Abstract
Biological processes involving environmental and genetic factors drive the interplay between age- and sex-regulating lipid profile. The relation between variations in the LPA gene with increasing the risk of coronary heart disease is dependent on population differences, sex, and age. The present study tried to do a gene candidate association analysis in people with myocardial infarction (MI) in a 22 year cohort family-based longitudinal cohort study, Tehran Cardiometabolic Genetic Study (TCGS). After adjusting p value by the FDR method, only the association of rs6415084 with the MI probability and the age-of-CHD-onset was significant in males in their middle age (p < 0.005). Surprisingly, a lack of association was observed for the rest of the markers (16 SNPs). These results revealed the moderator effects of age and sex on the association between the genetic variants (SNPs) of LPA and heart disease risk. Our observations may provide new insights into the biology that underlies lipid profile with age or the sexual dimorphism of Lp(a) metabolism. Finally, Lp(a) appears to be an independent risk factor; however, the role of sex and ethnicity is important.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13293-022-00413-7.
Keywords: TCGS, Lipoprotein (a), Lp(a), LPA locus, Single nucleotide polymorphism, Myocardial infarction (MI), Age-of-onset, Sex, Age
Introduction
Elevated lipoprotein (a) is associated with incidence and severity increasing of cardiovascular diseases [1, 2]. In the Chinese Han population, the association of five SNPs (rs1367211, rs3127596, rs9347438, rs6415085, and rs9364559) in the LPA gene with the development of coronary artery disease (CAD) were evaluated and also rs10455872 in predicting the risk of CHD events in statin therapy plays a significant role [3, 4]. Among these variants, rs6415085 was also associated with the increased LPA expression level and coronary artery disease (CAD) [5]. Rs3798220 and rs10455872 were associated with Lp(a) concentration and CAD prevalence [6, 7]. In assessing the prevalence of rs3798220 and rs10455872 polymorphisms of LPA gene in the subpopulation of patients with symptomatic and asymptomatic carotid stenosis, only a significant association was observed between rs3798220 polymorphism and carotid artery stenosis incidence. Contrastingly, no association was detected for rs3798220 and rs10455872 and atherogenic stroke [8]. A study in a large sample of Brazilian patients confirmed the association of rs10455872 with CAD development, while it showed a lack of association of the rs3798220 with this disease [9]. Heinz Nixdorf Recall’s study provided evidence for the association of LPA rs10455872 with higher Lp(a) and Coronary artery calcification (CAC), a well-proven marker for coronary artery disease and a risk factor for cardiovascular events. [10].
However, the investigation of tertiles Lipoprotein (a) concentration, rs10455872, and rs3798220 with all-cause mortality and cardiovascular mortality with the severity of disease in a large-scale study showed that lipoprotein (a) concentrations and the genetic variants have no associations with mortality in patients with prevalent coronary heart disease. The results showed that these variables are not useful risk factors to predict progression to death after coronary heart disease is established [11].
The Lv et al. study did not display any significant evidence of four SNPs' associations (rs2048327, rs3127599, rs7767084, and rs10755578) SLC22A3-LPAL2-LPA gene cluster with CAD in a large Chinese Han sample [12].
Lack of association between lipoprotein (a) genetic variants (rs6415084 and rs3798220) and subsequent cardiovascular events in Chinese Han patients with coronary artery disease after the percutaneous coronary intervention has also been reported [13].
To resolve ambiguity and investigate the between-population differences in Lp(a) levels, we refer to the Dumitrescu et al. study, which genotyped 19 LP(A) tag SNPs in 7159 participants from the Third National Health and Nutrition Examination Survey (NHANES III). Notably, there were more significant associations between Lp(a) and LP(A) SNPs in non-Hispanic blacks than non-Hispanic whites and Mexican Americans. Moreover, nearly, half of these associations were exclusive to non-Hispanic blacks [14]. LPA SNPs' prevalence and association with the size of apolipoprotein(a) isoforms, Lp(a), and OxPL-apoB levels are highly variable and ethnicity-specific. LPA SNP rs3798220 was most prevalent in Hispanics (42.38%), rs10455872 in whites (14.27%), and rs9457951 in blacks (32.92%). The correlation of each of these SNPs with the major apolipoprotein(a) isoform size was highly variable and in different directions among ethnic groups [15].
The current case–control study examines the association between 17 observed LPA polymorphisms with myocardial infarction (MI) risk across the Tehran Cardiometabolic Genetic Study (TCGS) participants. Our analysis considered the effect of age and sex in the process of evaluating this relationship.
Material and methods
In the present case–control study, 783 unrelated individuals with MII were selected and compared with the same number of controls. These individuals were assigned from the Tehran Cardiometabolic Genetic Study (TCGS), which is a large-scale family-based longitudinal cohort study [16] that is a subpopulation of the Tehran Lipid and Glucose Study (TLGS) [17]. TLGS is and community-based cohort study on fifteen thousand people launched in 1999 in the 13th district of Tehran. The first survey of the TLGS was initiated from 1999 to 2001 on 15,005 individuals aged 3 years, and subjects were genotyped and followed up to identify recently developed diseases every 3 years. The research council of the Endocrine Research Center of the Shahid Beheshti University of Medical Sciences approved the study.
At each survey of TLGS, participants signed a consent form. A standardized questionnaire collected information for age, sex, and history of using medication for diabetes, hypertension, and lipid disorders. Anthropometric measurements, including weight (kg), height (cm), and waist circumference (cm) recorded using standard protocols. Body mass index (BMI) is calculated as weight in kilograms divided by height in square meters. Systolic blood pressure (SBP), diastolic blood pressure (DBP) were measured as described previously [18]. A blood sample draws after 12–14 h overnight fasting. Samples were centrifuged within 30–45 min of collection, and the sera were used for biochemical measurements. Serum glucose, TC, TG, and HDL-C were measured using commercial kits using the enzymatic colorimetric method (Pars Azmoon, Tehran, Iran). Coefficients of variation (CV) for total cholesterol, HDL_C, and triglyceride measurements were below 5%. LDL_C concentrations were calculated using a modified Friedewald equation. Fasting plasma glucose (FPG), triglycerides (TG), total cholesterol (TC), and high-density lipoprotein cholesterol (HDL_C) levels were measured by Pars AzmunCo (Iran); also, coefficients of variation (CV) for total cholesterol, HDL_C and triglyceride measurements were below 5%. Non-HDL_C calculate by subtracting HDL_C from TC. LDL_C concentrations were computed using a modified Friedewald’s equation [19].
In this study, to evaluate the associations between these factors and genetic markers, the obtained values in the last measurement phase before the MI of each case (pre-MI phase) were considered.
Genetic analysis
Genomic samples were extracted from the buffy coat using the standard Proteinase K, the salting-out method. DNA samples were genotyped with HumanOmniExpress-24-v1-0 bead chips (containing 649,932 SNP loci with an average mean distance of 4 kb) at the deCODE genetics company (Iceland) according to the manufacturer’s specifications (Illumina Inc., San Diego, CA, USA). The PLINK program (V 1.07) [20] and R statistic (V 3.2) performed quality control procedures. The genotype information for 17 selected markers in LPA gene (rs7449650, rs11751605, rs7761293, rs6415084, rs9365171, rs7770628, rs6926458, rs6930542, rs13202636, rs7761377, rs10945682, rs7756317, rs1321196, rs1367211, rs9346833, rs783149, and rs1084651) was extracted for the studied population after performing quality control procedures.
The participants diagnosed with coronary heart disease until 2017 were selected as the case group (CHD) for the current study during the follow-up time [21]. We selected an unrelated participant of the same sex and age for each case by in-house python programming to control selection (non-CHD). The control participants also had no history of cardiovascular disease, diabetes mellitus, or metabolic syndrome. All of the controls and cases are unrelated, and they were not belong to the same big family since nearly all of these participants live in a specific region of the Tehran capital city, so they experienced the same environmental and pollution conditions.
Statistical analysis
Kolmogorov–Smirnov tests were used to determine deviations from the normal distribution for all continuous variables, and 0.05 was regarded as a significant level for this test. After that, continuous variables with normal distribution were expressed as mean ± standard deviation (SD) and compared two groups using the student's t-test. Deviation from Hardy–Weinberg equilibrium (HWE) and allele frequency were checked using PLINK (version 1.9) [20]. Linkage disequilibrium (LD) heatmap was made by the LDheatmap package [22]in the R software.
By python programming, Fisher exact test analysis for comparing the allele frequency and Cox analysis for assessing the age of MI of participants carrying out the different alleles was done. Four different age classes, early (20–45 years), middle (45–65 years), late (65–80 years), and (80 < years), and two different sexes for all 17 SNPs were investigated by considering four genetics models (Additive, Dominant, Recessive, and Overdominant). The adjusted odds ratio (OR) was calculated, and the FDR adjusted p value (or q value) of 0.05 was applied [23]. The statistical analyses were performed with python programming and SPSS 24.0 (SPSS, Chicago, IL, USA).
Results
Among TCGS study participants, 783 individuals (459 Men and 323 Women; 21 to 92 years old) had experienced MI (men: 62.7 ± 11.1 years; women: 62.2 ± 10.5 years). The descriptive table of demographics and biochemical characteristics of the case and control groups is presented in Table 1. Table 2 describes different age classes. The genomic (intron/exome) structure of the LPA locus and location of the investigated Rs are also presented in Fig. 1. Moreover, the frequencies of the different alleles of the SNPs are shown in Table 3 for males and females.
Table 1.
Baseline demographic and biochemical characteristics of the population
| Unrelated individuals | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Characteristics | Male (n = 918) | Female (n = 646) | All (n = 1564) | ||||||
| Non-CHD (n =) | CHD (n =) | P valueb | Non-CHD (n =) | CHD (n =) | P valueb | Non-CHD (n =) | CHD (n =) | P valueb | |
| Age (years) | 57 ± 11 | 57 ± 11 | 1 | 57 ± 9 | 57 ± 9 | 1 | 57 ± 10 | 57 ± 10 | 1 |
| SBP (mm Hg) | 122 ± 27 | 128 ± 30 | 0.001 | 123 ± 27 | 126 ± 30 | 0.26 | 122 ± 27 | 127 ± 30 | 0.001 |
| DBP (mm Hg) | 76 ± 15 | 79 ± 17 | 0.013 | 77 ± 15 | 79 ± 18 | 0.18 | 77 ± 15 | 79 ± 17 | 0.006 |
| BMI (kg/m2) | 25 ± 7 | 27 ± 6 | 0.002 | 27 ± 5 | 27 ± 6 | 0.86 | 26 ± 6 | 27 ± 6 | 0.01 |
| Cholesterol (mg/dl) | 220 ± 46 | 232 ± 48 | < 0.001 | 218 ± 40 | 234 ± 49 | < 0.001 | 219 ± 44 | 233 ± 49 | < 0.001 |
| Triglyceride (mg/dl) | 193 ± 130 | 228 ± 140 | 0.001 | 168 ± 91 | 224 ± 147 | < 0.001 | 183 ± 123 | 226 ± 156 | < 0.001 |
| LDL (mg/dl) | 141 ± 35 | 150 ± 39 | 0.001 | 138 ± 34 | 151 ± 41 | 0.001 | 140 ± 35 | 150 ± 39 | < 0.001 |
| HDL (mg/dl) | 42 ± 11 | 38 ± 9 | < 0.001 | 43 ± 10 | 40 ± 11 | < 0.001 | 42 ± 11 | 39 ± 10 | < 0.001 |
aCharacteristics based on Mean ± SE
bP value of t student or chi-square test between case and control groups, SBP, systolic blood pressure; DBP, diastolic blood pressure; BMI, body mass index; LDL, low-density lipoprotein cholesterol; HDL, high-density lipoprotein cholesterol. Indicates statistical significance (P < 0.05)
Table 2.
Different age class range and case and control frequencies in each class
| Age categories | Years old range | Male = 919 | Female = 646 | ||
|---|---|---|---|---|---|
| Case | Control | Case | Control | ||
| Early | 20–45 | 29 | 29 | 15 | 15 |
| Middle | 45–65 | 231 | 231 | 180 | 180 |
| Late | 65–80 | 175 | 175 | 120 | 120 |
| Old | > 80 | 25 | 24 | 8 | 8 |
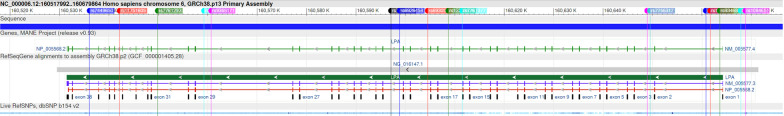
Fig. 1.
Genomic(intron/exome) structure of the LPA locus. Positions of the investigated Rs in this study are also marked on this locus
Table 3.
Investigated Rs in this study and case and control frequencies in male and female groups. All of these SNPs are intron_variant of the LPA gene
| Variation | Position | Ref Allele | Alt Allele | Male = 919 | Female = 646 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control = 459 | Case = 460 | Control = 323 | Case = 323 | ||||||||||||
| HomRef | Het | HomAlt | HomRef | Het | HomAlt | HomRef | Het | HomAlt | HomRef | Het | HomAlt | ||||
| rs7449650 | 6:160,536,082 | T | G | 52 | 199 | 208 | 59 | 186 | 214 | 50 | 128 | 145 | 33 | 140 | 150 |
| rs11751605 | 6:160,542,198 | T | C | 437 | 22 | 0 | 439 | 19 | 1 | 318 | 5 | 0 | 307 | 16 | 0 |
| rs7761293 | 6:160,549,931 | G | A | 148 | 242 | 69 | 162 | 211 | 87 | 124 | 147 | 52 | 104 | 166 | 53 |
| rs6415084 | 6:160,559,298 | T | C | 115 | 186 | 158 | 84 | 238 | 138 | 78 | 154 | 91 | 75 | 162 | 86 |
| rs9365171 | 6:160,560,704 | C | A | 156 | 204 | 99 | 142 | 230 | 88 | 121 | 141 | 61 | 116 | 156 | 50 |
| rs7770628 | 6:160,597,142 | C | T | 119 | 190 | 150 | 98 | 219 | 143 | 69 | 153 | 101 | 77 | 168 | 77 |
| rs6926458 | 6:160,598,834 | A | G | 237 | 189 | 33 | 220 | 196 | 44 | 160 | 126 | 37 | 167 | 134 | 21 |
| rs6930542 | 6:160,604,515 | T | C | 453 | 3 | 0 | 448 | 3 | 0 | 315 | 2 | 0 | 319 | 0 | 0 |
| rs13202636 | 6:160,608,696 | T | C | 234 | 189 | 33 | 219 | 196 | 44 | 160 | 126 | 37 | 167 | 134 | 21 |
| rs7761377 | 6:160,611,449 | A | G | 162 | 175 | 99 | 152 | 169 | 96 | 103 | 124 | 66 | 115 | 137 | 47 |
| rs10945682 | 6:160,648,909 | G | A | 166 | 195 | 95 | 158 | 210 | 92 | 109 | 148 | 65 | 115 | 161 | 47 |
| rs7756317 | 6:160,649,497 | C | T | 459 | 0 | 0 | 459 | 1 | 0 | 321 | 2 | 0 | 322 | 1 | 0 |
| rs1321196 | 6:160,660,810 | C | T | 94 | 205 | 160 | 92 | 215 | 153 | 66 | 145 | 112 | 47 | 163 | 113 |
| rs1367211 | 6:160,661,663 | T | C | 71 | 194 | 194 | 70 | 203 | 187 | 45 | 138 | 140 | 39 | 144 | 139 |
| rs9346833 | 6:160,663,610 | C | T | 102 | 218 | 138 | 109 | 219 | 131 | 76 | 152 | 95 | 64 | 159 | 99 |
| rs783149 | 6:160,667,886 | C | A | 374 | 80 | 5 | 365 | 91 | 4 | 254 | 62 | 6 | 259 | 61 | 3 |
| rs1084651 | 6:160,668,785 | G | A | 375 | 79 | 5 | 371 | 86 | 3 | 259 | 59 | 5 | 259 | 61 | 3 |
Table 4 shows the results of univariate analysis of the association between MI incidence with risk factors, including the allele frequencies of the variantrs6415084 in the OverDominant genetical model, sex, and age.
Table 4.
Univariate analysis results of the association between MI incidence with risk factors including the allele frequencies of the variant rs6415084 in OverDominant genetical model, sex, and age
| Variables in the Equation | ||||||
|---|---|---|---|---|---|---|
| B | SE | Wald | df | Sig | Exp(B) | |
| Sex | − 0.066 | 0.073 | 0.83 | 1 | 0.362 | 0.936 |
| AgeClass_In_selexted_phase | − 1.242 | 0.072 | 297.095 | 1 | 0 | 0.289 |
| rs6415084_OverDominant | 0.197 | 0.072 | 7.582 | 1 | 0.006 | 1.218 |
FDR adjusted p value (or q value) of 0.05 level of statistical significance put aside 16 SNPs. The only rs6415084 in the overdominant genetic model showed significant association with the MI probability. The statistical results of this association are reported in Table 5.
Table 5.
Fisher_exact statistical analysis of the association of the allele frequencies of the variant rs6415084 in OverDominant genetical model with a statistically significant impact on MI incidence
| AgeClass | HomoRef | Het | HomoAlt | p_value | Odds_Ratio (OR) | |
|---|---|---|---|---|---|---|
| Female | Early | 6 | 14 | 10 | 1.000 | 1 |
| Middle | 90 | 177 | 93 | 1.000 | 1.02248 | |
| Late | 48 | 119 | 73 | 0.606 | 1.18149 | |
| Old | 9 | 5 | 2 | 0.282 | 7 | |
| Male | Early | 12 | 25 | 21 | 1.000 | 0.86878 |
| Middle | 107 | 209 | 146 | 0.000 | 2.13976 | |
| Late | 70 | 169 | 111 | 0.134 | 1.41073 | |
| Old | 10 | 21 | 17 | 0.244 | 0.42308 |
Cox analysis results revealed that the same marker also showed a statistically significant association with age-of-CHD-onset. Our results showed an association between MI incidence and the rs6415084 variant in the overdominant genetic model. In addition, as presented in Table 6, there is a strong association between the frequency of different alleles of rs6415084 with age-of-CHD-onset and MI incidence in middle-aged men. Thus, this association depends on the age and sex of cases.
Table 6.
CoxPHFitter statistical analysis of the association of allele frequencies of the variant rs6415084 in OverDominant genetical model with the age of MI incidence
| rs: 6,415,084 Model: OverDominant |
CoxPHFitter | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Age Class |
Homo Ref |
Het | Homo Alt |
coef | exp(coef) | se(coef) | coef Lower 95% |
coef Upper 95% |
exp(coef) Lower 95% |
exp(coef) Upper 95% |
z | p | − log2(p) |
| Female | Early | 6 | 14 | 10 | 0.24 | 1.28 | 0.52 | − 0.78 | 1.26 | 0.46 | 3.54 | 0.47 | 0.64 | 0.64 |
| Middle | 90 | 177 | 93 | 0.02 | 1.02 | 0.15 | -0.27 | 0.31 | 0.76 | 1.36 | 0.12 | 0.91 | 0.14 | |
| Late | 48 | 119 | 73 | 0.08 | 1.08 | 0.18 | − 0.28 | 0.43 | 0.75 | 1.54 | 0.42 | 0.68 | 0.56 | |
| Old | 9 | 5 | 2 | 1.08 | 2.93 | 0.71 | − 0.32 | 2.48 | 0.72 | 11.89 | 1.51 | 0.13 | 2.92 | |
| Male | Early | 12 | 25 | 21 | 0.03 | 1.03 | 0.38 | − 0.71 | 0.77 | 0.49 | 2.15 | 0.07 | 0.94 | 0.09 |
| Middle | 107 | 209 | 146 | 0.5 | 1.64 | 0.13 | 0.24 | 0.76 | 1.27 | 2.13 | 3.76 | < 0.005 | 12.54 | |
| Late | 70 | 169 | 111 | 0.32 | 1.37 | 0.15 | 0.02 | 0.61 | 1.02 | 1.85 | 2.09 | 0.04 | 4.76 | |
| Old | 10 | 21 | 17 | − 0.52 | 0.6 | 0.43 | − 1.37 | 0.33 | 0.26 | 1.4 | − 1.19 | 0.23 | 2.1 | |
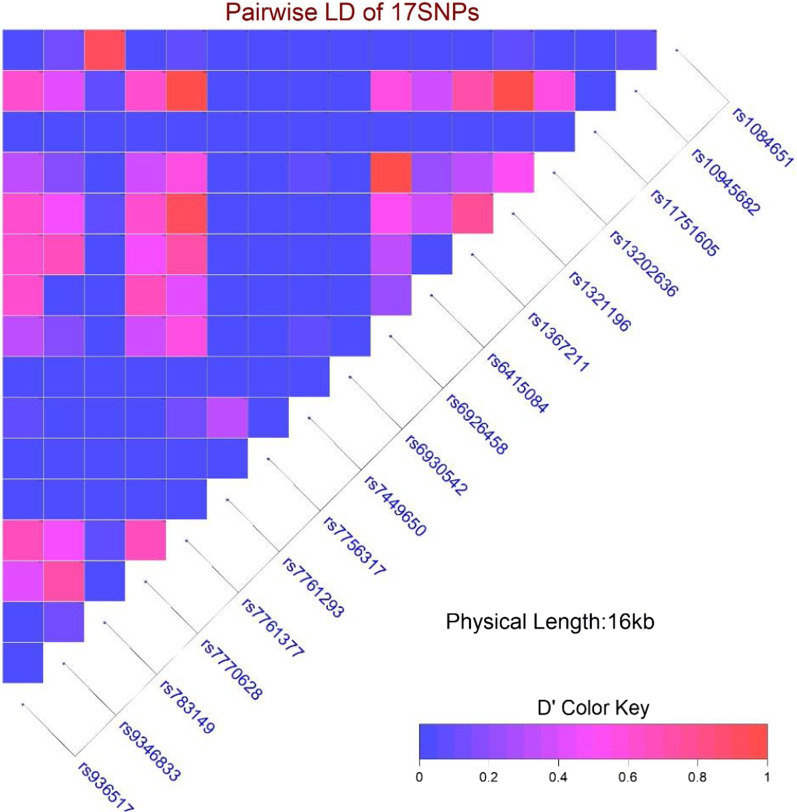
rs6415084 did not show any significant linkage disequilibrium (LD) with other studied markers. Figure 2 and Additional file 1: Table S1.
Fig. 2.
Linkage disequilibrium (LD) heatmap plot for all investigated SNPs in this study. This figure was produced by LDheatmap package in R software
To evaluate the associations between this genetic marker and other risk factors, univariate analysis results of the association between allele frequencies of the variant rs6415084 in OverDominant genetical model with risk factors including BMI, Cholesterol, TG, HDL, NHDL, and LDL. The parameters were obtained from the Pre-MI phase of each case.
Discussion
The influence of age on Lp(a) concentrations is controversial. In the other studies, both older age and female sex are independent significant predictors of higher plasma Lp(a) [24, 25].
Base on our results, even the rs6415084 variants in the males that showed association with the largest age category did not show association with the early age-class variables. Thus, it is not only a sex-dependent but also an age-dependent association. The frequencies of 16 variants (rs7449650, rs11751605, rs7761293, rs9365171, rs7770628, rs6926458, rs6930542, rs13202636, rs7761377, rs10945682, rs7756317, rs1321196, rs1367211, rs9346833, rs783149, and rs1084651) did not associate with any of examined age–sex classes. These results confirmed the inter-population difference in genetics markers in agreement with our previous findings on the other genes variation on the TCGS data.
The importance of rs10455872 and its association with Lp(a) level reported on pharmacogenetics, CAD development in Brazilian patients, familial hypercholesterolemia studies, and Coronary artery calcification (CAC) [4, 6, 7, 9, 10]. In addition, the association of LPA polymorphism and carotid artery stenosis incidence, a significant association was observed for rs3798220. Contrastingly, no associations were detected for rs3798220 and rs10455872 and atherogenic stroke [8], carotid artery stenosis incidence with rs10455872, and CAD in Brazilian patients with rs3798220 [8, 9]. In addition, in a large-scale study, rs10455872 and rs3798220, have shown no associations with mortality in patients with prevalent coronary heart disease [11]. Moreover, a lack of associations between rs2048327, rs3127599, rs7767084, and rs10755578 with CAD in Chinese Han samples has also been reported [12, 13]. Besides, the Dumitrescu et al.'s study, which genotyped 19 LPA tag SNPs in 7,159 participants from the Third National Health and Nutrition Examination Survey (NHANES III), showed significant associations between Lp(a) concentration and LPA SNPs in non-Hispanic blacks than non-Hispanic whites and Mexican Americans. [14]. Interestingly, a lack of association between rs6415084 and subsequent cardiovascular events after the percutaneous coronary intervention has also been reported [13] in Chinese Han patients with coronary artery disease.
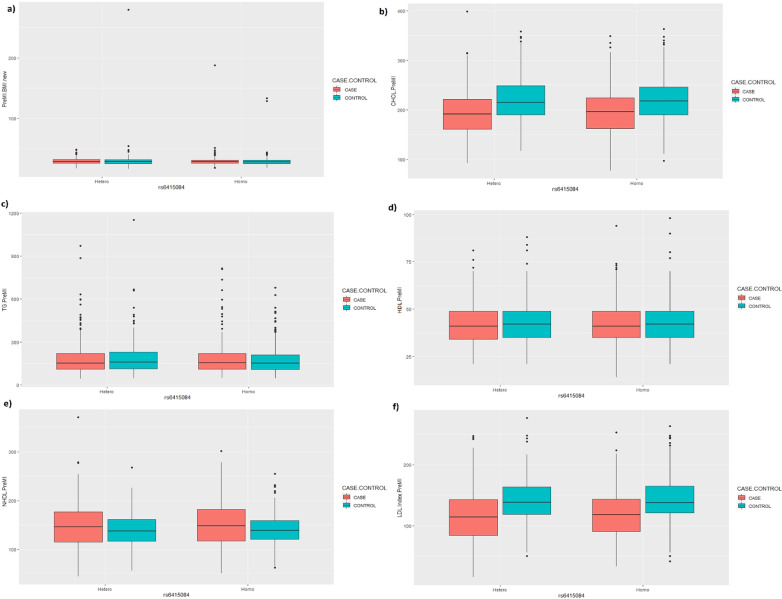
Genetic variation-based studies revealed the significant associations of variants in or near the LPA gene, with CHD risk incidence. Moreover, numerous case–control studies have confirmed that hyper-Lp(a) is a risk factor for premature cardiovascular disease [1, 2, 26]. Another study has shown that overweight and obesity are associated with significantly higher plasma Lp(a) [24]. Figure 3 shows the effect of the polymorphism on the plasma level of other risk factors such as obesity, LDL, HDL, etc. There was no statistically significant association between this polymorphism and these factors. This result proposes an independent relationship between the LP(a) factor and MI. This result is in agreement with the Paré et al. they also concluded that the LPA polymorphism association is independent of established MI risk factors, including diabetes mellitus, smoking, high blood pressure, and apolipoprotein B and A ratio. [27].
Fig. 3.
Univariate analysis results of the association between allele frequencies of the variant rs6415084 in OverDominant genetical model with risk factors, including a BMI, b Cholesterol, c TG, d HDL, e NHDL, and f LDL. There is a longitudinal study that every 3 years a new phase is started, so the above parameters were obtained from PreMI phase of each case
However, previous studies report the association of this polymorphism with the LP(a) level [28–30]; unfortunately, the Lp(a) level was not measured in this study.
These reports show a big controversy between the results of different studies and the importance of investigation of between-population differences in LPA genetic markers; in the same way, our results on the TCGS cohort also show specific association results; however, we found a sex and age dependency in the results.
Perspectives and significance
This study emphasizes the population, age, and sex dependency of the associations, and none of the 17 SNPs showed significant differences between the case and control groups independent of age and sex.
Conclusion
However, older patients are at an increased risk due to the human lifespan's natural limits. Still, our results show the genetic variation on the incidence of MI and the age-of-CHD-Onset. In addition, the population, age, and sex dependency of the association between the LPA variations and heart disease risk confirm that finding the effective variation in precision medicine is required to evaluate these factors in well-established cohorts. Thus, we propose the effects of these variants not only have differences among ethnic groups but also are sex and age-dependent.
Supplementary Information
Additional file 1: Table S1. The results of r2 calculation of Linkagedisequilibrium (LD) for all investigated SNPs in this study.
Acknowledgements
The authors would like to express their gratitude to the participants in the TCGS project. Also, special thanks for the scientific and financial support of the deCODE genetic company (Reykjavik, Iceland). This work was supported by the Research Institute for Endocrine Sciences of Shahid Beheshti University; Foundation under grant No: 240.
Authors' contributions
HL: Conceptualization, Data analysis, Python Programming, prepare the draft LNHB: Data analysis; MA: Statistical analysis; MM-J: Conceptualization, finalize the draft; AZ: Data preparation and Data preprocessing; SM: Data preparation and Data preprocessing; MSD: Conceptualization, finalize the draft. All authors read and approved the final manuscript.
Funding
The present study was funded by the Research Institute for Endocrine Sciences, Shahid Beheshti University of Medical Sciences (Tehran, Iran), and the scientific and financial support of the deCODE genetic company (Reykjavik, Iceland). Iranian molecular medicine network supported the genomic bank.
Availability of data and materials
The data is available as additional files, and if the journal requires additional information, it is possible to send more details.
Declarations
Ethics approval and consent to participate
All procedures were under the ethical standards of the ethics committee on human subject research at Research Institute for Endocrine Sciences, Shahid Beheshti University of Medical Sciences (code of“IR.SBMU.ENDOCRINE.REC.1395.362”) and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Consent for publication
As the corresponding author, I confirm that the manuscript has been read and approved for submission by all the named authors. We declare that this manuscript is original, has not been published before, and is not currently being considered for publication elsewhere.
Competing interests
We know of no conflicts of interest associated with this publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bermúdez V, Arráiz N, Aparicio D, Rojas E, Gotera D, Guerra X, et al. Lipoprotein(a): from molecules to therapeutics. Am J Ther. 2010 doi: 10.1097/MJT.0b013e3181e00bf1. [DOI] [PubMed] [Google Scholar]
- 2.Clarke R, Peden JF, Hopewell JC, Kyriakou T, Goel A, Heath SC, et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 3.Song ZK, Di WuH, Cao HY, Qin L. The association between the LPA gene polymorphism and coronary artery disease in chinese han population. Biomed Res Int. 2014 doi: 10.1155/2014/370670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei WQ, Li X, Feng Q, Kubo M, Kullo IJ, Peissig PL, et al. LPA variants are associated with residual cardiovascular risk in patients receiving statins. Circulation. 2018;138:1839–1849. doi: 10.1161/CIRCULATIONAHA.117.031356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song ZK, Cao HY, Di WH, Zhou LT, Qin L. LPA gene polymorphisms and gene expression associated with coronary artery disease. Biomed Res Int. 2017 doi: 10.1155/2017/4138376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paquette M, Bernard S, Thanassoulis G, Baass A. LPA genotype is associated with premature cardiovascular disease in familial hypercholesterolemia. J Clin Lipidol. 2019;13:627–633.e1. doi: 10.1016/j.jacl.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Page MM, Ellis KL, Pang J, Chan DC, Hooper AJ, Bell DA, et al. Coronary artery disease and the risk-associated LPA variants, rs3798220 and rs10455872, in patients with suspected familial hypercholesterolaemia. Clin Chim Acta. 2020;510:211–215. doi: 10.1016/j.cca.2020.07.029. [DOI] [PubMed] [Google Scholar]
- 8.Lasek-Bal A, Kula D, Urbanek T, Puz P, Szymszal J, Jarzab M, et al. The association of SNPs located in the CDKN2B-AS1 and LPA genes with carotid artery stenosis and atherogenic stroke. Front Neurol. 2019 doi: 10.3389/fneur.2019.01170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santos PCJL, Bueno CT, Lemos PA, Krieger JE, Pereira AC. LPA rs10455872 polymorphism is associated with coronary lesions in Brazilian patients submitted to coronary angiography. Lipids Health Dis. 2014 doi: 10.1186/1476-511X-13-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pechlivanis S, Mahabadi AA, Hoffmann P, Nöthen MM, Broecker-Preuss M, Erbel R, et al. Association between lipoprotein(a) (Lp(a)) levels and Lp(a) genetic variants with coronary artery calcification. BMC Med Genet. 2020 doi: 10.1186/s12881-020-01003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zewinger S, Kleber ME, Tragante V, McCubrey RO, Schmidt AF, Direk K, et al. Relations between lipoprotein(a) concentrations, LPA genetic variants, and the risk of mortality in patients with established coronary heart disease: a molecular and genetic association study. Lancet Diabetes Endocrinol. 2017;5:534–543. doi: 10.1016/S2213-8587(17)30096-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lv X, Zhang Y, Rao S, Liu F, Zuo X, Su D, et al. Lack of association between four SNPs in the SLC22A3-LPAL2-LPA gene cluster and coronary artery disease in a Chinese Han population: a case control study. Lipids Health Dis. 2012;11:128. doi: 10.1186/1476-511X-11-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li ZG, Li G, Zhou YL, Chen ZJ, Yang JQ, Zhang Y, et al. Lack of association between lipoprotein(a) genetic variants and subsequent cardiovascular events in Chinese Han patients with coronary artery disease after percutaneous coronary intervention. Lipids Health Dis. 2013;12:1–7. doi: 10.1186/1476-511X-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dumitrescu L, Glenn K, Brown-Gentry K, Shephard C, Wong M, Rieder MJ, et al. Variation in LPA is associated with Lp(a) levels in three populations from the third National Health and Nutrition Examination Survey. PLoS ONE. 2011;6:16604. doi: 10.1371/journal.pone.0016604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SR, Prasad A, Choi YS, Xing C, Clopton P, Witztum JL, et al. LPA gene, ethnicity, and cardiovascular events. Circulation. 2017;135:251–263. doi: 10.1161/CIRCULATIONAHA.116.024611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daneshpour MS, Fallah M-S, Sedaghati-Khayat B, Guity K, Khalili D, Hedayati M, et al. Rationale and design of a genetic study on cardiometabolic risk factors : protocol for the Tehran Cardiometabolic Genetic Study. JMIR Res Protoc. 2017;6:e28. doi: 10.2196/resprot.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azizi F, Rahmani M, Emami H, Mirmiran P, Hajipour R, Madjid M, et al. Cardiovascular risk factors in an Iranian urban population: Tehran lipid and glucose study (phase 1) Soz Praventivmed. 2002;47:408–426. doi: 10.1007/s000380200008. [DOI] [PubMed] [Google Scholar]
- 18.Azizi F, Ghanbarian A, Momenan AA, Hadaegh F, Mirmiran P, Hedayati M, et al. Prevention of non-communicable disease in a population in nutrition transition: Tehran Lipid and Glucose Study phase II. Trials. 2009;15:1–15. doi: 10.1186/1745-6215-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. doi: 10.1093/clinchem/18.6.499. [DOI] [PubMed] [Google Scholar]
- 20.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hadaegh F, Harati H, Ghanbarian A, Azizi F. Prevalence of coronary heart disease among Tehran adults: Tehran Lipid and Glucose Study ٢٠٠9 ،1 العدد ،عرش اخلامس املجلد ،العاملية الصحة منظمة ،املتوسط لرشق الصحية املجلة. East Mediterr Health J. 2009. 10.26719/2009.15.1.157 [PubMed]
- 22.LDheatmap—Graham & McNeney Labs—Simon Fraser University. http://stat.sfu.ca/statgen/research/ldheatmap.html. Accessed 9 May 2021.
- 23.Bland JM, Altman DG. The odds ratio. BMJ. 2000;320:1468. doi: 10.1136/bmj.320.7247.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michalczyk A, Budkowska M, Dołęgowska B, Chlubek D, Safranow K. Lysophosphatidic acid plasma concentrations in healthy subjects: circadian rhythm and associations with demographic, anthropometric and biochemical parameters. Lipids Health Dis. 2017 doi: 10.1186/s12944-017-0536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sunayama S, Daida H, Mokuno H, Miyano H, Yokoi H, Lee YJ, et al. Lack of increased coronary atherosclerotic risk due to elevated lipoprotein(a) in women ≥55 years of age. Circulation. 1996;94:1263–1268. doi: 10.1161/01.cir.94.6.1263. [DOI] [PubMed] [Google Scholar]
- 26.Schreiner PJ, Heiss G, Tyroler HA, Morrisett JD, Davis CE, Smith R. Race and gender differences in the association of Lp(a) with carotid artery wall thickness: The Atherosclerosis Risk in Communities (ARIC) Study. Arterioscler Thromb Vasc Biol. 1996;16:471–478. doi: 10.1161/01.ATV.16.3.471. [DOI] [PubMed] [Google Scholar]
- 27.Paré G, Çaku A, McQueen M, Anand SS, Enas E, Clarke R, et al. Lipoprotein(a) levels and the risk of myocardial infarction among 7 ethnic groups. Circulation. 2019;139:1472–1482. doi: 10.1161/CIRCULATIONAHA.118.034311. [DOI] [PubMed] [Google Scholar]
- 28.Dong H, Cong H, Wang J, Jiang Y, Liu C, Zhang Y, et al. Correlations between lipoprotein(a) gene polymorphisms and calcific aortic valve disease and coronary heart disease in Han Chinese. J Int Med Res. 2020 doi: 10.1177/0300060520965353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanktree MB, Anand SS, Yusuf S, Hegele RA. Comprehensive analysis of genomic variation in the LPA locus and its relationship to plasma lipoprotein(a) in South Asians, Chinese, and European Caucasians. Circ Cardiovasc Genet. 2010;3:39–46. doi: 10.1161/CIRCGENETICS.109.907642. [DOI] [PubMed] [Google Scholar]
- 30.Deo RC, Wilson JG, Xing C, Lawson K, Linda Kao WH, Reich D, et al. Single-nucleotide polymorphisms in LPA explain most of the ancestry-specific variation in Lp(a) levels in African Americans. PLoS ONE. 2011;6:14581. doi: 10.1371/journal.pone.0014581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The results of r2 calculation of Linkagedisequilibrium (LD) for all investigated SNPs in this study.
Data Availability Statement
The data is available as additional files, and if the journal requires additional information, it is possible to send more details.