Abstract
The incidence of tuberculosis (TB) in Madagascar is 150 cases per 100,000 people. Because of this endemicity, we studied the genetic diversity of Mycobacterium tuberculosis strains isolated in four big cities in 1994 to 1995 with the aim of monitoring TB transmission. Isolates from 316 cases of pulmonary TB (PTM+) were typed by Southern hybridization with genetic markers IS6110 and DR. Of the 316 PTM+ strains, 66 (20.8%) had a single IS6110 band and were differentiated by the DR marker into 33 profiles. Using both markers, 37.7% (119) of the patients were clustered, a proportion similar to that in countries with a high prevalence of TB. There was no significant difference between clustered and nonclustered patients in age, sex, Mycobacterium bovis BCG status, and drug susceptibility of strains. Clustering was significantly greater in the capital, Antananarivo, than in the other cities, suggesting a higher rate of transmission. However, most of the patients in clusters were living in different areas, and, within a distance of 0.7 km, we did not find epidemiologically unrelated strains with the same restriction fragment length polymorphism profile. Despite an apparently low polymorphism, genetic markers such as IS6110 are potentially valuable for monitoring TB transmission. However, the high proportion of Malagasy isolates with a single IS6110 copy makes this marker alone unsuitable for typing. Additional markers such as DR are necessary for the differentiation of the isolates and for epidemiological surveys.
Since the discovery of repetitive elements associated with polymorphism and their use as genetic markers for Mycobacterium tuberculosis, strain differentiation by DNA typing has greatly facilitated epidemiological studies (7, 8, 35). To type M. tuberculosis complex strains, the most extensively used method has been a standardized protocol of restriction fragment length polymorphism (RFLP) analysis with insertion sequence IS6110 as a marker (29, 31). This method has also been used successfully in confirming epidemics and nosocomial infections and in studies on transmission of tuberculosis (TB) in high-risk communities (5, 11, 13, 15, 17, 24). Another marker often used for discriminating strains is the direct repeat (DR) region, which contains multiple 36-bp DRs separated by variable spacers (31 to 45 bp in length), which are sources of diversity (21).
In Madagascar (about 13 million inhabitants), the incidence of pulmonary tuberculosis is very high (about 150 cases per 100,000 people) (9). In contrast, the prevalence of human immunodeficiency virus (HIV) is low, and between 1989 and 1994 only six of a total of 3,168 tuberculous patients tested were HIV seropositive (4). RFLP studies of clinical isolates in countries with a high incidence of TB describe limited polymorphism of TB genetic markers such as IS6110. To determine the usefulness of this marker to monitor transmission in these countries, we studied the diversity of M. tuberculosis complex strains from pulmonary patients (PTM+) in four big cities in Madagascar. We have correlated strain polymorphism with geographic areas. Such data will allow the detection of epidemics and an understanding of TB transmission in this country.
MATERIALS AND METHODS
Study population.
This study was carried out in the capital, Antananarivo, and in three other large cities, Antsirabe, Fianarantsoa, and Mahajanga, and was conducted jointly with a survey of M. tuberculosis primary resistance in 1994 to 1995 (10).
Between August 1994 and December 1995, of the 1,389 PTM+ patients identified in eight diagnostic and treatment centers, 1,108 new patients (81.5%) were included. For DNA fingerprinting, 316 patients were randomly sampled. They were representative of the eight centers: 153 patients from Antananarivo, 27 from Antsirabe, 64 from Fianarantsoa, and 72 from Mahajanga. The age of the patients was 11 to 74 years (mean age: 34 years). The male-to-female ratio was 1.53:1. Mycobacterium bovis BCG status was known for 306 patients, of whom 223 were vaccinated.
All patients included in this study were Malagasy and HIV seronegative.
Bacteriology.
All clinical specimens were cultured on standard Lowenstein-Jensen (LJ) medium (Diagnostics Pasteur, Paris, France) and on LJ medium without glycerol but supplemented with 0.5% pyruvate. Mycobacterial isolates were identified according to growth on LJ medium, colony morphology, and biochemical tests for the following: niacin production, catalase, urease, and nitrate reductase (20). Drug susceptibility of the PTM+ isolates to streptomycin, isoniazid, rifampin, and ethambutol was tested using the proportion method (6), as recommended by the Global Tuberculosis Programme of the World Health Organization and the International Union against Tuberculosis and Lung Disease (19).
RFLP typing.
Genomic DNA from only one strain from each PTM+ patient (316 isolates) was extracted according to the method described by van Soolingen et al. (36) and tested by RFLP analysis. DNA fingerprints were obtained using a standardized RFLP technique with the IS6110 insertion sequence (IS6110 pattern) (31). Briefly, chromosomal DNA was digested with restriction endonuclease PvuII (Pharmacia Biotech) and was hybridized with the 807-bp PvuII-XhoI fragment of IS6110 (29). For strains with one to four copies of IS6110, DNA was also digested with AluI (Pharmacia Biotech) and hybridized with the 36-bp DR sequence probe (5′GTCGTCAGACCCAAAACCCCGAGAGGGGACGGAAAC3′) (21). All probes were labeled with horseradish peroxidase and detected with the enhanced chemiluminescence system (ECL; Amersham).
Analysis of RFLP patterns.
The Taxotron software (P. A. D. Grimont, Institut Pasteur, Paris, France) was used for computer-assisted analysis of the RFLP patterns. Patterns were compared by the unweighted pair group clustering method of averages, and matching was further confirmed by visual examination. A cluster of M. tuberculosis strains was defined as two or more isolates with identical RFLP patterns. A cluster of patients was defined as two or more patients with identical strains. Epidemiological relationships between patients belonging to a cluster were also investigated. The geographical distribution of the clusters was assessed on the basis of the place of residence of the patients. The characteristics of clustered patients and nonclustered patients were compared. For assessing differences between percentages, we used the χ2 test or Fischer's exact test for values ≤5. Differences were considered significant if P values were <0.05.
RESULTS
DNA polymorphism of the M. tuberculosis complex strains isolated from PTM+ patients.
The IS6110 RFLP types for 316 PTM+ strains, a representative sample corresponding to 28.5% of the patients included (and 23% of the patients identified in the four cities for the 1994 to 1995 period), were determined. Of these strains, 312 were identified as M. tuberculosis and 4 were identified as M. bovis.
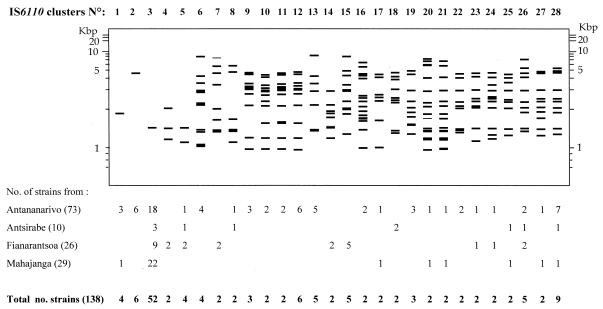
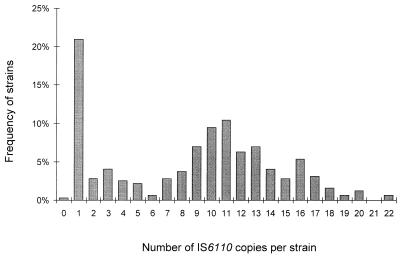
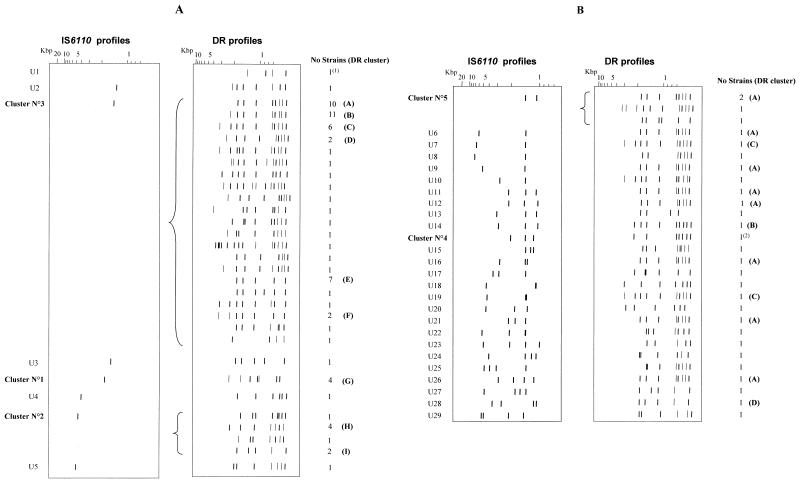
One strain from Antananarivo had no hybridizing band, suggesting that it did not contain the IS6110 element. One hundred and seventy-seven isolates (56%) had unique IS6110 patterns, and 138 (43.6%) were assigned to 28 IS6110 clusters of 2 to 52 strains (Fig. 1). The number of IS6110 copies per strain was between 1 and 22 (Fig. 2). The number of bands in the 177 isolates with unique banding patterns also varied from 1 to 22, and in the clustered isolates the number varied from 1 to 17. Sixty-six strains (20.8%) had a pattern with only one hybridizing band, suggesting that they contained a single IS6110 copy. The proportion of strains with a single IS6110 copy was the highest in Mahajanga (36.1%); the proportions were 17.6, 15.6, and 11.1% in Antananarivo, Fianarantsoa, and Antsirabe, respectively. Among these 66 strains, seven fragment sizes were observed: 1.35 (1 strain), 1.45 (52 strains), 1.56 (1 strain), 1.8 (4 strains identified as M. bovis), 4.1 (1 strain), 4.8 (6 strains), and 5.1 kb (1 strain). Moreover, 30 strains had a pattern with two to four bands containing IS6110. The strains with zero to four IS6110 copies were further genotyped with the DR marker (Fig. 3) (one strain of IS6110 cluster 4 was not viable and, therefore, could not be DR typed). Among those strains with zero to four IS6110 copies, from 4 to 14 restriction fragments were hybridized with the DR probe, producing 46 different DR patterns. Thirty-seven isolates had unique DR patterns, and 59 fell into nine clusters of 2 to 10 strains. There were isolates having the same DR pattern but having different IS6110 profiles (Table 1; Fig. 3). DR cluster A, for example, contained 19 isolates that could be differentiated into nine IS6110 profiles. However, for those strains with one IS6110 copy, strains with different IS6110 profiles also had different DR patterns (Table 2; Fig. 3A). Likewise, the same DR pattern was observed for all M. bovis strains, and it was different from the DR patterns of M. tuberculosis isolates. The strain without the IS6110 element had a DR profile different from that of all other strains.
FIG. 1.
IS6110 clusters of PTM+ isolates in Madagascar.
FIG. 2.
Numbers of IS6110 copies in 316 M. tuberculosis complex strains isolated from PTM+ patients in Madagascar.
FIG. 3.
(A) DR patterns of the isolates with zero or one IS6110 copy. (B) DR patterns of the isolates with two to four IS6110 copies. The numbers of IS6110 clusters are given on the left. U1 to U29, strains with unique IS6110 profiles; A through I, DR clusters; (1), M. bovis strains; (2), strain of IS6110 cluster 4 not viable and not available for DR typing.
TABLE 1.
Comparison of DR clusters and IS6110 patterns of the isolates with four or less IS6110 bands
| DR cluster | Total no. of strains | No. of IS6110 patterns
|
||
|---|---|---|---|---|
| Total | Unique | Clusters (no. of strains) | ||
| A | 19 | 9 | 7 | 2 (12) |
| B | 11 | 2 | 1 | 1 (10) |
| C | 8 | 3 | 2 | 1 (6) |
| D | 2 | 1 | 0 | 1 (2) |
| E | 8 | 2 | 1 | 1 (7) |
| F | 2 | 1 | 0 | 1 (2) |
| G | 4 | 1 | 0 | 1 (4) |
| H | 2 | 1 | 0 | 1 (2) |
| I | 3 | 2 | 1 | 1 (2) |
TABLE 2.
Number of DR patterns in 66 strains with one IS6110 copy
| Location (kb) of IS6110 band (cluster) | Total no. of strains | No. of DR patterns
|
||
|---|---|---|---|---|
| Total | Unique | Clustered (no. of strains) | ||
| 1.45 (3) | 52 | 21 | 15 | 6 (37) |
| 4.6 (2) | 6 | 4 | 2 | 2 (4) |
| 1.8 (1) | 4 | 1 | 0 | 1 (4a) |
| 1.35 | 1 | 1 | 1 | 0 |
| 5.1 | 1 | 1 | 1 | 0 |
| 1.56 | 1 | 1 | 1 | 0 |
| 4.1 | 1 | 1 | 1 | 0 |
| Total | 66 | 33 | 21 | 9 (45) |
M. bovis strains.
On the basis of both IS6110 and DR patterns, 197 PTM+ patients (62.3%) gave isolates that were not clustered and 119 (37.7%) gave isolates that were contained within 34 clusters of 2 to 10 isolates (Table 3). The 19 additional strains demonstrating a unique profile with two markers were composed of two groups: (i) 17 strains with one IS6110 band (Fig. 3A) and (ii) 2 strains with two IS6110 bands (Fig. 3B). Therefore, it appears that DR typing is especially useful for discrimination between isolates with one IS6110 copy. The small clusters were the most frequently observed group, accounting for 68% of the clusters but less than 50% of the isolates. These were further broken down as 21 clusters of two patients and 2 clusters of three patients. In comparing the characteristics of the clustered and the nonclustered patients (Table 4), we found that clusters of patients did not differ significantly in age, sex ratio, and BCG status and that the drug susceptibility patterns of the isolates also did not differ significantly. Similarly, no significant difference between the two groups within any of the individual cities was found (data not shown). Finally, a comparison of the RFLP profiles of drug-resistant and drug-susceptible strains revealed no correlation between drug resistance and RFLP pattern (36).
TABLE 3.
Degree of discrimination of IS6110 versus IS6110 plus DR
| Marker | Total no. of strains | No. of isolates from:
|
|
|---|---|---|---|
| Single strains with unique patterns | Clustered strains (no. of clusters) | ||
| IS6110 | 315 | 177 | 138 (28) |
| IS6110 + DR | 316 | 197a | 119 (34) |
Includes one strain with no IS6110 band.
TABLE 4.
Characteristics of clustered and nonclustered PTM+ patients
| Characteristic | Value for:
|
||
|---|---|---|---|
| Total (n = 316) | Clustered (n = 119) | Nonclustered (n = 197) | |
| Mean age (age range) (yr) | 34.01 (11–74) | 33.94 (13–70) | 34.08 (11–74) |
| Sex (no. of patients) | |||
| Female | 125 | 52 | 73 |
| Male | 191 | 67 | 124 |
| BCG status (no. of patients) | |||
| Vaccinated | 223 | 85 | 138 |
| Unvaccinated | 83 | 31 | 52 |
| Unknown | 10 | 3 | 7 |
| Susceptibility to S, H, R, and Ea (no. of patients) | |||
| Susceptible to all | 250 | 98 | 152 |
| Resistant to | |||
| One drug | 48 | 13 | 35 |
| Two drugs | 4 | 1 | 3 |
| Three drugs | 1 | 1 | 0 |
| H and R | 1 | 1 | 0 |
| ND | 13 | 6 | 7 |
S, streptomycin; H, isoniazid; R, rifampin; E, ethambutol; ND, not determined.
Geographical distribution of the clusters.
One hundred and thirty-eight strains (43.6% of the studied strains) were assigned to 28 IS6110 patterns. Thirteen of them (eight clusters of two patients and five with more than two patients) were recovered in at least two different cities (Fig. 1). One IS6110 pattern (two patients) was found only in Antsirabe, 4 were found only in Fianarantsoa, and 10 were found only in Antananarivo. An RFLP pattern with an IS6110 band at 1.45 kb (IS6110 cluster 3) was the only profile found in all four cities. Of the 46 DR patterns of the strains with one IS6110 copy, only 6 were found in two or more cities (data not shown).
When both the IS6110 and DR markers were used together, the proportion of clustered patients differed significantly between the cities. It was higher in Antananarivo (38.5%; 59 of 153) than in the three other cities (P = 0.003): 26.5% (17 of 64) in Fianarantsoa, 25% (18 of 72) in Mahajanga, and 7.4% (2 of 27) in Antsirabe. In Antsirabe, Fianarantsoa, and Mahajanga, the patients belonging to any single cluster lived in different areas of the city. In Antananarivo, except for two patients living at the same address and eight patients in Antananarivo prison, all the patients within clusters lived in different districts of the city. The two closest patients within the same cluster lived in different districts, within a distance of about 0.7 km (IS6110 cluster 3; DR pattern A). Although the polymorphism of strains in these cities seems relatively low, RFLP typing may be a valuable epidemiological tool for identifying transmission.
DISCUSSION
The present study is the first on DNA polymorphism of the M. tuberculosis complex strains in Madagascar, a country in which TB is hyperendemic (3). Four of the largest cities (Antananarivo, Antsirabe, Fianarantsoa, and Mahajanga) were chosen for this study. Using a two-genetic-marker typing method (IS6110 and DR), we found three IS6110 clusters containing strains that were differentiated using the second marker. There were two IS6110 clusters with isolates having one IS6110 band pattern (clusters 2 and 3) and one cluster with two-band-pattern isolates (cluster 5). Moreover, for some isolates having an IS6110 profile with two to four bands, some DR clusters contained isolates with different IS6110 patterns (Table 1). These results indicate that the changes in the DR and the IS6110 profiles are independent events and that only isolates with the same IS6110 and DR patterns can be considered identical strains. Our results can be favorably compared with those of Gillespie et al., who used the IS6110 and the PGRS markers for typing isolates from Tanzania with six or more band profiles (18). They found that only 1 of 13 IS6110 clusters contained isolates with different PGRS patterns. In this study, genotyping with a second marker was done only on isolates with patterns of less than five IS6110 bands. Thus, both our results and those of Gillespie et al. showed the usefulness of two-marker genotyping. An additional advantage of typing with the DR sequence that we chose as a second marker is that it can be performed using the PCR technique, avoiding large-scale cultures of M. tuberculosis (23).
The overall polymorphism of the strains as assessed by the number of clustered strains for the two genetic markers, IS6110 and DR, was low (37.7% for PTM+ strains). This is similar to results reported from other African and Asian countries but differs from what is observed in Europe or North America (16, 22, 35, 36,). The majority of the clusters (21 of 34) contained only two strains, probably due to the limited sample size. Only 42% (13 of 28) of the IS6110 patterns and 13% (6 of 46) of the DR patterns were found in more than one city, suggesting limited transmission between patients from different cities. Moreover, most patients contained within specific clusters lived in different areas in their city rather than in close proximity. The minimum distance between two clustered patients without any reported epidemiological link was 0.7 km. Despite the finding that the clustering of these patients was not related to risk, it is apparent that the polymorphism of markers used for M. tuberculosis strains is sufficient for detecting epidemics, for monitoring the transmission of resistant strains, and for tracking TB transmission in high-risk groups within particular areas (26).
Molecular epidemiology studies can lead to two hypotheses: first, that the degree of clustering can be used as a measure of recent and active transmission of TB (2, 28, 30), and second, that more young patients than old patients have recent infections and thus are more likely to belong to clusters. In our study, we found that the proportion of clustered patients was higher in Antananarivo than in the other cities, suggesting a more-active transmission of TB in the capital. However, ages of clustered and nonclustered patients were not significantly different. There are several possible explanations for this: (i) the population in the capital is highly mobile, (ii) the incubation time after an infection is highly variable, (iii) the prevalence of TB and the circulation of strains are so high that the risk of reinfection is the same throughout the population, and (iv) the reactivation of old infections is less frequent than expected (32). Another possibility is based on a recent study in Malaysia (12), where the authors reported an apparent increase in clustering with the age of the patients and suggested that strains with stable RFLP patterns may be in circulation for longer periods and thus may contribute to clustering. Differences in virulence among the strains could also explain the presence of some clusters. For example, a virulent strain would be isolated more frequently than a nonvirulent strain and thus would produce larger clusters.
Only one isolate did not contain the IS6110 genetic element. Zero-copy strains have previously been described, with slightly higher frequencies noted in patients from Vietnam, China, Thailand, and India (1, 14, 25, 36). One interesting characteristic of the Malagasy strains was the high proportion of single-IS6110-copy strains (21.6% of the PTM+ sampled strains) including seven different positions of the single-hybridizing band. These strains could be easily differentiated by using the DR marker. The strains having an RFLP pattern with a single-hybridizing band at 1.8 kb were all identified as M. bovis. This pattern is the same as that described for the majority of M. bovis strains elsewhere (27, 34). Several of the one-IS6110-band patterns have previously been found with similar frequencies in: Vietnam (12%) (37), India (30 to 40%) (14, 34), Thailand (20%) (25), and Malaysia (17.5%) (16). Thus, there appears to be a similarity between Asiatic and Malagasy strains (zero- or single-IS6110-band strains). This is not surprising because the Malagasy population is descended from African, Asian, and Arabic peoples who migrated to Madagascar several centuries ago, and there have been extensive economic and tourist contacts between countries of the Indian Ocean and Madagascar for centuries. It would be interesting to use other genetic markers or the spoligotyping method (23) to compare the Malagasy and Asiatic single-IS6110-copy strains and to look for the presence of other typical Asiatic patterns such as the Beijing and Nonthaburi groups (25, 33).
ACKNOWLEDGMENTS
We are grateful to Elie J. Vololonirina, T. Rasolonavalona, and P. Ravololonandriana for technical assistance and to F. Rakotomanana and M. Ratsitorahina for providing information about the patients. We also thank the physicians of the diagnostic and treatment centers of Antananarivo, Antsirabe, Fianarantsoa, and Mahajanga. We thank Isabelle Jeanne and the Bureau de I'Urbanisme d'Antananarivo for providing the Antananarivo city map.
The French National Reference Center for Mycobacteria (V. Vincent, Pasteur Institute, Paris, France) was the reference laboratory for the quality control of strain identification and drug susceptibility tests.
This study was supported by the French Cooperation (grant 93008) and the Raoul Follereau Foundation.
REFERENCES
- 1.Agasino C B, Ponce de Leon A, Jasmer R M, Small P M. Epidemiology of Mycobacterium tuberculosis strains in San Francisco that do not contain IS6110. Int J Tuberc Lung Dis. 1998;2:518–520. [PubMed] [Google Scholar]
- 2.Alland D, Kalkut G E, Moss A R, McAdam R A, Hahn J A, Bosworth W, Drucker E, Bloom B R. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N Engl J Med. 1994;17:1710–1716. doi: 10.1056/NEJM199406163302403. [DOI] [PubMed] [Google Scholar]
- 3.Aurégan G, Rakotondramarina D, Razafinimanana J, Ramarokoto H, Ratsirahonana O, Ralamboson M. Le Programme National de Lutte Anti-Tuberculeuse (PNLAT) à Madagascar. Arch Inst Pasteur Madagascar. 1995;62:4–12. [PubMed] [Google Scholar]
- 4.Aurégan G, Morvan J, Zeller H, Rasamindrakotroka A. SIDA et tuberculose: la situation à Madagascar. Arch Inst Pasteur Madagascar. 1995;62:24–25. [PubMed] [Google Scholar]
- 5.Bergmire-sweat D, Barnett B J, Harris S L, Taylor J P, Mazurek G H, Reddy V. Tuberculosis outbreak in a Texas prison, 1994. Epidemiol Infect. 1996;117:485–492. doi: 10.1017/s095026880005915x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canetti G, Rist N, Grosset J. Mesure de la sensibilité du bacille tuberculeux aux drogues antibacillaires par la méthode des proportions. Rev Tuberc Pneumol. 1963;27:217–272. [PubMed] [Google Scholar]
- 7.Cave M D, Eisenach K D, Templeton G, Salfinger M, Mazurek G, Bates J, Crawford J T. Stability of DNA fingerprint pattern produced with IS6110 in strains of Mycobacterium tuberculosis. J Clin Microbiol. 1994;32:262–266. doi: 10.1128/jcm.32.1.262-266.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cave M D, Eisenach K D, McDermott P F, Bates J H, Crawford J T. IS6110: conservation of sequence in the Mycobacterium tuberculosis complex and its utilization in DNA fingerprinting. Mol Cell Probes. 1991;5:73–80. doi: 10.1016/0890-8508(91)90040-q. [DOI] [PubMed] [Google Scholar]
- 9.Champetier de Ribes G, Ranaivoson G, Rakotoherisoa E, Andriamahefazafy B, Blanchy S. Le risque annuel d'infection tuberculeuse à Madagascar: étude réalisée de 1991 à 1994. Bull Soc Pathol Exot Fil. 1997;90:349–352. [PubMed] [Google Scholar]
- 10.Chanteau S, Rasolofo V, Ramarokoto H, Rasolonavalona T, Ratsirahonana O, Ratsitorahina M, Rakotomanana F, Boisier P, Cauchoix B, Aurégan G. Anti-tuberculosis drug resistance in Madagascar in 1994-95. Int J Tuberc Lung Dis. 1997;1:405–410. [PubMed] [Google Scholar]
- 11.Chaves F, Dronda F, Cave M D, Alonso-Sanz M, Gonzalez-Lopez A, Eisenach K D, Ortega A, Lopez-Cubero L, Fernandez-Martin I, Catalan S, Bates J H. A longitudinal study of transmission of tuberculosis in a large prison population. Am J Respir Crit Care Med. 1997;155:719–725. doi: 10.1164/ajrccm.155.2.9032218. [DOI] [PubMed] [Google Scholar]
- 12.Dale J W, Nor R M, Ramayah S, Tang T H, Zainudin Z F. Molecular epidemiology of tuberculosis in Malaysia. J Clin Microbiol. 1999;37:1265–1268. doi: 10.1128/jcm.37.5.1265-1268.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daley C L, Small P M, Schecter G F, Schoolnik K, McAdam R A, Jacobs W R, Hopewell P C. An outbreak of tuberculosis with accelerated progression among persons infected with human immunodeficiency virus. N Engl J Med. 1992;326:231–235. doi: 10.1056/NEJM199201233260404. [DOI] [PubMed] [Google Scholar]
- 14.Das S, Paramasivan C N, Lowrie D B, Prabhakar R, Narayanan P R. IS6110 restriction fragment length polymorphism typing of clinical isolates of Mycobacterium tuberculosis from patients with pulmonary tuberculosis in Madras, south India. Tuberc Lung Dis. 1995;76:550–554. doi: 10.1016/0962-8479(95)90533-2. [DOI] [PubMed] [Google Scholar]
- 15.Dwyer B, Jackson K, Raios K, Sievers A, Wilshire E, Ross B. DNA restriction fragment analysis to define an extended cluster of tuberculosis in homeless men and their associates. J Infect Dis. 1993;167:490–494. doi: 10.1093/infdis/167.2.490. [DOI] [PubMed] [Google Scholar]
- 16.Fomukong N G, Tang T H, Al-Maamary S, Ibrahim W A, Ramayah S, Yates M, Zainuddin F, Dale J W. Insertion typing of Mycobacterium tuberculosis: characterization of a widespread subtype with a single copy of IS6110. Tuberc Lung Dis. 1994;75:435–440. doi: 10.1016/0962-8479(94)90117-1. [DOI] [PubMed] [Google Scholar]
- 17.Genewein A, Telenti A, Bernasconi C, Mordasini C, Weiss S, Maurer A-M, Rieder H, Schopfer K, Bodmer T. Molecular approach to identifying route of transmission of tuberculosis in the community. Lancet. 1993;342:841–844. doi: 10.1016/0140-6736(93)92698-s. [DOI] [PubMed] [Google Scholar]
- 18.Gillespie S H, Dickens A, McHugh T D. False molecular clusters due to nonrandom association of IS6110 with Mycobacterium tuberculosis. J Clin Microbiol. 2000;38:2081–2086. doi: 10.1128/jcm.38.6.2081-2086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Global Tuberculosis Programme World Health Organisation (WHO) Geneva; International Union Against Tuberculosis and Lung Disease (IUATLD) Paris. Guidelines for surveillance of drug susceptibility resistance in tuberculosis. Int J Tuberc Lung Dis. 1997;2:72–89. [PubMed] [Google Scholar]
- 20.Helali N E, Vergez P. Identification des mycobactéries. Feuill Biol. 1993;190:5–18. [Google Scholar]
- 21.Hermans P W M, van Soolingen D, Bilk E M, de Haas P E W, Dale J W, van Embden J D A. Insertion element IS687 from Mycobacterium bovis is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59:2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hermans P W M, Messadj F, Guebrexabher H, van Soolingen D, de Haas P E W, Heersam H, de Neeling H, Ayoub A, Portaels F, Frommel D, Zribi M, van Embden J D A. Analysis of the population structure of Mycobacterium tuberculosis in Ethiopia, Tunisia, and The Netherlands: usefulness of DNA typing for global tuberculosis epidemiology. J Infect Dis. 1995;171:1504–1513. doi: 10.1093/infdis/171.6.1504. [DOI] [PubMed] [Google Scholar]
- 23.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J D A. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemaître N, Sougakoff W, Truffot-Pernot C, Cambeau E, Derenne J-P, Bricaire F. Use of DNA fingerprinting for primary surveillance of nosocomial tuberculosis in a large urban hospital: detection of outbreaks in homeless people and migrant workers. Int J Tuberc Lung Dis. 1998;2:390–396. [PubMed] [Google Scholar]
- 25.Palittapongarnpim P, Luangsook P, Tansuphaswadikul S, Chuchottaworn C, Prachaktam R, Sathapatayavongs B. Restriction fragment length polymorphism study of Mycobacterium tuberculosis in Thailand using IS6110 as probe. Int J Tuberc Lung Dis. 1997;1:370–376. [PubMed] [Google Scholar]
- 26.Rasolofo-Razanamparany V, Ménard D, Ratsitorahina M, Aurégan G, Gicquel B, Chanteau S. Transmission of tuberculosis in the prison of Antananarivo, Madagascar. Res Microbiol. 2000;151:785–795. doi: 10.1016/s0923-2508(00)01144-x. [DOI] [PubMed] [Google Scholar]
- 27.Skuce R A, Brittain D, Hughes M S, Beck L-A, Neill S D. Genomic fingerprinting of Mycobacterium bovis from cattle by restriction fragment length polymorphism analysis. J Clin Microbiol. 1994;32:2387–2392. doi: 10.1128/jcm.32.10.2387-2392.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Small P M, Hopewell P C, Singh S P, Paz A, Parsonnet J, Ruston D C, Schecter G F, Daley C L, Schoolnik G K. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N Engl J Med. 1994;330:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- 29.Thierry D, Brisson-Noël A, Vincent-Levy-Frebault V, Nguyen S, Guesdon J-L, Gicquel B. Characterization of a Mycobacterium tuberculosis insertion sequence, IS6110, and its application in diagnosis. J Clin Microbiol. 1990;28:2668–2673. doi: 10.1128/jcm.28.12.2668-2673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torrea G, Levée G, Grimont P, Martin C, Chanteau S, Gicquel B. Chromosomal DNA fingerprinting analysis using the insertion sequence IS6110 and the repetitive element DR as strain-specific markers for epidemiological study of tuberculosis in French Polynesia. J Clin Microbiol. 1995;33:1899–1904. doi: 10.1128/jcm.33.7.1899-1904.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P W M, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Rie A, Waren R, Ricardson M, Victor T C, Gie R P, Enarson D A, Beyers N, van Helden P D. Exogenous reinfection as a cause of recurrent tuberculosis after curative treatment. N Engl J Med. 1999;341:1174–1179. doi: 10.1056/NEJM199910143411602. [DOI] [PubMed] [Google Scholar]
- 33.van Soolingen D, Qian L, de Haas P E W, Douglas J T, Traore H, Portaels F, Qing H Z, Enkhsaikan D, Nymadawa P, van Embden J D A. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J Clin Microbiol. 1995;33:3234–3238. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Soolingen D, de Haas P E W, Haagsma J, Eger T, Hermans P W M, Ritacco V, Alito A, van Embden J D A. Use of various genetic markers in differentiation of Mycobacterium bovis strains from animals and humans and for studying epidemiology of bovine tuberculosis. J Clin Microbiol. 1994;32:2425–2433. doi: 10.1128/jcm.32.10.2425-2433.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Soolingen D, de Haas P E W, Hermans P W M, Groenen P M A, van Embden J D A. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Soolingen D, Hermans P W M, de Haas P E W, Soll D R, van Embden J D A. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependant DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuen L K W, Ross B C, Jackson K M, Dwyer B. Characterization of Mycobacterium tuberculosis strains from Vietnamese patients by Southern blot hybridization. J Clin Microbiol. 1993;31:1615–1618. doi: 10.1128/jcm.31.6.1615-1618.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]