2 and PROVEAN. Detailed scores predicted by the two programs are described in Figure 2A. In brief, the M132L SNP was estimated to be “benign” and “neutral” by PolyPhen-2 and PROVEAN, respectively. In addition, elk PrP with the M132 allele showed an amyloid propensity similar to that of elk PrP with the 132L allele (Figure 2B). We evaluated the electrostatic potential and 3D structure analysis according to the M132L SNP. Notably, we did not find significant changes in the electrostatic potential and 3D structure of elk PrP according to the M132L SNP (Figure 2C).
Figure 2.
In silico evaluation of the effect of the M132L SNP on elk prion protein (PrP). (A) Prediction of the functional effect of the M132L SNP on elk PrP by PolyPhen-2 and PROVEAN. (B) Prediction of the aggregation propensity of elk PrP according to alleles at codon 132 using AMYCO. (C) Electrostatic potential and 3D structure analysis of elk PrP according to the M132L SNP. Upper panel: electrostatic potential and 3D structure of elk PrP with the M132 allele. Lower panel: electrostatic potential and 3D structure of elk PrP with the L132 allele. Positive potentials are drawn in blue. Negative potentials are noted in red.
Discussion
In the present study, the M132L SNP of the PRNP gene was not associated with susceptibility to CWD in Korean elk (Table 1). Previous studies have reported that the genotype and allele frequencies of genetic polymorphisms of the PRNP gene in the Cervidae family can vary according to geographic areas of habitat (13, 16, 22). In addition, all Korean elk were raised on farms. Thus, the indigenous genetic features of Korean elk may be caused by geographically isolated habitats and breeding modes. In the present study, information on the age and sex of Korean elk was not available. Further analysis stratified by age and sex is highly desirable in the future.
We also integrated and evaluated the association of the M132L SNP of the PRNP gene with susceptibility to CWD in quantitatively synthesized elk populations by meta-analysis. Notably, we did not observe an association of the M132L SNP of this gene with susceptibility to CWD in any genetic models of meta-analyses (Table 2, Figure 1). Although previous experimental infections in elk and elk PrP transgenic mice showed the protective effect of the L132 allele, these results did not correspond with that of the present meta-analysis (14). In human prion diseases, the M129V SNP of the human PRNP gene plays a pivotal role in susceptibility to sporadic CJD (23, 24). However, the M129V SNP is not a solitary factor in the vulnerability of sporadic CJD. In the Asian population, the E219K SNP of the PRNP gene confers resistance to CJD (25–28). Although the overrepresented M129 allele of the PRNP gene was observed in Asian populations (>90%), including Korea and Japan, the incidence of sporadic CJD is similar and/or low compared to European populations, which have a relatively low frequency of the M129 allele (<70%). In addition, the G127V SNP of the PRNP gene was reported in Kuru-unaffected Papua New Guinean, and a follow-up study using transgenic mice carrying human PrP with the 127V allele confirmed that the V127 allele conferred potent resistance to several human prion diseases (29). Thus, given that the full amino acid sequences of the elk PrP of inoculation agents and recipient animals were not available in previous studies, further detailed studies applying other polymorphisms of elk PrP are highly desirable. With the exception of the M132L SNP of the elk PRNP gene, an association test for the entire amino acid sequence of elk PrP has not been performed. This association study is highly desirable in the future.
We also estimated the effect of the M132L SNP on elk PrP by in silico programs and did not find significant changes in structure, aggregation propensity or electrostatic potential according to the M132L SNP (Figure 2). In previous studies, human PrP with the 129M allele also showed similar characteristics to human PrP with the 129V allele. The protective effect of the 129V allele is thought to be associated with the inhibition of homotypic protein–protein interactions (28). In addition, a recent study using an in vitro conversion assay by RT-QuIC reported that conversion efficiency primarily depends on the correspondence of genotype at PRNP codon 132 of agent and recipient recombinant protein (15). However, our in silico analysis and previous in vitro and in vivo analyses did not reflect haplotypes of the elk PRNP gene, and further investigation of genetic polymorphisms and haplotypes of the elk PRNP gene is needed in the future.
Conclusion
We investigated the M132L SNP of the elk PRNP gene in Korean elk and performed association analysis between CWD-positive and CWD-negative elk for the first time. We did not observe a significant association between the M132L SNP of the PRNP gene and susceptibility to CWD in Korean elk. The meta-analysis also did not identify a strong association between the M132L SNP and susceptibility to CWD in quantitatively synthesized elk populations. Furthermore, we did not find significant changes in structure, amyloid propensity or electrostatic potential according to the M132L SNP in elk PrP using in silico analyses. To the best of our knowledge, this was the first report of an association analysis and meta-analysis in Korean elk and quantitatively synthesized elk populations, respectively.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank (Accession no. FJ590751.1).
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of Jeonbuk National University.
Author Contributions
I-SR, Y-CK, H-JS, and B-HJ conceived, designed the experiment, and wrote the paper. I-SR, Y-CK, S-YW, K-JP, H-CP, and J-YH performed the experiments. Y-CK, S-YW, H-EK, and B-HJ analyzed the data. All authors have read and approved the final manuscript.
Funding
This research was supported by the Basic Science Research Program and National Research Foundation (NRF) of Korea funded by the Ministry of Education (2017R1A6A1A03015876, 2021R1A6A3A010864), National Research Foundation of Korea (NRF) by the Korea government (2021R1A2C1013213), Ministry for Agriculture, Food and Rural Affairs (B-1543085-21-22-01), BK21 Plus Program in Department of Bioactive Material Sciences to Y-CK, and NRF (National Research Foundation of Korea) Grant funded by the Korean Government (NRF-2019-Fostering Core Leaders of the Future Basic Science Program/Global Ph.D Fellowship Program).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1.Orge L, Lima C, Machado C, Tavares P, Mendonca P, Carvalho P, et al. Neuropathology of animal prion diseases. Biomolecules. (2021) 11:466. 10.3390/biom11030466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim YC, Won SY, Jeong BH, Altered Altered expression of glymphatic system-related proteins in prion diseases: implications for the role of the glymphatic system in prion diseases. Cell Mol Immunol. (2021) 18:2281–3. 10.1038/s41423-021-00747-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim YC, Lee J, Lee DW, Jeong BH. Large-scale lipidomic profiling identifies novel potential biomarkers for prion diseases and highlights lipid raft-related pathways. Vet Res. (2021) 52:105. 10.1186/s13567-021-00975-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prusiner SB. The prion diseases. Brain Pathol. (1998) 8:499–513. 10.1111/j.1750-3639.1998.tb00171.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prusiner SB. Prions. Proc Natl Acad Sci USA. (1998) 95:13363–83. 10.1073/pnas.95.23.13363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim YC, Won SY, Jeong MJ, Jeong BH. Absence of proteinase K-resistant PrP in Korean Holstein cattle carrying potential bovine spongiform encephalopathy-related E211K somatic mutation. Transbound Emerg Dis. (2021) (in press). 10.1111/tbed.14053 [DOI] [PubMed] [Google Scholar]
- 7.Kim YC, Park KJ, Hwang JY, Park HC, Kang HE, Sohn HJ, et al. In-depth examination of PrP(Sc) in Holstein cattle carrying the E211K somatic mutation of the bovine prion protein gene (PRNP). Transbound Emerg Dis. (2021) (in press). 10.1111/tbed.14309 [DOI] [PubMed] [Google Scholar]
- 8.Arifin MI, Hannaoui S, Chang SC, Thapa S, Schatzl HM, Gilch S. Cervid prion protein polymorphisms: role in chronic wasting disease pathogenesis. Int J Mol Sci. (2021) 22:2271. 10.3390/ijms22052271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson SJ, Samuel MD, O'Rourke KI, Johnson CJ, The The role of genetics in chronic wasting disease of North American cervids. Prion. (2012) 6:153–62. 10.4161/pri.19640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guere ME, Vage J, Tharaldsen H, Kvie KS, Bardsen BJ, Benestad SL, et al. Chronic wasting disease in Norway-A survey of prion protein gene variation among cervids. Transbound Emerg Dis. (2021) (in press). 10.1111/tbed.14258 [DOI] [PubMed] [Google Scholar]
- 11.Roh IS, Kim YC, Kim HJ, Won SY, Jeong MJ, Hwang JY, et al. Polymorphisms of the prion-related protein gene are strongly associated with cervids' susceptibility to chronic wasting disease. Vet Rec. (2021) e940 (in press). 10.1002/vetr.940 [DOI] [PubMed] [Google Scholar]
- 12.Rivera NA, Brandt AL, Novakofski JE, Mateus-Pinilla NE. Mateus-pinilla, chronic wasting disease in cervids: prevalence, impact and management strategies. Vet Med. (2019) 10:123–39. 10.2147/VMRR.S197404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Rourke KI, Besser TE, Miller MW, Cline TF, Spraker TR, Jenny AL, et al. PrP genotypes of captive and free-ranging Rocky Mountain elk (Cervus elaphus nelsoni) with chronic wasting disease. J General Virol. (1999) 80:2765–9. 10.1099/0022-1317-80-10-2765 [DOI] [PubMed] [Google Scholar]
- 14.Green KM, Browning SR, Seward TS, Jewell JE, Ross DL, Green MA, et al. The elk PRNP codon 132 polymorphism controls cervid and scrapie prion propagation. J Gen Virol. (2008) 89:598–608. 10.1099/vir.0.83168-0 [DOI] [PubMed] [Google Scholar]
- 15.Moore SJ, Vrentas CE, Hwang S, West Greenlee MH, Nicholson EM, Greenlee JJ. Pathologic and biochemical characterization of PrP(Sc) from elk with PRNP polymorphisms at codon 132 after experimental infection with the chronic wasting disease agent. BMC Vet Res. (2018) 14:80. 10.1186/s12917-018-1400-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perucchini M, Griffin K, Miller MW, Goldmann W. PrP genotypes of free-ranging wapiti (Cervus elaphus nelsoni) with chronic wasting disease. J General Virol. (2008) 89:1324–8. 10.1099/vir.0.83424-0 [DOI] [PubMed] [Google Scholar]
- 17.Sohn HJ, Kim JH, Choi KS, Nah JJ, Joo YS, Jean YH, et al. A case of chronic wasting disease in an elk imported to Korea from Canada. J Vet Med Sci. (2002) 64:855–8. 10.1292/jvms.64.855 [DOI] [PubMed] [Google Scholar]
- 18.Otero A, Velasquez CD, Aiken J, McKenzie D. Chronic wasting disease: a cervid prion infection looming to spillover. Vet Res. (2021) 52:115. 10.1186/s13567-021-00986-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adzhubei I, Jordan DM, Sunyaev SR. Sunyaev, predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet Chapter. (2013) 7:Unit 7 20. 10.1002/0471142905.hg0720s76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. (2015) 31:2745–7. 10.1093/bioinformatics/btv195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. (2009) 30(Suppl. 1):S162–73. 10.1002/elps.200900140 [DOI] [PubMed] [Google Scholar]
- 22.White SN, Spraker TR, Reynolds JO, O'Rourke KI. Association analysis of PRNP gene region with chronic wasting disease in Rocky Mountain elk. BMC Res Notes. (2010) 3:314. 10.1186/1756-0500-3-314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovacs GG, Puopolo M, Ladogana A, Pocchiari M, Budka H, van Duijn C, et al. Genetic prion disease: the EUROCJD experience. Hum Genet. (2005) 118:166–74. 10.1007/s00439-005-0020-1 [DOI] [PubMed] [Google Scholar]
- 24.Lloyd S, Mead S, Collinge J. Collinge, Genetics of prion disease. Top Curr Chem. (2011) 305:1–22. 10.1007/128_2011_157 [DOI] [PubMed] [Google Scholar]
- 25.Jeong BH, Lee KH, Kim NH, Jin JK, Kim JI, Carp RI, et al. Association of sporadic Creutzfeldt-Jakob disease with homozygous genotypes at PRNP codons 129 and 219 in the Korean population. Neurogenetics. (2005) 6:229–32. 10.1007/s10048-005-0016-y [DOI] [PubMed] [Google Scholar]
- 26.Jeong BH, Kim YS. Genetic studies in human prion diseases. J Korean Med Sci. (2014) 29:623–32. 10.3346/jkms.2014.29.5.623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeong BH, Nam JH, Lee YJ, Lee KH, Jang MK, Carp RI, et al. Polymorphisms of the prion protein gene (PRNP) in a Korean population. J Hum Genet. (2004) 49:319–24. 10.1007/s10038-004-0150-7 [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi A, Teruya K, Matsuura Y, Shirai T, Nakamura Y, Yamada M, et al. The influence of PRNP polymorphisms on human prion disease susceptibility: an update. Acta Neuropathol. (2015) 130:159–70. 10.1007/s00401-015-1447-7 [DOI] [PubMed] [Google Scholar]
- 29.Asante EA, Smidak M, Grimshaw A, Houghton R, Tomlinson A, Jeelani A, et al. A naturally occurring variant of the human prion protein completely prevents prion disease. Nature. (2015) 522:478–81. 10.1038/nature14510 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank (Accession no. FJ590751.1).