Abstract
Hepatitis C virus (HCV) infections are emerging as one of the foremost challenges in healthcare owing to its chronicity and the virus’s quasispecies nature. Worldwide, over 170 million people are chronically infected with HCV, with an annual mortality of over 500,000 people across the world. The emerging pathophysiological evidence links HCV infections to a risk of developing liver diseases such as cirrhosis and hepatocellular carcinoma. Despite the great strides that have been made towards understanding the pathophysiology of disease progression, the tailored treatments of HCV infection remain to be established. The present review provides an update of the literature pertaining to evolving therapeutic approaches and prophylactic measures for the effective management of HCV infections. An extensive discussion of established and experimental immune prophylactic measures also sheds light on current developments in the design of vaccination strategies against HCV infection. We have also attempted to address the application of nanotechnology in formulating effective therapeutic interventions against HCV. Pointing out the limitations of the existing diagnostic methods and therapeutic approaches against HCV might inspire the design and development of novel, efficient, reliable, and cost-effective diagnostic technologies as well as novel therapeutic and immune prophylactic interventions for the effective management of HCV.
Introduction
Viral hepatitis can be caused by various viruses, including hepatitis A virus (HAV), hepatitis B virus (HBV), hepatitis C virus (HCV), hepatitis D virus (HDV), hepatitis E virus (HEV), and hepatitis G virus (HGV) [1]. Since its first detection in 1989 [2], it has become clear that HCV is a major cause of chronic hepatitis. HCV belongs to the genus Hepacivirus in the family Flaviviridae [3]. More than two decades since its discovery, HCV continues to be a major cause of concern and a huge burden on national public health systems worldwide. A recent report estimates that over 71 million people worldwide are chronically infected with HCV [4], and approximately 170 million people are estimated to be at risk of HCV infection. This virus has the highest prevalence in Africa and Asia, where it is four times more prevalent than HIV, and hence, HCV has the potential to cause the next pandemic, as the mortality has reached over 500,000 per annum [5]. The incubation period of HCV ranges from weeks to several months, with an average of around 7 weeks until symptoms occur [6]. HCV infection is often asymptomatic, making it very difficult to detect in its early stage. This is the main reason why patients do not receive early treatment, and HCV is often referred to as a “silent killer” [7]. This disease can have various outcomes, ranging from mild (minimal inflammation of the liver) to severe, and can lead to scar tissue formation. Chronic infection eventually causes cirrhosis, leading to hepatocellular carcinoma and, ultimately, death [8]. A report from the Centers for Disease Control and Prevention (CDC) states that HCV-related cirrhosis is observed in 5-20% of patients experiencing chronic HCV infections over a prolonged period of 20-30 years. Moreover, advanced cirrhosis is associated with end-stage liver disease, liver failure because of portal hypertension, hepatic encephalopathy, hepatorenal syndrome, and hepatocellular carcinoma [9]. As the most commonly observed type of liver cancer, research data from the last three decades suggest that hepatocellular carcinoma occurs in approximately 1-3% of HCV patients [10]. Of note, effective anti-HCV treatments can help to reduce the overall mortality rates in HCV patients with compensated cirrhosis [11].
HCV has a single-stranded positive-sense RNA genome of around 9.5 kb that encodes a single polyprotein precursor of 3011 to 3033 amino acids [12, 13]. Upon protease-mediated cleavage, the polyprotein precursor yields two distinct types of proteins that can be categorized as structural (Core, E1, E2, and p7) and non-structural (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins [14]. Owing to the importance of these proteins in the pathogenesis of HCV, we have reviewed recent advancements in the development of drugs targeting the structural and non-structural proteins of HCV. The evolving genetic heterogeneity of HCV serotypes is one of the important limiting factors in the effective management of HCV patients, and also in the development of novel pan-genotypic treatment modalities. The high genetic variability exhibited by HCV has resulted in the generation of seven to eight major genotypes. These genotypes appear to be quite diverse at the genetic level. For example, the identified genotypes show 30% difference in their genome sequences. Within these major genotypes, 84-86 subtypes have been identified that exhibit differences ranging from 15 to 25% at the nucleotide level [15, 16]. In the mainstream of HCV research, there are still questions waiting to be answered. For instance, it is not clear whether HCV genotypes originated from a single cross-species transmission and further diversified within the human population or whether it originated from separate zoonotic sources in different geographical regions [17].
In this review, we provide an update on advancements in the field of therapeutics and diagnosis of HCV. Previous reviews have mainly described diagnosis, therapy, or management of HCV infections [18–21] and have focused on individual genotypes [22, 23]. In contrast, in the present review, we provide an extensive literature survey on current diagnosis and therapeutic modalities for various genotypes of HCV and their associated shortfalls. Taking into consideration the current state of the art, the current review offers updated information on the development of novel drug strategies and factors limiting the effective management of HCV infections.
Attempts to develop new HCV therapeutic modalities: successes and failures
With the advent of the COVID-19 pandemic and the acknowledgment of the HCV research of Michael Houghton, Harvey Alter, and Charles Rice by the Nobel Committee in 2020, the search for novel and effective antiviral agents has accelerated, and some of these agents might also find use in the management of HCV. In HCV drug development programs, a sizable amount of human resources and capital is being invested in developing antiviral drugs. Although there is still no completely effective antiviral therapy tailored specifically for treatment of HCV infection, dozens of small-molecule drugs for stand-alone use or combination therapy have been approved and recommended by the FDA (Table 1), and others are in different phases of development, including clinical trials. These inhibitors mostly target viral nonstructural proteins such as NS3/4A, NS5A, and NS5B, or the internal ribosome entry site (IRES) [24]. Some of the most recently identified HCV antiviral agents are shown in Figure 1.
Table 1.
Summary of current anti-HCV therapeutic regimens
| Name of therapy | Year of FDA approval | Therapeutic target | Side effects and drawbacks associated with the therapy | Commercial source | Target Genotype | References |
|---|---|---|---|---|---|---|
| Victrelis (boceprevir + PegIFNα + ribavirin) | May 2011 | NS3/4A | Higher toxicity, adverse interaction with other drugs, triggering of decompensation, lack of safety at advanced stage of disease, high cost | Merck Inc., USA | Genotype 1 | [46] |
| Incivek (telaprevir + PegIFNα + ribavirin) | May 2011 | NS3 | Toxic epidermal necrolysis, skin reactions, adverse reaction with eosinophilia | Vertex Pharmaceuticals, USA | Genotype 1 | [45, 47] |
| Sovaldi (sofosbuvir+ PegIFNα + ribavirin) | December 2013 | NS5B | Anemia, nausea, fatigue, insomnia, and headache | Gilead Sciences Inc, USA | Genotype 1/4 | [45, 48] |
| Olysio (simeprevir + Sofosbuvir +PegIFNα/ribavirin) | November 2013 | NS3/4A | Photosensitivity reactions, nausea, rash, headache, pruritus, and fatigue | Medivir, Sweden, and Janssen Pharmaceuticals, Belgium | Genotype 1 | [4, 9, 45, 48] |
| Harvoni (ledipasvir + sofosbuvir) | October 2014 | NS5A and NS5B | Insomnia, nausea, headache, and fatigue especially in Afro-American populations | Gilead Sciences Inc, USA | Genotype 1, 4, 5/6 | [7, 45, 49, 50] |
| Viekira Pak (dasabuvir + paritaprevir + ritonavir + ombitasvir) | December 2014 | NS3/4A, NS5B, and NS5A | Asthenia, nausea, pruritus, skin rash, insomnia, and fatigue | AbbVie, USA | Genotype 1 | [45, 51] |
| Technivie (Ombitasvir + paritaprevir + ritonavir) | July 2015 | NS3/4A, NS5A | Fatigue, nausea, insomnia, and asthenia | AbbVie, USA | Genotype 4 | [45, 52] |
| Daklinza and Sovaldi (daclatasvir + sofosbuvir) | July 2015 | NS5A and NS5B | Fatigue and headache | Bristol Myers Squibb, USA, and Gilead Sciences Inc, USA, respectively | Genotype 1/3 | [45, 53] |
| Zepatier (grazoprevir + elbasvir) | January 2016 | NS3/4A and NS5A | Fatigue, headache, and nausea | Merck Inc., USA | Genotype 1 | [45, 54–56] |
| Epclusa (velpatasvir + sofosbuvir) | June 2016 | NS5A and NS5B | Diarrhea, insomnia, headache, fatigue, anemia, and nausea | Gilead Sciences Inc, USA | Pan-genotypic (1-6) | [45, 49] |
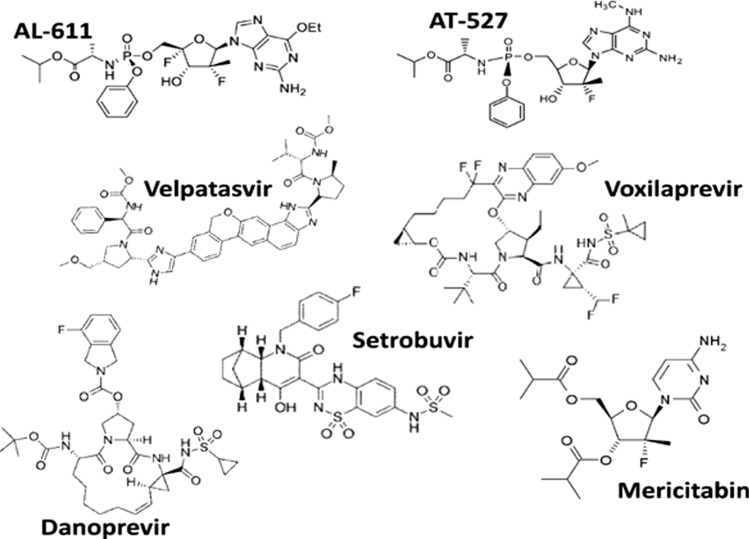
Fig. 1.
Chemical structures of newly identified HCV agents
Setrobuvir is a potent small-molecule, non-nucleoside inhibitor of the HCV genotype 1 NS5B RNA polymerase. This drug reached clinical trials at Roche Pharmaceuticals [25, 26]. However, it was removed from the development pipeline and from phase II clinical trials in 2015 due to adverse effects. Patients receiving setrobuvir monotherapy and those receiving setrobuvir in combination with other direct-acting antivirals developed rashes. Danoprevir is a macrocyclic, peptidomimetic, small-molecule compound that was developed at Roche Pharmaceuticals. This drug competitively inhibits the HCV NS3/4A protease [27–29]. After successfully completing phase III clinical trials, this drug was launched in the market in China in August 2018. Mericitabine is an oral, small-molecule nucleoside analogue that is phosphorylated intracellularly, resulting in two distinct active triphosphate forms that target the NS5B RNA-dependent RNA polymerase of HCV, affecting the viral replication [29–31]. However, this drug was discontinued in phase II clinical trials in 2016 because of an inadequate number of HCV patients participating in the trials [32]. Iridoids (such as the aglycones of shanzhiside methyl ester loganin and verbenalin) and amphipathic DNA polymers are novel antiviral molecules that were designed to inhibit entry of HCV into cells [33]. The drug telaprevir (VX-950), which targets the NS3-4A protease, showed some success in clinical trials [34]. It is being marketed worldwide by Janssen R&D Ireland. Different analogues of ribavirin, namely 4-iodo-1-beta-ribofuranosylpyrazole-3-carboxamide, 4-propynyl-1-beta-ribofuranosylpyrazole-3-carboxamide, and 4-phenylethynyl-1-beta-ribofuranosylpyrazole-3-carboxamide, are now being considered as alternative candidate drugs [35].
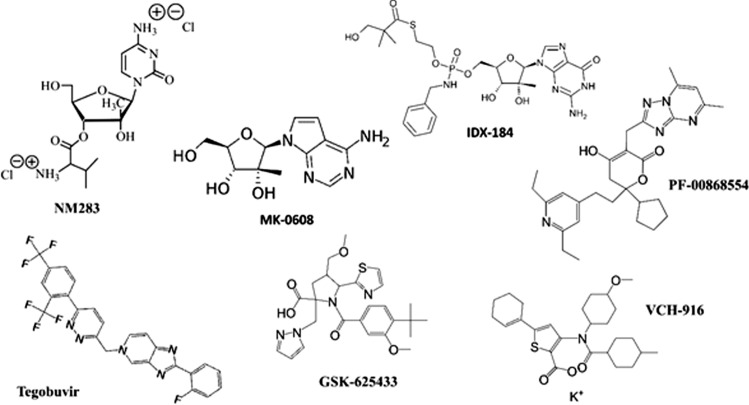
BMS-824393 is a small molecule that targets the NS5A protein [36–38], and it was discontinued in phase II clinical trials due to severe adverse effects. Zinc meso-porphyrin holds immense potential as a novel drug to treat HCV infection [39]. In the last decade, the NS5B protein has been shown to be a good target for developing candidate drugs against HCV, and various NS5B inhibitors, such as NM-283 [40], IDX-184, and MK-0608 have been described. NM-283, developed by Idenix Pharmaceuticals, was found to be associated with severe gastrointestinal toxicity and hence discontinued from the phase II clinical trials. Despite initially promising results, the development of IDX-184 has been kept on clinical hold since February 2013, and no further update is available for this agent. MK-0608, despite encouraging results in animal model studies, was discontinued in 2011 due to unsuccessful clinical trials. Pfizer's PF-00868554 was discontinued from phase II clinical trials in February 2013 due to adverse effects, including headache, fatigue, insomnia, and nausea. GS-9190 (tegobuvir), produced by Gilead Corporation, is an NS5B protease inhibitor that was discontinued due to various adverse effects in phase II clinical trials, including flu-like syndrome, pyrexia, headache, and myalgia [41]. Several other molecules, such as VCH-916 (Viro Chem Pharma) [42], were discontinued from phase II clinical trials due to gastrointestinal disorders, throat irritation, and nausea. GSK-625433 (GlaxoSmithKline) [43] reached phase I but was later discontinued (Fig. 2). ABT-333/dasabuvir (developed by Abbott Laboratories) is currently being successfully marketed in Israel, the UK, Switzerland, and the USA, and, after completing phase III trials, was approved for monotherapy as well as combination therapy with ribavirin [44].
Fig. 2.
Chemical structures of newly identified HCV agents that are in early clinical trials
Evolving approaches in HCV therapeutics
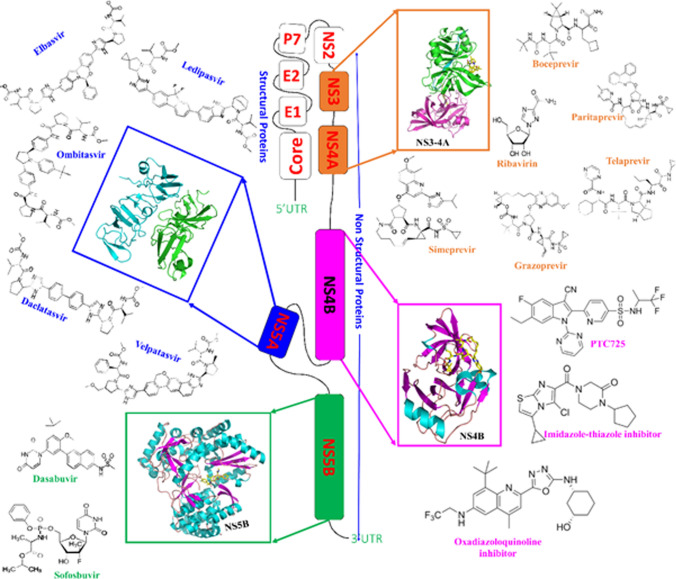
The scientific community and researchers at various pharmaceutical companies are continuously working on developing novel and effective therapeutic interventions against HCV. In the past five years, novel lead compounds with improved efficacy and safety have been discovered [45], and a series of anti-HCV candidate drugs have been proposed. The following section attempts to summarize these new candidates and their respective drug targets. NS3/4A has been found to be an attractive target for series of therapeutic agents, including danoprevir, voxilaprevir, vedroprevir, MK-8831, faldaprevir, ABT493, BMS-605339, and BMS-890068. NS4B has also proven to be a good drug target, as various drugs, including PTC725, 2-oxadiazoloquinoline derivative, imidazo[2,1-b]thiazole derivative, and piperazinone have been proposed based on modulation of NS4B activity. NS5A also appears to be a promising drug target, and various drugs, including ABT-530, ruzasvir, ravidasvir, GSK2336805, EDP-239, samatasvir, biphenylimidazole analogues, and disulfiram target this protein. NS5B has been described as an effective therapeutic target, and various chemical agents, including beclabuvir, GS-9669, thiophene carboxylate allosteric inhibitor, mericitabine, deleobuvir, DAPN-PD1, TMC647055, filibuvir/VX-222, GS-6620, tegobuvir, JNJ-54257099, 6-aminoquinolone derivative, and pyrazolobenzothiazines have been proposed as inhibitors of NS5B. In addition, the structural proteins E1/E2 and p7 are being explored as therapeutic targets for drugs such as benzimidazole derivative, cynaropicrin/grosheimol, saikosaponin b2, chlorcyclizine derivative, flunarizine, monoclonal antibodies and adamantane/rimantadine. In addition to direct-acting antivirals, immunostimulators such as alisporivir, bis-amide derivative, NIM258, phenylepyrrolidine derivative, isothiazolo[5,4-b] pyridine, ITX-5061, MA026, and soraphen A are also under consideration for repurposing as anti-HCV agents. These drugs have been reviewed extensively by Li and De Clercq [45]. Table 1 summarizes the therapies that are now commonly offered to patients with HCV infection, and Fig. 3 provides a brief overview of the location of the genes for these drug targets in the HCV genome. Table 2 summarizes the profile of clinical trials of promising anti-HCV candidate drugs that are in the early stages of development and has enormous potential to act as effective antivirals.
Fig. 3.
Map of the HCV genome, showing the positions of genes encoding viral proteins that are targets for antiviral therapy. The drugs are color coded according to their target: NS5B (green, PDB 4KAI), NS4B (magenta, PDB 6NZV), NS4A (blue, PDB 4CL1), NS3-4A (orange, PDB 3LOX).
Table 2.
Summary of clinical trials of promising anti-HCV drugs
| Name of the drug | Sponsor/company | Therapeutic target | Highest clinical trial stage reached | Outcome or special comments | Clinical trial | Reference/clinical trials ID |
|---|---|---|---|---|---|---|
| Velpatasvir | Radboud University, University of Pittsburgh, USA | NS3/4A protease | Phase I | Combination therapy with sofosbuvir/velpatasvir/Voxilaprevir was generally well tolerated, with mild or moderate side effects. Common adverse effects included nausea, fatigue, diarrhea, and headache | Phase 1 B multiple-dose, double-blind, randomized trial involving 103 participants. This drug demonstrated improved and pan-genotypic activity, even against preexisting resistant variants. | NCT03513393, NCT04382404, [57–60] |
| Holybuvir (SH229) | Nanjing Sanhome Pharmaceutical, China | NS5B | Phase II/III | In phase II/III data from the combination therapy, this drug was observed to be safe, with mild to moderate adverse effects | In a phase II/III randomized clinical trial, 440 participants with chronic HCV were treated with the drug for 12 weeks and showed a favourable SVR response. | NCT03748745, NCT03588923 [61, 62] |
| CDI-31244 | Cocrystal Pharma, Inc. USA | NS5B polymerase | Phase II | Combination therapy with CDI-31244 and sofosbuvir/velpatasvir resulted in a sustained virologic response after 12 and 24 weeks. Side effects included headache in 42% of subjects | 12 participants were involved in this open-label single-center trial, and 67% achieved post-treatment SVR at both 12 and 42 weeks. | NCT02760758 [63, 64] |
| MK-1075 | Merck Sharp and Dohme, USA | NS3/4a protease | Phase I | A phase I clinical study in 2015 showed that more than 92% of patient from multiple cohorts achieved SVR12 when given this drug | 12 subjects participated in an open-label, randomized clinical trial, and a 3 log10 reduction in HCV RNA was observed in 70% of patients | NCT03588923, NCT02461563, NCT02392494 [59, 65] |
| Voxilaprevir (GS 9857) | Gilead Sciences, USA | NS3/4A protease | Phase II | Triple combination therapy with voxilaprevir, sofosbuvir, and velpatasvir showed high efficacy and was well tolerated, with no serious adverse effects. The adverse effects included mainly headache, fatigue, and nausea | In a clinical trial including 273 participants, 88% achieved SVR12 after treatment for 6 weeks | NCT02397707, NCT02402452 [29, 59, 66] |
| AT-527 | Atea Pharmaceuticals, Inc., USA | NS5B polymerase | Phase II | Well tolerated with a pan-genotypic antiviral effect | Results from two triple randomized, sequential assignment clinical trials involving 88 and 10 participants are yet to be disclosed | NCT03219957, NCT04309734 [59, 64, 67–69] |
| HEC74647PA | Sunshine Lake Pharma, China | Not known | Phase I | Phase I clinical trials were completed in China. No further clinical data are available | Two separate clinical trials with 105 and 28 subjects were initiated, and the data are expected to be made available soon. | NCT04201275, NCT03673696 |
| AL-611 | AliosBioPharma, USA | NS5B polymerase | Phase I | Development was terminated at phase I. No further data are available | 24 subjects participated in this placebo-controlled, double-blind, randomized clinical trial. | NCT03253471 [59, 70] |
| MB-110 | Microbio Co Ltd., Taiwan | NS5A protein | Phase I | Phase I clinical trials are being conducted in Taiwan and the USA | A randomized double-blind clinical trial was initiated with 40 participants. The results of this study are awaited. | NCT02617615 [59] |
| EDP-494 | Enanta Pharmaceuticals, USA | Cyclophilin | Phase I | The drug targets human cyclophilin, which plays an important role in the HCV replicative cycle. Phase I clinical trials were terminated due to gastrointestinal complications, venipuncture bruises, and headache in subjects | Out of 82 subjects that participated in this trial, it was reported that 6.3% showed eye disorders, and 40.6% suffered from gastrointestinal disorders. | NCT02652377 [59, 71] |
| Ravidasvir/ ASC16 Tablets | XTL Biopharmaceuticals, Israel | NS5A protein | Phase II/III | No serious treatment-related adverse effects have been noted so far | This drug demonstrated a 99% rate of sustained virological response after 12 weeks of treatment | NCT03288636, NCT03430830 [59, 72] |
| AT-777 | Atea Pharmaceuticals, USA | NS5A protein | Phase II | Phase II clinical trials are being conducted in Belgium with no clinical data available yet | This triple-blind randomized trial is being conducted on 26 participants, and results are expected to be available in September 2022 | NCT04309734 [59] |
| AL-704 | Janssen Pharmaceutical, Belgium | NS5B polymerase | Phase I | Discontinued in phase I clinical trials | A clinical trial with 42 participants was initiated in 2015 and terminated in 2017 | NCT02510248 [59] |
Current immunoprophylactic approaches for the management of HCV infections
The growing incidence of HCV infections and related public-health complications prompted the World Health Organization to establish a program to eliminate HCV by 2030. To achieve this goal, there is a need for an effective, safe, and affordable vaccine against HCV.
Since the discovery of HCV more than 30 years ago, there have been significant advances in understanding the molecular aspects of HCV pathophysiology, the molecular immunological mechanisms involved in spontaneous viral clearance, and the rational design of vaccines. There is a need to develop effective prophylactic vaccines against HCV, as current HCV treatment approaches are not effective in preventing recurrent infections and are not affordable in economically challenged parts of the world restricting their clinical use to developed countries [73, 74]. In general, antiviral therapies require long-term treatment, are expensive and toxic, and are effective only in about 50% of patients against the majority of the HCV genotypes [75]. There have been a number of reports indicating progress toward the development of therapeutic vaccines for the management of HCV infections. For example, IC41, a synthetic peptide vaccine containing HCV T-cell epitopes has been shown recently to be safe and is currently undergoing a phase I clinical trial. IC41 can induce HCV-specific INF-gamma-secreting CD4+ and CD8+ T cells in healthy individuals. It has been shown to induce HCV-specific Th1/Tc1 responses in a subset of difficult-to-treat HCV non-responder patients [76]. Another interesting candidate for a peptide vaccine is the HCV-specific HLA-A2-restricted epitope NS3-1073 [77]. Table 3 summarizes the promising candidate vaccines against HCV that are in clinical trials, and Table 4 lists the important CD8 and CD4 epitopes of HCV recognized by T cells that have been identified in the past five years [78]. The epitopes identified before this period have been reviewed by Yu and Chiang [79]. These epitopes have potential for use in peptide-based subunit vaccine formulations.
Table 3.
Profile of promising candidate HCV vaccines in clinical trials
| Vaccine | Sponsor/company | Antigen targeted | Highest clinical trial stage reached | Outcome or special comments | References/clinical trials ID |
|---|---|---|---|---|---|
| GLS-6150 (hepatitis C vaccine—Inovio/VGX; INO-8000; VGX-6150) | Originated at the University of Pennsylvania and developed at Inovio Pharmaceuticals and GeneOne Life Science, Inc | DNA Vaccine | Phase I clinical trials | The vaccine works by increasing Treg cell frequency, thereby enhancing the virus-specific T-cell response with no apparent side effects | NCT03674125 [63, 78, 80] |
| ChAd3-hliNSmut and MVA-hliNSmut | Originated at Oxford University and developed at ReiThera Srl, European Commission, and GlaxoSmithKline | No data available | Phase I | These two biologicals are termed ‘NSmut' vaccines. Biological ChAd3-hliNSmut is a biological vaccine developed using a chimpanzee adenovirus (ChAd) vector, while MVA-hliNSmut was constructed using the modified vaccinia Ankara (MVA) vector. Phase I clinical trials are being conducted in the United Kingdom | NCT03688061 [62] |
| HCVax | GeneCure Biotechnologies | No data available | Phase I | HCVax was designed by combining several HCV antigens and is vectored using a replication-defective simian immunodeficiency virus using the ‘SimVec technology’ | NCT04318379 [63] |
| Electroporation-mediated plasmid DNA vaccine | Developed in collaboration between National Cancer Institute (NCI) and Inovio Pharmaceuticals | DNA vaccine | Phase I | This vaccine was designed by encoding interleukin-12 in a DNA plasmid and INO-9012. This vaccine may work by boosting the production of antigen-specific T cells | NCT02772003 [63] |
| Autologous DC-vaccines (autologous dendritic cell vaccine) | Russian Academy of Medical Sciences | Core and NS3 proteins | Phase II | Dendritic cells are major players in mounting and subsequently sustaining the T-cell response against an antigen that is derived from a pathogen. This vaccine is expected to induce an immune response against HCV NS3/core protein and thereby reduce the viral load in individuals suffering from chronic HCV infections | NCT03119025 [63, 78] |
| Mesenchymal stem cells | Gulhane Military Medical Academy Ankara, Turkey | Mesenchymal stem cells | Phase II | Liver cirrhosis usually develops in chronic HCV patients. This technique involves the transplantation of mesenchymal stem cells, which are either autologous or obtained from adipose tissues, to patients that have developed advanced cirrhosis. This technique is thus an effort to establish an alternative to expensive liver transplant procedures | NCT02705742 [63] |
Table 4.
Important CD4 and CD8 epitopes of HCV recognized by T cells
| Epitope type | HCV antigen/protein | Epitope (start position-amino acid sequence-end position) | HLA restriction allele | Functional properties | References |
|---|---|---|---|---|---|
| CD8 epitopes | E2 | 607-CLIDYPYRL-615 | A*02 | An effective CD8 T-cell response was observed. | [81] |
| E2 | 610-DYPYRLWHF-618 | A*24 | |||
| NS2 | 838-WLARGLWWI-846 | A*02 | |||
| NS2 | 954-TPMSDWPPY-962 | B*35 | |||
| NS3 | 1367-LPTTGEIPF-1375 | B*35 | |||
| NS5A | 2143-DEVSFSVGL-2151 | B*18 | |||
| NS5A | 2285-LPIWARPDY-2293 | B*35 | |||
| NS5B | 2990-RYLLLCLLI-2998 | A*24 | |||
| CD4 epitopes | NS2 | 923-LLRICALARKMAGGHYVQMA-942 | DRB1*1104 | Effective IFN gamma and TNF alpha responses were observed. | [82] |
| NS2 | 975-HNGLRDLAVAVEPVVFSQME-994 | DRB1*1301 | |||
| NS3 | 1266-TLGFGAYMSKAHGIDPNIRT-1285 | DRB1*1101 | |||
| NS4B | 1964-LHQWLSSECTTPCSG-1978 | DRB1*0401 | |||
| NS5A | 2020-YRGVWRGDGIMHTRCHCGAE-2039 | DRB1*0301 | |||
| NS5B | 2609-LPVAVMGSSYGFQYSPGQRV-2629 | DRB1*0401 | |||
| NS5B | 2674-ARVAIRSLTERLYVGGPLTN-2694 | DRB1*1301 | |||
| NS5B | 2852-ILMTHFFSVLIARDQLEQAL | DRB1*1301 |
Challenges in HCV disease management
Challenges in vaccine design
The major challenge in the development of a vaccine against HCV infection is the unusual genetic diversity of the virus [83–85]. There are seven or eight known genotypes and more than 84-86 subtypes, making this virus even more diverse than human immunodeficiency virus [15, 16, 85–87]. HCV has 30% genetic diversity at the genotype level and 15% at the strain level. The error-prone polymerase of HCV and immune selection generate related but genetically diverse virus variants, even within the same infected individual, thereby posing the problem of resistance to antibody and T-cell-mediated immune responses [88–91]. The use of live-attenuated whole virus particles is a common strategy for vaccine development. However, in the case of HCV, the feasibility of live vaccines is uncertain, as in vitro-grown strains of HCV have a high rate of adaptive mutation, which eventually leads to the generation of highly efficient replicative strains [92–94]. However, for unknown reasons, the infectivity of such strains in primate cell lines and primate animal models is very low [95–97]. Adaptive mutations in such highly replicative strains of HCV are linked to the nature of viral genotypes [87, 93]. Another setback in the field of HCV vaccine development is the lack of immunocompetent animal models. Although chimpanzees can be used as an alternative natural host for HCV, factors such as low chronicity and a lack of clinical signs of infection, limit their utility as an animal model [98]. Other primates such as rhesus macaques (Macaca mulatta), doguera baboons (Papio anubis), chacma baboons (Papio ursinus), Japanese macaques (Macaca fuscata), green monkeys (Chlorocebus sabaeus), and New World monkeys (Saguinus mystax, Saguinus labiatus, and Saguinus oedipus) develop asymptomatic HCV infections [99–102]. Although HCV-like viruses have been found in rats, allowing them to be used as a small-animal model for HCV-like infection, the genetic diversity of rat hepacivirus limits its usefulness in vaccine testing.
Surface protein modification through mutations in various regions of the envelope proteins E1 and E2 is one of the ways in which HCV escapes from adaptive immune responses of the host. Mutations in hypervariable region 1 (HVR1) subvert the neutralizing antibody response by masking conserved epitopes. Furthermore, post-translation processes such as glycosylation have been reported to cause steric interference with the binding of neutralizing antibodies [103]. Various experimental studies of the effects of glycans at different positions have demonstrated that changes in glycosylation can lead to antibody resistance [104, 105]. This is supported by studies showing that natural and site-directed mutations at various glycosylation sites on the E2 protein decrease the sensitivity of cell-culture-grown HCV (HCVcc) and HCV pseudoparticles (HCVpp) to neutralizing antibodies [105].
Some attributes of the host also help HCV to escape from antibody neutralization. High-density lipoproteins in human serum are known to interact with scavenger receptor class B type I (SR-BI). Such interactions increase the rate of virus entry into the cell and reduce the time window for neutralizing antibodies to act [106]. Apolipoprotein E is functionally involved in viral transport and was also demonstrated recently to help mature HCV particles to escape neutralization by masking their epitopes [107]. A recent study on HCV-infected cell cultures showed that E2-protein-coated exosomes were able to make HCV less sensitive to neutralizing antibodies. These findings may find applications in the development of an HCV vaccine [108].
Challenges in diagnostics
Serological methods are commonly used for the diagnosis of most bacterial and viral infections. In the case of HCV, ELISA, recombinant immunoblot assay (RIBA), and rapid assays are the most commonly used serological detection methods. These methods have the disadvantage that they can give false-negative results when testing immunocompromised patients and when used during the window period of HCV infection. False-positive results can also occur due to cross-reactivity in individuals with lupus or rheumatoid arthritis. NAAT (nucleic acid amplification testing) is considered the gold standard for diagnosis of various diseases, especially viral diseases, and it is generally found to be advantageous over traditional serological tests. PCR and transcription-mediated amplification are the two prominent technologies currently employed to detect viral genomes. NAAT has the advantage that it can discriminate between actively infected and recovered patients and thus provides an additional safety layer for preventing transfusion-transmitted infections (TTIs). It also has the important advantage that it allows a diagnosis to be made at an earlier stage of infection. This is important because early treatment of an acute HCV infection improves the likelihood of eliminating the virus and preventing the establishment of a chronic infection. Using NAAT, viral RNA can be detected in an HCV-infected sample within one week of infection [109]. The high sensitivity and specificity of NAAT make it more reliable than serological tests, with fewer false-negative results [110]. In a survey published in 2007, 486,676 seronegative blood samples were screened by NAAT, with detection limits of 1:97,000, 1:490,000, and 1:2800 for samples containing HIV, HCV, and HBV, respectively [111]. Although NAAT is a highly sensitive and specific method for HCV detection, its high cost and requirement for skilled workers and sophisticated instrumentation limits its usage on a mass scale in resource-constrained countries. To make the method more affordable, institutions are using a mini-pool model of NAAT (MP-NAAT) for the detection of virus contamination. Moreover, highly sensitive immunological assays are being developed that can ensure blood safety at an affordable cost [112].
Other possible challenges
Another major challenge in the management of HCV-related disease is the large number of undiagnosed and therefore untreated cases of HCV infection in the population. In the USA, about 1.8 million people are unaware that they are HCV positive [113]. This is partially due to healthcare providers not taking time to ask appropriate questions when examining patients, as recommended in the guidelines of AASLD-IDSA (American Association for the Study of Liver Diseases and the Infectious Diseases Society of America). Furthermore, risk-assessment strategies sometimes fail to identify HCV-infected patients, and some infected patients do not want to disclose their risk factors. Furthermore, the cost of modern HCV therapies such as direct-acting antivirals (DAAs) is a hurdle in HCV disease management. In the USA, the initial price of the drug sofosbuvir is about 1000 USD for a single tablet, and a standard 12-week treatment costs approximately 10,000 USD [114]. At the global level, the cost issue is a major obstacle to the HCV eradication program.
Traditional treatment regimens: past and present
In the 1990s, interferon combined with the nucleotide analogue ribavirin was the only therapy available and was the sole standard care for HCV infections [115]. Subsequently, the introduction of pegylated interferons (alfa-2a and alfa-2b) in combination with ribavirin improved the sustained virological response (SVR) up to 40% to 50% for genotype 1. Similarly, the SVR was observed to improve to 70% to 80% for genotypes 2 and 3, which was 7 to 12% higher than the previously used interferon-ribavirin combination [115, 116]. However, the use of pegylated interferon is associated with psychological, autoimmune, and blood-related side effects. Moreover, it fails to cure chronic cirrhosis caused by HCV infections [18]. It has been reported that, in some cases, complications such as ischemia and neuropsychiatric emergencies do not completely resolve even after withdrawal of the interferon medication [117]. Interferon treatment has been found to be associated with the suppression of bone marrow, leading to a decline in major blood cell lines [117]. Moreover, medications used in interferon therapy are supplied in the form of prefilled syringes that need to be injected subcutaneously, which can be intimidating for patients in the initial stages of the treatment, thereby requiring trained professionals to remain accessible during the entire course of treatment. Ribavirin can also cause side effects, such as teratogenesis, hemolysis, flu-like symptoms, and rashes on the skin [116]. For these reasons, patients need to be closely monitored throughout their treatment, and there is a need for very careful counseling regarding the risks associated with their medications. These side effects of pegylated interferon-ribavirin (PIR) therapy make it unsuitable for standard care of HCV infections.
In 2011, the FDA approved two direct-acting antiviral DAAs, boceprevir and telaprevir, which were marketed by two different companies. SVR was observed to increase to 70% when PIR therapy was combined with either of these DAAs. However, complex dosages, the need for a selective diet, and adverse effects remain major impediments to the success of this therapeutic regimen. DAAs are grouped into three classes based on their target. These include NS3 protease inhibitors, NS5A inhibitors, and NS5B polymerase inhibitors [118]. The NS3 protease is involved in the cleavage and processing of structural and non-structural proteins of HCV. NS5A is a target for drugs due to its important role in the modulation of the interferon response. NS5B is the viral RNA-dependent RNA polymerase. RNA polymerase inhibitors are further classified as nucleotide analogue inhibitors and non-nucleotide analogue inhibitors boceprevir and telaprevir are first-generation drugs, and grazoprevir and voxilaprevir are second-generation NS3 protease inhibitors. The use of first-generation NS3 protease inhibitors is believed to contribute in the emergence of resistant strains in non-responder patients [119]. During monotherapy with telaprevir, although the HCV RNA level decreases significantly in the early phase of treatment, the development of resistance is observed in a large proportion of cases, which has been associated with four mutations in the NS3 region [120]. Similarly, six mutations have been observed to be associated with resistance to boceprevir treatment [121]. Frequent treatment failures due to the acquisition of resistance mutations due to boceprevir and telaprevir therapy have resulted in a shift to triple therapy primarily consisting of PIR and telaprevir. Despite the good results of triple therapy in clinical trials, 96.17% of patients experienced adverse effects, and 34.98% experienced severe adverse effects, including renal failure, skin-related problems, and anaemia [120]. This failure necessitated the quest for an alternative therapy, and a breakthrough came in 2013 with the approval of an NS3 protease inhibitor, simeprevir [122], and an NS5B inhibitor, sofosbuvir (Sovaldi, Gilead Sciences) [122, 123]. The combination of these two drugs, with or without PIR, resulted in an increase in SVR to 90%, with a well-tolerated daily dose [18]. Different combinations of sofosbuvir and simeprevir are associated with various adverse effects, including fatigue, headache, nausea, insomnia, anaemia, and decreased appetite. In addition, treatment regimens including sofosbuvir and simeprevir have been reported to generate drug resistance-associated virus variants [124–126]. Further research in the field of HCV drug development has added well-tolerated, orally active DAAs that are effect without interferon treatment, and combinations of these can be used to treat patients with comorbidities and chronic HCV infections with various genotypes [18, 127]. Clinical trials have been conducted on four DAA drugs (paritaprevir/ritonavir/ombitasvir plus dasabuvir) in patients who had been treated previously with pegylated interferon and in untreated patients. The resulting SVR scores were in the range of 90.2 to 100% for HCV genotypes 1a and 1b [128]. In a meta-analysis of published data on the four-DAA drug regimen, among the 20 cohorts tested in 12 countries, SVR was 96.8% for genotype 1 and 98.9% for genotype 4. The same study reported a 1.3% virologic relapse rate among 5158 patients included in the analysis [129]. Thus, DAA treatments show promise, with improved SVR, good tolerability, and a very narrow range of drug resistance when prescribed in combination. However, combination therapy is also associated with various mild, moderate, and severe adverse effects. Furthermore, virological response failures due to mutations in the target proteins have been reported [130].
Future perspectives
DAAs and other therapies can be successful in the primary stage of HCV infection, provided that the infection is diagnosed at an early stage, and in this regard, the methods currently available for HCV diagnosis are not up to the mark in terms of early diagnosis. The potential of nanotechnology in disease diagnosis is gaining attention because it is simple, reliable, durable, and inexpensive. Gold nanoparticles (AuNP) are a well-studied nanosystem that has advantages over other types of nanoparticles. AuNP can be electrostatically surface-functionalized with various biomolecules including antibody peptides and nucleic acids [131–133]. In addition, AuNPs are highly sensitive to surface plasmon resonance (SPR), showing a prominent color switch from red to blue while transforming from the dispersed state to the aggregate state [134]. Acetylcholinesterase E (AChE) enzyme-conjugated AuNP have been found to be highly efficient for the detection of HCV antibodies. RT-PCR can be used for virus detection, but it requires highly skilled personnel and may not be available in many places. Colorimetric detection of HCV RNA in patient samples without amplification using AuNP has been suggested to be a useful approach. [135, 136]. In a recent study, 27-nucleotide-long RNA probes specific for the 5’UTR of HCV RNA were used for the detection of HCV RNA, either assembled on metallic nanoparticles or without nanoparticles. If a sample is positive, the RNA probes hybridize to the viral RNA and aggregate upon addition of unmodified AuNP, turning the solution blue [135]. Other nanosystems have been used to detect HCV RNA. For example, chemically labelled HCV-RNA-specific probes conjugated with iron oxide nanoparticles can detect HCV in samples from patients, without interference from cellular DNA and RNA [137].
Cytotoxicity and adverse effects of interferon and pegylated interferon can be minimized by site-directed delivery. Hyaluronic acid (HA) is one of the abundant extracellular polysaccharides that naturally target the liver by binding to cluster determinant 44 (CD 44/HA receptors) and get internalized into the cell. HA-decorated AuNPs combined with interferon through hydrophobic and electrostatic forces have been reported to be delivered efficiently to the liver [80]. Furthermore, the polyanionic nature of HA prevents the adsorption of serum proteins onto the nanoparticle-bound interferon. Using this delivery system, no side effects of interferon or pegylated interferon were observed [138]. Targeted delivery of ribavirin can be used to alter its distribution pattern, thereby reducing its cytotoxicity to erythrocytes. The use of polymeric nanoparticles consisting of poly-D, L-lactic acid (PLA) and arabinogalactan poly-L-lysine (AG-PLL) conjugated with ribavirin has been shown not only to reduce cytotoxicity but also to result in targeted delivery to liver cells [138, 139].
Despite the success of DAA therapy, there has been a significant decrease in the popularity of DAAs due to the issues of affordability, resistance, and genotype selectivity. This situation is motivating the development of new therapeutic options. Peptide C5A, derived from the NS5A protein of HCV, has demonstrated potent toxicity to HCV in Huh 7.5 cells [141]. Another peptide, from the N-terminal region of the NS5A protein has amphipathic properties and has been observed to lyse lipid vesicles in a model system for lysis of the virus [141]. Peptides from the NS5A protein are highly toxic for HCV at sub-micromolar concentrations. Furthermore, these peptides were found to be non-toxic to the host in vitro and in vivo at doses 100-fold higher than those required for antiviral activity [142].
Nano-vaccines offer several advantages over traditional vaccines, including rapid delivery and penetration across the membrane of targeted cells. Recombinant core and NS3 fusion proteins conjugated with polylactide-co-glycolide nanoparticles (rC–N/PLGA NPs) have been shown to induce a T-cell-mediated immune response [143]. The shape, size, and polydispersity index of rC–N/PLGA NPs determine their penetration capacity. Triblock copolymers (e.g., poly (D, L-lactic acid)-co-poly(ethylene glycol)-co-poly(D, L-lactic acid [PLA–PEG–PLA] and poly(D, L-lactic-co-glycolic acid)-b-poly(ethylene glycol)-b-poly(D, L-lactic-co-glycolic acid [PLGA–PEG–PLGA]) have been found to be good carriers of DNA encoding multiple viral epitopes [144]. Furthermore, these copolymers increase both cell-mediated and humoral immune responses of such multiepitope gene constructs compared to their naked forms [145]. Controlled release of multiepitope antigen genes from such micellar systems is expected to provide immunological benefits in a single-dose immunization [146].
A strong and long-lasting immune response is one of the ideal characteristics of an efficient vaccine. A natural adjuvant (heat shock protein 27) and cell penetration protein (CPP- Caddy2 and HR9) co-expressed with NS3 protein have been shown to increase the cellular Th1 immune response [147]. This study also showed an elevated cytokine profile of IgG2a, IgG2b, and IFN-γ and increased granzyme B secretion when using this system. A stronger immune response was observed in a mouse model when an NS3-encoding genetic vaccine and an NS3 protein vaccine were administered together than when administered separately [148]. A potent immune response had been reported to be induced by double-stranded RNA encoding the NS3 protein. In mice immunized with recombinant NS3 protein (rNS3) and poly(I:C) emulsified in montanide ISA 720 (M720), the CD8 T cell immune response was found to persist for 7 months [149]. These studies collectively show that such formulations can induce a strong and persistent cellular immune response against HCV.
Conclusion
The overall objective of the current review was to provide a bird-eye view of recent advancements in the field of HCV therapeutics and prophylaxis, giving a summary of existing HCV therapies and their limitations. We initially summarized recent efforts to develop new HCV therapeutic modalities, discussing a few examples of successful drugs and failed attempts. We also attempted to summarize the features of existing therapies and their inherent drawbacks to pinpoint the opportunities and challenges to consider when developing novel treatments. We also summarized the features of drugs that are in the early stages of clinical trials, providing their clinical trial identifiers for effective monitoring of new HCV therapies. We also covered the discussion on emerging immune prophylactic measures, including new vaccine candidates and important biologicals that are in early clinical trials, with special emphasis on newly identified CD4 and CD8 epitopes that can be included in peptide-based subunit vaccines. We also discussed the hurdles that the scientific community is facing in terms of effective disease diagnosis and challenges in existing HCV therapies. Finally, we attempted to provide a nanotech perspective on vaccine design. In summary, although we have tried to paint a positive picture regarding the development of novel HCV therapeutics, recent therapies are still far from perfect, and further improvements are still needed.
Author contributions
All three authors wrote the paper, RNG conceived the idea and supervised the progress of the project, and RJM and GHK prepared figures and tables.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rohan Janardhan Meshram and Gunderao Hanumantrao Kathwate contributed equally for this work.
References
- 1.Keikha M, Eslami M, Yousefi B, Ali-Hassanzadeh M, Kamali A, Yousefi M, Karbalaei M. HCV genotypes and their determinative role in hepatitis C treatment. Virus Dis. 2020;31(3):235–240. doi: 10.1007/s13337-020-00592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farci P, Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome [Science 1989; 244:359–362] J Hepatol. 2002;36(5):582–585. doi: 10.1016/s0168-8278(02). [DOI] [PubMed] [Google Scholar]
- 3.Alzahrani N, Wu MJ, Shanmugam S, Yi M. Delayed by design: role of suboptimal signal peptidase processing of viral structural protein precursors in Flaviviridae virus assembly. Viruses. 2020 doi: 10.3390/v12101090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stasi C, Silvestri C, Voller F. Update on hepatitis C epidemiology: unaware and untreated infected population could be the key to elimination. SN Compr Clin Med. 2020;2:2808–2815. doi: 10.1007/s42399-020-00588-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukhopadhya A. HCV: the Indian scenario. Trop Gastroenterol. 2006;27(3):105–110. [PubMed] [Google Scholar]
- 6.Hashem M, Zaghla H, Zakaria Z, Allam WR, Sameea EA, Mikhail NN, Sobhy M, Galal IF, Mokhtar Y, Hamdy S, Galal G, Abdelwahab SF, Waked I. High spontaneous clearance of symptomatic iatrogenic acute hepatitis C genotype 4 infection. J Med Virol. 2018;90(12):1841–1847. doi: 10.1002/jmv.25270. [DOI] [PubMed] [Google Scholar]
- 7.Yu ML, Chen PJ, Dai CY, Hu TH, Huang CF, Huang YH, Hung CH, Lin CY, Liu CH, Liu CJ, Peng CY, Lin HC, Kao JH, Chuang WL. Taiwan consensus statement on the management of hepatitis C: part (II) special populations. J Formos Med Assoc Taiwan Yi Zhi. 2020;119(7):1135–1157. doi: 10.1016/j.jfma.2020.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Wang W, Huang X, Fan X, Yan J, Luan J. Progress in evaluating the status of hepatitis C infection based on the functional changes of hepatic stellate cells [review] Mol Med Rep. 2020;22(5):4116–4124. doi: 10.3892/mmr.2020.11516. [DOI] [PubMed] [Google Scholar]
- 9.Ge PS, Runyon BA. Treatment of patients with cirrhosis. N Engl J Med. 2016;375(8):767–777. doi: 10.1056/NEJMra1504367. [DOI] [PubMed] [Google Scholar]
- 10.El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142(6):1264–1273.e1261. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nahon P, Bourcier V, Layese R, Audureau E, Cagnot C, Marcellin P, Guyader D, Fontaine H, Larrey D, De Lédinghen V, Ouzan D, Zoulim F, Roulot D, Tran A, Bronowicki JP, Zarski JP, Leroy V, Riachi G, Calès P, Péron JM, Alric L, Bourlière M, Mathurin P, Dharancy S, Blanc JF, Abergel A, Serfaty L, Mallat A, Grangé JD, Attali P, Bacq Y, Wartelle C, Dao T, Benhamou Y, Pilette C, Silvain C, Christidis C, Capron D, Bernard-Chabert B, Zucman D, Di Martino V, Thibaut V, Salmon D, Ziol M, Sutton A, Pol S, Roudot-Thoraval F. Eradication of hepatitis C virus infection in patients with cirrhosis reduces risk of liver and non-liver complications. Gastroenterology. 2017;152(1):142–156.e142. doi: 10.1053/j.gastro.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Houghton M, Weiner A, Han J, Kuo G, Choo QL. Molecular biology of the hepatitis C viruses: implications for diagnosis, development and control of viral disease. Hepatology. 1991;14(2):381–388. doi: 10.1002/hep.1840140227. [DOI] [PubMed] [Google Scholar]
- 13.Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65(3):1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang H, Grisé H. Cellular and molecular biology of HCV infection and hepatitis. Clin Sci (Lond, Engl). 1979;117(2):49–65. doi: 10.1042/cs20080631]:2009. [DOI] [PubMed] [Google Scholar]
- 15.Palladino C, Ezeonwumelu IJ, Marcelino R, Briz V, Moranguinho I, Serejo F, Velosa JF, Marinho RT, Borrego P, Taveira N. Epidemic history of hepatitis C virus genotypes and subtypes in Portugal. Sci Rep. 2018;8(1):12266. doi: 10.1038/s41598-018-30528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borgia SM, Hedskog C, Parhy B, Hyland RH, Stamm LM, Brainard DM, Subramanian MG, McHutchison JG, Mo H, Svarovskaia E, Shafran SD. Identification of a novel hepatitis C virus genotype from Punjab, India: expanding classification of hepatitis C virus into 8 genotypes. J Infect Dis. 2018;218(11):1722–1729. doi: 10.1093/infdis/jiy401. [DOI] [PubMed] [Google Scholar]
- 17.Pybus OG, Thézé J. Hepacivirus cross-species transmission and the origins of the hepatitis C virus. Curr Opin Virol. 2016;16:1–7. doi: 10.1016/j.coviro.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Kish T, Aziz A, Sorio M. Hepatitis C in a new era: a review of current therapies. P T Peer-Rev J Formul Manag. 2017;42(5):316–329. [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamoto S, Kanda T, Shirasawa H, Yokosuka O. Antiviral therapies for chronic hepatitis C virus infection with cirrhosis. World J Hepatol. 2015;7(8):1133–1141. doi: 10.4254/wjh.v7.i8.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta E, Bajpai M, Choudhary A. Hepatitis C virus: Screening, diagnosis, and interpretation of laboratory assays. Asian J Transfus Sci. 2014;8(1):19–25. doi: 10.4103/0973-6247.126683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukherjee R, Burns A, Rodden D, et al. Diagnosis and management of hepatitis C virus infection. J Lab Autom. 2015;20(5):519–538. doi: 10.1177/2211068214563794. [DOI] [PubMed] [Google Scholar]
- 22.Dalgard O, Weiland O, Noraberg G, Karlsen L, Heggelund L, Färkkilâ M, Balslev U, Belard E, Øvrehus A, Skalshøi Kjær M, Krarup H. Sofosbuvir based treatment of chronic hepatitis C genotype 3 infections—a Scandinavian real-life study. PLoS ONE. 2017;12(7):e0179764. doi: 10.1371/journal.pone.0179764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Biagio A, Taramasso L, Cenderello G. Treatment of hepatitis C virus genotype 4 in the DAA era. Virol J. 2018;15(1):1–8. doi: 10.1186/s12985-018-1094-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganta NM, Gedda G, Rathnakar B, Satyanarayana M, Yamajala B, Ahsan MJ, Jadav SS, Balaraju T. A review on HCV inhibitors: significance of non-structural polyproteins. Eur J Med Chem. 2019;164:576–601. doi: 10.1016/j.ejmech.2018.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Clercq E. Current race in the development of DAAs (direct-acting antivirals) against HCV. Biochem Pharmacol. 2014;89(4):441–452. doi: 10.1016/j.bcp.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Tong X, Le Pogam S, Li L, Haines K, Piso K, Baronas V, Yan JM, So SS, Klumpp K, Nájera I. In vivo emergence of a novel mutant L159F/L320F in the NS5B polymerase confers low-level resistance to the HCV polymerase inhibitors mericitabine and sofosbuvir. J Infect Dis. 2014;209(5):668–675. doi: 10.1093/infdis/jit562. [DOI] [PubMed] [Google Scholar]
- 27.Chowdhury AY, Tavis JE, George SL. Human Pegivirus (GB virus C) NS3 protease activity inhibits induction of the type I interferon response and is not inhibited by HCV NS3 protease inhibitors. Virology. 2014;456–457:300–309. doi: 10.1016/j.virol.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 28.Cheng R, Tu T, Shackel N, McCaughan GW. Advances in and the future of treatments for hepatitis C. Expert Rev Gastroenterol Hepatol. 2014;8(6):633–647. doi: 10.1586/17474124.2014.909725. [DOI] [PubMed] [Google Scholar]
- 29.Gane EJ, Pockros PJ, Zeuzem S, Marcellin P, Shikhman A, Bernaards C, Zhou J, Yetzer ES, Ballester R, Dwyer C, Tong X, Nájera I, Bertasso A, Hammond J, Kindrick A, Morcos PN, Smith P, Stancic S, Shulman NS. Mericitabine and ritonavir-boosted danoprevir with or without ribavirin in treatment-naive HCV genotype 1 patients: INFORM-SVR study. Liver. 2015;35(1):79–89. doi: 10.1111/liv.12588. [DOI] [PubMed] [Google Scholar]
- 30.Gentile I, Coppola N, Buonomo AR, Zappulo E, Borgia G. Investigational nucleoside and nucleotide polymerase inhibitors and their use in treating hepatitis C virus. Expert Opin Investig Drugs. 2014;23(9):1211–1223. doi: 10.1517/13543784.2014.921680. [DOI] [PubMed] [Google Scholar]
- 31.Haznedar J, Moreira S, Marbury T, Robson R, Smith W, Kulkarni R, Munson ML, Thommes JA, Lemenuel-Diot A, Washington C, Smith P, Chen YC. The effect of mild to moderate renal impairment on the pharmacokinetics of the nucleoside analog hepatitis C virus polymerase inhibitor mericitabine. Drug Dev Res. 2014;75(2):107–113. doi: 10.1002/ddr.21166. [DOI] [PubMed] [Google Scholar]
- 32.Jensen DM, Brunda M, Elston R, Gane EJ, George J, Glavini K, Hammond JM, Le Pogam S, Nájera I, Passe S, Piekarska A, Rodriguez I, Zeuzem S, Chu T. Interferon-free regimens containing setrobuvir for patients with genotype 1 chronic hepatitis C: a randomized, multicenter study. Liver. 2016;36(4):505–514. doi: 10.1111/liv.12997. [DOI] [PubMed] [Google Scholar]
- 33.Zhang P, Zhong L, Struble EB, Watanabe H, Kachko A, Mihalik K, Virata-Theimer ML, Alter HJ, Feinstone S, Major M. Depletion of interfering antibodies in chronic hepatitis C patients and vaccinated chimpanzees reveals broad cross-genotype neutralizing activity. Proc Natl Acad Sci USA. 2009;106(18):7537–7541. doi: 10.1073/PNAS;0609027491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferreon JC, Ferreon AC, Li K, Lemon SM. Molecular determinants of TRIF proteolysis mediated by the hepatitis C virus NS3/4A protease. J Biol Chem. 2005;280(21):20483–20492. doi: 10.1074/jbc.M500422200. [DOI] [PubMed] [Google Scholar]
- 35.Moriyama K, Suzuki T, Negishi K, Graci JD, Thompson CN, Cameron CE, Watanabe M. Effects of introduction of hydrophobic group on ribavirin base on mutation induction and anti-RNA viral activity. J Med Chem. 2008;51(1):159–166. doi: 10.1021/jm7009952. [DOI] [PubMed] [Google Scholar]
- 36.Lemm JA, Leet JE, O’Boyle DR, 2nd, Romine JL, Huang XS, Schroeder DR, Alberts J, Cantone JL, Sun JH, Nower PT, Martin SW, Serrano-Wu MH, Meanwell NA, Snyder LB, Gao M. Discovery of potent hepatitis C virus NS5A inhibitors with dimeric structures. Antimicrob Agents Chemother. 2011;55(8):3795–3802. doi: 10.1128/AAC.00146-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemm JA, O’Boyle D, 2nd, Liu M, Nower PT, Colonno R, Deshpande MS, Snyder LB, Martin SW, St-Laurent DR, Serrano-Wu MH, Romine JL, Meanwell NA, Gao M. Identification of hepatitis C virus NS5A inhibitors. J Virol. 2010;84(1):482–491. doi: 10.1128/jvi.01360-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romine JL, St-Laurent DR, Leet JE, Martin SW, Serrano-Wu MH, Yang F, Gao M, O’Boyle DR, 2nd, Lemm JA, Sun JH, Nower PT, Huang XS, Deshpande MS, Meanwell NA, Snyder LB. Inhibitors of HCV NS5A: from Iminothiazolidinones to symmetrical stilbenes. ACS Med Chem Lett. 2011;2(3):224–229. doi: 10.1021/ml1002647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou W, Tian Q, Zheng J, Bonkovsky HL. Zinc mesoporphyrin induces rapid proteasomal degradation of hepatitis C nonstructural 5A protein in human hepatoma cells. Gastroenterology. 2010;138(5):1909–1919. doi: 10.1053/j.gastro.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gardelli C, Attenni B, Donghi M, Meppen M, Pacini B, Harper S, Di Marco A, Fiore F, Giuliano C, Pucci V, Laufer R, Gennari N, Marcucci I, Leone JF, Olsen DB, MacCoss M, Rowley M, Narjes F. Phosphoramidate prodrugs of 2′-C-methylcytidine for therapy of hepatitis C virus infection. J Med Chem. 2009;52(17):5394–5407. doi: 10.1021/jm900447q. [DOI] [PubMed] [Google Scholar]
- 41.Pockros PJ. Non-nucleoside analogue polymerase inhibitors in development. Clin Liver Dis. 2013;17(1):123–128. doi: 10.1016/j.cld.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 42.De Clercq E, Li G. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev. 2016;29(3):695–747. doi: 10.1128/cmr.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burton JR, Jr, Everson GT. HCV NS5B polymerase inhibitors. Clin Liver Dis. 2009;13(3):453–465. doi: 10.1016/j.cld.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 44.Cloherty G, Cohen D, Sarrazin C, Wedemeyer H, Chevaliez S, Herman C, Bernstein B, Pawlotsky JM. HCV RNA assay sensitivity impacts the management of patients treated with direct-acting antivirals. Antivir Ther. 2015;20(2):177–183. doi: 10.3851/imp2810. [DOI] [PubMed] [Google Scholar]
- 45.Li G, De Clercq E. Current therapy for chronic hepatitis C: the role of direct-acting antivirals. Antiviral Res. 2017;142:83–122. doi: 10.1016/j.antiviral.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.González-Grande R, Jiménez-Pérez M, González Arjona C, Mostazo TJ. New approaches in the treatment of hepatitis C. World J Gastroenterol. 2016;22(4):1421–1432. doi: 10.3748/wjg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwong AD, Kauffman RS, Hurter P, Mueller P. Discovery and development of telaprevir: an NS3-4A protease inhibitor for treating genotype 1 chronic hepatitis C virus. Nat Biotechnol. 2011;29(11):993–1003. doi: 10.1038/nbt.2020. [DOI] [PubMed] [Google Scholar]
- 48.Sofia MJ, Bao D, Chang W, Du J, Nagarathnam D, Rachakonda S, Reddy PG, Ross BS, Wang P, Zhang HR, Bansal S, Espiritu C, Keilman M, Lam AM, Steuer HM, Niu C, Otto MJ, Furman PA. Discovery of a β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J Med Chem. 2010;53(19):7202–7218. doi: 10.1021/jm100863x. [DOI] [PubMed] [Google Scholar]
- 49.Rosenquist Å, Samuelsson B, Johansson PO, Cummings MD, Lenz O, Raboisson P, Simmen K, Vendeville S, de Kock H, Nilsson M, Horvath A, Kalmeijer R, de la Rosa G, Beumont-Mauviel M. Discovery and development of simeprevir (TMC435), a HCV NS3/4A protease inhibitor. J Med Chem. 2014;57(5):1673–1693. doi: 10.1021/jm401507s. [DOI] [PubMed] [Google Scholar]
- 50.Lin TI, Lenz O, Fanning G, Verbinnen T, Delouvroy F, Scholliers A, Vermeiren K, Rosenquist A, Edlund M, Samuelsson B, Vrang L, de Kock H, Wigerinck P, Raboisson P, Simmen K. In vitro activity and preclinical profile of TMC435350, a potent hepatitis C virus protease inhibitor. Antimicrob Agents Chemother. 2009;53(4):1377–1385. doi: 10.1128/aac.01058-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Link JO, Taylor JG, Xu L, Mitchell M, Guo H, Liu H, Kato D, Kirschberg T, Sun J, Squires N, Parrish J, Kellar T, Yang ZY, Yang C, Matles M, Wang Y, Wang K, Cheng G, Tian Y, Mogalian E, Mondou E, Cornpropst M, Perry J, Desai MC. Discovery of ledipasvir (GS-5885): a potent, once-daily oral NS5A inhibitor for the treatment of hepatitis C virus infection. J Med Chem. 2014;57(5):2033–2046. doi: 10.1021/jm401499g. [DOI] [PubMed] [Google Scholar]
- 52.Wilder JM, Jeffers LJ, Ravendhran N, Shiffman ML, Poulos J, Sulkowski MS, Gitlin N, Workowski K, Zhu Y, Yang JC, Pang PS, McHutchison JG, Muir AJ, Howell C, Kowdley K, Afdhal N, Reddy KR. Safety and efficacy of ledipasvir-sofosbuvir in black patients with hepatitis C virus infection: a retrospective analysis of phase 3 data. Hepatology. 2016;63(2):437–444. doi: 10.1002/hep.28334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klibanov OM, Gale SE. Ombitasvir SB/paritaprevir/ritonavir and dasabuvir tablets for hepatitis C virus genotype 1 infection. Ann Pharmacother. 2015;49(5):566–581. doi: 10.1177/1060028015570729. [DOI] [PubMed] [Google Scholar]
- 54.Keating GM. Ombitasvir/paritaprevir/ritonavir: a review in chronic HCV Genotype 4 infection. Drugs. 2016;76(12):1203–1211. doi: 10.1007/s40265-016-0612-1. [DOI] [PubMed] [Google Scholar]
- 55.Belema M, Meanwell NA. Discovery of daclatasvir, a pan-genotypic hepatitis C virus NS5A replication complex inhibitor with potent clinical effect. J Med Chem. 2014;57(12):5057–5071. doi: 10.1021/jm500335h. [DOI] [PubMed] [Google Scholar]
- 56.Harper S, McCauley JA, Rudd MT, Ferrara M, DiFilippo M, Crescenzi B, Koch U, Petrocchi A, Holloway MK, Butcher JW, Romano JJ, Bush KJ, Gilbert KF, McIntyre CJ, Nguyen KT, Nizi E, Carroll SS, Ludmerer SW, Burlein C, DiMuzio JM, Graham DJ, McHale CM, Stahlhut MW, Olsen DB, Monteagudo E, Cianetti S, Giuliano C, Pucci V, Trainor N, Fandozzi CM, Rowley M, Coleman PJ, Vacca JP, Summa V, Liverton NJ. Discovery of MK 5172, a macrocyclic hepatitis C virus NS3/4A protease inhibitor. ACS Med Chem Lett. 2012;3(4):332–336. doi: 10.1021/ml300017p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahmed H, Abushouk AI, Menshawy A, Attia A, Mohamed A, Negida A, Abdel-Daim MM. Meta-analysis of grazoprevir plus elbasvir for treatment of hepatitis C virus Genotype 1 infection. Ann Hepatol. 2018;17(1):18–32. doi: 10.5604/01.3001.0010.7532. [DOI] [PubMed] [Google Scholar]
- 58.Karaoui LR, Mansour H, Chahine EB. Elbasvir-grazoprevir: a new direct-acting antiviral combination for hepatitis C. Am J Health Syst Pharm AJHP. 2017;74(19):1533–1540. doi: 10.2146/ajhp160558. [DOI] [PubMed] [Google Scholar]
- 59.Heo YA, Deeks ED. Sofosbuvir/Velpatasvir/Voxilaprevir: a review in chronic hepatitis C. Drugs. 2018;78(5):577–587. doi: 10.1007/s40265-018-0895-5. [DOI] [PubMed] [Google Scholar]
- 60.Lawitz EJ, Dvory-Sobol H, Doehle BP, et al. Clinical resistance to velpatasvir (GS-5816), a novel pan-genotypic inhibitor of the hepatitis C virus NS5A protein. Antimicrob Agents Chemother. 2016;60(9):5368–5378. doi: 10.1128/AAC.00763-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Greig SL. Sofosbuvir/velpatasvir: a review in chronic hepatitis C. Drugs. 2016;76(16):1567–1578. doi: 10.1007/s40265-016-0648-2. [DOI] [PubMed] [Google Scholar]
- 62.Lin L, Li H. Analysis of clinical trials of new drugs for liver diseases in China. Drug Des Devel Ther. 2021;20(15):3181–3191. doi: 10.2147/DDDT.S309964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Merck S, Dohme (2021) MalaCards integrated aliases for hepatitis C. https://www.malacards.org/card/hepatitis_c. Accessed 01 Dec 2021
- 64.Chua JV, Ntem-Mensah A, Abutaleb A, Husson J, Mutumbi L, Lam KW, Ghosh A, Romani S, Poonia B, Lee S, Luz Pascual M, Frumkin LR, Kottilil S. Short-duration treatment with the novel non-nucleoside inhibitor CDI-31244 plus sofosbuvir/velpatasvir for chronic hepatitis C: an open-label study. J Med Virol. 2021;93(6):3752–3760. doi: 10.1002/jmv.26652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lawitz E, Gane E, Pearlman B, et al. Efficacy and safety of 12 weeks versus 18 weeks of treatment with grazoprevir (MK-5172) and elbasvir (MK-8742) with or without ribavirin for hepatitis C virus genotype 1 infection in previously untreated patients with cirrhosis and patients with previous null response with or without cirrhosis (C-WORTHY): a randomised, open-label phase 2 trial. Lancet. 2015;385(9973):1075–1086. doi: 10.1016/S0140-6736(14)61795-5. [DOI] [PubMed] [Google Scholar]
- 66.Nanjing Sanhome Pharmaceutical C, Ltd. SH229 tablets combined with daclatasvir dihydrochloride tablets in treatment adult patients with chronic hepatitis C
- 67.Holmes JA, Chung RT. New approaches in viraemic organ transplantation and antiviral therapies. Nat Rev Gastroenterol Hepatol. 2020;17(2):78–79. doi: 10.1038/s41575-019-0257-0. [DOI] [PubMed] [Google Scholar]
- 68.Good SS, Moussa A, Zhou XJ, Pietropaolo K, Sommadossi JP. Preclinical evaluation of AT-527, a novel guanosine nucleotide prodrug with potent, pan-genotypic activity against hepatitis C virus. PLoS ONE. 2020;15(1):e0227104. doi: 10.1371/journal.pone.0227104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berliba E, Bogus M, Vanhoutte F, Berghmans PJ, Good SS, Moussa A, Pietropaolo K, Murphy RL, Zhou XJ, Sommadossi JP. Safety, pharmacokinetics and antiviral activity of AT-527, a novel purine nucleotide prodrug, in HCV-infected subjects with and without cirrhosis. Antimicrob Agents Chemother. 2019 doi: 10.1128/aac.01201-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang G, Dyatkina N, Prhavc M, Williams C, Serebryany V, Hu Y, Huang Y, Wu X, Chen T, Huang W, Rajwanshi VK, Deval J, Fung A, Jin Z, Stoycheva A, Shaw K, Gupta K, Tam Y, Jekle A, Smith DB, Beigelman L. Synthesis and anti-HCV activity of sugar-modified guanosine analogues: discovery of AL-611 as an HCV NS5B polymerase inhibitor for the treatment of chronic hepatitis C. J Med Chem. 2020;63(18):10380–10395. doi: 10.1021/acs.jmedchem.0c00935. [DOI] [PubMed] [Google Scholar]
- 71.Künnemann K (2016) The liver meeting. Drugs Today, 67th Annual Meeting. American Association for the Study of Liver Diseases; 2016–Boston, Massachusetts, USA). p. 673–80 10.1358/dot.2016.52.12.2571703(PMID: 28276539 DOI: (Barcelona. Spain: 1998); 52(12)) [DOI] [PubMed]
- 72.Xu X, Feng B, Guan Y, Zheng S, Sheng J, Yang X, Ma Y, Huang Y, Kang Y, Wen X, Li J, Tan Y, He Q, Xie Q, Wang M, An P, Gong G, Liu H, Ning Q, Hua R, Ning B, Xie W, Zhang J, Huang W, Yang Y, Lin M, Zhao Y, Yu Y, Jia J, Yang D, Chen L, Ye Y, Nan Y, Gong Z, Zhang Q, Hu P, Wang F, Li Y, Li D, Jia Z, Hou J, Chen C, Wu JJ, Wei L. Efficacy and safety of all-oral, 12-week ravidasvir plus ritonavir-boosted danoprevir and ribavirin in treatment-naïve noncirrhotic HCV genotype 1 patients: results from a phase 2/3 clinical trial in China. J Clin Transl Hepatol. 2019;7(3):213–220. doi: 10.14218/jcth.2019.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ahlén G, Frelin L. Methods to evaluate novel hepatitis C virus vaccines. Methods Mol Biol. 2016;1403:221–244. doi: 10.1007/978-1-4939-3387-7_11. [DOI] [PubMed] [Google Scholar]
- 74.Duncan JD, Urbanowicz RA, Tarr AW, Ball JK. Hepatitis C virus vaccine: challenges and prospects. Vaccines. 2020 doi: 10.3390/Vaccines:8010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Manns MP, Wedemeyer H, Cornberg M. Treating viral hepatitis C: efficacy, side effects, and complications. Gut. 2006;55(9):1350–1359. doi: 10.1136/gut.2005.076646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wedemeyer H, Schuller E, Schlaphoff V, Stauber RE, Wiegand J, Schiefke I, Firbas C, Jilma B, Thursz M, Zeuzem S, Hofmann WP, Hinrichsen H, Tauber E, Manns MP, Klade CS. Therapeutic vaccine IC41 as late add-on to standard treatment in patients with chronic hepatitis C. Vaccine. 2009;27(37):5142–5151. doi: 10.1016/j.vaccine.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 77.Fytili P, Dalekos GN, Schlaphoff V, Suneetha PV, Sarrazin C, Zauner W, Zachou K, Berg T, Manns MP, Klade CS, Cornberg M, Wedemeyer H. Cross-genotype-reactivity of the immunodominant HCV CD8 T cell epitope NS3-1073. Vaccine. 2008;26(31):3818–3826. doi: 10.1016/j.vaccine.2008.05.045. [DOI] [PubMed] [Google Scholar]
- 78.Oлeйник EA, Лeплинa O, Tыpинoвa T, Tиxoнoвa M, Пыpинoвa Г, Ocтaнин A, Cтapocтинa H, Чepныx E. Bлияниe peкoмбинaнтныx бeлкoв Gore и NS3 виpyca гeпaтитa c нa coзpeвaниe и фyнкции дeндpитныx клeтoк, гeнepиpyeмыx in vitro в пpиcyтcтвии интepфepoнa-a. Иммyнoлoгия. 2016;37(5).
- 79.Yu CI, Chiang BL. A new insight into hepatitis C vaccine development. J Biomed Biotechnol. 2010 doi: 10.1155/2010/548280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Han JW, Sung PS, Hong SH, Lee H, Koh JY, Lee H, White S, Maslow JN, Weiner DB, Park SH, Jeong M, Heo J, Ahn SH, Shin EC. IFNL3-adjuvanted HCV DNA vaccine reduces regulatory T cell frequency and increases virus-specific T cell responses. J Hepatol. 2020;73(1):72–83. doi: 10.1016/j.jhep.2020.02.009. [DOI] [PubMed] [Google Scholar]
- 81.Luxenburger H, Graß F, Baermann J, Boettler T, Marget M, Emmerich F, Panning M, Thimme R, Nitschke K, Neumann-Haefelin C. Differential virus-specific CD8(+) T cell epitope repertoire in hepatitis C virus genotype 1 versus 4. J Viral Hepat. 2018;25(7):779–790. doi: 10.1111/jvh.12874. [DOI] [PubMed] [Google Scholar]
- 82.Lucas M, Deshpande P, James I, Rauch A, Pfafferott K, Gaylard E, Merani S, Plauzolles A, Lucas A, McDonnell W, Kalams S, Pilkinton M, Chastain C, Barnett L, Prosser A, Mallal S, Fitzmaurice K, Drummer H, Ansari MA, Pedergnana V, Barnes E, John M, Kelleher D, Klenerman P, Gaudieri S. Evidence of CD4(+) T cell-mediated immune pressure on the hepatitis C virus genome. Sci Rep. 2018;8(1):7224. doi: 10.1038/s41598-018-25559-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Manos MM, Shvachko VA, Murphy RC, Arduino JM, Shire NJ. Distribution of hepatitis C virus genotypes in a diverse US integrated health care population. J Med Virol. 2012;84(11):1744–1750. doi: 10.1002/jmv.23399. [DOI] [PubMed] [Google Scholar]
- 84.Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61(1):77–87. doi: 10.1002/hep.27259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Petruzziello A, Marigliano S, Loquercio G, Cozzolino A, Cacciapuoti C. Global epidemiology of hepatitis C virus infection: an up-date of the distribution and circulation of hepatitis C virus genotypes. World J Gastroenterol. 2016;22(34):7824–7840. doi: 10.3748/wjg.v22.i34.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Timm J, Roggendorf M. Sequence diversity of hepatitis C virus: implications for immune control and therapy. World J Gastroenterol. 2007;13(36):4808–4817. doi: 10.3748/wjg.v13.i36.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bailey JR, Barnes E, Cox AL. Approaches, progress, and challenges to hepatitis C vaccine development. Gastroenterology. 2019;156(2):418–430. doi: 10.1053/j.gastro.2018.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Davis GLJ. Hepatitis C virus genotypes and quasispecies. Am J Med. 1999;107(6B):21S–S26. doi: 10.1016/s0002-9343(99)00376-9. [DOI] [PubMed] [Google Scholar]
- 89.Martell M, Esteban JI, Quer J, Genescà J, Weiner A, Esteban R, Guardia J, Gómez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66(5):3225–3229. doi: 10.1128/JVI.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Timm J, Lauer GM, Kavanagh DG, Sheridan I, Kim AY, Lucas M, Pillay T, Ouchi K, Reyor LL, Schulze-zur-Wiesch JS, Gandhi RT, Chung RT, Bhardwaj N, Klenerman P, Walker BD, Allen TM. CD8 epitope escape and reversion in acute HCV infection. J Exp Med. 2004;200(12):1593–604. doi: 10.1084/jem.20041006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bailey JR, Wasilewski LN, Snider AE, El-Diwany R, Osburn WO, Keck Z, Foung SK, Ray SC. Naturally selected hepatitis C virus polymorphisms confer broad neutralizing antibody resistance. J Clin Invest. 2015;125(1):437–447. doi: 10.1172/JCI78794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Q, Li Y, Liu S-A, Xie W, Cheng J. Cell culture-adaptive mutations in hepatitis C virus promote viral production by enhancing viral replication and release. World J Gastroenterol. 2018;24(12):1299–311. doi: 10.3748/wjg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chung A, Jin B, Han KH, Ahn SH, Kim S. Cell culture-adaptive mutations of NS5A affect replication of hepatitis C virus differentially depending on the viral genotypes. J Med Virol. 2017;89(1):146–152. doi: 10.1002/jmv.24591. [DOI] [PubMed] [Google Scholar]
- 94.Mori KI, Matsumoto A, Maki N, Ichikawa Y, Tanaka E, Yagi S. Production of infectious HCV genotype 1b virus in cell culture using a novel Set of adaptive mutations. BMC Microbiol. 2016;16(1):224. doi: 10.1186/s12866-016-0846-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bukh J, Pietschmann T, Lohmann V, Krieger N, Faulk K, Engle RE, Govindarajan S, Shapiro M, St Claire M, Bartenschlager R. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc Natl Acad Sci U S A. 2002;99(22):14416–14421. doi: 10.1073/pnas.212532699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sourisseau M, Goldman O, He W, Gori JL, Kiem HP, Gouon-Evans V, Evans MJ. Hepatic cells derived from induced pluripotent stem cells of pigtail macaques support hepatitis C virus infection. Gastroenterology. 2013;145(5):966–969.e7. doi: 10.1053/j.gastro.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Berggren KA, Suzuki S, Ploss AJI. Animal models used in hepatitis C virus research. Int J Mol Sci. 2020;21(11):3869. doi: 10.3390/ijms21113869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guo X, Zhong JY, Li JW. Hepatitis C virus infection and vaccine development. J Clin Exp Hepatol. 2018;8(2):195–204. doi: 10.1016/j.jceh.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Garson JA, Whitby K, Watkins P, Morgan AJ. Lack of susceptibility of the cottontop tamarin to hepatitis C infection. J Med Virol. 1997;52(3):286–288. doi: 10.1002/(sici)1096-9071(199707)52:3<286::Aid-jmv9>3.0.Co;2-z. [DOI] [PubMed] [Google Scholar]
- 100.Tabor E, Seeff LB, Gerety RJ. Lack of susceptibility of marmosets to human non-A, non-B hepatitis. J Infect Dis. 1979;140(5):794–797. doi: 10.1093/infdis/140.5.794. [DOI] [PubMed] [Google Scholar]
- 101.Abe K, Kurata T, Teramoto Y, Shiga J, Shikata T. Lack of susceptibility of various primates and woodchucks to hepatitis C virus. J Med Primatol. 1993;22(7–8):433–434. doi: 10.1111/j.1600-0684.1993.tb00697.x. [DOI] [PubMed] [Google Scholar]
- 102.Sithebe NP, Kew MC, Mphahlele MJ, Paterson AC, Lecatsas G, Kramvis A, de Klerk W. Lack of susceptibility of Chacma baboons (Papio ursinus orientalis) to hepatitis C virus infection. J Med Virol. 2002;66(4):468–71. doi: 10.1002/jmv.2167. [DOI] [PubMed] [Google Scholar]
- 103.Lavie M, Hanoulle X, Dubuisson J. Glycan shielding and modulation of hepatitis C virus neutralizing antibodies. Front Immunol. 2018;9:910. doi: 10.3389/fimmu.2018.00910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Prentoe J, Velázquez-Moctezuma R, Augestad EH, Galli A, Wang R, Law M, Alter H, Bukh J. Hypervariable region 1 and N-linked glycans of hepatitis C regulate virion neutralization by modulating envelope conformations. Proc Natl Acad Sci U S A. 2019;116(20):10039–10047. doi: 10.1073/pnas.1822002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pantua H, Diao J, Ultsch M, Hazen M, Mathieu M, McCutcheon K, Takeda K, Date S, Cheung TK, Phung Q, Hass P, Arnott D, Hongo JA, Matthews DJ, Brown A, Patel AH, Kelley RF, Eigenbrot C, Kapadia SB. Glycan shifting on hepatitis C virus (HCV) E2 glycoprotein is a mechanism for escape from broadly neutralizing antibodies. J Mol Biol. 2013;425(11):1899–1914. doi: 10.1016/j.jmb.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 106.Voisset C, de Beeck AO, Horellou P, Dreux M, Gustot T, Duverlie G, Cosset FL, Vu- Dac N, Dubuisson J. High-density lipoproteins reduce the neutralizing effect of hepatitis C virus (HCV)-infected patient antibodies by promoting HCV entry. J Gen Virol. 2006;87(9):2577–81. doi: 10.1099/vir.0.81932-0. [DOI] [PubMed] [Google Scholar]
- 107.Fauvelle C, Felmlee DJ, Crouchet E, Lee J, Heydmann L, Lefèvre M, Magri A, Hiet MS, Fofana I, Habersetzer F, Foung SK, Milne R, Patel AH, Vercauteren K, Meuleman P, Zeisel MB, Bartenschlager R, Schuster C, Baumert TF. Apolipoprotein E mediates evasion from hepatitis C virus neutralizing antibodies. Gastroenterology. 2016;150(1):206–217.e204. doi: 10.1053/j.gastro.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 108.Deng L, Jiang W, Wang X, Merz A, Hiet MS, Chen Y, Pan X, Jiu Y, Yang Y, Yu B, He Y, Tu Z, Niu J, Bartenschlager R, Long G. Syntenin regulates hepatitis C virus sensitivity to neutralizing antibody by promoting E2 secretion through exosomes. J Hepatol. 2019;71(1):52–61. doi: 10.1016/j.jhep.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 109.Picciotto A, Torre F, Brizzolara R, Campo N, Giusto R, Sinelli N, Lantieri BP, Risso D, Celle G. Chronic hepatitis C new therapeutic strategies. Minerva Gastroenterol Dietol. 1999;45(3):169–72. [PubMed] [Google Scholar]
- 110.Yaseen SG, Ahmed SA, Johan MF, Kiron R, Daher AM. Evaluation of serological transfusion-transmitted viral diseases and mutliplex nucleic acid testing in Malaysian blood donors. Transfus Apher Sci. 2013;49(3):647–51. doi: 10.1016/j.transci.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 111.Phikulsod S, Oota S, Tirawatnapong T, Sakuldamrongpanich T, Chalermchan W, Louisirirotchanakul S, Tanprasert S, Chongkolwatana V, Kitpoka P, Phanuphak P, Wasi C, Nuchprayoon C. One-year experience of nucleic acid technology testing for human immunodeficiency virus Type 1 Hepatitis C virus, and hepatitis B virus in Thai blood donations. Transfusion. 2009;49(6):1126–35. doi: 10.1111/j.1537-2995.2009.02176.x. [DOI] [PubMed] [Google Scholar]
- 112.Chandrashekar S. Half a decade of mini-pool nucleic acid testing: cost-effective way for improving blood safety in India. Asian J Transfus Sci. 2014;8(1):35–8. doi: 10.4103/0973-6247.126688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Denniston MM, Klevens RM, McQuillan GM, Jiles RB. Awareness of infection, knowledge of hepatitis C, and medical follow-up among individuals testing positive for hepatitis C: National Health and Nutrition Examination Survey 2001–2008. Hepatology. 2012;55(6):1652–1661. doi: 10.1002/hep.25556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Terrault NA. Hepatitis C elimination: challenges with under-diagnosis and undertreatment. F1000Research. 2019 doi: 10.12688/f1000research.15892.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Carrat F, Bani-Sadr F, Pol S, Rosenthal E, Lunel-Fabiani F, Benzekri A, Morand P, Goujard C, Pialoux G, Piroth L, Salmon-Céron D, Degott C, Cacoub P, Perronne C. Pegylated interferon alfa-2b vs standard interferon alfa-2b, plus ribavirin, for chronic hepatitis C in HIV-infected patients: a randomized controlled trial. JAMA. 2004;292(23):2839–48. doi: 10.1001/JAMA:292.23.2839. [DOI] [PubMed] [Google Scholar]
- 116.Yau AH, Yoshida EM. Hepatitis C drugs: the end of the pegylated interferon era and the emergence of all-oral interferon-free antiviral regimens: a concise review. Can J Gastroenterol Hepatol. 2014;28(8):445–51. doi: 10.1155/2014/549624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Geddawy A, Ibrahim YF, Elbahie NM, Ibrahim MA. Direct acting anti-hepatitis C virus drugs: clinical pharmacology and future direction. J Transl Intern Med. 2017;5(1):8–17. doi: 10.1515/jtim-2017-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ward RP, Kugelmas M. Using pegylated interferon and ribavirin to treat patients with chronic hepatitis C. Am Fam Physician. 2005;72(4):655–662. [PubMed] [Google Scholar]
- 119.Takehara T. Hepatitis C virus treatment. Springer; 2017. Features of the second wave of the first generation protease inhibitors: effect and tolerance; pp. 9–16. [Google Scholar]
- 120.Karino Y. Hepatitis C virus treatment. Springer; 2017. Treatment of chronic hepatitis C with the first-generation protease inhibitor telaprevir: its efficacy and resistance mutations; pp. 1–8. [Google Scholar]
- 121.Susser S, Welsch C, Wang Y, Zettler M, Domingues FS, Karey U, Hughes E, Ralston R, Tong X, Herrmann E, Zeuzem S, Sarrazin C. Characterization of resistance to the protease inhibitor Boceprevir in hepatitis C virus-infected patients. Hepatology. 2009;50(6):1709–18. doi: 10.1002/hep.23192. [DOI] [PubMed] [Google Scholar]
- 122.Vaidya A, Perry CM. Simeprevir: first global approval. Drugs. 2013;73(18):2093–2106. doi: 10.1007/s40265-013-0153-9. [DOI] [PubMed] [Google Scholar]
- 123.Lin MV, Chung R. Recent FDA approval of sofosbuvir and simeprevir. Implications for current HCV treatment. Clin Liver Dis. 2014;3(3):65–8. doi: 10.1002/cld.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shepherd SJ, Abdelrahman T, MacLean AR, Thomson EC, Aitken C, Gunson RN. Prevalence of HCV NS3 pre-treatment resistance associated amino acid variants within a Scottish cohort. J Clin Virol. 2015;65:50–3. doi: 10.1016/j.jcv.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hsu SJ, Enkhzaya S, Lin YY, Tseng TC, Khosbayar T, Tsai CH, Wang TS, Enkhtuya D, Ivshinkhorol D, Naranzul N, Jargalsaikhan B, Amarsanaa J, Baatarkhuu O, Kao JH. Resistance-associated substitution and ledipasvir/sofosbuvir therapy in Mongolian chronic hepatitis C patients. J Formos Med Assoc. 2020;119(3):712–719. doi: 10.1016/j.jfma.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 126.Newsum AM, Molenkamp R, van der Meer JT, Rebers SP, Prins M, van der Valk M, Schinkel J. Persistence of NS5B-S282T, a sofosbuvir resistance-associated substitution, in a HIV/HCV-coinfected MSM with risk of onward transmission. J Hepatol. 2018;69(4):968–970. doi: 10.1016/j.jhep.2018.06.021. [DOI] [PubMed] [Google Scholar]
- 127.Curry MP, O’Leary JG, Bzowej N, Muir AJ, Korenblat KM, Fenkel JM, Reddy KR, Lawitz E, Flamm SL, Schiano T, Teperman L, Fontana R, Schiff E, Fried M, Doehle B, An D, McNally J, Osinusi A, Brainard DM, McHutchison JG, Brown RS, Charlton M. Sofosbuvir and Velpatasvir for HCV in Patients with Decompensated Cirrhosis. N Engl J Med. 2015;373(27):2618–2628. doi: 10.1056/NEJMoa1512614. [DOI] [PubMed] [Google Scholar]
- 128.Paritaprevir/ritonavir/ombitasvir plus dasabuvir with ribavirin for chronic hepatitis C. Aust Prescr. 2016;39(4):141–3 10.18773/austprescr.2016.045(PMID: 27756980 PMCID: PMC4993703) [DOI] [PMC free article] [PubMed]
- 129.Wedemeyer H, Craxí A, Zuckerman E, Dieterich D, Flisiak R, Roberts SK, Pangerl A, Zhang Z, Martinez M, Bao Y, Calleja JL. Real-world effectiveness of ombitasvir/paritaprevir/ritonavir±dasabuvir±ribavirin in patients with hepatitis C virus genotype 1 or 4 infection: a meta-analysis. J Viral Hepat. 2017;24(11):936–943. doi: 10.1111/jvh.12722. [DOI] [PubMed] [Google Scholar]
- 130.Degasperi E, Aghemo A. Sofosbuvir for the treatment of chronic hepatitis C: between current evidence and future perspectives. Hepatic Med Evid Res. 2014;6:25–33. doi: 10.2147/hmer.vol.S44375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zong J, Cobb SL, Cameron NR. Peptide-functionalized gold nanoparticles: versatile biomaterials for diagnostic and therapeutic applications. Biomater Sci. 2017;5(5):872–886. doi: 10.1039/c7bm00006e. [DOI] [PubMed] [Google Scholar]
- 132.Emami F, Banstola A, Vatanara A, Lee S, Kim JO, Yook J-H, SJMP, Doxorubicin and anti-PD-L1 antibody conjugated gold nanoparticles for colorectal cancer photochemotherapy. Mol Pharm. 2019;16(3):1184–99. doi: 10.1021/acs.molpharmaceut.8b01157. [DOI] [PubMed] [Google Scholar]
- 133.Sánchez-Visedo A, Gallego B, Royo LJ, Soldado A, Valledor M, Ferrero FJ, Campo JC, Costa-Fernández JM, Fernández-Argüelles MT. Visual detection of microRNA146a by using RNA-functionalized gold nanoparticles. Mikrochim Acta. 2020;187(3):192. doi: 10.1007/s00604-020-4148-4. [DOI] [PubMed] [Google Scholar]
- 134.Kateshiya MR, George G, Rohit JV, Malek NI, Kumar KS. Ractopamine as a novel reagent for the fabrication of gold nanoparticles: colorimetric sensing of cysteine and Hg2+ ion with different spectral characteristics. Microchem J. 2020;158:0026–265X. doi: 10.1016/j.microc.2020.105212. [DOI] [Google Scholar]
- 135.Shawky SM, Bald D, Azzazy HM. Direct detection of unamplified hepatitis C virus RNA using unmodified gold nanoparticles. Clin Biochem. 2010;43(13–14):1163–1168. doi: 10.1016/j.clinbiochem.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 136.Shawky SM, Awad AM, Allam W, Alkordi MH, El-Khamisy SF. Gold aggregating gold: A novel nanoparticle biosensor approach for the direct quantification of hepatitis C virus RNA in clinical samples. Biosens Bioelectron. 2017;92:349–356. doi: 10.1016/j.bios.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shawky SM, Guirgis BS, Azzazy HME. Detection of unamplified HCV RNA in serum using a novel two metallic nanoparticle platform. Clin Chem Lab Med (CCLM). 2014;52(4):565–572. doi: 10.1515/cclm-2013-0521. [DOI] [PubMed] [Google Scholar]
- 138.Di Stefano G, Colonna FP, Bongini A, Busi C, Mattioli A, Fiume L. Ribavirin conjugated with lactosaminated poly-L-lysine: selective delivery to the liver and increased antiviral activity in mice with viral hepatitis. Biochem Pharmacol. 1997;54(3):357–63. doi: 10.1016/s0006-2952. [DOI] [PubMed] [Google Scholar]
- 139.Pathak A, Vyas SP, Gupta KC. Nano-vectors for efficient liver specific gene transfer. Int J Nanomed. 2008;3(1):31–49. doi: 10.2217/17435889.3.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.El-Shenawy R, Tabll A, Bader-El-Din NG, El-Abd Y, Mashaly M, Abdel-Malak CA, Dawood R, El-Awady M. Antiviral activity of virocidal peptide derived from NS5A against two different HCV genotypes: an in vitro study. J Immunoassay Immunochem. 2015;36(1):63–79. doi: 10.1080/15321819.2014.896264. [DOI] [PubMed] [Google Scholar]
- 141.Cho NJ, Dvory-Sobol H, Xiong A, Cho SJ, Frank CW, Glenn JS. Mechanism of an amphipathic alpha-helical peptide’s antiviral activity involves size-dependent virus particle lysis. ACS Chem Biol. 2009;4(12):1061–7. doi: 10.1021/cb900149b. [DOI] [PubMed] [Google Scholar]
- 142.Cheng G, Montero A, Gastaminza P, Whitten-Bauer C, Wieland SF, Isogawa M, Fredericksen B, Selvarajah S, Gallay PA, Ghadiri MR, Chisari FV. A virocidal amphipathic{alpha}-helical peptide that inhibits hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2008;105(8):3088–93. doi: 10.1073/pnas.0712380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hekmat S, Siadat SD, Aghasadeghi MR, Sadat SM, Bahramali G, Aslani MM, Mahdavi M, Shahbazi S. From in-silico immunogenicity verification to in vitro expression of recombinant Core-NS3 fusion protein of HCV. Bratisl Lek Listy. 2017;118(4):189–95. doi: 10.4149/bll_2017_038. [DOI] [PubMed] [Google Scholar]
- 144.Semete B, Booysen LIJ, Kalombo L, Venter JD, Katata L, Ramalapa B, Verschoor JA, Swai H. In vivo uptake and acute immune response to orally administered chitosan and PEG coated PLGA nanoparticles. Toxicol Appl Pharmacol. 2010;249(2):158–165. doi: 10.1016/j.taap.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 145.Rietscher R, Schröder M, Janke J, Czaplewska J, Gottschaldt M, Scherließ R, Hanefeld A, Schubert US, Schneider M, Knolle PA, Lehr CM. Antigen delivery via hydrophilic PEG-b-PAGE-b-PLGA nanoparticles boosts vaccination induced T cell immunity. Eur J Pharm Biopharm. 2016;102:20–31. doi: 10.1016/j.ejpb.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 146.Lu Y, Wu F, Duan W, Mu X, Fang S, Lu N, Zhou X, Kong W. Engineering a ”PEGg-PEI/DNA nanoparticle-in-PLGA microsphere” hybrid controlled release system to enhance immunogenicity of DNA vaccine. Mater Sci Eng C Mater Biol Appl. 2020;106:110294. doi: 10.1016/j.msec.2019.110294. [DOI] [PubMed] [Google Scholar]
- 147.Alizadeh S, Irani S, Bolhassani A, Sadat SM. Simultaneous use of natural adjuvants and cell penetrating peptides improves HCV NS3 antigen-specific immune responses. Immunol Lett. 2019;212:70–80. doi: 10.1016/j.imlet.2019.06.011. [DOI] [PubMed] [Google Scholar]
- 148.Masalova OV, Lesnova EI, Shingarova LN, Tunitskaia VL, Ulanova TI, Burkov AN, Kushch AA. The combined application of nucleotide and amino acid sequences of NS3 hepatitis C virus protein, DNA encoding granulocyte macrophage colony-stimulating factor and inhibitor of regulatory T cells induces effective immune response against hepatitis C virus. Mol biol (Mosk) 2012;46(3):525–534. doi: 10.1134/S0026893312030077. [DOI] [PubMed] [Google Scholar]
- 149.Jin B, Wang RY, Qiu Q, Sugauchi F, Grandinetti T, Alter HJ, Shih JW. Induction of potent cellular immune response in mice by hepatitis C virus NS3 protein with double stranded RNA. Immunology. 2007;122(1):15–27. doi: 10.1111/j.1365-2567.2007.02607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]