Abstract
Motivation
Adverse outcome pathways (AOPs) are a conceptual framework developed to support the use of alternative toxicology approaches in the risk assessment. AOPs are structured linear organizations of existing knowledge illustrating causal pathways from the initial molecular perturbation triggered by various stressors, through key events (KEs) at different levels of biology, to the ultimate health or ecotoxicological adverse outcome.
Results
Artificial intelligence can be used to systematically explore available toxicological data that can be parsed in the scientific literature. Recently, a tool called AOP-helpFinder was developed to identify associations between stressors and KEs supporting thus documentation of AOPs. To facilitate the utilization of this advanced bioinformatics tool by the scientific and the regulatory community, a webserver was created. The proposed AOP-helpFinder webserver uses better performing version of the tool which reduces the need for manual curation of the obtained results. As an example, the server was successfully applied to explore relationships of a set of endocrine disruptors with metabolic-related events. The AOP-helpFinder webserver assists in a rapid evaluation of existing knowledge stored in the PubMed database, a global resource of scientific information, to build AOPs and Adverse Outcome Networks supporting the chemical risk assessment.
Availability and implementation
AOP-helpFinder is available at http://aop-helpfinder.u-paris-sciences.fr/index.php
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Structured organization of toxicological and ecotoxicological data is now feasible using the adverse outcome pathways (AOP) framework (Ankley et al., 2010). An AOP is defined by a linear combination of biological events, started from a molecular initiating event (MIE) triggered by stressors (pollutants, ionizing radiations, nanomaterials or climate stressors) connected through a series of key events (KEs) occurring at various levels of the biological organization, to an adverse outcome (AO). Biological events (MIE, KE and AO) are not linked to a unique AOP, but can be shared, allowing the establishment of Adverse Outcome Network (AON) that reflect better the true complexity of the biology. Combined with new approach methodologies (Parish et al., 2020), AOPs and AONs are extremely useful in establishing integrated approaches to testing and assessment (IATA) for environmental and risk assessment, and they aid to the development of novel nonanimal toxicity testing strategies (Delrue et al., 2016).
With advances in technologies, huge amounts of data have become available, compiled in well-structured toxicological databases (e.g. CTD, CompTox), in AOP-oriented webservers (AOP-wiki, sAOP, AOP4EUpest) and scientific publications (Williams et al., 2017). Innovative data mining tools are needed to identify sparse but complementary data such as Abstract Sifter allowing to have a view of the toxicological information landscape for a set of entities as chemicals (Baker et al., 2017) or ComptoxAI (https://comptox.ai/index.html). Artificial intelligence (AI) technology, that uses natural language processing (NLP), is an interesting way to facilitate the identification of links between relevant information that can be used to build novel AOPs (Song et al., 2020), and identify knowledge gaps and research needs (Zgheib et al., 2021). Several tools use text mining (TM), an AI method to transform unstructured into structured text. For example, Limtox provides a biomedical search for adverse hepatobiliary reactions (Cañada et al., 2017). Recently, the AOP-helpFinder tool, based on TM and graph theory was proposed to identify stressor-KE relationships by examining large collections of scientific abstracts, and was applied to bisphenol A substituents and pesticides (Carvaillo et al., 2019; Jornod et al., 2020; Rugard et al., 2020).
Here, we present the AOP-helpFinder webserver, which uses an updated version of the tool, to provide an easy but effective resource for identifying and compiling existing knowledge from the scientific literature. The main optimized features are (i) the capability to choose to search in full abstracts or without considering the introductory parts, (ii) the possibility to perform a refined search using machine learning and (iii) an automatic update of the PubMed database before each search. A case study with endocrine disruptors (ED) and metabolism-related events is provided, illustrating the capacity of the tool to collate quickly an overview of the existing information.
2 Materials and methods
2.1 The AOP-helpFinder webserver
The proposed webserver is easy to use, and requires only the user email to access the upload page and to receive information when the results are available for download. This simple procedure is in line with digital sobriety that aims to reduce the environmental impact by limiting computing use. Two input files are needed: one with the stressors of interest and the second with biological events (i.e. MIE, KE and/or AO). Before running the tool to identify if knowledge connecting stressors and biological events exists, the user can choose between two options: reduce search and refinement filter (see the following section and Supplementary Material), as well as the output format (date, title, PMID, etc.).
2.2 The AOP-helpFinder tool
To increase the performance of the previously developed version, several methods were tested using a set on ED and biological events related to metabolism, and two were kept through the process (see Supplementary Material, https://github.com/jornod/aop-helpFinder):(i) ‘reduced search’: searches are performed in the full abstracts or without considering the introductory part, which appears to be covered usually by the first 20% of the abstracts. This option allows avoiding too many false positives, as the introduction often reflects a working hypothesis instead of the conclusions of the publication and (ii) ‘refinement filter’: after the preprocessing that uses a stemming process (Carvaillo et al., 2019), the tool can refine the searches by combining a deletion of sentences containing context words with a lemmatization process. Lemmatization is a machine learning method for text normalization used in NLP that considers the context and converts the word to its meaningful base form. This option is very useful when terms have common stems (e.g. tests, testis □ test) leading to incorrect meanings and spelling errors. Further, an automatic daily update of the PubMed database was newly implemented using the NCBI API to screen the full existing knowledge.
The current version of the AI tool mined the PubMed database, that is a global source for scientific literature. Nevertheless, the developed method screens text-based knowledge, and therefore the AOP-helpFinder server could be improved for mining multiple sources (databases, literature), including studies reporting negative findings, to accelerate information gathering when data are limited and present in diverse sources (Carvaillo et al., 2019).
The advantage of the proposed method, is its capacity to be adapted for literature searches in general, independent of AOP development, in order to identify interconnections between the query keywords, as it was successfully done to decipher nonvalidated test methods for ED (Zgheib et al., 2021).
2.3 Case study on endocrine disruptors and metabolism
The AOP-helpFinder webserver was used for a case study aiming at automatically identifying existing relationships and knowledge gaps between 10 ED (Supplementary Table S1) and 294 biological events related to metabolism (Supplementary Table S2). The webserver was launched using ‘reduced search’ (omitting searches in the first 20% of the abstracts) and ‘refinement filter’. Among the 83 970 abstracts retrieved in the PubMed database as of May 10, 2021 related to at least one ED (Supplementary Table S1), a total of 4622 were retained (comentioning ED and event). Among the 294 events, 108 were identified as comentioned with at least one ED (Supplementary Table S2). Figure 1 illustrates the large disparity of knowledge for the 10 selected ED in the area of metabolism (see Supplementary Fig. S1 for all results). For example, cadmium, bisphenol A and di(2-ethylhexyl) phthalate (DEHP) are well studied chemicals as the webserver retrieved scientific articles for almost all biological events of interest. Other chemicals (bisphenol F, bisphenol S, butyl-paraben) appear to be less studied (Supplementary Table S1), and the information were essentially identified for extensively studied biological events such as oxidative stress or obesity.
Fig. 1.
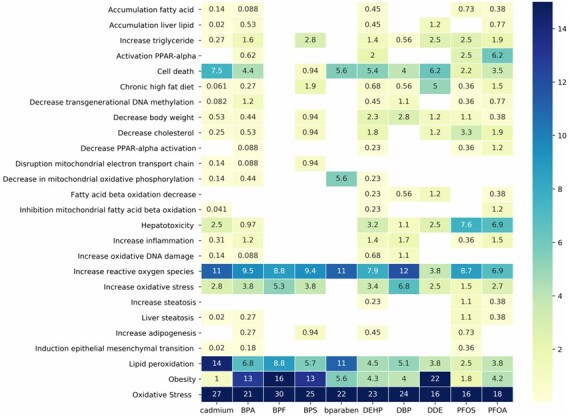
Example of ED comentioned in PubMed scientific abstracts with biological events related to metabolism, identified by the AOP-helpFinder webserver. The numbers correspond to the % of retrieved abstracts mentioning both the stressor (column) and the event (line) among all identified abstracts (the colors are according to the percentage for better visualization). For example, among all identified abstracts that comentioned bisphenol S (BPS) and at least one event from the list, 13% of the abstracts were comentioning BPS (fourth column) and obesity (the second line from the bottom)
3 Conclusion
The AOP-helpFinder webserver uses an automatic AI screening to rapidly retrieve existing knowledge on links between stressors and biological events to build AOPs and AONs. This webserver allows highly effective searches in PubMed as it considerably reduces the time of finding relevant scientific articles. The comprehensive AI-based analyses of existing literature support various needs of the risk assessment such as establishment of causality between chemicals and AOs through AOPs and AONs, identification of gaps or prioritization and design of future experimental and epidemiological studies.
Supplementary Material
Acknowledgements
The authors would like to acknowledge Inserm and the Université de Paris for supporting the work.
Funding
This work was supported by the European Union’s Horizon 2020 Research and Innovation Programme OBERON [https://oberon-4eu.com, Grant 825712] and HBM4EU [https://www.hbm4eu.eu/, Grant 733032].
Conflict of Interest: none declared.
Contributor Information
Florence Jornod, Université de Paris, T3S, Inserm UMR-S1124, Paris F-75006, France.
Thomas Jaylet, Université de Paris, T3S, Inserm UMR-S1124, Paris F-75006, France.
Ludek Blaha, RECETOX, Faculty of Science, Masaryk University, Brno CZ62500, Czech Republic.
Denis Sarigiannis, HERACLES Research Center on the Exposome and Health, Aristotle University of Thessaloniki, Center for Interdiciplinary Research and Innovation, Thessaloniki 57001, Greece.
Luc Tamisier, Université de Paris, SPPIN CNRS UMR 8003,Paris F-75006, France.
Karine Audouze, Université de Paris, T3S, Inserm UMR-S1124, Paris F-75006, France.
References
- Ankley G.T. et al. (2010) Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem., 29, 730–741. [DOI] [PubMed] [Google Scholar]
- Baker N. et al. (2017) Abstract Sifter: a comprehensive front-end system to PubMed. F1000Research, 6, 2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañada A. et al. (2017) LimTox: a web tool for applied text mining of adverse event and toxicity associations of compounds, drugs and genes. Nucleic Acids Res., 45, W484–W489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvaillo J.-C. et al. (2019) Linking bisphenol S to adverse outcome pathways using a combined text mining and systems biology approach. Environ. Health Perspect., 127, 47005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delrue N. et al. (2016) The adverse outcome pathway concept: a basis for developing regulatory decision-making tools. Altern. Lab. Anim., 44, 417–429. [DOI] [PubMed] [Google Scholar]
- Jornod F. et al. (2020) AOP4EUpest: mapping of pesticides in Adverse Outcome Pathways using a text mining tool. Bioinformatics, 36, 4379–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish S.T. et al. (2020) An evaluation framework for new approach methodologies (NAMs) for human health safety assessment. Regul. Toxicol. Pharmacol., 112, 104592. [DOI] [PubMed] [Google Scholar]
- Rugard M. et al. (2020) Deciphering adverse outcome pathway network linked to bisphenol F using text mining and systems toxicology approaches. Toxicol. Sci., 173, 32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. et al. (2020) Upregulation of angiotensin converting enzyme 2 by shear stress reduced inflammation and proliferation in vascular endothelial cells. Biochem. Biophys. Res. Commun., 525, 812–818. [DOI] [PubMed] [Google Scholar]
- Williams A.J. et al. (2017) The CompTox chemistry dashboard: a community data resource for environmental chemistry. J. Cheminform., 9, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgheib E. et al. (2021) Identification of non-validated endocrine disrupting chemical characterization methods by screening of the literature using artificial intelligence and by database exploration. Environ. Int., 154, 106574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


