Abstract
We assessed the reproducibility of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) and protease sequencing using cryopreserved plasma aliquots obtained from 46 heavily treated HIV-1-infected individuals in two laboratories using dideoxynucleotide sequencing. The rates of complete sequence concordance between the two laboratories were 99.1% for the protease sequence and 99.0% for the RT sequence. Approximately 90% of the discordances were partial, defined as one laboratory detecting a mixture and the second laboratory detecting only one of the mixture's components. Only 0.1% of the nucleotides were completely discordant between the two laboratories, and these were significantly more likely to occur in plasma samples with lower plasma HIV-1 RNA levels. Nucleotide mixtures were detected at approximately 1% of the nucleotide positions, and in every case in which one laboratory detected a mixture, the second laboratory either detected the same mixture or detected one of the mixture's components. The high rate of concordance in detecting mixtures and the fact that most discordances between the two laboratories were partial suggest that most discordances were caused by variation in sampling of the HIV-1 quasispecies by PCR rather than by technical errors in the sequencing process itself.
Mutations in the human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) and protease enzymes are responsible for resistance to current antiretroviral drugs. The prognostic value and clinical utility of HIV-1 gene sequencing for resistance testing are supported by both retrospective and prospective clinical trials (1, 4, 7, 12). Within the past 2 years, assays for the sequencing of HIV-1 RT and protease genes have become commercially available, and in early 2000, two expert panels recommended resistance testing to help with the selection of the appropriate antiretroviral drugs in certain clinical situations (2, 14, 20).
The reproducibility of detection of drug resistance mutations by RT and protease sequencing is critical not only because HIV-1 RT and protease sequencing is one of the first applications of gene sequencing for clinical purposes but also because many positions in these two genes have been linked to drug resistance (10). The evaluation of tests used for sequencing of HIV-1 is complicated by the fact that HIV-1 exists in vivo as a quasispecies: a mixture of genetically distinct viral variants that evolve from the initial virus inoculum. Previous studies in which both clonal and population-based sequencing have been done with the same samples have shown that samples with approximately equal mixtures of alleles at a given position lead to double-peaked electropherograms (9, 17, 22, 25; J. Martinez-Picado, M. P. DePasquale, L. Ruiz, V. Miller, A. V. Savara, L. Sutton, B. Clotet, and R. T. D'Aquila, Antivir. Ther. 4(Suppl. 1):50, abstr. 72, 1999) but that the potential for missing mutations is high when one of the alleles is present at a low proportion (22, 25). Suboptimal reproducibility therefore can result from either technical artifacts or variability in the detection of minor viral variants.
To determine the reproducibility of sequencing for assessment of HIV-1 drug resistance in clinical samples, we compared the RT and protease sequencing results of two different laboratories testing replicate cryopreserved plasma aliquots obtained from 46 heavily treated HIV-1-infected individuals. We characterized each of the discordant positions as partial or complete and synonymous or nonsynonymous. Additionally, we correlated the numbers and types of discordances with the HIV-1 RNA level in the samples.
MATERIALS AND METHODS
Patients and samples.
The study population consisted of 46 patients described in a previously published report (29). Most patients had been heavily treated, receiving a median of two protease inhibitors and four RT inhibitors. Replicate plasma samples for the present study were available for 46 of the 54 patients in the original study. HIV-1 RT and protease sequences from this group of patients are in GenBank under the accession numbers AF085086 to AF085139 and AF088078 to AF088131. The patients in this study have previously been referred to as PCC1 to PCC3, PCC5 to PCC7, PCC9, PCC10, PCC12 to PCC18, PCC20 to PCC27, PCC29 to PCC32, PCC35, and PCC37 to PCC54 (29).
Laboratory methods.
At laboratory A (Stanford University Hospital Diagnostic Virology Laboratory, Stanford, Calif.), samples were initially obtained from patients between November 1996 and March 1998 for plasma HIV-1 RNA testing. Excess plasma from these samples was aliquoted and stored at −70°C. Between December 1997 and March 1998, laboratory A thawed plasma samples and sequenced RT and protease for the study described above (29). In May 1999, the 46 available replicate aliquots were sent to laboratory B (VIRCO Belgium, Mechelen, Belgium), which was blinded to the original sequencing results.
Both laboratories sequenced the complete protease gene. Laboratory A sequenced RT codons 1 to 250. Laboratory B sequenced RT codons 1 to 400. Each laboratory performed plasma HIV-1 RNA extraction, reverse transcription to create HIV-1 cDNA, nested PCR, and direct PCR (population-based) dideoxynucleotide sequencing. In both laboratories, overlapping sequencing reactions were performed in both directions and were resolved electrophoretically on an ABI 377 sequencer (Applied Biosystems Incorporated [ABI], Foster City, Calif.).
In laboratory A, RNA was extracted from 0.2 ml of plasma with the guanidine thiocyanate lysis reagent in the AMPLICOR HIV Monitor test kit (Roche Diagnostic Systems, Branchburg, N.J.). Reverse-strand cDNA was generated from viral RNA, and first-round PCR was performed by using Superscript One-Step RT-PCR (Life Technologies, Rockville, Md.). A 1.3-kb product encompassing the protease gene and the first 300 residues of the RT gene was then amplified with nested PCR primers. Direct PCR (population-based) cycle sequencing was performed with AmpliTaq DNA FS polymerase and dRhodamine terminators (ABI). The primers used for amplification and sequencing are described in a previous publication (25). Electropherograms were created with Sequence Analysis, version 3.0, software (ABI), and the sequences were assembled with the manufacturer's FACTURA and AUTOASSEMBLER sequence analysis software (ABI). The FACTURA program flagged heterozygous positions (positions with signals or peaks indicative of at least two nucleic acids, with the minor peak being ≥30% of the major peak).
In laboratory B, RNA was extracted from 0.2 ml of plasma with the QIAamp Viral RNA Extraction kit (Qiagen, Hilden Germany), and protease and RT cDNAs were created with Expanded Reverse Transcriptase (Boehringer, Mannheim, Germany). A 2.2-kb PCR product was amplified with nested PCR primers, and direct PCR (population-based) cycle sequencing was performed with AmpliTaq DNA FS polymerase and BigDye (ABI). The primers used for amplification and sequencing are described in a previous publication (13). Electropherograms were created with Sequence Analysis, version 3.0, software (ABI), and sequences were assembled with the manufacturer's FACTURA and AUTOASSEMBLER sequence analysis software (ABI). The FACTURA program flagged heterozygous positions in which the minor peak was estimated to be ≥20% of the major peak.
Both laboratories used standard physical precautions to prevent sample contamination with DNA from other sources (16) and sequence analysis techniques to detect the possibility of contamination with other samples studied during the same time period (18). The sequence analysis techniques included comparison of each sequence to other recently generated sequences to look for unexpected high levels of similarity and the creation of neighbor-joining trees to visually detect unexpectedly similar isolates. Neighbor-joining trees were created with the PHYLIP package (8).
Definitions.
A partial nucleotide discordance was considered to be present when one laboratory reported a nucleotide mixture and the other laboratory reported one of the mixture's components (e.g., laboratory A reported Y, the International Union of Biochemistry and Molecular Biology code for C/T, and laboratory B reported C). A complete nucleotide discordance was considered to be present if each laboratory reported a different nonambiguous nucleotide at the same position for a sample (e.g., laboratory A reported C and laboratory B reported T).
Mutations were defined as amino acid differences between a patient sequence and the HIV-1 subtype B consensus sequence (15). Mutations were considered to be present if they were detected as part of a mixture (together with a wild-type allele) or in pure form. Protease inhibitor resistance mutations were defined as mutations at codons 10, 20, 24, 30, 32, 36, 46, 47, 48, 50, 53, 54, 60, 63, 71, 73, 77, 82, 84, 88, 90, or 93 and included all differences from the consensus B sequence at position 82 except V82I. RT inhibitor resistance mutations were defined as mutations at codons 41, 62, 65, 67, 69 (including insertions at this position), 70, 74, 75, 77, 115, 116, 151, 184, 210, 215, or 219 for the nucleoside RT inhibitors and at codons 98 (G not S), 100, 101, 103 (N not R), 106, 108, 179 (D not I), 181, 188, 190, 225, or 236 for the nonnucleoside RT inhibitors. This list of mutations is not meant to be complete, and several accessory and recently identified protease and RT mutations are not listed.
RESULTS
Nucleic acid sequences.
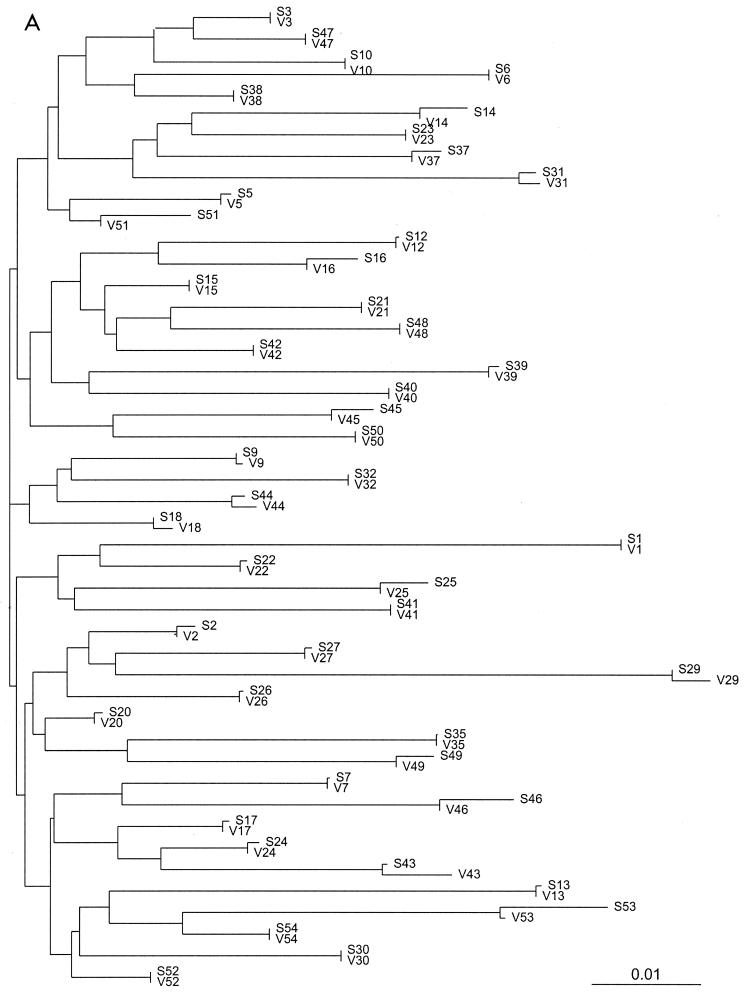
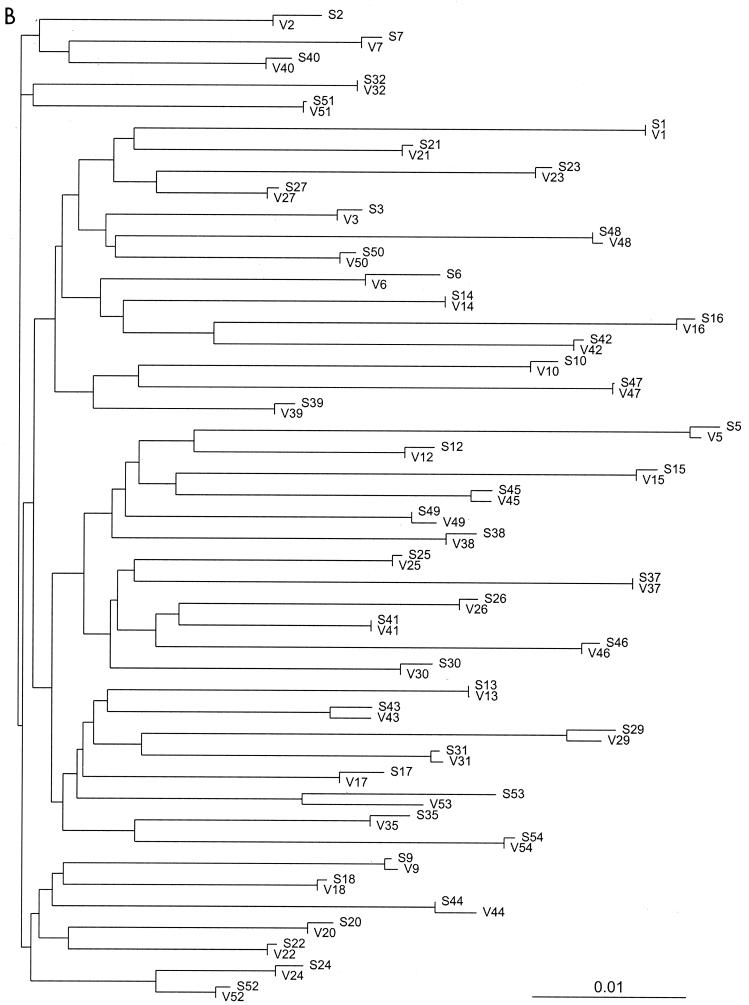
Sequencing data were obtained for all 46 samples in both laboratories. Neighbor-joining trees of the 46 pairs of protease and RT sequences based on the uncorrected distances between the sequences confirm the absence of cross-contamination or sample mix-up; the paired sequences obtained for each isolate are more similar to one another than to the sequences of any other isolate (Fig. 1). The rates of complete sequence concordance between the two laboratories were 99.1% at 13,662 protease nucleotide positions and 99.0% at 34,398 RT nucleotide positions (Table 1). Of the 123 discordant protease nucleotides, 112 (91%) were partially discordant (one laboratory reported a mixture and the other reported only one component of the mixture). Of the 347 discordant RT nucleotides, 311 (90%) were partially discordant.
FIG. 1.
Unrooted neighbor-joining trees of the 46 protease (A) and RT (B) sequence pairs. The sequence name consists of a letter indicating the laboratory (S, Stanford [laboratory A]; V, VIRCO [laboratory B]). In all cases the paired sequences were closer to one another than to any other sequence in the data set.
TABLE 1.
Comparison of protease and RT sequencing at two laboratories: concordance of nucleotide, amino acid, and drug resistance mutationsa
| HIV-1 enzyme | Nucleotide concordance
|
Amino acid concordance
|
Drug resistance mutation concordance
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total NA concordance (%) | Total no. of NAs | No. of complete NA differences | No. of partial NA differences | Total AA concordance (%) | Total no. of AAs | No. of complete AA differences | No. of partial AA differences | Total no. of DRMs | No. of DRMs with complete match | No. of complete DRM mismatches | No. of partial DRM mismatches | |
| Protease | 99.1 | 13,662 | 11 | 112 | 98.4 | 4,554 | 6 | 65 | 229 | 197 | 3 | 29 |
| RT | 99.0 | 34,398 | 36 | 311 | 99.0 | 11,466 | 13 | 107 | 228 | 204 | 6 | 18 |
NA, nucleotide; AA, amino acid; DRM, drug resistance mutation. Mutation was a change at one of the sites in the protease sequence (position 10, 20, 32, 36, 46, 48, 50, 53, 54, 60, 63, 71, 73, 77, 82, 84, 88, 90, or 93) and at one of the sites in the RT sequence (41, 62, 65, 67, 69, 70, 74, 75, 77, 100, 101, 103, 106, 108, 115, 116, 151, 181, 184, 188, 190, 210, 215, 219, 225, or 236). A partial mismatch or difference was considered to be present when one laboratory reported a nucleotide mixture and the other laboratory reported one of the mixture's components. A complete mismatch or difference was considered to be present if each laboratory reported a different nonambiguous nucleotide at the same position. In two cases, laboratory A sequenced codons 1 to 235 and 1 to 231 (rather than codons 1 to 250). Therefore, the number of RT nucleic acids is 102 (34 × 3) less than 34,500 (46 × 750).
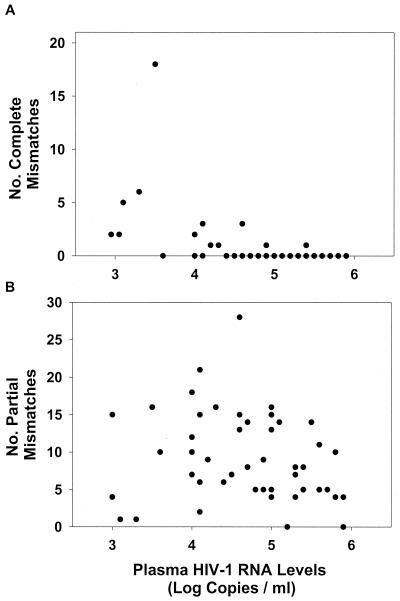
Complete interlaboratory discordances occurred at 47 positions in 12 plasma samples. These 12 plasma samples with complete discordances had significantly lower mean HIV-1 RNA levels compared to those for the 34 samples without complete discordances (3.9 versus 4.9 log copies/ml [P < 0.001, Student's t test]) (Fig. 2A). A single sample with a plasma HIV-1 RNA level of 3.0 log copies/ml accounted for 18 of the 47 discordances. There was no correlation between plasma HIV-1 RNA levels and the presence of partial discordances (Fig. 2B).
FIG. 2.
(A) Twelve samples with one or more complete nucleotide discordances had significantly lower mean plasma HIV-1 RNA levels than the 34 samples without a complete nucleotide mismatch (3.9 versus 4.9 log copies/ml; P < 0.001). Panel A appears to have less than 46 points because 34 of the samples had no complete mismatches and these overlap with one another along the x axis. (B)There was no statistically significant relationship between the number of partial nucleotide mismatches and plasma HIV-1 RNA levels in the 46 samples. Each panel has 46 points, one for each plasma sample. The position along the x axis is based on the HIV-1 RNA level of the plasma sample. The position along the y axis is the number of nucleotide mismatches between laboratories A and B upon testing of the plasma sample.
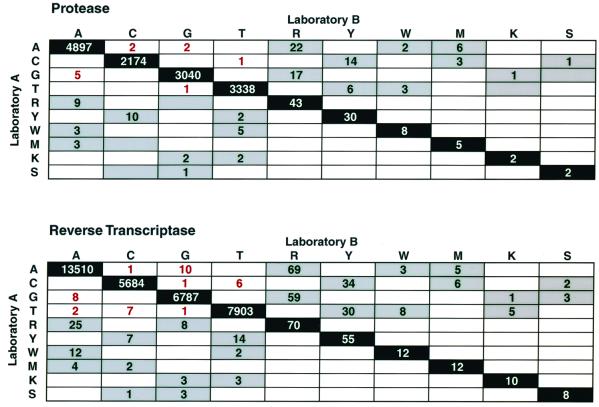
Most of the completely discordant positions (39 of 47 [83%]) and most of the partially discordant nucleotides (326 of 421 [77%]) comprised nucleotide pairs resulting from transitions (R = A/G, Y = C/T) rather than transversions (W = A/T, M = A/C, K = G/T, S = C/G) (Fig. 3). The predilection of both complete and partial discordances for transitions was statistically significant when one considers the fact that transversions would be expected to occur twice as frequently as transitions (P < 0.001; by the binomial test for comparison of two proportions, expected proportion = 0.33).
FIG. 3.
Matrices showing the exact numbers of nucleotide concordances and discordances between laboratories A (vertical, left) and B (horizontal, top). Exact matches are shown along the diagonal. The numbers of partial discordances are written in black on a grey background, and the numbers of complete discordances are written in red on a white background. R (A/G) and Y (C/T) represent transitions. M (A/C), W (A/T), K (G/T), and S (C/G) represent transversions. Data from the protease sequencing are shown at the top, and data from the RT sequencing are shown below. One RT sequence had a B and another had an H (data not shown). There were no N's or other highly ambiguous nucleotides.
Ambiguous nucleotides (mixtures) were detected at approximately 1% of the nucleotide positions (0.7% for laboratory A and 1.1% for laboratory B). In every case in which one laboratory detected a mixture, the other laboratory also detected either the mixture (40%) or one of the mixture's components (60%). For example, if one laboratory detected a Y, the other laboratory detected Y, C, or T (Fig. 3). If one laboratory detected an R, the other laboratory detected R, A, or G (Fig. 3).
Amino acid sequences.
Among the 123 discordant protease nucleotide positions, 71 (58%) resulted in the two laboratories detecting different amino acids (nonsynonymous differences). Among the 347 discordant RT positions, 120 (35%) were nonsynonymous. Most of the nonsynonymous changes (65 of 71 in the protease sequence and 107 of 120 in the RT sequence) represented partial amino acid discordances. Only 6 of 4,554 assignments in the protease sequence and 13 of 11,466 assignments in the RT sequence were completely discordant.
Of the 71 nonsynonymous protease discordances, 32 occurred at positions known to be associated with protease inhibitor resistance, including 9 partial discordances at positions strongly associated with drug resistance (positions 48, 82, 84, and 90), 3 partial discordances in the protease flap (positions 46, 47, and 54), and 20 discordances at accessory drug resistance positions (positions 10, 20, 36, 63, 71, 77, and 93), of which 17 were partial. Of 120 nonsynonymous RT discordances, 24 occurred at positions known to be related to drug resistance (positions 41, 62, 67, 74, 103, 106, 179, 181, 188, 190, 210, 215, and 219), of which 18 were partial. Forty-eight discordances occurred at several other polymorphic positions (positions 39, 43, 49, 64, 118, 122, 135, 142, 178, 200, 208, 211, and 245) (24).
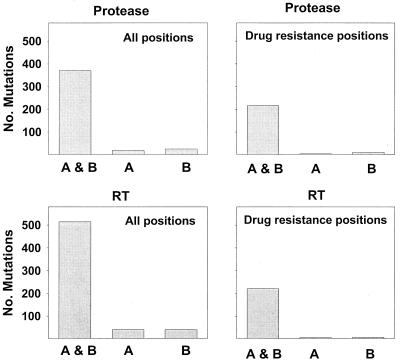
Figure 4 shows the numbers of protease and RT mutations detected by each laboratory. Laboratory A detected 388 mutations in the protease sequence (mean, 8.4 per sequence), including 219 drug resistance mutations (mean, 4.9 per sequence). Laboratory B detected a total of 395 protease mutations (mean, 8.6 per sequence), including 226 resistance mutations (mean, 5.0 per sequence). The laboratories agreed in the detection of 370 total mutations (mean, 8.2 per sequence) and 216 drug resistance mutations. Laboratory A detected 18 mutations (including 3 protease inhibitor resistance mutations) that were not detected by laboratory B, and laboratory B detected 25 mutations (including 10 protease inhibitor resistance mutations) that were not detected by laboratory A.
FIG. 4.
Each panel shows the mutations detected in common between the laboratories (A & B) as well as those mutations detected by laboratory A but not laboratory B (A) and those detected by laboratory B but not laboratory A (B). The panels on the left show the total number of mutations (differences from consensus B) for protease and RT. The panels on the right show the number of drug resistance mutations for protease and RT.
In the RT sequence, laboratory A detected 554 mutations (mean, 11.4 per sequence), including 221 drug resistance mutations. Laboratory B detected a total of 554 RT mutations (mean, 11.4 per sequence), including 223 drug resistance mutations. The laboratories agreed in the detection of 514 total mutations and 216 drug resistance mutations. Laboratory A detected an additional 40 mutations (including 5 drug resistance mutations) that were not detected by laboratory B, and laboratory B detected 40 mutations (including 7 drug resistance mutations) that were not detected by laboratory A.
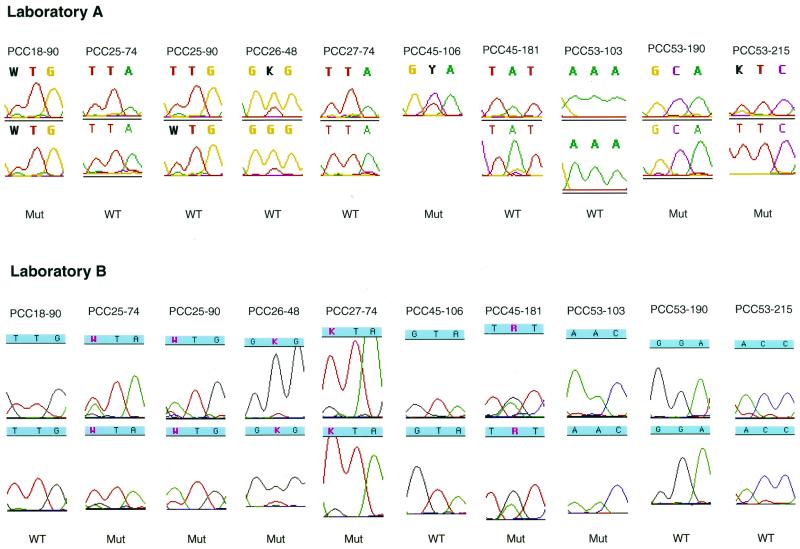
There were 10 instances in which a primary resistance mutation was detected by just one of the two laboratories (four by laboratory A and six by laboratory B) (Fig. 5). Seven of these instances represented partial mismatches in that the laboratory detecting the mutation detected it as part of a mixture (for the protease sequence, in patients PCC18 [L90L/M], PCC25 [L90L/M], and PCC26 [G48G/V]; for the RT sequence, in patients PCC25 [L74L/I], PCC27 [L74L/V], and PCC45 [V106V/A, Y181Y/C]). Three instances represented complete mismatches, and each of these complete mismatches occurred upon testing of the plasma sample from patient PCC53. Of the six mutations detected only by laboratory B, three were found in retrospect by laboratory A to have small mutant peaks, but these positions were not reported as mutations either because the mutation was not flagged by the FACTURA program (patient PCC45, Y181YC) or because a minor mutation was found in only one of the two sequencing reactions (patient PCC25, L90L/M; patient PCC26, G48G/V).
FIG. 5.
Chromatographic tracings showing the automated DNA sequence analysis of the 10 instances in which a primary resistance mutation was detected by just one laboratory (the PCC numbers at the tops of the tracings indicate patient code-codon number). In seven instances, the laboratory detecting the mutation detected it as part of a mixture, and in three instances (for patient PCC53) there was a complete mismatch between the two laboratories. The tracings for the forward and reverse sequences of each laboratory are shown (the reverse sequence at RT codon 106 for patient PCC45 at laboratory A is missing). Nucleotides with mixtures are shown in bold for laboratory A and in red for laboratory B. Laboratory A reported protease codon 90 for patient PCC25 and protease codon 48 for patient PCC26 as being of the wild type (WT) because the mutant (Mut) peak was detected in only one of the sequencing reactions. Laboratory A's reverse sequence of RT codon 181 for patient PCC45 shows a mixture of TAT and TGT. However, the minor G peak was not flagged by the ABI FACTURA program at the then-recommended cutoff of 30%. For three of the mixtures (for patient PCC25, codon 90; patient PCC45, codon 106; and patient PCC45, codon 181), the predominant nucleotide (having the larger peak) was different between the two laboratories.
DISCUSSION
We assessed the reproducibility of HIV-1 RT and protease sequencing using cryopreserved plasma aliquots obtained from 46 heavily treated HIV-1-infected individuals. Sequencing was performed in two full-service clinical diagnostic laboratories, each of which used different protocols for plasma HIV-1 RNA extraction, reverse transcription, nested PCR, and automated dideoxynucleotide cycle sequencing. This study differs from previous interlaboratory sequence comparisons, as it is the first to examine the reproducibility of sequencing of HIV-1 isolates in clinical plasma samples.
The rates of sequence concordance between the two laboratories were 99.1% for the protease sequence and 99.0% for the RT sequence. Because 90% of the discordances were partial (defined as one laboratory detecting a mixture and the second laboratory detecting just one of the mixture's components), only 0.1% of the total nucleotides were completely discordant. Nucleotide mixtures were detected at approximately 1% of the nucleotide positions, and in every case in which one laboratory detected a mixture, the second laboratory detected either the same mixture or one of the mixture's components. Laboratory B detected mixtures more frequently than laboratory A, in part because laboratory B used BigDye terminators (28) and in part because laboratory B used more aggressive measures to flag and call mixtures than laboratory A.
Several lines of evidence suggest that most discordances resulted from differences in sampling of the HIV-1 plasma population rather than from technical errors. The high proportion of partial mismatches and the 40% agreement in identifying mixtures would not be expected if mismatches were unrelated to the in vivo distribution of HIV-1 variants in plasma. The predominance of transitions at discordant positions also likely reflects the predominance of transitions within the HIV-1 quasispecies (19). In contrast, technical errors, with the exception of those caused by Taq polymerase, would be expected to cause a significantly higher proportion of transversions. The fact that complete discordances were significantly more likely to occur when plasma samples with lower HIV-1 RNA levels were tested is also consistent with sampling variation.
The high rates of concordance in this study may be related in part to the similar methods for sequencing used by laboratories A and B and to the large volume of HIV-1 sequencing regularly performed at each laboratory. A high rate of nucleotide concordance (99.7%) was also previously reported in a multicenter study of dideoxynucleotide sequencing of cultured cell pellets (5). However, the extent of nucleotide concordance was lower in two previous multicenter comparisons that involved sequencing of isolates from mixtures of plasmid clones and spiked plasma samples (22; R. Schuurman, D. J. Brambilla, T. De Groot, and C. Boucher, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother. abstr. 1168, 1999). In these multicenter comparisons, some laboratories failed to detect mutant nucleotides that were present in 100% of the clones and nearly one-half of the laboratories failed to detect mutations present as 25 and 50% mixtures (22; Schuurmann et al., 39th ICAAC). Lower rates of concordance have also been reported in studies that compared different sequencing technologies (9, 11, 27). The previous studies have underscored the need for ongoing quality assurance and have spurred the development of standardized assay kits.
The inability to detect minor drug-resistant HIV-1 variants is a recognized limitation of clinical HIV-1 drug susceptibility testing when one is using either genotypic or phenotypic methods (3, 21). Moreover minor variants are more likely to be missed in samples with lower RNA levels because plasma HIV-1 RNA extraction and reverse transcription of large RNA fragments are inefficient procedures, with recovery rates ranging between 1 and 10% (9, 23). In samples with low RNA levels, ultracentrifugation of larger volumes of plasma (e.g., ≥1.0 ml) prior to RNA extraction would theoretically improve both the rate of detection of minor variants and sequencing reproducibility. Nonetheless, the possibility that an undetected mutant virus is present should be considered when interpreting the results of drug susceptibility tests, particularly for patients with complicated antiretroviral treatment histories or patients who have discontinued one or more antiretroviral drugs (6, 26). Further studies are also needed to identify those patients who are at greater risk of having minor mutant drug-resistant viral populations and to develop sequencing methods with increased sensitivities for these variants.
ACKNOWLEDGMENTS
We thank Caroline Tolman, Chris Imazumi, Christina Trevino, and Cheryl Ikemoto for doing the gene sequencing at Stanford. We thank Christina Trevino and Joanne Dileanos (ABI) for critical review of the manuscript. We thank Mathew Gonzales for collating the chromatograms and preparing Fig. 5.
REFERENCES
- 1.Baxter J D, Mayers D L, Wentworth D N, Neaton J D, Hoover M L, Winters M A, Mannheimer S B, Thompson M A, Abrams D I, Brizz B J, Ioannidis J P, Merigan T C. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. CPCRA 046 Study Team for the Terry Beirn Community Programs for Clinical Research on AIDS. AIDS. 2000;14:F83–F93. doi: 10.1097/00002030-200006160-00001. [DOI] [PubMed] [Google Scholar]
- 2.Carpenter C C, Cooper D A, Fischl M A, Gatell J M, Gazzard B G, Hammer S M, Hirsch M S, Jacobsen D M, Katzenstein D A, Montaner J S, Richman D D, Saag M S, Schechter M, Schooley R T, Thompson M A, Vella S, Yeni P G, Volberding P A. Antiretroviral therapy in adults: updated recommendations of the International AIDS Society-USA Panel. JAMA. 2000;283:381–390. doi: 10.1001/jama.283.3.381. [DOI] [PubMed] [Google Scholar]
- 3.D'Aquila R T. Limits of resistance testing. Antivir Ther. 2000;5:71–76. [PubMed] [Google Scholar]
- 4.DeGruttola V, Dix L, D'Aquila R, Holder D, Phillips A, Ait-Khaled M, Baxter J, Clevenbergh P, Hammer S, Harrigan R, Katzenstein D, Lanier R, Miller M, Para M, Yerly S, Zolopa A, Murray J, Patick A, Miller V, Castillo S, Pedneault L, Mellors J. The relation between baseline HIV drug resistance and response to antiretroviral therapy: re-analysis of retrospective and prospective studies using a standardized data analysis plan. Antivir Ther. 2000;5:41–48. doi: 10.1177/135965350000500112. [DOI] [PubMed] [Google Scholar]
- 5.Demeter L M, D'Aquila R, Weislow O, Lorenzo E, Erice A, Fitzgibbon J, Shafer R, Richman D, Howard T M, Zhao Y, Fisher E, Huang D, Mayers D, Sylvester S, Arens M, Sannerud K, Rasheed S, Johnson V, Kuritzkes D, Reichelderfer P, Japour A. Interlaboratory concordance of DNA sequence analysis to detect reverse transcriptase mutations in HIV-1 proviral DNA. ACTG Sequencing Working Group. AIDS Clinical Trials Group. J Virol Methods. 1998;75:93–104. doi: 10.1016/s0166-0934(98)00100-1. [DOI] [PubMed] [Google Scholar]
- 6.Devereux H L, Youle M, Johnson M A, Loveday C. Rapid decline in detectability of HIV-1 drug resistance mutations after stopping therapy. AIDS. 1999;13:F123–F127. [PubMed] [Google Scholar]
- 7.Durant J, Clevenbergh P, Halfon P, Delgiudice P, Porsin S, Simonet P, Montagne N, Boucher C A, Schapiro J M, Dellamonica P. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomised controlled trial. Lancet. 1999;353:2195–2199. doi: 10.1016/s0140-6736(98)12291-2. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein J. PHYLIP (Phylogeny Inference Package), version 3.5c. Seattle: University of Washington; 1993. [Google Scholar]
- 9.Gunthard H F, Wong J K, Ignacio C C, Havlir D, Richman D D. Comparative performance of high-density oligonucleotide sequencing and dideoxynucleotide sequencing of type 1 pol from clinical samples. AIDS Res Hum Retrovir. 1998;14:869–876. doi: 10.1089/aid.1998.14.869. [DOI] [PubMed] [Google Scholar]
- 10.Hammond J, Calef C, Larder B, Schinazi R, Mellors J W. Mutations in retroviral genes associated with drug resistance. In: Korber B T, Kuiken C, Foley B, Hahn B, McCutchan F, Mellors J, Sodrowski J, editors. Human retroviruses and AIDS: a compilation and analysis of nucleic and amino acid sequences. Los Alamos, N.M: Theoretical Biology and Biophysics, Los Alamos National Laboratory; 1998. [Google Scholar]
- 11.Hanna G J, Johnson V A, Kuritzkes D R, Richman D D, Martinez-Picado J, Sutton L, Hazelwood J D, D'Aquila R T. Comparison of sequencing by hybridization and cycle sequencing for genotyping of human immunodeficiency virus type 1 reverse transcriptase. J Clin Microbiol. 2000;38:2715–2721. doi: 10.1128/jcm.38.7.2715-2721.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrigan P R, Cote H C. Clinical utility of testing human immunodeficiency virus for drug resistance. Clin Infect Dis. 2000;30:S117–S122. doi: 10.1086/313861. [DOI] [PubMed] [Google Scholar]
- 13.Hertogs K, de Bethune M P, Miller V, Ivens T, Schel P, Van Cauwenberge A, Van Den Eynde C, Van Gerwen V, Azijn H, Van Houtte M, Peeters F, Staszewski S, Conant M, Bloor S, Kemp S, Larder B, Pauwels R. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob Agents Chemother. 1998;42:269–276. doi: 10.1128/aac.42.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirsch M S, Brun-Vezinet F, D'Aquila R T, Hammer S M, Johnson V A, Kuritzkes D R, Loveday C, Mellors J W, Clotet B, Conway B, Demeter L M, Vella S, Jacobsen D M, Richman D D. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA Panel. JAMA. 2000;283:2417–2426. doi: 10.1001/jama.283.18.2417. [DOI] [PubMed] [Google Scholar]
- 15.Korber B, Kuiken C, Foley B, Hahn B, McCutchan F E, Mellors J, Sodroski J. Human retroviruses and AIDS: a compilation and analysis of nucleic and amino acid sequences. Los Alamos, N.M: Theoretical Biology and Biophysics, Los Alamos National Laboratory; 1998. [Google Scholar]
- 16.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature (London) 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 17.Larder B A, Kohli A, Kellam P, Kemp S D, Kronick M, Henfry R D. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature. 1993;365:671–673. doi: 10.1038/365671a0. [DOI] [PubMed] [Google Scholar]
- 18.Learn G H, Korbert B T M, Foley B, Hahn B H, Wolinsky S M, Mullins J I. Maintaining the integrity of human immunodeficiency virus sequence databases. J Virol. 1996;70:5720–5730. doi: 10.1128/jvi.70.8.5720-5730.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mansky L M. Retrovirus mutation rates and their role in genetic variation. J Gen Virol. 1998;79:1337–1345. doi: 10.1099/0022-1317-79-6-1337. [DOI] [PubMed] [Google Scholar]
- 20.Panel on Clinical Practices for Treatment of HIV Infection, U.S. Department of Health and Human Services. 2000. Guidelines for the use of antiretroviral agents in HIV-infected adults adolescents. [Online.] www.hivatis.org/trtgdlns.html.
- 21.Richman D D. Principles of HIV resistance testing and overview of assay performance characteristics. Antivir Ther. 2000;5:27–31. [PubMed] [Google Scholar]
- 22.Schuurman R, Demeter L, Reichelderfer P, Tijnagel J, de Groot T, Boucher C. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J Clin Microbiol. 1999;37:2291–2296. doi: 10.1128/jcm.37.7.2291-2296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shafer R W, Levee D J, Winters M A, Richmond K L, Huang D, Merigan T C. Comparison of QIAamp HCV kit spin columns, silica beads, and phenol-chloroform for recovering human immunodeficiency virus type 1 RNA from plasma. J Clin Microbiol. 1997;35:520–522. doi: 10.1128/jcm.35.2.520-522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shafer R W, Stevenson D, Chan B. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 1999;28:346–348. doi: 10.1093/nar/27.1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shafer R W, Warford A, Winters M A, Gonzales M J. Reproducibility of human immunodeficiency virus type 1 (HIV-1) protease and reverse transcriptase sequencing of plasma samples from heavily treated HIV-1-infected individuals. J Virol Methods. 2000;86:143–153. doi: 10.1016/s0166-0934(00)00144-0. [DOI] [PubMed] [Google Scholar]
- 26.Verhofstede C, Wanzeele F V, Van Der Gucht B, De Cabooter N, Plum J. Interruption of reverse transcriptase inhibitors or a switch from reverse transcriptase to protease inhibitors resulted in a fast reappearance of virus strains with a reverse transcriptase inhibitor-sensitive genotype. AIDS. 1999;13:2541–2546. doi: 10.1097/00002030-199912240-00007. [DOI] [PubMed] [Google Scholar]
- 27.Wilson J W, Bean P, Robins T, Graziano F, Persing D H. Comparative evaluation of three human immunodeficiency virus genotyping systems: the HIV-GenotypR Method, the HIV PRT GeneChip Assay, and the HIV-1 RT Line Probe Assay. J Clin Microbiol. 2000;38:3022–3028. doi: 10.1128/jcm.38.8.3022-3028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zakeri H, Amparo G, Chen S-M, Spurgeon S, Kwok P-Y. Peak height pattern in dichlororhodamine and energy transfer dye terminator sequencing. BioTechniques. 1998;25:406–414. doi: 10.2144/98253st01. [DOI] [PubMed] [Google Scholar]
- 29.Zolopa A R, Shafer R W, Warford A, Montoya J G, Hsu P, Katzenstein D, Merigan T C, Efron B. HIV-1 genotypic resistance patterns predict response to saquinavir- ritonavir therapy in patients in whom previous protease inhibitor therapy had failed. Ann Intern Med. 1999;131:813–821. doi: 10.7326/0003-4819-131-11-199912070-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]