Key Points
Question
What is the global diversity in childhood acute lymphoblastic leukemia (ALL) subtypes, and how is genetic ancestry associated with ALL treatment outcomes?
Findings
This genetic association study comprehensively analyzed the molecular subtypes of ALL in 2428 patients from racially and ethnically diverse populations and described differences in leukemia somatic genomics across ancestries. Genetic ancestry was also associated with treatment outcomes, with Native American and African ancestries associated with poorer prognosis even with contemporary ALL therapy and after adjusting for ALL subtypes.
Meaning
This study suggests that the associations of genetic ancestry with ALL subtype and prognosis may underscore the biological basis of the racial and ethnic disparities in ALL, pointing to the need for molecular subtype–driven treatment individualization to help address racial and ethnic gaps in ALL survival.
Abstract
Importance
Racial and ethnic disparities persist in the incidence and treatment outcomes of childhood acute lymphoblastic leukemia (ALL). However, there is a paucity of data describing the genetic basis of these disparities, especially in association with modern ALL molecular taxonomy and in the context of contemporary treatment regimens.
Objective
To evaluate the association of genetic ancestry with childhood ALL molecular subtypes and outcomes of modern ALL therapy.
Design, Setting, and Participants
This multinational, multicenter genetic association study was conducted from March 1, 2000, to November 20, 2020, among 2428 children and adolescents with ALL enrolled in frontline trials from the United States, South East Asia (Singapore and Malaysia), and Latin America (Guatemala), representing diverse populations of European, African, Native American, East Asian, and South Asian descent. Statistical analysis was conducted from February 3, 2020, to April 19, 2021.
Main Outcomes and Measures
Molecular subtypes of ALL and genetic ancestry were comprehensively characterized by performing RNA sequencing. Associations of genetic ancestries with ALL molecular subtypes and treatment outcomes were then evaluated.
Results
Among the participants in the study, 1340 of 2318 (57.8%) were male, and the mean (SD) age was 7.8 (5.3) years. Of 21 ALL subtypes identified, 8 were associated with ancestry. East Asian ancestry was positively associated with the frequency of somatic DUX4 (odds ratio [OR], 1.30 [95% CI, 1.16-1.45]; P < .001) and ZNF384 (OR, 1.40 [95% CI, 1.18-1.66]; P < .001) gene rearrangements and negatively associated with BCR-ABL1–like ALL (OR, 0.79 [95% CI, 0.66-0.92]; P = .002) and T-cell ALL (OR, 0.80 [95% CI, 0.71-0.90]; P < .001). By contrast, occurrence of CRLF2 rearrangements was associated with Native American ancestry (OR, 1.48 [95% CI, 1.29-1.69]; P < .001). When the percentage of Native American ancestry increased, ETV6-RUNX1 fusion became less frequent (OR, 0.80 [95% CI, 0.70-0.91]; P < .001), with the opposite trend observed for ETV6-RUNX1–like ALL. There was a marked preponderance of T-cell ALL in children of African descent compared with those with a high percentage of Native American ancestry (African: OR, 1.22 [95% CI, 1.07-1.37]; P = .003; Native American: OR, 0.53 [95% CI, 0.40-0.67]; P < .001). African ancestry was also positively associated with the prevalence of TCF3-PBX1 (OR, 1.49 [95% CI, 1.25-1.76]; P < .001) and negatively associated with DUX4 rearrangements (OR, 0.70 [95% CI, 0.48-0.93]; P = .01) and hyperdiploidy (OR, 0.77 [95% CI, 0.68-0.86]; P < .001). African and Native American ancestries as continuous variables were both associated with poorer event-free survival (for every 25% increase in ancestry: hazard ratio [HR], 1.2; 95% CI, 1.1-1.4; P = .001 for African ancestry; HR, 1.3; 95% CI, 1.0-1.6; P = .04 for Native American ancestry) and overall survival (for every 25% increase in ancestry: HR, 1.2; 95% CI, 1.1-1.5; P = .01 for African ancestry; HR, 1.4; 95% CI, 1.0-1.8; P = .03 for Native American ancestry). Even after adjusting for biological subtypes and clinical features, Native American and African ancestries remained associated with poor prognosis.
Conclusions and Relevance
This study suggests that ALL molecular subtypes and prognosis are associated with genetic ancestry, potentially pointing to a genetic basis for some of the racial and ethnic disparities in ALL. Therefore, molecular subtype–driven treatment individualization is needed to help address racial and ethnic gaps in outcomes.
This genomic association study evaluates the association of genetic ancestry with childhood acute lymphoblastic leukemia (ALL) molecular subtypes and outcomes of modern acute lymphoblastic leukemia therapy.
Introduction
Acute lymphoblastic leukemia (ALL) is the most common cancer in children,1,2 and cure rates have improved markedly around the world with contemporary combination chemotherapy and refined risk stratification.3,4 However, not all children have benefitted equally from this progress; stark racial and ethnic disparities persist in both disease susceptibility and treatment outcomes.1,5,6,7 In the United States, Hispanic children have the highest incidence of ALL but one of the lowest survival rates. In addition, African American children are least likely to develop ALL,8 but they tend to fare worse in survival compared with those of European and Asian descent.5,6 The underlying reasons for the racial and ethnic disparities in ALL are undoubtedly multifactorial and remain incompletely understood.
To our knowledge, there is a paucity of data describing the biological differences of ALL by ancestry. First, recent genomic profiling studies of ALL have led to markedly revised taxonomies of this cancer and have identified many novel subtypes previously loosely classified as “B other.”9 The prevalence of these newly reported subtypes in racially and ethnically diverse populations is largely unknown. Second, prior studies of ALL disparities predominantly focused on treatment regimens developed in previous decades,1,5,6,7,10 and it is not known how they correlate with outcomes after contemporary, risk-directed therapies. This is important because incremental treatment intensification adopted in recent trials can abrogate racial and ethnic differences in outcomes.11 Third, with few exceptions, racial and ethnic comparison of ALL subtypes and prognosis relied mostly on self-reported race and ethnicity,5 with significant heterogeneity occurring within each population. This is particularly true for Hispanic patients, for whom there can be varying degrees of admixture, and therefore a wide cline in the composition of European, African, and Native American ancestries.12 Similarly, within Asian populations, East Asian and South Asian individuals are often grouped together, although they represent 2 distinctly separate genetic superpopulations. Fourth, most previous reports focused on US populations (ie, Black, Hispanic, and White populations),5,10,13 with significant deficiencies in the characterization of Asian ancestry in ALL. The lack of data describing ALL subtypes in South and East Asian populations is particularly notable, given the fact that Asian individuals represent more than 50% of the global burden of childhood cancer.14,15 Overall, the true global diversity of ALL still remains unclear and consequently represents a significant barrier to the development of molecular subtype–based therapeutic interventions to eliminate racial and ethnic gaps in this cancer.
In this study, we sought to comprehensively characterize population differences in the new taxonomy of childhood ALL subtypes and to examine the relevance of genetic ancestry to ALL outcomes in the context of contemporary therapy. To that end, we quantitatively defined the ancestral composition of 2428 children, adolescents, and young adults treated in a set of national and international frontline ALL trials with wide racial and ethnic diversity. We then systematically evaluated the associations of genetic ancestries with clinical characteristics, biological subtypes, and survival outcomes of ALL.
Methods
The participants included in this study, conducted from March 1, 2000, to November 20, 2020, were enrolled in the following frontline ALL trials in the US, Singapore, Malaysia, and Guatemala: (1) St Jude Children’s Research Hospital Total XV (N = 460)16 and XVI (N = 552)17 protocols; (2) the Children’s Oncology Group P9906 (N = 35),18 AALL0232 (N = 442),19 AALL0331 (N = 134),20 AALL0932 (N = 82),21 and AALL1131 (N = 121)22 protocols; (3) the Malaysia-Singapore (Ma-Spore) group MS2003 (N = 208)23 and MS2010 (N = 206)24 studies; and (4) the Guatemala LLAG-0707 study (N = 188).25 Patients were selected for this analysis based only on genomic data availability. This study was approved by the St Jude Children’s Research Hospital institutional review board. The clinical trials were approved by the respective institutional review boards, and written informed consents for trial enrollment and banking of specimens for future research were obtained from parents, guardians, and/or patients, as appropriate. This study followed the Strengthening the Reporting of Genetic Association Studies (STREGA) reporting guideline.
RNA Sequencing and Subtype Calling
We performed RNA sequencing to derive molecular subtypes based on gene expression profile and fusion genes, which have been described previously.9,26 Full details of RNA sequencing and subtype calling are described in the eMethods in the Supplement.
Determining Genetic Ancestry
For each individual, the admixture fraction was estimated using the iAdmix program,27 and allele frequencies from the 1000 Genomes Project reference populations (European, African, Native American, East Asian, and South Asian) were used as reference.28 The overall genetic ancestral composition for each single individual was derived based on comparison of allele frequencies between patient and reference genomes. Full details of the determination of genetic ancestry and the categorization of patients into racial and ethnic groups are detailed in the eMethods and eFigure 1 in the Supplement.
Statistical Analysis
Statistical analysis was conducted from February 3, 2020, to April 19, 2021. Disease outcomes were overall survival (OS), event-free survival (EFS), and cumulative incidence of any relapse (CIR). Overall survival was measured from date of initial diagnosis of ALL to date of death from any cause or date of last contact. Event-free survival was calculated as the interval from the date of diagnosis until the date of first treatment failure (including induction failure, relapse, second malignant neoplasm, and death from any cause). Cumulative incidence of any relapse was calculated from the date of diagnosis and included any form of relapse (bone marrow, central nervous system, other, or combined). For those who did not experience events, OS and EFS were the time to last contact. Survival probabilities were calculated separately for each of the 7 categories of genetically defined races and ethnicities (Black, East Asian, Hispanic, South Asian, White, other-Asian [dominant admixture with East Asian or South Asian ancestry], and other-US [primarily admixed with European or African ancestry]). We evaluated associations between genetically defined race and ethnicity and EFS or OS using the Mantel log-rank test and associations with CIR using the Fine-Gray competing risk regression model.29 Multivariable analysis of EFS and OS was performed with the Cox proportional hazards regression model,30 and CIR was assessed using competing risks regression, for both genetically defined racial and ethnic category and percentage genetic ancestry. For all univariable and multivariable models assessing genetic ancestry, 4 non-European ancestries were included in the model, leaving out European as the reference. We evaluated ancestries in intervals of 25% increase (eg, hazard ratio [HR] of relapse associated with a 25% increase in African ancestry refers to the elevation of relapse risk when African ancestry increases 25% with a concurrent decrease of European ancestry by 25% while other ancestries [Native American, East Asian, and South Asian] stay constant). Prognostic presenting features (ie, biological subtypes, age at diagnosis [<1, 1 to <10 years, or ≥10 years], and leukocyte count at diagnosis [<50 000 vs ≥50 000 cells/µL; to convert to cells ×109/L, multiply by 0.001]) were included in the multivariable analysis, together with race and ethnicity or percentage genetic ancestry. We included treatment protocol (Ma-Spore MS2003, Ma-Spore MS2010, St Jude Children’s Research Hospital Total XV, St Jude Children’s Research Hospital Total XVI, and Guatemala LLAG-0707) as a covariate for all outcome analyses. Because each cohort and site had their own ALL protocol, protocols adjusted for potential confounding effects arising from differences in treatment and other factors across sites were included. Associations among categorical values were examined using the χ2 test.
Associations of biological subtypes with genetic ancestries were evaluated using a 2-step procedure. In step 1, we first asked if there was an overall ancestry-related difference in the proportion of a given subtype. We performed an overall likelihood ratio test—a χ2 test comparing a logistic regression model without any ancestry variable with a model including all 4 ancestries as continuous variables (leaving out European ancestry as the reference). Because we tested each of the 18 ALL subtypes, the P value cutoff for significance was set as .003 using Bonferroni adjustment for multiple testing. In step 2, we focused on subtypes that reached this significance threshold and evaluated the association of each ancestry with each subtype. This association was assessed using multiple logistic regression comparing the proportion of a given subtype in each of the 4 ancestries (Native American, African, East Asian, and South Asian) with European ancestry as the reference. Here, we applied a Bonferroni-corrected threshold of P = .01. Similar to assessment of HRs, odds ratios (ORs) here represent non-European ancestry increment intervals of 25%, with a concurrent 25% decrease of European ancestry and remaining ancestries fixed.
Linearity of ancestry in the logistic regression models was confirmed by adding quadratic and cubic polynomial transformations of each ancestry to the regression models, with P > .05 from the likelihood ratio test for all nonlinear terms. Analyses were performed by using Prism, version 9.0 (GraphPad Software) and R, version 4.0.3 (R Group for Statistical Computing).
Results
Study Population and Clinical Characteristics Across Genetic Ancestries
A total of 2428 patients (1340 of 2318 male participants [57.8%]; mean [SD] age, 7.8 [5.3] years) were included in this study, with 814 participants from the Children’s Oncology Group, 414 from Ma-Spore, 188 from the LLAG-0707 study in Guatemala, and 1012 from the St Jude frontline ALL trial (Table 1). The distribution of genetically defined racial and ethnic categories of the study population was 8.2% Black (n = 199), 9.6% East Asian (n = 232), 21.4% Hispanic (n = 520), 2.8% South Asian (n = 69), 46.5% White (n = 1128), and 11.6% other (n = 280). In the other category, 162 participants (6.7% of entire cohort) had a dominant admixture with East Asian or South Asian ancestry (other-Asian), and 118 (4.9% of the entire cohort) were primarily admixed with European or African ancestry (other-US). The genetic ancestral composition of all 2428 children is shown in eFigure 2A in the Supplement, and the presenting features of the study cohort are shown in Table 1.
Table 1. Clinical Characteristics of Entire Study Cohort Across Genetically Defined Racial Categories.
| Clinical characteristic | Patients, No./total No. (%) | P valueb | |||||||
|---|---|---|---|---|---|---|---|---|---|
| All | Genetically defined race and ethnicitya | ||||||||
| Black | East Asian | Hispanic | South Asian | White | Other-Asian | Other-US | |||
| Age at diagnosis, y | |||||||||
| <1 | 27/2428 (1.1) | 0/199 | 6/232 (2.6) | 2/520 (0.4) | 2/69 (2.9) | 7/1128 (0.6) | 7/162 (4.3) | 3/118 (2.5) | <.001 |
| ≥1 to <10 | 1704/2428 (70.2) | 131/199 (65.8) | 175/232 (75.4) | 333/520 (64.0) | 58/69 (84.1) | 790/1128 (70.0) | 137/162 (84.6) | 80/118 (67.8) | |
| ≥10 | 697/2428 (28.7) | 68/199 (34.2) | 51/232 (22.0) | 185/520 (35.6) | 9/69 (13.0) | 331/1128 (29.3) | 18/162 (11.1) | 35/118 (29.7) | |
| Sex | |||||||||
| Male | 1340/2318 (57.8) | 112/195 (57.4) | 146/230 (63.5) | 270/488 (55.3) | 42/68 (61.8) | 610/1065 (57.3) | 102/162 (63.0) | 58/110 (52.7) | .26 |
| Female | 978/2318 (42.2) | 83/195 (42.6) | 84/230 (36.5) | 218/488 (44.7) | 26/68 (38.2) | 455/1065 (42.7) | 60/162 (37.0) | 52/110 (47.3) | |
| Leukocyte count at diagnosis | |||||||||
| <50 000 cells/µL | 1646/2326 (70.8) | 132/195 (67.7) | 163/228 (71.5) | 365/494 (73.9) | 43/68 (63.2) | 753/1069 (70.4) | 116/161 (72.0) | 74/111 (66.7) | .40 |
| ≥50 000 cells/µL | 680/2326 (29.2) | 63/195 (32.3) | 65/228 (28.5) | 129/494 (26.1) | 25/68 (36.8) | 316/1069 (29.6) | 45/161 (28.0) | 37/111 (33.3) | |
| Study cohort | |||||||||
| COG | 814/2428 (33.5) | 52/199 (26.1) | 21/232 (9.1) | 215/520 (41.3) | 11/69 (15.9) | 455/1128 (40.3) | 3/162 (1.9) | 57/118 (48.3) | NA |
| Ma-Spore | 414/2428 (17.1) | NA | 202/232 (87.0) | NA | 50/69 (72.5) | 2/1128 (0.2) | 156/162 (96.3) | 4/118 (3.4) | NA |
| St Jude | 1012/2428 (41.7) | 147/199 (73.9) | 9/232 (3.9) | 117/520 (22.5) | 8/69 (11.6) | 671/1128 (59.5) | 3/162 (1.9) | 57/118 (48.3) | NA |
| Guatemala | 188/2428 (7.7) | NA | NA | 188/520 (36.2) | NA | NA | NA | NA | NA |
Abbreviations: ALL, acute lymphoblastic leukemia; COG, Children’s Oncology Group; Ma-Spore, Malaysia-Singapore ALL Study Group; NA, not applicable; St Jude, St Jude Children’s Research Hospital.
SI conversion factor: To convert leukocytes to cells ×109/L, multiply by 0.001.
Criteria for defining racial and ethnic categories are as described in the eMethods in the Supplement.
Determined by use of the χ2 test.
Association of Percent Genetic Ancestry With ALL Biological Subtypes
A total of 21 ALL biological subtypes were identified in the entire study cohort, based on fusion gene, expression profile, point mutation, and karyotype as appropriate. Eighteen subtypes (CRLF2 [OMIM 300357] rearrangement, DUX4 [OMIM 606009] rearrangement, ETV6-RUNX1 [OMIM 600618] fusion, ETV6-RUNX1–like, hyperdiploidy, KMT2A [OMIM 159555] fusion, low hypodiploidy, MEF2D [OMIM 600663] fusion, near haploidy, NUTM1 [OMIM 608963] fusion, PAX5 [OMIM 167414] P80R variation, PAX5 alteration, BCR-ABL1 [OMIM 151410] fusion, BCR-ABL1–like, TCF3-PBX1 [OMIM 147141] fusion, ZNF384 [OMIM 609951] fusion, B other, T-cell ALL [T-ALL]) were included in subsequent analysis, whereas 3 subtypes (HLF [OMIM 142385] fusion, BCL2/MYC [OMIM 151430] rearrangement, and IKZF1 [OMIM 603023] N159Y variation) were grouped under B other because there were fewer than 10 patients with these subtypes in the entire cohort. Overall, biological subtypes varied substantially across genetic ancestries (Table 2) and genetically defined racial categories (eFigure 2B and 2C, and eTable 1 in the Supplement). The exact distribution of subtypes by race and ethnicity in each cohort is shown in eTable 2 and eFigure 3 in the Supplement.
Table 2. Association of Genetic Ancestries (25% Interval) as Continuous Variables With Biological Subtypes.
| Subtype | Overall P valuea | Percentage African | Percentage Native Americanb | Percentage East Asianb | Percentage South Asianb | ||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI)c | P valuea | OR (95% CI) | P valuea | OR (95% CI)c | P valuea | OR (95% CI)c | P valuea | ||
| B-ALL | |||||||||
| Hyperdiploid | <.001d | 0.77 (0.68-0.86) | <.001d | 0.86 (0.78-0.94) | <.001d | 0.95 (0.88-1.02) | .20 | 0.84 (0.70-0.98) | .02 |
| ETV6-RUNX1 | .001d | 1.07 (0.96-1.20) | .22 | 0.80 (0.70-0.91) | <.001d | 0.98 (0.89-1.06) | .59 | 1.10 (0.94-1.27) | .24 |
| B other | <.001d | 0.94 (0.78-1.10) | .45 | 1.32 (1.18-1.47) | <.001d | 0.96 (0.85-1.08) | .51 | 0.88 (0.64-1.11) | .30 |
| BCR-ABL1–like | <.001d | 0.84 (0.68-1.02) | .08 | 1.12 (0.98-1.26) | .10 | 0.79 (0.66-0.92) | .002d | 0.81 (0.55-1.07) | .16 |
| PAX5alt | .09 | 1.15 (0.97-1.35) | .11 | 1.10 (0.93-1.27) | .26 | 1.15 (1.02-1.30) | .02 | 1.19 (0.94-1.45) | .14 |
| DUX4 | <.001d | 0.70 (0.48-0.93) | .01d | 1.02 (0.85-1.2) | .83 | 1.30 (1.16-1.45) | <.001d | 1.26 (1.01-1.52) | .04 |
| CRLF2 | <.001d | 0.78 (0.54-1.04) | .09 | 1.48 (1.29-1.69) | <.001d | 0.81 (0.64-0.99) | .04 | 1.18 (0.88-1.49) | .25 |
| TCF3-PBX1 | <.001d | 1.49 (1.25-1.76) | <.001d | 1.30 (1.09-1.53) | .004d | 1.19 (1.01-1.38) | .03 | 1.02 (0.67-1.38) | .91 |
| KMT2A | .47 | 1.09 (0.85-1.36) | .47 | 0.86 (0.64-1.10) | .25 | 0.94 (0.76-1.13) | .52 | 1.18 (0.85-1.52) | .29 |
| BCR-ABL1 | .79 | 0.96 (0.71-1.24) | .79 | 0.89 (0.66-1.13) | .34 | 1.03 (0.84-1.22) | .79 | 0.92 (0.54-1.30) | .69 |
| ETV6-RUNX1–like | .06 | 0.70 (0.38-1.04) | .08 | 1.22 (0.99-1.48) | .06 | 0.98 (0.78-1.20) | .87 | 1.18 (0.81-1.56) | .35 |
| ZNF384 | .004d | 1.08 (0.76-1.45) | .63 | 1.09 (0.81-1.40) | .55 | 1.40 (1.18-1.66) | <.001d | 1.15 (0.73-1.60) | .49 |
| MEF2D | .004d | 1.37 (1.06-1.74) | .02 | 1.14 (0.85-1.47) | .36 | 1.08 (0.79-1.37) | .59 | 0.04 (2.45E-07-1.04) | .06 |
| Near haploid | .09 | 1.05 (0.74-1.41) | .76 | 0.56 (0.24-0.93) | .02 | 0.84 (0.58-1.10) | .22 | 1.08 (0.62-1.54) | .74 |
| Low hypodiploid | .48 | 1.30 (0.91-1.77) | .14 | 1.09 (0.74-1.51) | .63 | 0.95 (0.62-1.31) | .77 | 0.92 (0.20-1.59) | .82 |
| PAX5 P80R | .17 | 0.85 (0.40-1.36) | .55 | 1.24 (0.89-1.66) | .19 | 0.70 (0.32-1.08) | .12 | 1.25 (0.69-1.84) | .38 |
| NUTM1 | .67 | 1.42 (0.89-2.12) | .13 | 1.09 (0.61-1.71) | .73 | 1.20 (0.82-1.69) | .33 | 1.39 (0.68-2.19) | .29 |
| T-ALLe | <.001d | 1.22 (1.07-1.37) | .003d | 0.53 (0.40-0.67) | <.001d | 0.80 (0.71-0.90) | <.001d | 0.97 (0.79-1.16) | .76 |
Abbreviations: OR, odds ratio; B-ALL, B-cell acute lymphoblastic leukemia; T-ALL, T-cell acute lymphoblastic leukemia.
Derived from the Firth multiple logistic regression model. Overall subtype P values were determined by likelihood ratio test, and the Bonferroni-adjusted P value threshold is .003. For individual subtype-ancestry assessment, P values were determined by the Wald test, and the Bonferroni-adjusted threshold for significance is .01.
Percentage genetic ancestry assessed as a continuous variable, with each ancestry varying from 0% to 100%. The 4 non-European ancestries were included in the model, leaving European ancestry out as the reference.
ORs represent intervals of every 25% increase; for example, for every 25% increase in Native American ancestry with corresponding 25% decrease in European ancestry (and remainder of ancestries fixed), there is a 1.48-fold increase in the odds of developing CRLF2-rearranged ALL.
Significant after correction for multiple testing.
T-ALL evaluated within Ma-Spore, St Jude, and Guatemala cohorts only, as these were unselected cohorts that treated both B-ALL and T-ALL.
Because we quantified the ancestral composition in each patient, we first evaluated the percentage of genetic ancestry as a continuous variable for association with ALL subtype. Because the sum of 5 ancestries is 100% for each individual, we tested African, Native American, East Asian, and South Asian ancestries, leaving European ancestry out of the model as the reference (Table 2). First, we tested the overall differences in the proportion of each ALL subtype across ancestries. After Bonferroni adjustment for multiple testing (with an adjusted P value threshold of .003), we identified 8 subtypes that exhibited overall ancestry-related differences: hyperdiploid, ETV6-RUNX1, B other, BCR-ABL1–like, DUX4, CRLF2, TCF3-PBX1, and T-ALL, with ZNF384 and MEF2D approaching this significance cutoff. Because our defined ALL subtypes are mutually exclusive, we also confirmed their independent association with ancestry using stepwise conditional analyses (eTable 3 in the Supplement). Focusing on the subtypes with cross-ancestry differences, we then examined which specific ancestries were associated with a given subtype. With European ancestry as the reference, we then quantified the associations of increasing each non-European ancestry in 25% increments. That is, we estimated ORs for every 25% increase in a given non-European ancestry, with a corresponding 25% decrease in European ancestry and other ancestries fixed. We found that percentage of African ancestry showed the strongest association with TCF3-PBX1 (OR, 1.49 [95% CI, 1.25-1.76]; P < .001) and was highly associated with T-ALL (OR, 1.22 [95% CI, 1.07-1.37]; P = .003), whereas it was negatively associated with the frequency of DUX4 rearrangement (OR, 0.70 [95% CI, 0.48-0.93]; P = .01) and hyperdiploidy (OR, 0.77 [95% CI, 0.68-0.86]; P < .001) (Table 2). Percentage of Native American ancestry was strongly associated with an increased occurrence of CRLF2 rearrangement (OR, 1.48 [95% CI, 1.29-1.69]; P < .001) and TCF3-PBX1 (OR, 1.30 [95% CI, 1.09-1.53]; P = .004), whereas it was negatively associated with the frequency of hyperdiploid ALL (OR, 0.86 [95% CI, 0.78-0.94]; P < .001), ETV6-RUNX1 (OR, 0.80 [95% CI, 0.70-0.91]; P < .001), and T-ALL (OR, 0.53 [95% CI, 0.40-0.67]; P < .001). We further examined this association within the Guatemala cohort, which was enriched with individuals with a high percentage of Native American ancestry. In this subset, the preponderance of CRLF2 rearrangement and underrepresentation of ETV6-RUNX1 became even more pronounced with increasing percentage of Native American ancestry (eFigure 3 and eFigure 4 in the Supplement). Percentage of East Asian ancestry was associated with a higher frequency of DUX4 rearrangement (OR, 1.30 [95% CI, 1.16-1.45]; P < .001) and ZNF384 fusion (OR, 1.40 [95% CI, 1.18-1.66]; P < .001), while it was protective against the occurrence of BCR-ABL1–like ALL (OR, 0.79 [95% CI, 0.66-0.92]; P = .002) and T-ALL (OR, 0.80 [95% CI, 0.71-0.90]; P < .001). South Asian ancestry remained the most similar to European ancestry in terms of association with molecular subtypes. We also examined the distribution of subtypes across genetically defined racial and ethnic categories and obtained similar results (detailed in the eResults and eTable 1 in the Supplement).
Association of Genetic Ancestry With Treatment Outcomes
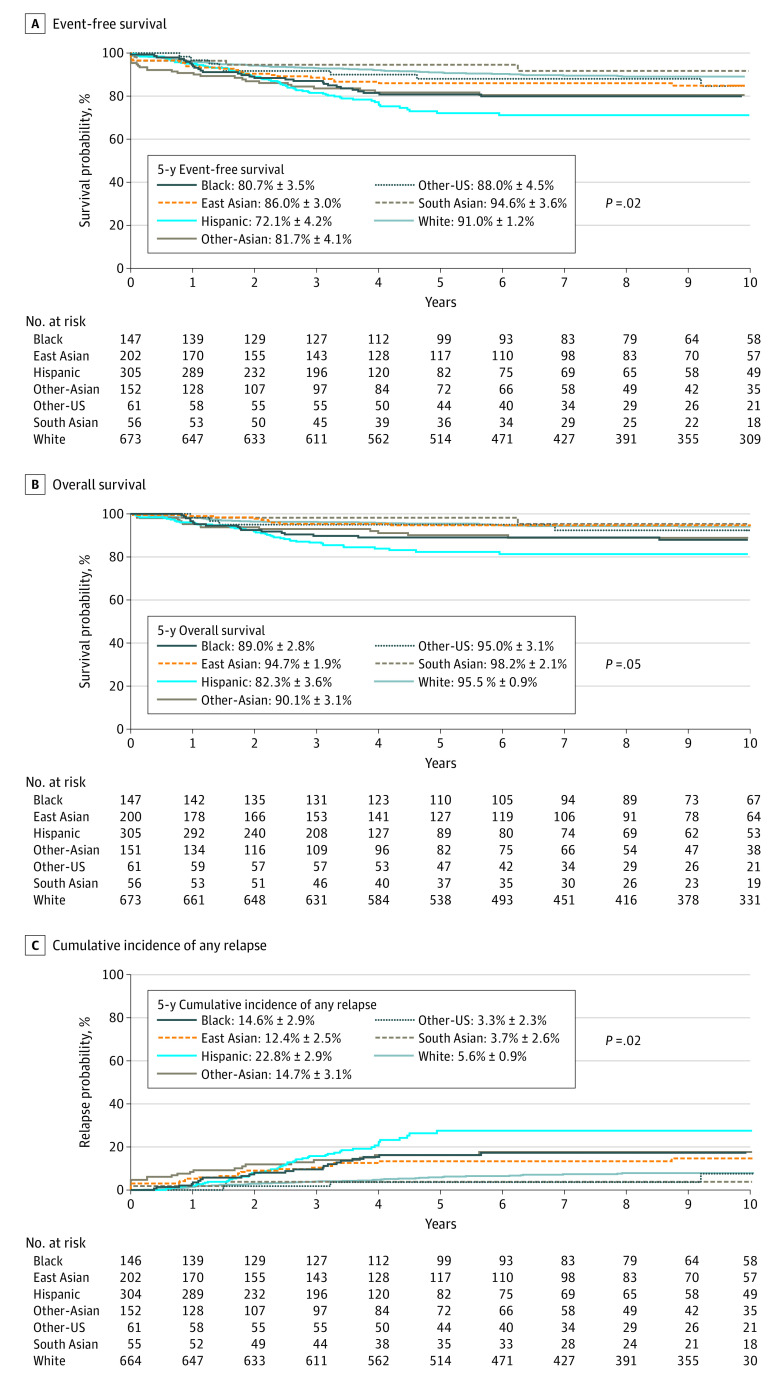
To evaluate the association of ancestry with ALL survival, we elected to focus on frontline protocols with nonselected patients representing the full range of risk groups (ie, St Jude, Ma-Spore, and Guatemala cohorts [n = 1596]). Event-free survival, OS, and CIR were all significantly associated with ancestry (Figure). We analyzed genetic ancestry as a continuous variable, and HRs were estimated for every 25% increase in a given non-European ancestry, with a corresponding 25% decrease in European ancestry and other ancestries fixed. We found that African ancestry was associated with poorer EFS (HR, 1.2; 95% CI, 1.1-1.4; P = .001), OS (HR, 1.2; 95% CI, 1.1-1.5; P = .01), and relapse (HR, 1.3; 95% CI, 1.1-1.5; P = .002), whereas Native American ancestry was associated with worse EFS (HR, 1.3; 95% CI, 1.0-1.6; P = .04) and OS (HR, 1.4; 95% CI, 1.0-1.8; P = .03). Evaluation of outcomes by genetically defined racial and ethnic categories are described in the eResults in the Supplement.
Figure. Kaplan-Meier Survival Curves Comparing All 7 Genetically Defined Racial and Ethnic Categories for 1596 Children, Adolescents, and Young Adults With Acute Lymphoblastic Leukemia.
Criteria for defining racial and ethnic categories are as described in the eMethods in the Supplement. Event-free survival (A), overall survival (B), and cumulative incidence of any relapse (C) were worse for Hispanic and Black patients, with South Asian patients having the most favorable outcomes. P values were obtained after adjusting for treatment protocol.
To explore the hypothesis of whether genetic ancestry was independently associated with treatment outcomes even beyond the associations with biological subtypes and presenting features, we performed multivariable regression analyses adjusting for all biological subtypes, age, leukocyte count at diagnosis, and treatment protocol. We first evaluated percentage genetic ancestry as a continuous variable for association with outcomes (Table 3). With EFS as the outcome of interest, percentage of Native American ancestry and percentage of African ancestry were both associated with a poorer prognosis (HR, 1.3; 95% CI, 1.0-1.6; P = .04 for percentage of Native American ancestry; HR, 1.2; 95% CI, 1.1-1.4; P = .005 for percentage of African ancestry). This finding was similar for OS, for which a higher percentage of Native American ancestry and a higher percentage of African ancestry were both associated with poorer outcomes (HR, 1.4; 95% CI, 1.0-1.9; P = .04 for percentage of Native American ancestry; HR, 1.2; 95% CI, 1.1-1.4; P = .04 for percentage of African ancestry). Assessing relapse, only percentage of African ancestry remained independently associated (HR, 1.3; 95% CI, 1.1-1.5; P = .006). Even in the Guatemala cohort, higher percentage of Native American ancestry continued to be associated with poorer outcome after accounting for subtype and presenting features (EFS: HR, 3.2 [95% CI, 1.0-8.9]; P = .04; CIR: HR, 5.7 [95% CI, 1.6-19.8]; P = .001; eTable 4 in the Supplement). We performed similar multivariable analyses within each of 3 patient cohorts (eTable 5 and eTable 6 in the Supplement) and with ancestry-defined racial and ethnic groups as categorical variables (eTable 7 in the Supplement) and observed largely similar results.
Table 3. Multivariable Analysis of Genetic Ancestries for Association With Survival and Relapse Outcomes for the Entire Cohort.
| Prognostic factor | Event-free survival | Overall survival | Cumulative incidence of relapse | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | P valuea | HR (95% CI) | P valuea | HR (95% CI) | P valuea | |
| Age at diagnosis, y | ||||||
| <1 | 2.4 (1.1-5.0) | .06 | 4.3 (1.6-11.6) | .001 | 1.0 (0.4-2.6) | .93 |
| ≥1 to <10 | 1 [Reference] | 1 [Reference] | 1 [Reference] | |||
| ≥10 | 1.2 (0.9-1.6) | 1.7 (1.2-2.6) | 1.0 (0.7-1.4) | |||
| Leukocyte count at diagnosis, cells/µL | ||||||
| <50 000 | 1 [Reference] | .006 | 1 [Reference] | .04 | 1 [Reference] | .007 |
| ≥50 000 | 1.5 (1.1-2.1) | 1.5 (1.0-2.3) | 1.6 (1.1-2.4) | |||
| Biological subtypesb | ||||||
| ETV6-RUNX1 | 1 [Reference] | <.001 | 1 [Reference] | <.001 | 1 [Reference] | <.001 |
| DUX4 | 1.8 (0.7-4.4) | 1.5 (0.4-6.6) | 2.2 (0.8-6.2) | |||
| Hyperdiploid | 2.0 (1.0-3.8) | 1.9 (0.7-5.4) | 2.1 (1-4.6) | |||
| ZNF384 | 2.9 (0.9-8.9) | 3.4 (0.6-17.9) | 2.9 (0.8-10.9) | |||
| B other | 3.4 (1.7-6.8) | 3.8 (1.3-10.7) | 4.7 (2.1-10.7) | |||
| CRLF2 | 7.2 (3.4-15.0) | 8.8 (3.0-26.2) | 5.4 (2.2-13.5) | |||
| ETV6-RUNX1–like | 6.3 (2.7-14.9) | 9.4 (2.8-31.1) | 5.2 (1.9-14.4) | |||
| KMT2A | 6.5 (2.9-14.7) | 6.7 (2.0-22.5) | 9.9 (3.9-24.8) | |||
| Low hypodiploid | 10.0 (3.7-27.5) | 17.4 (5.0-60.4) | 20.0 (6.9-58.0) | |||
| MEF2D | 4.5 (1.3-15.8) | 6.8 (1.3-35.7) | 3.4 (0.7-16.9) | |||
| Near haploid | 12.2 (3.9-38.1) | 35.0 (9.0-136.0) | 8.7 (1.9-39.9) | |||
| PAX5alt | 3.5 (1.6-7.5) | 2.0 (0.5-7.7) | 5.1 (2.2-12.2) | |||
| BCR-ABL1 | 7.4 (3.3-16.5) | 7.4 (2.3-24.1) | 7.5 (2.8-20.1) | |||
| BCR-ABL1–like | 6.9 (3.1-15.4) | 7.3 (2.3-23.5) | 6.7 (2.6-17.2) | |||
| T-ALL | 4.8 (2.5-9.0) | 6.3 (2.4-16.9) | 4.0 (1.8-9) | |||
| TCF3-PBX1 | 2.3 (1.0-5.3) | 3.5 (1.0-11.5) | 2.4 (0.9-6.4) | |||
| Percentage genetic ancestryc | ||||||
| European | 1 [Reference] | 1 [Reference] | 1 [Reference] | |||
| African (every 25% increase) | 1.2 (1.1-1.4) | .005 | 1.2 (1.01-1.4) | .04 | 1.3 (1.1-1.5) | .006 |
| Native American (every 25% increase) | 1.3 (1.01-1.6) | .04 | 1.4 (1.01-1.9) | .04 | 1.2 (0.9-1.6) | .16 |
| East Asian (every 25% increase) | 1.0 (0.7-1.4) | .87 | 1.0 (0.7-1.6) | .88 | 1.0 (0.7-1.5) | .96 |
| South Asian (every 25% increase) | 0.9 (0.7-1.3) | .70 | 1.0 (0.7-1.6) | .98 | 0.8 (0.5-1.3) | .42 |
Abbreviations: ALL, acute lymphoblastic leukemia; HR, hazard ratio; T-ALL, T-cell ALL.
SI conversion factor: To convert leukocytes to cells × 109/L, multiply by 0.001.
P value for event-free survival and overall survival determined by Cox proportional hazards regression test and P value for cumulative incidence of any relapse determined by Fine-Gray competing risks regression. Treatment protocol was included as a covariate for all analyses. Because each cohort and site had their own ALL protocol, this adjusted for potential confounding effects arising from differences in treatment and other factors across sites.
Subtypes with fewer than 10 patients (ie, NUTM1 and PAX5 P80R) in the whole outcomes cohort were combined into B other.
Percentage genetic ancestry assessed as a continuous variable with each ancestry varying from 0% to 100%. The 4 non-European ancestries were included in the model, leaving European ancestry out as a reference. Hazard ratios are represented for every 25% increase in ancestry; for example, for every 25% increase in African ancestry with corresponding 25% decrease in European ancestry (and remaining ancestries fixed), there is a 1.2-fold increase in risk of event and 1.3-fold increase in risk of relapse.
Discussion
Racial and ethnic disparity among patients with ALL manifests not only as differences in disease molecular subtype and treatment outcomes but also in clinical and genomic research of this cancer. Although more than 90% of childhood ALL cases occur in low- and middle-income countries in Asia and Africa, most ALL genomic studies are restricted to high-income countries, such as the US and European countries.14,15 The racial and ethnic composition of patients with ALL with available genomic data are thus substantially different from the actual global burden in diverse populations, leading to potential bias in our understanding of the molecular subtypes of ALL. Also, contemporary ALL treatment regimens have been developed largely based on patients of European ancestry. Extrapolating these therapeutic approaches to global populations without considering the racial and ethnic diversity of the patients with ALL can no longer be justified, especially with the growing implementation of genomics-based precision medicine. Our study comprising genetically diverse patient cohorts described the full spectrum of ALL molecular subtypes across ancestries and the association of ancestry with ALL prognosis, highlighting a genetic basis for the racial and ethnic disparities in this childhood cancer.
Our study cohort is enriched in Asian children, which has therefore enabled us to examine several new subtypes occurring more frequently in this population, such as DUX4 and ZNF384. Importantly, we delineated the differences between East Asian and South Asian children. It is already generally known that Asian children have excellent survival outcomes, comparable to those of White children.1,6 Here, we identify that further within this previously loosely grouped “Asian” racial and ethnic category, South Asian patients have exceedingly favorable survival outcomes when treated with modern ALL therapy. The exact reasons for their generally superior outcomes remain unclear, but it is plausible that there are germline pharmacogenomic variants unique in this population with a significant association with antileukemic drug disposition and treatment response or toxic effects.31 Pharmacogenomic variants with prominent racial and ethnic differences in frequency have already been described (eg, TPMT and NUDT15 variants associated with thiopurine myelosuppression).11,32,33,34,35,36,37
The toxic effects of several other anti-ALL drugs also associated with substantial racial and ethnic disparities.38,39,40 Hence, deintensification of treatment in this group to mitigate toxic effects could be a future consideration. Racial and ethnic disparity in ALL outcomes is in part associated with the differences in the incidence of biologic subtypes across ancestries. For example, ETV6-RUNX1 and hyperdiploidy, which are associated with outstanding outcomes, collectively occurred in 44.2% of White participants, 32.2% of Black participants, and 29.2% of Hispanic participants; we estimated that 64.5% of the ancestry-related differences in EFS were explained by ALL subtypes. Despite this, genetic ancestry remained associated with outcome in multivariable analysis, beyond the associations with biological subtypes. In fact, these subtype-independent associations of ancestry accounted for another 2.2% of the variance in EFS beyond the 8.4% explained by subtype alone. Therefore, evaluating ancestry in addition to molecular subtype may improve ALL outcomes by an appreciable margin (ie, an improvement of approximately 26%). Also, the interaction between ancestry and ALL therapy is likely to be highly complex, and completely disentangling their association with outcome is challenging. This is particularly true in our study because our ALL cohorts were from diverse geographical regions, each with their own ALL treatment regimen and distinctive ancestral background. Differences in patient ascertainment and/or ALL therapy delivered (which are also associated with ancestry) could confound the analyses associating ancestry with treatment outcomes. Therefore, we deliberately included the ALL treatment protocol as a covariate in all of our outcome analyses, and genetic ancestries remained significant in these multivariable models. Similarly, when we analyzed each ALL cohort separately (St Jude, Ma-Spore, and Guatemala), ancestry exhibited a consistent association with outcomes. That said, we cannot completely rule out variance in patient ascertainment and treatment delivered. Larger trials or population-level studies might be needed in the future to definitively define the association of these factors with ALL prognosis and to formally evaluate the possibility of incorporating ancestry into ALL risk stratification.
Limitations
This study has some limitations. One is that nonbiological factors, such as socioeconomic status, access to care, and treatment adherence, were not evaluated. It has been shown that equal access to effective contemporary treatment can result in Black children with ALL faring as well as White children.10,13 On the other hand, when studying the association of medication adherence and parental education with relapse rates, Bhatia et al41 found that, although these social factors may partly explain the racial and ethnic differences in ALL relapse, Hispanic individuals with good adherence rates continued to demonstrate a higher risk of relapse, suggesting that race and ethnicity–related biological factors beyond adherence may play a role. Granular analysis of individual-level socioeconomic status, access to care, and adherence factors should be explored in future studies and may inform the development of holistic approaches to address racial and ethnic disparities and improve care for all patients.
Another caveat of the present analyses arose from our RNA sequencing–based ALL subtyping. Although RNA sequencing is highly efficient in identifying fusion genes and expression profile and thus enables the identification of most known ALL subtypes, this approach is suboptimal for calling subtypes defined by sequence mutations or those without a characteristic expression signature (eg, ALL with TP53 mutations). Future studies using genome sequencing (both germline and somatic) are warranted to obtain the complete picture of ancestry-related differences in ALL genomics.
Last, although our study is one of the first, to our knowledge, to describe ALL genomics in globally diverse populations, our sample size is still relatively limited, especially for individuals of Asian descent. Plausibly for this reason, a number of ancestry-subtype associations did not withstand stringent Bonferroni correction. Therefore, caution should be exercised when interpreting these results (either positively or negatively), and further validation is needed.
Conclusions
Our study provides a comprehensive characterization of ancestry-related differences in childhood ALL in globally diverse populations, and our findings provide a rationale for the development of molecular subtype–driven treatment protocols to help address the racial and ethnic gaps in ALL survival. Further large-scale pharmacogenomic studies are also needed to identify the exact genomic variation and the mechanisms explaining the racial and ethnic differences in drug response and leukemia molecular subtypes.
eMethods.
eResults.
eReferences.
eTable 1. Distribution of ALL Biological Subtypes Across Genetically Defined Racial Categories
eTable 2. Distribution of ALL Biological Subtypes by Individual Trial Group Across all Racial Categories
eTable 3. Stepwise Conditional Analyses of Association of Molecular Subtypes With Genetic Ancestries
eTable 4. Multivariable Analysis of Genetic Ancestries for Association With Survival and Relapse Outcomes for the Guatemala Cohort
eTable 5. Multivariable Analysis of Genetic Ancestry for Association With Survival and Relapse Outcomes in the Ma-Spore Cohort
eTable 6. Multivariable Analysis of Genetic Ancestry for Association With Survival and Relapse Outcomes in the St Jude Cohort
eTable 7. Multivariable Analysis of Genetically Defined Racial Categories for Association With Survival and Relapse Outcomes for the Entire Cohort
eFigure 1. Correlation of Ancestry Estimates Between Leukemia RNA-seq and Germline SNP Array
eFigure 2. Genetic Ancestral Composition of 2428 Children and Adolescents With ALL and Subtype Distribution Across Genetically Defined Racial Categories
eFigure 3. Distribution of ALL Subtypes Across Various Racial Categories in the (A) COG Cohort, (B) Ma-Spore Cohort, (C) St Jude Cohort and (D) Guatemala Cohort
eFigure 4. Native American Ancestry and Prognosis Within Guatemala Cohort
References
- 1.Kadan-Lottick NS, Ness KK, Bhatia S, Gurney JG. Survival variability by race and ethnicity in childhood acute lymphoblastic leukemia. JAMA. 2003;290(15):2008-2014. doi: 10.1001/jama.290.15.2008 [DOI] [PubMed] [Google Scholar]
- 2.Inaba H, Greaves M, Mullighan CG. Acute lymphoblastic leukaemia. Lancet. 2013;381(9881):1943-1955. doi: 10.1016/S0140-6736(12)62187-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354(2):166-178. doi: 10.1056/NEJMra052603 [DOI] [PubMed] [Google Scholar]
- 4.Hunger SP, Lu X, Devidas M, et al. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the Children’s Oncology Group. J Clin Oncol. 2012;30(14):1663-1669. doi: 10.1200/JCO.2011.37.8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pollock BH, DeBaun MR, Camitta BM, et al. Racial differences in the survival of childhood B-precursor acute lymphoblastic leukemia: a Pediatric Oncology Group Study. J Clin Oncol. 2000;18(4):813-823. doi: 10.1200/JCO.2000.18.4.813 [DOI] [PubMed] [Google Scholar]
- 6.Bhatia S, Sather HN, Heerema NA, Trigg ME, Gaynon PS, Robison LL. Racial and ethnic differences in survival of children with acute lymphoblastic leukemia. Blood. 2002;100(6):1957-1964. doi: 10.1182/blood-2002-02-0395 [DOI] [PubMed] [Google Scholar]
- 7.Pui CH, Sandlund JT, Pei D, et al. Results of therapy for acute lymphoblastic leukemia in Black and White children. JAMA. 2003;290(15):2001-2007. doi: 10.1001/jama.290.15.2001 [DOI] [PubMed] [Google Scholar]
- 8.McNeil DE, Coté TR, Clegg L, Mauer A. SEER update of incidence and trends in pediatric malignancies: acute lymphoblastic leukemia. Med Pediatr Oncol. 2002;39(6):554-557. doi: 10.1002/mpo.10161 [DOI] [PubMed] [Google Scholar]
- 9.Gu Z, Churchman ML, Roberts KG, et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet. 2019;51(2):296-307. doi: 10.1038/s41588-018-0315-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pui CH, Boyett JM, Hancock ML, Pratt CB, Meyer WH, Crist WM. Outcome of treatment for childhood cancer in Black as compared with White children: the St Jude Children’s Research Hospital experience, 1962 through 1992. JAMA. 1995;273(8):633-637. doi: 10.1001/jama.1995.03520320043039 [DOI] [PubMed] [Google Scholar]
- 11.Yang JJ, Cheng C, Devidas M, et al. Ancestry and pharmacogenomics of relapse in acute lymphoblastic leukemia. Nat Genet. 2011;43(3):237-241. doi: 10.1038/ng.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, Lewis CM, Jakobsson M, et al. Genetic variation and population structure in native Americans. PLoS Genet. 2007;3(11):e185. doi: 10.1371/journal.pgen.0030185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pui CH, Pei D, Pappo AS, et al. Treatment outcomes in Black and White children with cancer: results from the SEER database and St Jude Children’s Research Hospital, 1992 through 2007. J Clin Oncol. 2012;30(16):2005-2012. doi: 10.1200/JCO.2011.40.8617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhakta N, Force LM, Allemani C, et al. Childhood cancer burden: a review of global estimates. Lancet Oncol. 2019;20(1):e42-e53. doi: 10.1016/S1470-2045(18)30761-7 [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Galindo C, Friedrich P, Alcasabas P, et al. Toward the cure of all children with cancer through collaborative efforts: pediatric oncology as a global challenge. J Clin Oncol. 2015;33(27):3065-3073. doi: 10.1200/JCO.2014.60.6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pui CH, Campana D, Pei D, et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med. 2009;360(26):2730-2741. doi: 10.1056/NEJMoa0900386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeha S, Pei D, Choi J, et al. Improved CNS control of childhood acute lymphoblastic leukemia without cranial irradiation: St Jude Total Therapy Study 16. J Clin Oncol. 2019;37(35):3377-3391. doi: 10.1200/JCO.19.01692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowman WP, Larsen EL, Devidas M, et al. Augmented therapy improves outcome for pediatric high risk acute lymphocytic leukemia: results of Children’s Oncology Group trial P9906. Pediatr Blood Cancer. 2011;57(4):569-577. doi: 10.1002/pbc.22944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsen EC, Devidas M, Chen S, et al. Dexamethasone and high-dose methotrexate improve outcome for children and young adults with high-risk B-acute lymphoblastic leukemia: a report from Children’s Oncology Group study AALL0232. J Clin Oncol. 2016;34(20):2380-2388. doi: 10.1200/JCO.2015.62.4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maloney KW, Devidas M, Wang C, et al. Outcome in children with standard-risk B-cell acute lymphoblastic leukemia: results of Children’s Oncology Group Trial AALL0331. J Clin Oncol. 2020;38(6):602-612. doi: 10.1200/JCO.19.01086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angiolillo AL, Schore RJ, Kairalla JA, et al. Excellent outcomes with reduced frequency of vincristine and dexamethasone pulses in standard-risk B-lymphoblastic leukemia: results from Children’s Oncology Group AALL0932. J Clin Oncol. 2021;39(13):1437-1447. doi: 10.1200/JCO.20.00494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burke MJ, Salzer WL, Devidas M, et al. Replacing cyclophosphamide/cytarabine/mercaptopurine with cyclophosphamide/etoposide during consolidation/delayed intensification does not improve outcome for pediatric B-cell acute lymphoblastic leukemia: a report from the COG. Haematologica. 2019;104(5):986-992. doi: 10.3324/haematol.2018.204545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeoh AE, Ariffin H, Chai EL, et al. Minimal residual disease-guided treatment deintensification for children with acute lymphoblastic leukemia: results from the Malaysia-Singapore Acute Lymphoblastic Leukemia 2003 Study. J Clin Oncol. 2012;30(19):2384-2392. doi: 10.1200/JCO.2011.40.5936 [DOI] [PubMed] [Google Scholar]
- 24.Yeoh AEJ, Lu Y, Chin WHN, et al. Intensifying treatment of childhood B-lymphoblastic leukemia with IKZF1 deletion reduces relapse and improves overall survival: results of Malaysia-Singapore ALL 2010 Study. J Clin Oncol. 2018;36(26):2726-2735. doi: 10.1200/JCO.2018.78.3050 [DOI] [PubMed] [Google Scholar]
- 25.Antillón FG, Blanco JG, Valverde PD, et al. The treatment of childhood acute lymphoblastic leukemia in Guatemala: biologic features, treatment hurdles, and results. Cancer. 2017;123(3):436-448. doi: 10.1002/cncr.30257 [DOI] [PubMed] [Google Scholar]
- 26.Qian M, Zhang H, Kham SK, et al. Whole-transcriptome sequencing identifies a distinct subtype of acute lymphoblastic leukemia with predominant genomic abnormalities of EP300 and CREBBP. Genome Res. 2017;27(2):185-195. doi: 10.1101/gr.209163.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bansal V, Libiger O. Fast individual ancestry inference from DNA sequence data leveraging allele frequencies for multiple populations. BMC Bioinformatics. 2015;16:4. doi: 10.1186/s12859-014-0418-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auton A, Brooks LD, Durbin RM, et al. ; 1000 Genomes Project Consortium . A global reference for human genetic variation. Nature. 2015;526(7571):68-74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94(446):496-509. [Google Scholar]
- 30.Cox DR. Regression models and life-tables. J R Stat Soc Series B Stat Methodol. 1972;34(2):187-202. [Google Scholar]
- 31.Hariprakash JM, Vellarikkal SK, Verma A, et al. SAGE: a comprehensive resource of genetic variants integrating South Asian whole genomes and exomes. Database (Oxford). 2018;2018:1-10. doi: 10.1093/database/bay080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper SC, Ford LT, Berg JD, Lewis MJ. Ethnic variation of thiopurine S-methyltransferase activity: a large, prospective population study. Pharmacogenomics. 2008;9(3):303-309. doi: 10.2217/14622416.9.3.303 [DOI] [PubMed] [Google Scholar]
- 33.Jack J, Havener TM, McLeod HL, Motsinger-Reif AA, Foster M. Evaluating the role of admixture in cancer therapy via in vitro drug response and multivariate genome-wide associations. Pharmacogenomics. 2015;16(13):1451-1463. doi: 10.2217/PGS.15.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Liu AP, Devidas M, et al. Association of GATA3 polymorphisms with minimal residual disease and relapse risk in childhood acute lymphoblastic leukemia. J Natl Cancer Inst. 2020;113(4):408-417. doi: 10.1093/jnci/djaa138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang W, Treviño LR, Yang JJ, et al. ARID5B SNP rs10821936 is associated with risk of childhood acute lymphoblastic leukemia in Blacks and contributes to racial differences in leukemia incidence. Leukemia. 2010;24(4):894-896. doi: 10.1038/leu.2009.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moriyama T, Nishii R, Perez-Andreu V, et al. NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat Genet. 2016;48(4):367-373. doi: 10.1038/ng.3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang JJ, Landier W, Yang W, et al. Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J Clin Oncol. 2015;33(11):1235-1242. doi: 10.1200/JCO.2014.59.4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor OA, Brown AL, Brackett J, et al. Disparities in neurotoxicity risk and outcomes among pediatric acute lymphoblastic leukemia patients. Clin Cancer Res. 2018;24(20):5012-5017. doi: 10.1158/1078-0432.CCR-18-0939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu C, Yang W, Devidas M, et al. Clinical and genetic risk factors for acute pancreatitis in patients with acute lymphoblastic leukemia. J Clin Oncol. 2016;34(18):2133-2140. doi: 10.1200/JCO.2015.64.5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karol SE, Yang W, Van Driest SL, et al. Genetics of glucocorticoid-associated osteonecrosis in children with acute lymphoblastic leukemia. Blood. 2015;126(15):1770-1776. doi: 10.1182/blood-2015-05-643601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhatia S, Landier W, Shangguan M, et al. Nonadherence to oral mercaptopurine and risk of relapse in Hispanic and non-Hispanic White children with acute lymphoblastic leukemia: a report from the Children’s Oncology Group. J Clin Oncol. 2012;30(17):2094-2101. doi: 10.1200/JCO.2011.38.9924 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods.
eResults.
eReferences.
eTable 1. Distribution of ALL Biological Subtypes Across Genetically Defined Racial Categories
eTable 2. Distribution of ALL Biological Subtypes by Individual Trial Group Across all Racial Categories
eTable 3. Stepwise Conditional Analyses of Association of Molecular Subtypes With Genetic Ancestries
eTable 4. Multivariable Analysis of Genetic Ancestries for Association With Survival and Relapse Outcomes for the Guatemala Cohort
eTable 5. Multivariable Analysis of Genetic Ancestry for Association With Survival and Relapse Outcomes in the Ma-Spore Cohort
eTable 6. Multivariable Analysis of Genetic Ancestry for Association With Survival and Relapse Outcomes in the St Jude Cohort
eTable 7. Multivariable Analysis of Genetically Defined Racial Categories for Association With Survival and Relapse Outcomes for the Entire Cohort
eFigure 1. Correlation of Ancestry Estimates Between Leukemia RNA-seq and Germline SNP Array
eFigure 2. Genetic Ancestral Composition of 2428 Children and Adolescents With ALL and Subtype Distribution Across Genetically Defined Racial Categories
eFigure 3. Distribution of ALL Subtypes Across Various Racial Categories in the (A) COG Cohort, (B) Ma-Spore Cohort, (C) St Jude Cohort and (D) Guatemala Cohort
eFigure 4. Native American Ancestry and Prognosis Within Guatemala Cohort