Abstract
Motivation
Recent advancements in fluorescence in situ hybridization (FISH) techniques enable them to concurrently obtain information on the location and gene expression of single cells. A key question in the initial analysis of such spatial transcriptomics data is the assignment of cell types. To date, most studies used methods that only rely on the expression levels of the genes in each cell for such assignments. To fully utilize the data and to improve the ability to identify novel sub-types, we developed a new method, FICT, which combines both expression and neighborhood information when assigning cell types.
Results
FICT optimizes a probabilistic function that we formalize and for which we provide learning and inference algorithms. We used FICT to analyze both simulated and several real spatial transcriptomics data. As we show, FICT can accurately identify cell types and sub-types, improving on expression only methods and other methods proposed for clustering spatial transcriptomics data. Some of the spatial sub-types identified by FICT provide novel hypotheses about the new functions for excitatory and inhibitory neurons.
Availability and implementation
FICT is available at: https://github.com/haotianteng/FICT.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
A number of different technologies have been recently developed for spatial transcriptomics. In contrast to single-cell RNA-Seq most spatial transcriptomics platforms rely on image analysis by extending Fluorescence in situ hybridization (FISH) methods. This enables the quantification of expression levels for several genes at a single-cell resolution while still recording the location of each of the cells in the sample. Examples of platforms for spatial transcriptomics include MERFISH (Chen et al., 2015; Moffitt et al., 2016; Moffitt and Zhuang, 2016), seqFISH (Eng et al., 2017; Lubeck et al., 2014), seqFISH+ (Eng et al., 2019), osmFISH (Codeluppi et al., 2018) and the 3D transcriptomics record (STARmap) (Wang et al., 2018). Spatial transcriptomics techniques have now been applied to study several different organs and tissues including lung (Schiller et al., 2019), kidney (Park et al., 2019) and brain (Codeluppi et al., 2018; Eng et al., 2017, 2019; Moffitt et al., 2018; Wang et al., 2018). These studies have led to new insights about the set of cell types in these regions, their location and their interactions (Li et al., 2020; Partel and Wählby, 2021; Yuan and Bar-Joseph, 2020).
A key question in the analysis of single-cell expression data (both for scRNA-Seq and for spatial transcriptomics) is the assignment of cell types. This is often the essential task performed in any analysis of such data and downstream analysis often relies on these assignments (for example, when studying cell–cell interactions (Arnol et al., 2019; Yuan and Bar-Joseph, 2020)). Several packages have been developed to aid in such clustering for single-cell expression data (Abdelaal et al., 2019). These methods often start by clustering cells (usually in low-dimensional space). Next, clusters are assigned to known or new cell types based on the expression of a subset of marker genes. Most spatial transcriptomics studies have also relied on similar methods for cell-type assignment. For example, in the osmFISH paper, hierarchical clustering of the gene expression profiles is used to assign cell types (Codeluppi et al., 2018). For the MERFISH data, cell-type assignment is performed by Louvain community detection applied to a neighborhood graph which is constructed using low-dimension representation of gene expression profiles (Pandey et al., 2018; Shekhar et al., 2016).
While using gene expression levels often leads to successful assignments, relying on scRNA-Seq cell assignment methods, for example the Seurat (Stuart et al., 2019) or other clustering methods, for spatial transcriptomics may not fully utilize the available location information. Specifically, the set of neighboring cells which is known in spatial transcriptomics studies may provide valuable information about the likely cell type of a specific cell. In many cases, specific cell types are known to reside together (Xia et al., 2019) or next to other types of cells (Stoltzfus et al., 2020). Knowledge of the cell types of neighboring cells may thus provide information on the correct assignment of the cell itself. In other cases, such knowledge can lead to the identification of new cell types based on their neighborhood profiles. Recently a method termed smfishHmrf was developed to utilize spatial information when assigning cell types (Zhu et al., 2018). smfishHmrf starts with an initial cell-type assignment using a support vector machine classifier, which is trained using annotated expression data. Next, some assignments are updated based on a neighborhood affinity score which takes into account the fraction of cells assigned to the same cluster. While smfishHmrf utilizes some the spatial information, it only assumes that cells of the same type reside in close proximity and does not look at the overall distribution of cell types in the neighborhood of each cell. Thus, important information about the neighborhood of the cell may not be fully utilized which can lead to decrease in assignment accuracy.
To enable the use of both expression and spatial information for cell-type assignment, we developed FICT (FISH Iterative Cell-Type assignment). FICT maximizes a joint probabilistic likelihood function that takes into account both the expression of the genes in each cell and the joint multi-variate spatial distribution of cell types. We discuss how to formulate the likelihood function and present a method for learning and inference in this model.
We applied FICT to both simulated and real spatial transcriptomics datasets. As we show using the simulation data FICT can correctly determine both expression and parameters that provide information on the distribution of neighboring cell types for each cell for different cell types, improving on generative and discriminative methods that rely only on expression levels and on methods that do not take into account the complete neighborhood of each cell. For the real data, we show that the models learned by FICT for different animals for the same tissue are in good agreement, that it can indeed use the spatial information to correct errors resulting from noise in the expression values and that it can be used to identify spatially different cell sub-types even when their expression profiles are similar.
2 Materials and methods
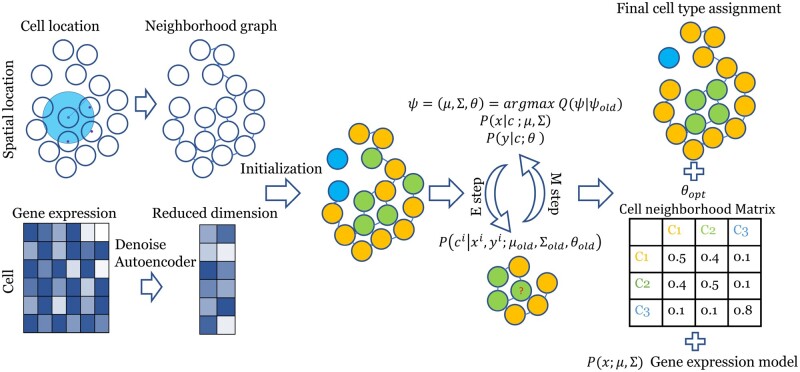
Our goal is to cluster spatial transcriptomics data using both gene expression levels and cell location. A generative mixture model is defined first: each cell is assigned a cell type given its neighborhood, and then the dimension-reduced representation of gene expression levels are drawn from cell-type specific distribution. We next learn the parameters of this generative model by maximizing the joint likelihood of gene expression and cell location (Fig. 1). The cell type is then inferred by the posterior distribution of this generative model given the gene expression level and cell location.
Fig. 1.
FICT pipeline. A reduced dimension expression profile is generated using a Denoising Autoencoder (Vincent et al., 2008), and an undirected graph is constructed according to the spatial locations information. Cells are initially clustered using an expression only GMM. Next, the model is iteratively optimized using an EM algorithm to improve the joint likelihood of the expression and neighborhood models given both the gene expression representation and the spatial graph. The final output is an assignment of cells to clusters, a Gaussian gene expression model and a Multinomial neighborhood model for each class
2.1 A generative model for spatial transcriptomics data
We assume an undirected, weighted graph G representing cell neighborhoods. Each node in G is a cell. We assume a total of M cell types in the dataset. We denote by Z the cell type assignments for nodes in G where zi is the cell type of cell i and denote by the gene expression matrix. Here, xi is the gene expression levels vector for cell i and X is the gene expression matrix for the expression of all cells. Finally, we define the neighbors of cell i in G using . Neighborhood is either defined using the k-nearest neighbors (we used k = 10 in this article) or a cutoff on the distance between i and other nodes in G (a cutoff on the edge weight). Using these definitions, we assume the following generative model for a single-cell transcriptomics dataset.: (i) First, a cell type is selected according to , in which is the conditional distribution for the assignment of cell i given its neighborhood capturing the relationship between neighboring cells in G. (ii) Next, expression levels X are generated according to a cell type specific probability distribution .
Given this model the likelihood of a dataset with a set of gene expression levels X and cell locations (G) is:
| (1) |
We use a multinomial distribution to model the relationship with neighborhood cells and so the product of the conditional probability can be written as:
| (2) |
where yi is a vector summarizing the cell-type assignments for neighbors of i. Specifically, yi is of dimensions M (number of cell types) and each entry j demotes the number of neighbors of cell i assigned to cell type j. Combined, the overall likelihood function is:
| (3) |
where X is the dimension-reduced gene expression matrix and Y is the neighborhood cell type count matrix for each cell, m denotes the mth cell type, we also change the order of product and sum as y is now treated as a property of the cells. We assume that follows a Gaussian distribution and follows a multinomial distribution.
2.2 Inferring cell types (E-step)
We use an Expectation Maximization (EM) approach to learn the parameters of the model. EM iterates between the expectation (E) and maximization (M) steps. Given the generative model, to infer cell types, we need to calculate the posterior probability . However, computing these assignments is challenging since changing the assignment of a specific cell type (i.e. changes to Z’) also change the neighborhood count Y for other cells. Thus, we perform an iterative procedure as follows: In the first phase, Y is treated as a fixed vector for each cell, and is used to calculate the posterior distribution of cell i given the gene expression matrix xi and current neighborhood count yi by setting:
| (4) |
In which is a multi-variate Gaussian distribution with mean and covariance matrix , and is a Multinomial distribution with as the frequency parameter, and we use to denote all the model parameters. We next use the posterior distribution calculations to update cell-type assignments for a subset of the cells. Specifically, we randomly select a set of non-adjacent cells in the adjacency graph G and update their types by the posterior probability. Next, the neighborhood count matrix for all cells, Y, is updated, and is used in the next iteration. We continue with this iterative process until convergence. This method extends the well-known Iterative Condition Modes (ICM) update method (Besag, 1986) by updating multiple cells in each iteration instead of a single one. However, since we only update non-adjacent cells, those updated cells still have the same neighborhood after each round of updates guaranteeing convergence due to the monotonical increase in overall likelihood.
2.3 Learning model parameters (M-step)
For M-step, we have:
| (5) |
When conditioning on the cell type, the values observed for the gene expression xi and neighborhood for a cell become independent. Thus, we can write:
| (6) |
| (7) |
So as mentioned earlier (Section 2.2), the posterior distribution is calculated using an alternated ICM algorithm, in which follows a multivariate Gaussian distribution , and the neighborhood vector for each cell follows a Multi-Nominal distribution . We set , where M is the number of cell types, k is the number of neighborhood cells, is the neighborhood frequency of cell type j given the current cell type i, and is row-wise normalized so that , where is the mth row of . is the prior distribution for cell types.
With , then by maximizing the given Q function, we can obtain the parameters:
| (8) |
The above likelihood function assumes equal weight for each term in the two types of data (expression and neighborhood). However, there are often much more genes than cell types which can lead to over reliance on the expression data. We use two ways to address this problem, first our model is using the dimensional-reduced gene expression as input, instead of the raw expression profile. But the dimension of this input can still be high, e.g. 20, compared with the typical cell type number to be clustered, for example 7, thus then we include a weight term that balances the contribution of the gene and spatial components, named power factor (see Supplementary Section SA1.1). And also during EM training, the neighborhood count is calculated in term of the assigned probability (a soft update), while usual multinomial distribution is defined in , so we expand the scope of the multinomial distribution to to address this. See Supplementary Appendix SA1.2 for details.
2.4 Dimensionality reduction using denoising autoencoder
A dimension-reduced representation of the original gene expression data is used as the input to our model. While the original gene expression data usually does not follow a Gaussian distribution, using a denoising autoencoder, we can transform the data to better fit such model (Vincent et al., 2008). We use a single-layer linear neural network for the auto-encoder though it is possible to adapt the method to use multi-layered networks if the outcome does not fit the required Gaussian distribution. We note that when comparing FICT to the expression only GMM method, we use the same reduced dimension data as input to both. Thus, the only difference between the GMM model and FICT is the use of the spatial information.
2.5 Generating simulated data for testing the method
We tested our method on both real and simulated data. While it is not trivial to simulate data for these experiments, simulation data provide an opportunity to test methods against ground truth which is hard to do with real data.
To perform simulations, we first selected the location of 2000 cells from one of the MERFISH datasets (Supplementary Fig. SA4). We used the cell-type assignments as in the original paper and three other cell groupings (shown in Results, Fig. 2) to assign cell type to each cell. We next generated expression distributions for each cell type. For this, we simulate 1000 genes. Of these genes, 100 are cell type specific (i.e. cell-type-specific mean and variance) and the others have the same distribution in all cell types. Finally, we sample for each cell, a 1000 gene expression profile from the distribution parameters for the cell type to which the cell is assigned to. We also performed a second simulation in which we set both the expression and location of the cells to test the robustness of the method to other spatial cell-type neighborhoods not seen in the MERFISH data. For these, we first generate a neighborhood graph, then generate a cell-type assignment on the neighborhood graph which gives the desired neighborhood frequency, and finally we sample expression data for each cell based on its type. See Supplementary Methods SA1.4 for detail.
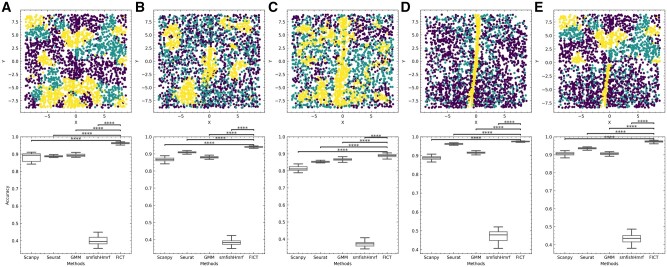
Fig. 2.
Evaluation using simulated data. Top: Simulated ground truth cell-type assignments. Cells locations are from the MERFISH dataset (see Supplementary Fig. SA4 for selected cells). Four neighborhood frequency configurations were simulated: (A) Addictive configuration where cells prefer to aggregate with cells from same type. (B) Exclusive configuration where type 1 and type 2 cells are mixed (green and purple cells) while type 3 cells (yellow cells) cluster together. (C) Consecutive configuration where, type 1 cells surround type 2 cells but not type 3 cells. (D) Cell-type assignments from the MERFISH paper (yellow—Ependymal cells, green—Excitatory cells and purple—inhibitory cells). (E) A mixture model where neighborhood distribution for each cell type is a mixture of the distributions in A and D. Bottom: performance of the five methods we tested on simulated datasets. Accuracy for each method is averaged from 50 random expression assignment (Section 2). P value is calculated using paired samples t-test. ****P<0.0001 (Color version of this figure is available at Bioinformatics online.)
2.6 Software used for comparison to other methods
We used Seurat 4.0.3, Scanpy 1.7.2 and smfishHmrf 0.1 to perform the comparison. Seurat and Scanpy use the Leiden clustering and we used the recommended value for the resolution number (0.9 for coarse cluster assignment and 1.1 for sub cluster analysis).
3 Results
We developed a joint expression and location clustering method to infer cell types in spatial transcriptomics studies. To test the method, we used both simulated and real single-cell spatial transcriptomics data.
3.1 Evaluation using simulated data
While a number of spatial transcriptomics datasets exist, we do not have ground truth information about cell types in these studies. Thus, we first tested our method using simulated data where we can assign both expression and cell type and test if the method can correctly recover the cell types. As noted in Section 2, generating simulated data for such analysis is not trivial since the data needs to satisfy both expression and location constraints. To enable a realistic setting for simulation analysis, we used the spatial information from a real dataset (subset of the MERFISH dataset (Supplementary Fig. SA4). (See Section 2 for details about the simulation setup.) We used the simulated data to test FICT and to compare it with four prior generative and discriminative methods that have been previously used to assign cell types in spatial transcriptomics data. Three of these [ GMM (Tian et al., 2019; Xie et al., 2016), scanpy (Traag et al., 2019; Wolf et al., 2018) and Seurat (Blondel et al., 2008; Butler et al., 2018; Stuart et al., 2019)] only use expression data for clustering while the fourth, smfishHmrf combines gene expression data with cell location and neighborhood information. However, unlike FICT smfishHmrf only considers neighboring cells of the same type (similar to only manually setting the diagonal values in the FICT cell neighborhood matrix and ignoring the off diagonal elements).
In addition to using the cell-type assignments from the original paper, we also simulated four other cell-type assignment settings. Results are presented in Figure 2. As can be seen, for all simulation settings, FICT is the best-performing method followed by Seurat. FICT obtains almost perfect accuracy on all settings, significantly improving upon Seurat and all other methods, we compared with (P < 0.0001 using paired samples t-test) A1. Cluster assignment examples for all methods can be found in Supplementary Figure SA5.
We also compared FICT and the other methods using simulated location and expression data (Section 2). Again, FICT significantly outperformed all other methods (Supplementary Fig. SA1 and Supplementary Table SA2). We also tested the robustness of FICT and determined that it was robust to random initialization and to a wide range of values for determining the set of neighbors for each cell (Supplementary Figs SA15 and SA14).
3.2 Performance on the MERFISH dataset
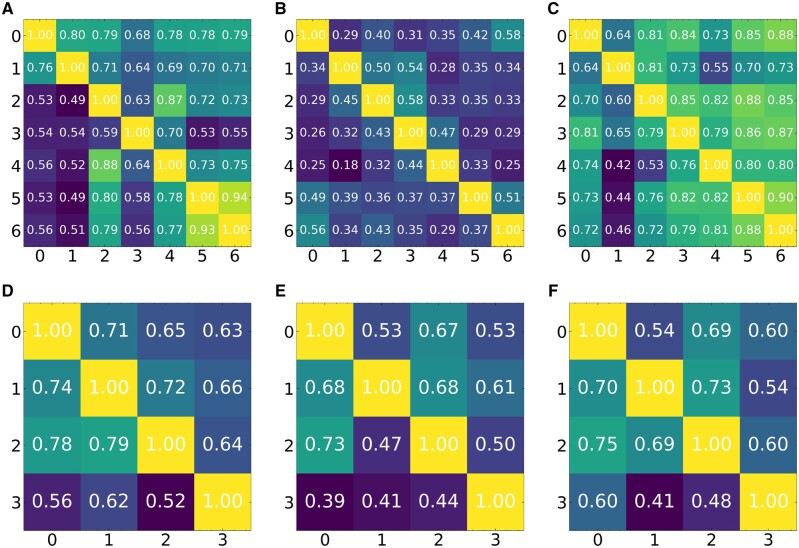
We next tested FICT using real single-cell spatial transcriptomics data. We first focused on mouse hypothalamus data generated by the multiplexed error-robust fluorescence in situ hybridization (MERFISH) method (Moffitt et al., 2018). The MERFISH data profiles the expression of 258 genes in 480 000 cells from 11 animals (4 females and 7 males). Since there is no ground truth for this data, we used a different approach to compare the different clustering methods. For all gender pairs (i.e. 21 male pairs and 6 female pairs), we performed the following analysis. Let A and B be a pair of animals from the same gender. We first train FICT on A and use the parameters learned for the model trained on A to assign cells in B. We next learn a FICT model for B. We then compare the Adjusted Rand Index (ARI) of the clustering results for the two animals. Higher ARIs mean that the results are more consistent between animals indicating better fit to the underlying biology. Note that, this process is not symmetric and so results for training on A and testing on B would be different from those trained on B and tested on A.
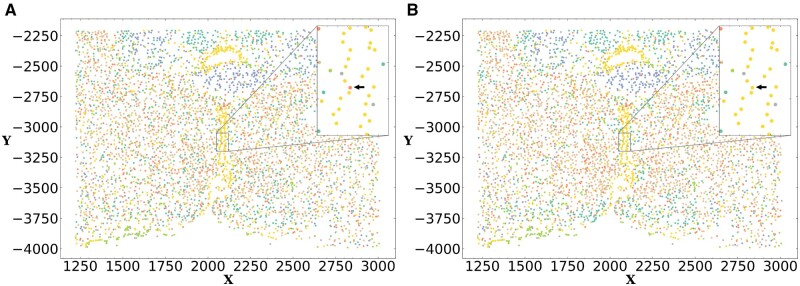
Results for this comparison are presented in Figure 3 for both female and male animals. Note that, since both Seurat and scanpy are not generative methods the models they learn on one dataset cannot be directly applied to another. Thus, for the real data, we compared FICT with smfishHmrf and GMM. Results show that for 32 of the 54 pairs (59%) FICT is more consistent than GMM. The result for the larger dataset of male pairs is (29/42, 69%). The improvement upon smfishHmrf is even larger than that and FICT is more consistent in 52 of the 54 pairs (96.3%). We also tried to compare Seurat and scanpy by learning a classifier using the clustering of one animal and comparing the assignments of the learned classifier to the unsupervised clustering using Seurat and scanpy on another animal. As expected, results indicate that performance of such supervised/unsupervised comparisons is inferior to the results of the generative models as we show in Supplementary Figure SA12. We note that based on prior studies that indicated that gene expression and cell distribution differ based on gender (Dewing et al., 2003; McCarthy and Arnold, 2011), the above analysis was performed by only testing models learned from male animals on male animals and from female animals on female animals. An example of the difference in assignments between expression only GMM clustering and FICT is presented in Figure 4. As can be seen, the yellow cells (Ependymal cells) are spatially clustered in the center of the hypothalamus tile profiled. However, due to small variations in gene expression, GMM assigns some cells in that cluster as OD Immature cells. In contrast FICT is able to correctly assign these cells as shown in the inset.
Fig. 3.
Mean Adjusted Rand index (ARI) based on cross-validation analysis of the MERFISH dataset. Results presented for expression only GMM, smfishHmrf and FICT. Each entry (i, j) in the matrix represents the ARI of the two cluster assignments (one learned on animal A and applied to animal B and the other learned directly on B). (A–C) Results for the 7 Male animals (A) GMM, (B) smfishHmrf and (C) FICT. (D–F) Results for the 4 Females (D) GMM, (E) smfishHmrf and (F) FICT. The x and y axes are the index of the dataset being cross validated on
Fig. 4.
FICT can correct expression noise. Cell-type assignments using expression only GMM (left) and FICT (right). Using the spatial information FICT correctly assigns Ependymal cells along the periventricular hypothalamic nucleus. In contrast, the GMM method mistakenly classified the cell as OD Immature Cell
3.2.1 Sub-type clustering
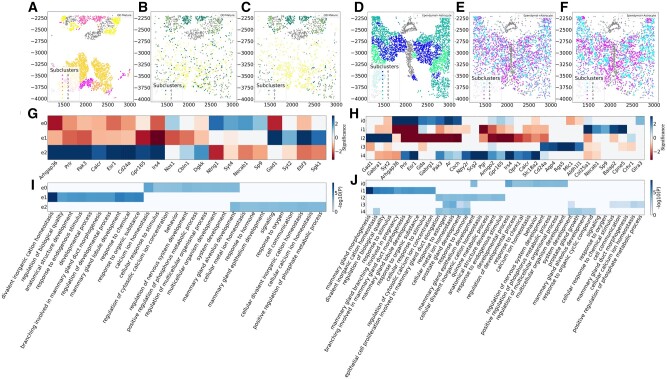
An important question in the analysis of brain single-cell data is the identification of new sub-types of various neuronal cells (Lake et al., 2016). We thus examined the assignments to see if FICT can identify new subtypes of neurons. For this, we focused on the subset of excitatory neurons identified in the MERFISH dataset. FICT identified three sub-types of cells that were all determined to be excitatory in the original analysis but displayed different spatial patterns (Fig. 5). To determine if the three sub-clusters are indeed different, we performed differential expression (DE) analysis for each of the sub-clusters. While, as expected, their overall expression profiles are similar (leading to their similar assignment by the expression only method), we were able to identify a number of distinct genes for each of these sub-types using MAST (Finak et al., 2015). We next performed GO enrichment analysis (Ashburner et al., 2000; Mi et al., 2017; The Gene Ontology Consortium, 2019) on the significant DE genes in each sub-clusters. Results are presented in Figure 5. As can be seen, some unique functional terms are associated with each of the three sub-clusters. For example, the first sub-cluster (e0) seems to be mainly related to response to chemicals. The second (e1) seems to be related to signaling and regulation of calcium homeostasis while the third (e2) is linked to responses to activity changes and behavior. Thus, while all share similar expression profiles and act as excitatory neurons, each of the sub-clusters may have a further specific function as predicted by the spatial clustering. We performed similar sub-clustering analysis using the other methods we compared with. Results are presented in Supplementary Figures SA16–SA20 and indicate that FICT finds both, relevant GO terms such as ‘behavior’ that are not identified by other methods for this data and more significant enrichment for GO categories related to cell and synapse signaling. We performed similar sub-clustering analysis for inhibitory neurons and obtained similar results both in terms of the more coherent placing of cells from different sub-types and in terms of the unique genes and functions assigned to each of the sub-types identified by FICT (Fig. 5C and D).
Fig. 5.
Cell sub-type clustering on MERFISH data from animal 1. We used smfishHmrf (A and D), expression only GMM (B and E) and FICT (C and F) to sub-cluster excitatory neurons cells (A, B and C) and inhibitory neuron cells (D, E and F). As can be seen, for both types of neurons FICT assignments are better spatially conserved creating a central core for sub-cluster 2 surrounded by cells assigned to sub-cluster 0. In contrast, the expression only assignment mixes cells from different sub-types much more. smfishHmrf with Potts model only assigns affinity score between the same cell types making it harder to infer more complex structures of synergistic activity. (E) DE genes for the three FICT sub-clusters from the excitatory neurons and (F) inhibitory neurons. As can be seen, even though the sub-clusters are overall similar in terms of their expression profiles, some genes can be identified for each of the sub-clusters. (G) GO enrichment analysis identifies unique functions for each of the sub-clusters on excitatory neurons and (H) inhibitory neurons. Significance of the differential expressed genes is measured by the log of gene enrichment fold change
3.3 Performance on osmFISH and seqFISH
To demonstrate the generality of our method, we further tested it on two other datasets from two additional spatial transcriptomics platforms: osmFISH (Codeluppi et al., 2018) and seqFISH (Zhu et al., 2018). The osmFISH dataset profiled 6470 cells in the mouse somatosensory cortex. The seqFISH dataset profiled 1597 cells in the mouse visual cortex. Since both datasets only profiled a single animal we performed the cross validation by manually splitting each dataset into 4 smaller regions with approximately the same number of cells. Results for these analyses are presented in Figure 6. As can be seen, FICT was able to successfully cluster cells not only just based on type but also based on their layer, whereas clustering using only the expression data, as was performed in the original study, cannot separate layers as well. We also performed cross-validation analysis, as we did for the MERFISH data. Given the small number of cells for each dataset, we see a drop in performance for all generative model methods. As the figure shows, smfishHmrf was unable to identify more than a single cell type for many of the cross-validation runs resulting in errors. As for GMM and FICT while both were able to successfully assign cells in the cross validation runs for the osmFISH and seqFISH datasets, results were not as good as the MERFISH results presented earlier. Still, even though FICT fits more parameters than the expression only model we observe comparable performance on these smaller datasets suggesting that there is no downside to using the joint expression-spatial assignment A8.
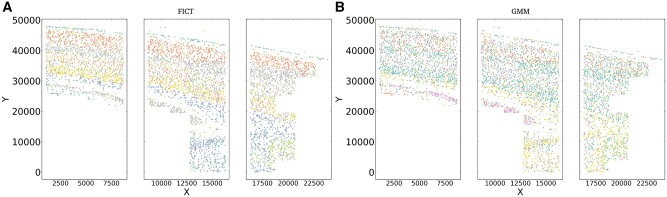
Fig. 6.
Cluster assignment scatter plot for osmFISH dataset. (A) Clusters generated by FICT and (B) clusters based on using expression data only as was done in the original paper. As can be seen, FICT correctly distinguishes between neurons in different layers of the brain, whereas expression only clustering mixes cells from different brain layers
4 Discussion
Spatial transcriptomics has emerged as a valuable tool for the analysis of single-cell expression data. Similar to scRNA-Seq, this technology provides information on the expression of genes at the single-cell resolution. In addition, it also provides information on the location of each of the cells and their spatial relationships which can help understand cell–cell interactions, the organization of cells in specific regions and tissues and how changes in such organization impact development and disease.
A key question in spatial transcriptomics analysis is the assignment of types to the cells profiled. To date, most studies relied on the profiled expression levels for such assignment using tools and techniques originally developed for the analysis of scRNA-Seq data. While such methods work well, they do not fully utilize the information obtained in spatial transcriptomics studies. Specifically, information about the location of cells and their neighbors is usually not used in such assignments even though in several cases cell types are known to co-locate with other cells from the same or different types. To enable the use of the spatial information in cell assignments, we developed FICT which uses an EM method to learn both expression and spatial distribution models. We presented a likelihood optimization function and learning and inference methods for FICT and used it to assign cell types in both simulated and real datasets.
As we have shown, for both simulated and large real datasets FICT improves on both, gene expression only methods and methods that only use part of the spatial information when assigning cell types. Since FICT estimates more parameters than expression only assignment methods its performance suffers when applied to smaller datasets. Still, even for the smallest datasets, we tested on (seqFISH, which profiled only 1500 cells) FICT performance was comparable to expression only methods making it a reasonable alternative for such methods. Since more recent studies often profile more cells, FICT is likely to generalize better to future datasets.
In addition to improved accuracy FICT can also identify cell sub-types that are similar in terms of their expression while differ in their spatial organization. As we have shown, FICT divided the set of excitatory neuron cells into three sub-types based on other cells in their neighborhood. Analysis of DE genes between these spatial clusters identified a number of biological functions that differ between the clusters indicating that each sub-type may indeed serve a different goal as predicted by FICT. The use of spatial information can also improve assignment and analysis for larger regions in the brain. As we have shown, FICT can improve the identification of layer-specific cells in the brain which is useful for both, segmenting various regions based on the cells present and identifying specific markers for sub-populations of cell.
While FICT worked well for most of the datasets we tested on, there are still a number of ways in which it can be improved. We would like to improve its run-time since it currently takes one hour to perform the joint expression and spatial cell-type assignment on a single animal MERFISH dataset (∼100K cells). As we noted, parts of FICT learning resemble HMRFs and so methods used to speed up HMRF inference including belief propagation can be incorporated to further improve in FICT (Yedidia et al. 2001). In addition, we would like to extend FICT so that it could also be used for cell-type assignment of data generated using the Visium (Mantri et al. 2021) or nanostring (Lewis et al. 2021) platforms. Unlike the data analyzed in this article, these platforms do not generate single-cell level data and so to extend FICT for them we would need to combine deconvolution with clustering in an iterative manner.
FICT is implemented in Python and both data and an open source version of the software are available in https://github.com/haotianteng/FICT. Given the results presented in this article, we hope that it can be used to improve the analysis of the increasing number of studies that rely on spatial transcriptomics profiling.
Funding
This work was partially supported by National Institutes of Health [1R01GM122096, OT2OD026682, 1U54AG075931, 1U24CA268108] and a C3.ai DTI Research Award to Z.B.-J.
Conflict of Interest: none declared.
Supplementary Material
Contributor Information
Haotian Teng, Department of Computational Biology, School of Computer Science, Carnegie Mellon University, Pittsburgh, PA 15213, USA.
Ye Yuan, Institute of Image Processing and Pattern Recognition, Shanghai Jiao Tong University, and Key Laboratory of System Control and Information Processing, Ministry of Education of China, Shanghai 200240, China.
Ziv Bar-Joseph, Department of Computational Biology, School of Computer Science, Carnegie Mellon University, Pittsburgh, PA 15213, USA.
References
- Abdelaal T. et al. (2019) A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol., 20, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnol D. et al. (2019) Modeling cell-cell interactions from spatial molecular data with spatial variance component analysis. Cell Rep., 29, 202–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. et al. (2000) Gene ontology: tool for the unification of biology. Nat. Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besag J. (1986) On the statistical analysis of dirty pictures. J. R. Stat. Soc. Ser. B (Methodological), 48, 259–279. [Google Scholar]
- Blondel V.D. et al. (2008) Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp., 2008, P10008. [Google Scholar]
- Butler A. et al. (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol., 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.H. et al. (2015) Spatially resolved, highly multiplexed RNA profiling in single cells. Science, 348, aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codeluppi S. et al. (2018) Spatial organization of the somatosensory cortex revealed by osmfish. Nat. Methods, 15, 932–935. [DOI] [PubMed] [Google Scholar]
- Dewing P. et al. (2003) Sexually dimorphic gene expression in mouse brain precedes gonadal differentiation. Mol. Brain Res., 118, 82–90. [DOI] [PubMed] [Google Scholar]
- Eng C.-H.L. et al. (2017) Profiling the transcriptome with RNA spots. Nat. Methods, 14, 1153–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng C.-H.L. et al. (2019) Transcriptome-scale super-resolved imaging in tissues by RNA seqfish. Nature, 568, 235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G. et al. (2015) Mast: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol., 16, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake B.B. et al. (2016) Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science, 352, 1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S.M. et al. (2021) Spatial omics and multiplexed imaging to explore cancer biology. Nat. Methods, 18, 997–1012. [DOI] [PubMed] [Google Scholar]
- Li D. et al. (2020) Identifying signaling genes in spatial single cell expression data. Bioinformatics, 37, 968–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck E. et al. (2014) Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods, 11, 360–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantri M. et al. (2021) Spatiotemporal single-cell RNA sequencing of developing chicken hearts identifies interplay between cellular differentiation and morphogenesis. Nat. Commun., 12, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy M.M., Arnold A.P. (2011) Reframing sexual differentiation of the brain. Nat. Neurosci., 14, 677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H. et al. (2017) Panther version 11: expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res., 45, D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt J.R., Zhuang X. (2016) RNA imaging with multiplexed error-robust fluorescence in situ hybridization (merfish). Methods Enzymol., 572, 1–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt J.R. et al. (2016) High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc. Natl. Acad. Sci. USA, 113, 11046–11051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt J.R. et al. (2018) Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science, 362, eaau5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S. et al. (2018) Comprehensive identification and spatial mapping of habenular neuronal types using single-cell RNA-seq. Curr. Biol., 28, 1052–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. et al. (2019) Understanding the kidney one cell at a time. Kidney Int., 96, 862–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partel G., Wählby C. (2021) Spage2vec: unsupervised detection of spatial gene expression constellations. FEBS J., 288, 1859. [DOI] [PMC free article] [PubMed]
- Schiller H.B. et al. (2019) The human lung cell atlas: a high-resolution reference map of the human lung in health and disease. Am. J. Respir. Cell Mol. Biol., 61, 31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K. et al. (2016) Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell, 166, 1308–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus C.R. et al. (2020) Cytomap: a spatial analysis toolbox reveals features of myeloid cell organization in lymphoid tissues. Cell Rep., 31, 107523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T. et al. (2019) Comprehensive integration of single-cell data. Cell, 177, 1888–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. (2019) The gene ontology resource: 20 years and still going strong. Nucleic Acids Res., 47, D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T. et al. (2019) Clustering single-cell RNA-seq data with a model-based deep learning approach. Nat. Mach. Intell., 1, 191–198. [Google Scholar]
- Traag V.A. et al. (2019) From louvain to leiden: guaranteeing well-connected communities. Sci. Rep., 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent P. et al. (2008) Extracting and composing robust features with denoising autoencoders. In: Proceedings of the 25th international conference on Machine learning, Helsinki, Finland, pp. 1096–1103.
- Wang X. et al. (2018) Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science, 361, eaat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F.A. et al. (2018) Scanpy: large-scale single-cell gene expression data analysis. Genome Biol., 19, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C. et al. (2019) Spatial transcriptome profiling by merfish reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc. Natl. Acad. Sci. USA, 116, 19490–19499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J. et al. (2016) Unsupervised deep embedding for clustering analysis. In International Conference on Machine Learning, New York City, NY, USA, pp. 478–487.
- Xu C., Su Z. (2015) Identification of cell types from single-cell transcriptomes using a novel clustering method. Bioinformatics, 31, 1974–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedidia J.S. et al. (2001) Generalized belief propagation. In: Advances in Neural Information Processing Systems, Vancouver, Canada, pp. 689–695.
- Yuan Y., Bar-Joseph Z. (2020) GCNG: graph convolutional networks for inferring cell–cell interactions. Genome Biol., 21, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappia L. et al. (2017) Splatter: simulation of single-cell RNA sequencing data. Genome Biol., 18, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q. et al. (2018) Identification of spatially associated subpopulations by combining scrnaseq and sequential fluorescence in situ hybridization data. Nat. Biotechnol., 36, 1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.