Abstract
Extracellular vesicles (EVs) are nano-sized lipid bilayer vesicles released by virtually every cell type. EVs have diverse biological activities, ranging from roles in development and homeostasis to cancer progression, which has spurred the development of EVs as disease biomarkers and drug nanovehicles. Owing to the small size of EVs, however, most studies have relied on isolation and biochemical analysis of bulk EVs separated from biofluids. Although informative, these approaches do not capture the dynamics of EV release, biodistribution, and other contributions to pathophysiology. Recent advances in live and high-resolution microscopy techniques, combined with innovative EV labeling strategies and reporter systems, provide new tools to study EVs in vivo in their physiological environment and at the single-vesicle level. Here we critically review the latest advances and challenges in EV imaging, and identify urgent, outstanding questions in our quest to unravel EV biology and therapeutic applications.
Knowledge of EV biogenesis pathways and biological activities has grown rapidly in the past decade1 (Fig. 1a,b). EVs are membrane-enclosed structures that are released into the extracellular milieu by all organisms and cell types studied so far. EVs are a diverse family in which subtypes have been defined based on subcellular origin, size, and composition: endosome-derived vesicles (including multivesicular endosome-derived exosomes with a diameter of 50–150 nm and secretory autophagosome-derived EVs); ectosomes and other microvesicles that bud from the plasma membrane (PM) as small as exosomes or up to several μm in size; midbody remnants released by dividing cells (Box 1); migrasomes trailing behind migrating cells2,3; apoptotic bodies dislodged from dying and disintegrating cells; and large oncosomes released by transformed cells with exaggerated membrane plasticity (Fig. 2a and Table 1). Recent discoveries reveal additional subclasses of microparticles and nanoparticles, such as exophers4,5, exomeres6, supramolecular attack particles7, and elongated particles8. Initial discoveries implicated EVs in cellular adherence (as ‘adherons’)9 and clearance10 in the early 1980s, and in immune regulation in the mid-1990s11. EVs also play crucial roles in neurodegenerative diseases, cancer progression, metabolic homeostasis, angiogenesis, inflammation, neuronal plasticity, migration, trophic support, and pathogenic infections12–15. These roles are primarily supported by the capacity of EVs to shuttle molecules from one cell to another.
Fig. 1 |. Timeline of EV imaging milestones and broad overview of microscopy techniques to resolve EVs at different scales.
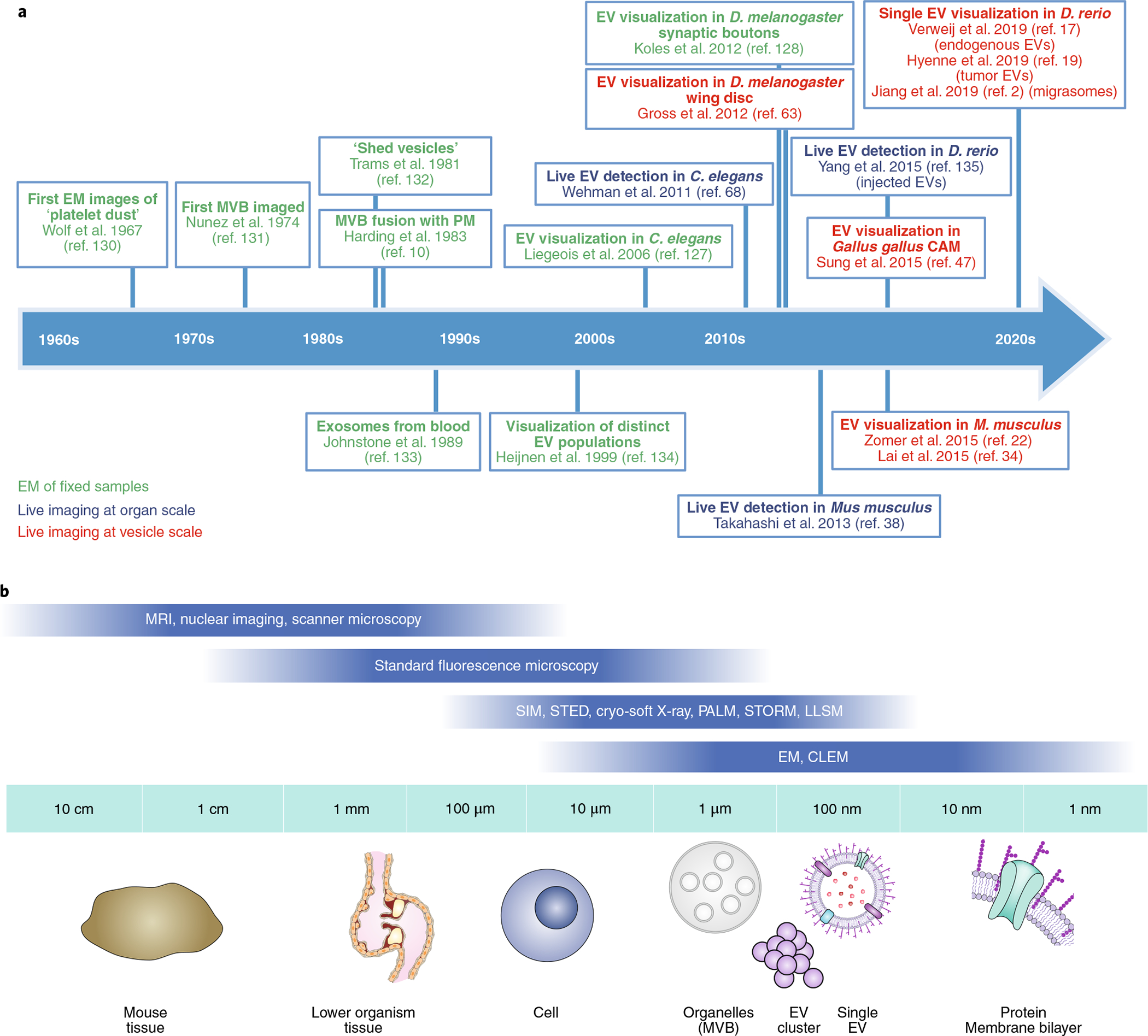
a, Timeline of imaging milestones in EV research. EM, electron microscopy. Refs. 2,10,17,19,22,34,38,47,63,68,128,129,131–136. b, Schematic of the resolution range of different microscopic approaches to resolve EVs at increasing resolution.
Box 1 |. Glossary.
Glycan extended trees.
Protein modification involving attached polymerized glycans that possess structural and/or modulatory function (for example, ligand binding).
Lipid membrane dye.
Lipophilic fluorescent dye that integrates in lipid membranes.
EV subtype.
EV with specific subcellular origin, size, and/or composition (Table 1).
Tetraspanin.
Family of membrane proteins with four transmembrane domains enriched in EVs.
Inner leaflet.
Cytosol- or EV-lumen-facing layer of a lipid bilayer.
Fluorescent complementation.
A technology used to validate protein interactions through the association of complementary fluorescent protein fragments attached to components of the same macromolecular complex.
Steric hindrance.
Here, spatial extent of an exogenous label preventing native interaction(s) of the labeled protein.
EV cargo.
Any molecule (lipid, protein, metabolite, genetic material) shuttled within or on EVs.
CLEM.
Imaging technique to correlate (live) light microscopy with ultrastructural information obtained on the same sample after fixation.
Diffraction limit.
Theoretical limit of optical microscopes to distinguish objects separated by a lateral distance less than half of the wavelength used.
Photobleaching.
Photon-induced alteration of a fluorophore that causes it to permanently lose its ability to fluoresce.
Phototoxicity.
Photon-induced damage to cellular macromolecules that impairs sample physiology.
Intraluminal vesicles.
Vesicles formed inside endosomes and precursors of canonical exosomes (Table 1).
3D microenvironment.
Local environment surrounding a cell, consisting of ECM, soluble factors, and other cells.
Gene traps.
Here, insertion of fluorescent tag such that the labelled protein is expressed under its endogenous promoter.
Lectins.
Saccharide binding proteins.
Midbody remnants.
Condensed membrane structure derived from the intercellular bridge that is left over after cell division.
V-ATPase.
Transmembrane proton pump functioning to acidify intracellular compartments.
Back-fusion.
Process in which ILVs or internalized EVs fuse with the late-endosomal limiting membrane, exposing their lumen to the cytosol and delivering their luminal content to the cytoplasm of recipient cells.
Fig. 2 |. Tagging strategies to image EV production.
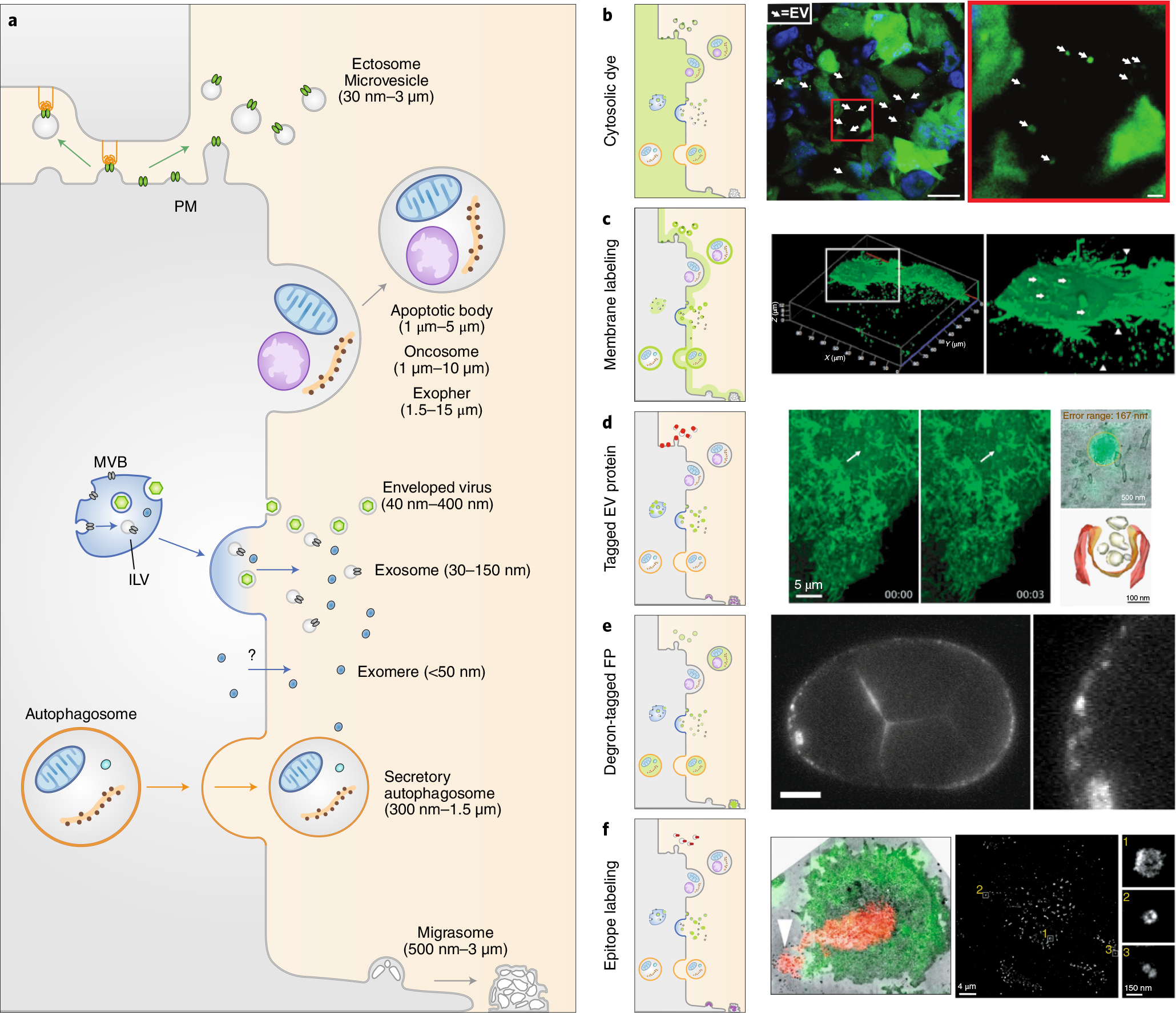
a, EVs are diverse double-leaflet membrane-enclosed structures generated from the PM (microvesicles, apoptotic bodies, oncosomes, exophers, enveloped viruses, and migrasomes), from endosomal compartments (exosomes and enveloped retroviruses), and from autophagic compartments (secretory autophagosomes). The origin of exomeres is still uncertain. b–d, Tagging strategies to image EVs. Cytoplasmic labeling facilitates pan-EV tagging by labeling the cell cytosol and the lumen of any EVs (b). Right, large EVs released from MDA-MB-231 cells expressing Dendra2 in mice mammary glands22. Arrows, EVs. Scale bars: 10 mm (left image), 1 mm (right image). Membrane labeling tags multiple EV subtypes (c). Right, confocal microscopy of live PalmGFP-expressing 293 T cells releasing EVs34. Arrows, bud-like structure from the surface; arrowheads, processes extending from cells. Expressing tagged cargo proteins allows the tracking of EV subtypes (d). Right, live imaging of a burst of CD63-pHluorin fluorescence at the HeLa cell surface (arrows, fusion event), overlaid using CLEM (top right image) to observe an MVB fusing with the PM to release exosomes (bottom right image)49. e, Expression of degron-tagged fluorescent proteins allows EV tagging while cytosolic fluorescence in the source cell is degraded. Right, PH::CTPD-labeled EVs released from the unlabeled PM in C. elegans51. Scale bar, 10 μm. f, Targeting of EV surface proteins by antibodies. Right, optical-EM correlation of M. musculus T cell that released EVs (red)53. Arrowhead, released microvesicles. Single EV imaging by dSTORM analysis of antibody staining54 (right image and insets).
Table 1 |.
EVs and particles
| Name | Size | Acronyms and other names | origin | Features |
|---|---|---|---|---|
| Exosomes | 30 nm–150 nm | Tolerosomes, prostasomes | MVBs, late or recycling endosomes, amphisomes | Lipid bilayer; contains proteins, genetic material, metabolites |
| Microvesicles | 50 nm–5μm | MVs; ectosomes, microparticles, synaptosomes, myelosomes, prostasomes, prominosomes | PM, microvilli | Lipid bilayer; contains proteins, genetic material, metabolites |
| Apoptotic bodies | 1 μm–5 μm | Apoptotic blebs | PM | Lipid bilayer; contains proteins, cytosolic components, organelles, nuclear fragments |
| Oncosomes | 100 nm–400 nm | None | PM | Lipid bilayer; contains oncoproteins, genetic material, oncometabolites |
| Large oncosomes | 1 μm–>10 μm | LO | PM | Lipid bilayer; contains peculiar cancer cell metabolism related enzymes |
| Enveloped viruses | 40 nm–400 nm | Miscellaneous | Endosomes, PM | Lipid bilayer, virion, viral proteins, viral genetic material |
| Exomeres | <50 nm | None | ND | Might lack a lipid bilayer; contains proteins such as argonaute and APP, lipids, and nucleic acids |
| Exophers | 1.5–15 μm | None | PM | Lipid bilayer; contains metabolic waste, protein aggregates, organelles |
| Secretory autophagosomes | 300 nm–1.5 μm | Mitovesicles? | Autophagic pathway | Lipid bilayer; contains cytoplasmic contents, excess or damaged proteins, organelles, microorganisms |
| Migrasomes | 500 nm–3 μm | None | PM-derived retraction fibers | Lipid bilayer; cytoplasmic content |
| Supramolecular attack particles | 120 nm | SMAPs | ND, cytotoxic granules | No lipid bilayer; cytotoxic core surrounded by thrombospondin-1 shell |
| Elongated particles | 1.9 μm–112 μm | Shear-derived particles, SDP | PM | Lipid bilayer; shear-derived particle, observed in rolling neutrophils |
EVs comprise a heterogeneous population of membrane vesicles. Their sizes vary between <50 nm and >5 μm. They can originate from the PM, or the endosomal or autophagic pathways. ND, not determined.
Despite the clear importance of EV biology, EV research faces challenges imposed by the small size and heterogeneity of EVs. Most studies have used bulk separation and characterization of heterogeneous populations of EVs from biological fluids or extended, large-scale in vitro cell cultures. These approaches allow robust characterization16 at the population level—for example, size and molecular profiles—but removing EVs from their context precludes insight into subcellular origin, release and uptake dynamics, and half-life. Separation can also disrupt fragile components such as branched glycans (Box 1), potentially altering EV functionality. Furthermore, studies in two-dimensional (2D) monocultures do not necessarily reflect what occurs in vivo.
Recent advances in live and high-resolution microscopy, combined with novel EV labeling strategies, now allow us to interrogate the composition and behavior of EVs at the single-vesicle level in living organisms17–20 (Fig. 1a). Functional transfer of EV proteins and RNA can also be assessed using novel reporters in vivo21,22 and in vitro23. These developments open new vistas in EV biology, providing the means to examine previously intractable issues such as assessing the lifespan of EVs in vivo. Here we review the state-of-the-art in EV labeling and tracking in animal model systems. We identify pitfalls and propose solutions and best practices. Finally, we discuss how recent advances in imaging can address open questions in EV biology, from biogenesis to uptake and function, thereby enhancing the development of EV therapeutics.
Tagging strategies, microscopy technology, and animal models
Labeling strategies that allow imaging with subcellular resolution are necessary for imaging EVs. In recent years, several such strategies or applications have been developed, ranging from novel lipid dyes (Box 1) to luminal dyes and genetic labeling (Table 2 and Fig. 2).
Table 2 |.
Tagging strategies for EVs
| Strategy | Biogenesis | Secretion | Transfer | Biodistribution | uptake | Functional transfer | Subcellular resolution | Body-wide resolution | Microscopy techniques | Modality |
|---|---|---|---|---|---|---|---|---|---|---|
| Lipid dyes | ||||||||||
| PKH, MemBright, DiI, DiO, DiR | − | −/+ | + | + | + | − | + | −/+ | CM, SDM, BFM | Live, fixed |
| Radiolabels and metabolic labels | ||||||||||
| Radioisotopes (that is, 99mTc) | − | − | − | ++ | − | − | − | ++ | SPECT, PET | Live |
| Metabolic labeling (for example, glycan) | − | − | − | ++ | + | − | −/+ | ++ | CM, SDM, BFM | Live |
| Genetic labeling strategies | ||||||||||
| Protein fused to fluorescent protein (for example, TSPAN–XFP) | + | ++ | + | + | + | − | + | −/+ | IEM, CM, SDM, TIRFM, BFM | Live, fixed |
| Degron tagging | − | + | + | + | + | − | + | −/+ | CM, SDM, BFM | Live, fixed |
| Cre/loxP | − | − | − | −/+ | −/+ | ++ | − | + | CM, SDM, BFM | Live, fixed |
| APEX | + | − | − | − | + | + | + | − | EM | Fixed |
| Nanoluciferase | − | + | − | + | + | − | + | ++ | BLIM, IEM | Live, fixed |
Different labeling strategies are suitable for visualizing EV (subtype) biogenesis, secretion, transfer, biodistribution, uptake, and functional (cargo) transfer, as well as for live or fixed imaging at subcellular or body-wide resolution. −, unsuitable; −/+, low suitability; +, suitable; ++, highly suitable. BFM, bright-field microscopy; BLIM, bioluminescence imaging microscopy; CM, confocal microscopy; IEM, immuno-electron microscopy; PET, positron emission tomography; SDM, spinning disk microscopy; SPECT, single-photon emission computed tomography; TIRFM, total internal reflection fluorescence microscopy.
Lipid dyes.
Lipid dyes (for example, PKH67, DiR/DiD, MemGlow) have been widely applied to label EVs with various excitation and emission wavelengths24, including the infrared range for greater penetration through tissues for in vivo studies. However, the application of lipophilic dyes to study EVs is complicated by unbound dye, aggregate and micelle formation, promiscuous labeling of non-EV particles, and the long half-life25. Labeling protocols should therefore limit dye concentrations during labeling, remove free dye after labeling, include appropriate controls (for example, ‘dye only’ control in EV solvent), and consider using multiple differentially stained EV populations to demonstrate absence of dye transfer or vesicle aggregation after co-isolation26. Recently, MemGlow27 was reported to be brighter and less prone to aggregate formation compared with traditional lipid dyes19.
Lipid dyes can be applied directly to producer cells followed by EV isolation19. However, it is unknown whether cell labeling affects EV release or function, or equally labels EV subtypes. Lipid dyes might also affect membrane–membrane fusion, fluidity of membrane proteins, membrane stiffness and EV size28. As the half-life of lipid dyes greatly exceeds that of EVs29,30, EV degradation after cellular uptake can be masked by recycling and redistribution of fluorescent dye. Therefore, lipophilic dye labeling of EVs may be best suited for short-term studies31.
Dye labeling of the EV lumen.
Dyes such as carboxyfluorescein diacetate succinimidyl ester (CFDA-SE) and calcein-AM label proteins in the EV lumen30,32. Their dependence on luminal esterases for conversion into a fluorescent product may produce fewer false-positive EV signals than lipophilic dyes but probably restricts labeling to a subpopulation of esterase-containing EVs33.
Fluorescent and bioluminescent protein EV reporters.
Various genetically encoded reporters have been developed to label all EVs or subtypes (Box 1) using fluorescence or bioluminescence. Labeled proteins expressed in the cytosol can be shuttled into the lumen of both exosomes and ectosomes (Fig. 2b)22. Addition of a palmitoylation signal associates the reporter with the inner leaflet (Box 1) of PM-derived EVs in vivo (Fig. 2c)34. For labeling of specific EV subtypes, reporters (including GFPs, RFPs, and the bioluminescent ThermoLuc) can be attached to EV cargos (for example, syntenin or tetraspanin (TSPAN; Box 1) family members (TSPAN4, CD63, CD81 and CD9)2,17,19,26,35, of which CD63 is most widely used). Alternative scaffolds and double labeling strategies36 can be considered to permit subtype detection. In contrast to fluorescent proteins, bioluminescent proteins emit signal after substrate addition with a high signal-to-noise ratio but comparatively lower spatiotemporal resolution37. Therefore, bioluminescence-based reporters (gLuc-lactadherin, GlucB) are predominantly used in small animal models to track EV biodistribution at whole-animal and organ scales38,39 (Table 2). More recently, a third category of EV reporter using bioluminescence resonance energy transfer (BRET) has been described (PalmGRET), allowing EV biodistribution analysis and in vivo quantification from whole animal to super-resolution without requiring multiple reporters40.
Excitingly, genetic labeling allows access to the entire fluorescent protein toolbox, including photoswitching and photoactivation, biosensors and bimolecular fluorescent complementation (Box 1). However, genetic labeling also comes with challenges. Labeling transmembrane proteins might disrupt conformation or cause steric hindrance (Box 1) of ligand–receptor interaction and organotropism41–43. EV surface-associated reporters may also be prone to proteolytic cleavage44, removing the signal45. Reporter overexpression may affect cellular signaling, EV cargo loading, or endogenous EV production and trafficking. Although a recent study demonstrated that CD63-GFP labeling of EVs only minimally perturbed the EV proteome26, other studies reported alterations in endolysosomal trafficking46, suggesting context-specific effects. Overexpression may also misdirect the reporter protein to unintended EV subtypes. Moreover, the amount of fluorescence emitted by the producing cell will ordinarily overpower the fluorescent signal of small EVs (approximately one millionth of the cell volume) in the immediate vicinity. One solution is the use of pH-sensitive fluorophores (for example, pHluorin), which are quenched in acidic cellular organelles but detected upon EV release, as successfully applied in vitro47–50 and in vivo17 (Fig. 2d). A second strategy is degron tagging, whereby cytosolic signal in the producing cell is degraded, while the signal in EVs persists51 (Fig. 2e).
Epitope targeting of EV surface proteins.
EV-enriched surface proteins and glycans can be targeted to visualize and characterize EVs in live and fixed cells (Fig. 2f). Pre-labeling of glycans on the PM with fluorescent hyaluronic acid binding complex (fHABC) allows live visualization of EV budding and fission from the cell surface52. Fluorescently labelled antibody fragments, such as nanobodies or fragment antigen-binding (Fab) domains, can also target EV-enriched proteins, with the advantages of eliminating the need for a secondary antibody and their smaller size compared to intact immunoglobulins. These strategies are compatible with most microscopy approaches53 and allow imaging at single-EV resolution54. With these tags, imaging EVs near the producing cells can be difficult if the epitope is present on both EVs and the PM. Depending on the resolving power of the imaging modality, the use of EV capture55 or immobilization strategies56 may be necessary.
A ‘one size fits all’ EV reporter does not exist (yet), and a particular reporter should be chosen based on the biological question and available imaging equipment. The specificity of the strategies to label EVs should preferably be validated with super-resolution and/or ultrastructural techniques. Along these lines, several recent studies have used combinations of correlative light and electron microscopy (CLEM; Box 1), immuno-electron microscopy (IEM), and/or scanning electron microscopy (SEM) to validate in vitro and in vivo approaches17,19,48,49 (Table 2).
Microscopy.
Apart from successful labeling, live imaging of EVs in vivo also requires a dedicated imaging set-up. Ideally, the set-up is suitable for deep tissue imaging while being resolutive and sensitive enough to observe EVs without inducing phototoxicity (Fig. 1b). This means relying on fast but often diffraction-limited systems (Box 1). Super-resolution microscopy (SRM)—for example, stochastic optical reconstruction microscopy (STORM) and photoactivated localization microscopy (PALM)—improve resolution to the nanometer scale, but often require fixation and are time-consuming. Other SRM approaches better suited for live-cell imaging of EV uptake and processing are structured illumination microscopy (SIM) and stimulated emission depletion microscopy (STED). All SRM techniques depend on high photon intensities, complicating detection of smaller EVs and increasing the risk of photobleaching and phototoxicity (Box 1), especially when imaging larger volumes in vivo over time. This renders some of the current SRM techniques incompatible with robust live imaging of EVs in vivo.
What is the best fluorescence microscopy system to study EV biology in vivo? The answer depends on the specific research question and the physiological and pathological context (Table 3). Confocal laser scanning microscopy (CLSM) can detect EVs in the sub-200-nm range, track their uptake by living cells, and their dynamic intracellular distribution on a time scale of seconds. However, EVs will appear as a small set of pixels by light microscopy and insufficient structural detail is attained to determine EV diameter and distinguish single EVs from EV clusters, dye aggregates, or dye-labeled protein aggregates and other particles. In addition, tracking of rapidly moving EVs (for example, in circulation17,19) and/or longer time-lapses require high-speed imaging with systems such as spinning disk microscopy and selective plane illumination microscopy (SPIM). These set-ups allow fast acquisition of EV movement, image larger volumes in vivo, and limit photobleaching and phototoxicity57. However, cells might be negatively affected by illumination even before they start to display morphological changes such as membrane blebbing57,58. Subtle impacts of prolonged imaging (for example, on cellular metabolic state) must be kept in mind, as they may affect EV release quantitatively and/or qualitatively. Emerging techniques, including lattice light-sheet microscopy (LLSM), could prove instrumental to enable sustained high-resolution live imaging with minimal photobleaching and phototoxicity59.
Table 3 |.
Microscopy methods
| Modalities | Resolution (XY) | Resolution (Z) | Illumination | Probes | Acquisition time | Post-acquisition processing | Live or fixed |
|---|---|---|---|---|---|---|---|
| Standard fluorescence microscopy | 250 nm | 500 nm | Epi, confocal, TIRF | Conventional fluorescent probes | Seconds | Live, fixed | |
| SIM, airyscan | 80 nm–150 nm | 250 nm–350 nm | Widefield (epi and TIRF) | Conventional fluorescent probes | Seconds | Yes, FTT | Live, fixed |
| STED | 30 nm–80 nm | 150 nm | Laser scanning | Limited selection of probes (match depletion laser) | Seconds | No | Live, fixed; optimal for fixed |
| Cryo-soft X-ray tomography | 25 nm–40 nm | 30 nm | Widefield | none | No | Fixed (near-native state vitrification) | |
| PALM | 20 nm | 50 nm | Widefield (epi and TIRF) | Photoactivatible fluorescent proteins | Minutes | Yes (PSF mapping) | Live, fixed |
| STORM | 20 nm | 50 nm | Widefield (epi and TIRF) | Photoswitchable dyes | Minutes | Yes (PSF mapping) | Live, fixed |
| LLSM | 100 nm–200 nm | 400 nm | Multi-Bessel beam plane illumination | Conventional fluorescent probes | Seconds, minutes or hours | Not necessary, but often tracking dynamic processes | Live, fixed; optimal for live |
| TEM | <1 nm | 70 nma | Electron beam | Contrast reagent, immunochemistry | Seconds | Yes | Fixed |
| CLEM | <1 nm/150 nmb | 5 nmc | Electron beam, widefield | Contrast reagent, nanodots, and fluorescent proteins | Minutes | Yes (aligning) | Live and/or fixed |
Characteristics of imaging methods used to visualize EVs. epi, epifluorescence; FTT, fast Fourier transform; PSF, point spread function; TEM, transmission electron microscopy.
Resolution corresponding to the thickness of the section.
Resolution gap between electron microscopy and light microscopy data, respectively.
Tomography from double-tilted 250 nm sections.
IEM and CLEM allow validation of EV-labeling approaches; for example, to confirm proper association with intraluminal vesicles (ILVs; Box 1)17,19,48,49. These approaches can be used in in vitro cultures and in vivo models to study aspects of the EV lifespan like extracellular fate post-secretion or subcellular distribution in receiving cells17,19,60. Importantly, EM provides ultrastructural resolution and label-free visualization of EVs in their native environment. In addition, immunolabeling detects proteins at the single-EV or single-ILV level. However, IEM with CLEM is restricted to a posteriori imaging of fixed samples.
Model organisms.
Molecular processes involved in EV biogenesis, secretion, and uptake can be studied as isolated processes using in vitro approaches. However, the physiological quantities, content, release dynamics, natural targets, and stability of EVs are likely to be affected by the 3D microenvironment (Box 1). Particularly when studying EVs in the context of intercellular communication, one of the main paradigms in the field, a relevant context is essential. The use of primary cell sources and 3D models is therefore arguably a much-needed step to provide more physiological relevance compared to 2D monocultures of immortalized cell lines in vitro.
Drosophila melanogaster is an attractive model system for studying EVs in tissue organization, development, and systemic crosstalk61,62. Wnt and Hh-containing EVs have been observed ex vivo in D. melanogaster wing imaginal discs63–65. In addition, D. melanogaster has been used to study EV biology during mating behavior and in adaptive immunity66. Recently, an EV subpopulation from Rab11-positive multivesicular bodies (MVBs) was shown to be evolutionarily conserved in flies and human cells46. The worm Caenorhabditis elegans is also an interesting model organism to study inter-animal EV communication with fluorescently labeled EVs67 and EV biogenesis mechanisms using the ultrastructural resolution of EM68,69.
Imaging of more complex tissues, like those from vertebrates, comes with additional restraints (Table 4 and Fig. 3). The smaller the observed particle, the more important optical accessibility of the surrounding tissue becomes to reduce noise. For instance, a chorioallantoic membrane (CAM) model system allows the visualization of CD63-positive and CD44-positive EVs in vivo48,70. The zebrafish (Danio rerio), as a transparent vertebrate model, allows continuous live imaging of the blood flow of endogenous EVs and EVs exogenously administered throughout the embryo17,19. This model has permitted the exploration of EV biology in unprecedented detail71 (Fig. 3c,d), revealing correlates of EV characteristics and function43. In mice, functional EV cargo transfer from immune to neuronal cells and between tumor cells has been observed21,22, as well as stroma–glioblastoma interactions, including microRNA (miRNA) transfer18,72. Still, live imaging of EVs in mice is currently restricted to tissues immediately adjacent to the imaging window or to larger EVs, as small EVs probably escape detection in these models18,22,73 (Fig. 3b). Imaging less accessible areas or across organs often requires organ extraction and ex vivo (post-fixation) analysis74, and is possible only with sufficient EV accumulation over time. Moreover, sites of accumulation might not equate with sites of function. These considerations have complicated efforts to understand EV physiology in mammalian models.
Table 4 |.
Model systems for EV imaging
| Model system | Biogenesis | Secretion | Transfer | Biodistribution | uptake | Functional cargo transfer | Subcellular resolution | Relevance to human physiology | Cost | Throughput |
|---|---|---|---|---|---|---|---|---|---|---|
| In vitro (2D) | ++ | ++ | − | − | + | + | ++ | −/+ | Low | High |
| In vitro (3D; for example, organoids) | + | + | −/+ | − | + | + | ++ | ++ | Low | High |
| G. gallus CAM47,70 | + | + | + | −/+ | + | ++ | ++ | + | Low | Medium or low |
| C. elegans67–69,128 | + | + | + | + | ++ | ++ | ++ | −/+ | Low | Medium |
| D. melanogaster 63–65,129,130 | + | + | + | + | ++ | ++ | ++ | −/+ | Low | Medium |
| D. rerio 2,17,19 | + | + | ++ | ++ | ++ | ++ | ++ | + | Medium | Medium |
| M. musculus, Rattus norvegicus 21,22,34,118 | − | −/+ | + | ++ | + | ++ | −/+ | ++ | High | Low |
The suitability and relevance of different model systems for EV imaging to visualize disparate aspects of EV biology at different scales. −, unsuitable; −/+, low suitability; +, suitable; ++, highly suitable.
Fig. 3 |. Imaging EV propagation in vivo.
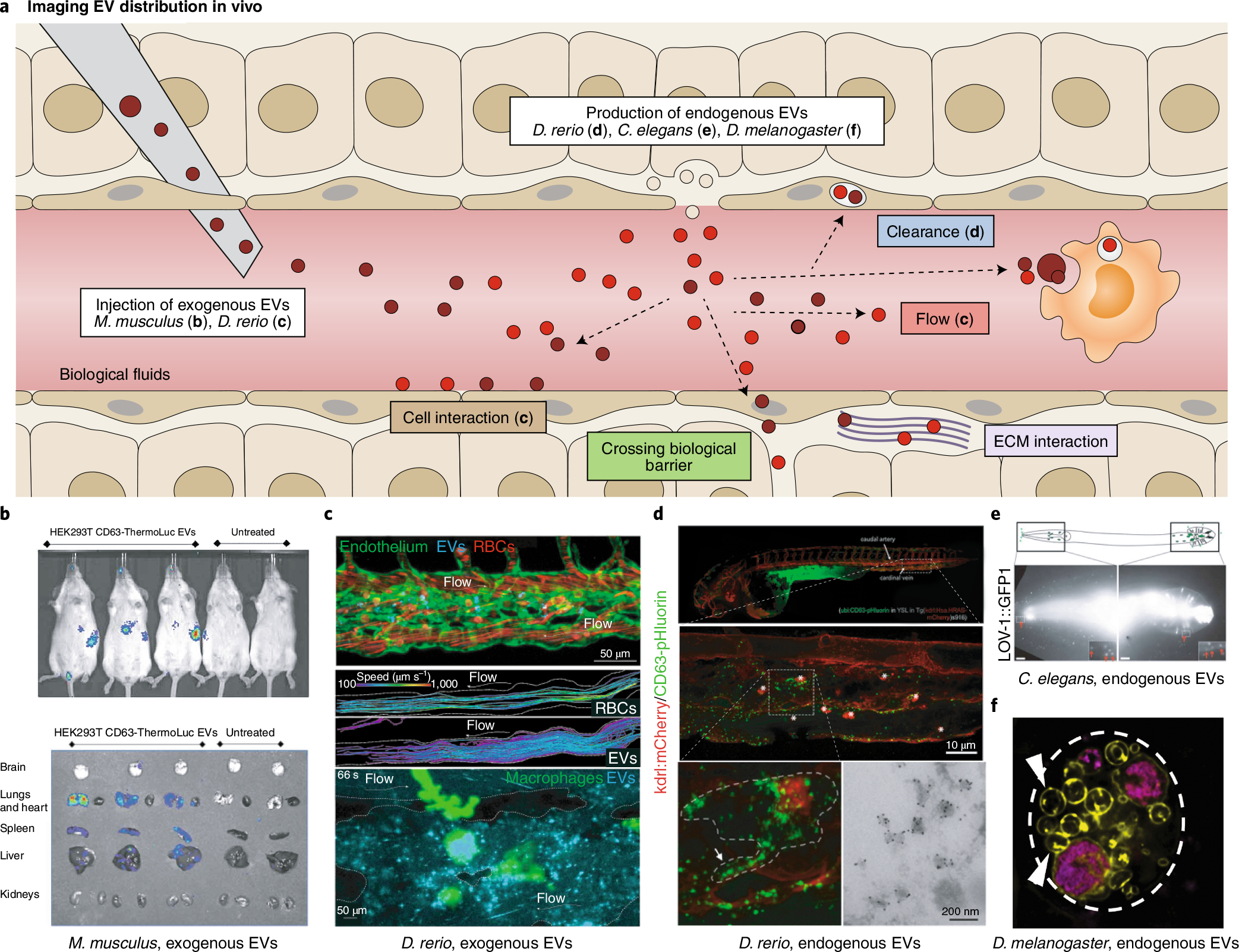
a, EV biodistribution can be mapped in the complex architecture of an organism after injection of labeled exogenous EVs or tagging endogenous EVs in situ. The in vivo fates of EVs (white boxes) are shown. b–d, Imaging using injected or endogenous EVs in live animals. b, EV accumulation tracked at the organ scale using CD63-ThermoLuciferase in mice73. c, EVs interacting with endothelial cells (green, top) or macrophages (green, bottom) tracked live in transparent zebrafish D. rerio; EV circulation in comparison to red blood cells (RBCs, red, middle)19. d, Endogenous EV clearance by scavenger endothelial cells and macrophages in D. rerio (top panels); inset of macrophage internalizing EVs captured from blood flow by SEC (bottom left); and IEM confirms the vesicular nature of the CD63-pHluorin signal in situ (bottom right)17. Arrow, macrophage protrusion; asterisks, macrophages. e, Fluorescently tagged EV cargo proteins track released EVs in C. elegans67. Red arrows, EVs surrounding the head and tail. Scale bars, 10 μm. f, Fluorescently tagged EV cargo proteins track EVs released from giant secretory MVB-like compartments in D. melanogaster130.
Thus, each model organism has its own strengths and weaknesses. Live imaging of single EVs in D. rerio, CAM, D. melanogaster, and C. elegans is highly realistic (Fig. 3e,f) in contrast with murine models. Although mice models display a higher degree of relevance to human physiology, the applicability of non-mammalian model systems to study human pathologies remains considerable: 82% of all disease-related genes are conserved in D. rerio, 75% in D. melanogaster, and >65% in C. elegans75–77. For example, disease-related models of neurodegenerative pathologies and tumor development have been introduced over the past decade77,78. Therefore, while considerations regarding relevance to human pathophysiology are important for any model system, these considerations should not block the access to a superior level of insight in smaller model organisms or preclude important questions from being addressed. D. melanogaster, C. elegans and D. rerio allow fundamental investigations in cell biology and development and are often vastly superior to murine models with regard to optical accessibility, genetic amenability, costs, and suitability for medium- or high-throughput approaches (Table 4). For example, exogenous tagging of proteins and tissue-specific expression using gene traps (Box 1) is well established using the GAL4–UAS system79. Various CRISPR–Cas9 and CRISPR–Cas12a systems are available for functional studies in vivo80,81, allowing loss- and gain-of-function studies and endogenous tagging and live imaging of proteins at endogenous expression levels (although these levels may not be sufficient to reliably follow small-sized particles such as EVs). Additionally, these models can be used as ‘pre-mouse’ models, where mice are subsequently deployed for key validation steps. Such strategies are consistent with the ‘3R’ principles in animal research. The choice of model system should therefore depend on the research question, the necessary level of resolution (single versus bulk EVs), and the required throughput (Table 4).
Imaging EV biogenesis, release, and distribution
In vitro studies revealed that most cells release EVs continuously and/or adapt release in response to triggers49,82,83. Similarly, most cells can take up EVs. Bulk EV isolation from culture media thereby neglects the subset of EVs that has been released and recaptured or does not spread beyond cell–cell interfaces. Moreover, culture media components and 2D versus 3D culture methods significantly affect EV release and EV composition84–89. Furthermore, little is known about bulk or subtype EV release dynamics or its dependence on characteristics of specific tissues and conditions (growth, homeostasis, pathology, specific triggers). Live imaging techniques now let us grasp these temporal, spatial, and conditional EV dynamics.
Imaging EV biogenesis and release.
EVs have two main subcellular origins: intracellular compartments and the PM. Although biogenesis at the PM is synonymous with release, EV release from intracellular compartments requires multiple steps, from ILV or autophagic vacuole biogenesis to organelle fusion with the cell surface (Fig. 2a).
Recent developments have enabled live visualization of PM-generated EVs by various approaches. Direct budding and fission of EVs into the extracellular milieu has been visualized in living cells after PM labeling with fHABC (fluorescent hyaluronic acid binding complex) in various cell types52 (Fig. 2c). Lectins (Box 1) such as wheat germ agglutinin (WGA) have also been used to label the surface of migrating cells and detect the formation of migrasomes on retraction fibers90. Alternative approaches have exploited migrasome-enriched transmembrane proteins such as TSPAN4 to track migrasome formation live in migrating cultured cells and during embryonic development in D. rerio2,3,91. Fluorescently tagged cytosolic proteins enriched in PM-derived EVs, such as midbody remnants, can also be harnessed to track biogenesis and uptake20,92. Immune cell synaptic microvesicle release can be studied on planar-supported lipid bilayers containing fluorescently labelled triggers of cargo loading into EVs via CLEM and STORM techniques53,54. These approaches may allow study of the molecular machinery of EV generation in an ideal setting for super-resolution microscopy.
To visualize exosome release, one successful approach is to image MVB–PM fusion. The acidic late-endosomal pH underlies the reason that PM fusion results in a burst of fluorescence from (super ecliptic) CD63-pHluorin93, which can be observed by live microscopy approaches, including total internal reflection fluorescence (TIRF) and spinning-disk microscopy47,49,50. This approach depends on fast acquisition times or dynamic CLEM to distinguish full MVB–PM fusion from rapid kiss-and-run motions that are inefficient in exosome release49 (Fig. 2d). CD63-pHluorin provides single-cell spatial information of release with high temporal resolution47,49,50. But this approach is best suited for flat surfaces (for example, the basolateral side of cells) and shorter time acquisitions at single-cell level, and hence less suitable than luciferase-coupled CD63 for medium- and high-throughput screens of EV biogenesis modulators94. Dual-color microscopy of dual-tagged reporters (pHluorin-CD63-mScarlet) allows MVBs to be tracked before fusion48, while other reporter combinations can unravel the molecular identity of MVBs that fuse with the PM50. However, using CD63-pHluorin to visualize MVB–PM fusion remains challenging in vivo due to the lack of high-speed and high-resolution modalities with limited phototoxicity17.
Imaging ILV formation in MVBs to study putative exosome biogenesis processes is equally challenging, as most live approaches lack single-vesicle resolution. The induction of enlarged endosomes by overexpressing GTPase-defective Rab5 improves resolution, but alters MVB maturation and function95. Moreover, MVBs may be destined for lysosomal degradation rather than EV secretion, limiting their relevance for exosome biogenesis. The giant secretory MVB-like compartments from D. melanogaster accessory glands allow unperturbed confocal and super-resolution visualization of intracellular sorting events and colocalization analysis of fluorescently labelled cargo proteins on ILVs in vivo46, but these processes may be specific to specialized cells.
Future developments are needed to combine measurements of ILV generation, exosome release, and PM budding simultaneously; for example, using high-speed 3D imaging. A clever approach to visualize protein trafficking has already revealed differences in endosome- and PM-derived EV proteomes96. Understanding these processes in further detail will let us interfere with formation and/or release of EV subclasses and provide an invaluable asset in our quest to attribute specific functions to EV subtypes in vivo.
Imaging EV distribution.
After EV release in vivo, the microenvironment plays a major role in EV distribution and function. Apart from EV-intrinsic factors (for example, adhesion molecules), the local 3D architecture, extracellular matrix (ECM)97 and biological barriers between organs affect EV diffusion and influence the physiological role of EVs (Fig. 3a). As these constraints determine local retention47,98 versus distant transport and may not be fully recapitulated in vitro, the need for realistic in vivo models of EV distribution is clear (Fig. 3).
Although murine studies are limited mostly to organ scale26 and disclose only the ‘final destination’ of EVs, smaller, transparent organisms allow subcellular resolution19 and live tracking of EV diffusion and transport (Table 4). Bioluminescent labeling, radiolabeling, and metabolic labeling are compatible with the former strategy, whereas the latter typically employs fluorescent protein- and lipid-labeling strategies.
Compared with studying endogenous EVs, isolation and injection of exogenous EVs permits fine control of engineering and dosing for optimal half-life and functional43 studies. Such studies have suggested rapid removal by tissue and cell types with sustained phagocytic capacity, even within 5 min after injection99. Although EV injection does not recapitulate the earliest aspects of the EV lifespan, two recent in vivo studies demonstrated that pre-labeled injected tumor EVs did not deviate considerably in fate from physiological EVs that are endogenously released in the blood flow17,19 (Fig. 3c,d).
Yet, it is not clear whether these examples are sufficient to warrant a generalized verdict concerning all EVs and all aspects of EV biology, especially regarding mRNA transfer100. Indeed, exogenous administration incompletely mimics physiological EV release levels (unless approximated by sustained delivery methods101), and physiological and pathological factors that might influence endogenous EV subset(s) might be absent in vitro84–89. EV subtypes isolated from in vitro cultures, some of which would normally act locally, would also artificially reach non-physiological sites upon injection in vivo. For example, EVs involved in ECM deposition and modulation47,102 might normally act near the cell of origin, as would EVs released at immunological or neurological synapses35,53,103,104. In addition, anatomical differences in vascular permeability (for example, liver versus brain), pathological conditions affecting endothelial barrier function, or antiviral mechanisms restricting EV diffusion could alter the efficiency of EV propagation and uptake99,105. Imaging the release and biodistribution of endogenous EV subsets in vivo under various conditions will reveal how EVs cross biological barriers under physiological conditions, for which only indirect proof is currently available; for example, intravenously injected EVs in the brain106,107. Ultimately, comparative studies of both endogenous and exogenous EV administration are needed. Studying endogenous EVs will show physiological concentrations and dynamics of EV release and biodistribution that highlight the best sites and frequencies of injection. This will help us to interpret exogenous EV studies and will permit finer control of certain EV-intrinsic variables. Together, these comparisons will inform EV targeting approaches for therapeutics.
Imaging interaction and uptake of EVs by recipient cells and related functions
The EV lifespan is often depicted as cell A releasing EVs that reach cell B, where endocytosis and (intraluminal) cargo delivery trigger a phenotypic response. Although this communication paradigm is exciting and supported by literature, EVs can also act in an autocrine fashion or have other ‘delivery-independent’ extracellular functions such as ECM modulation, PM receptor engagement or transfer of EV-resident proteins to recipient cells108–110 (Fig. 4a).
Fig. 4 |. Tagging strategies to image EV interaction, uptake, and fate.
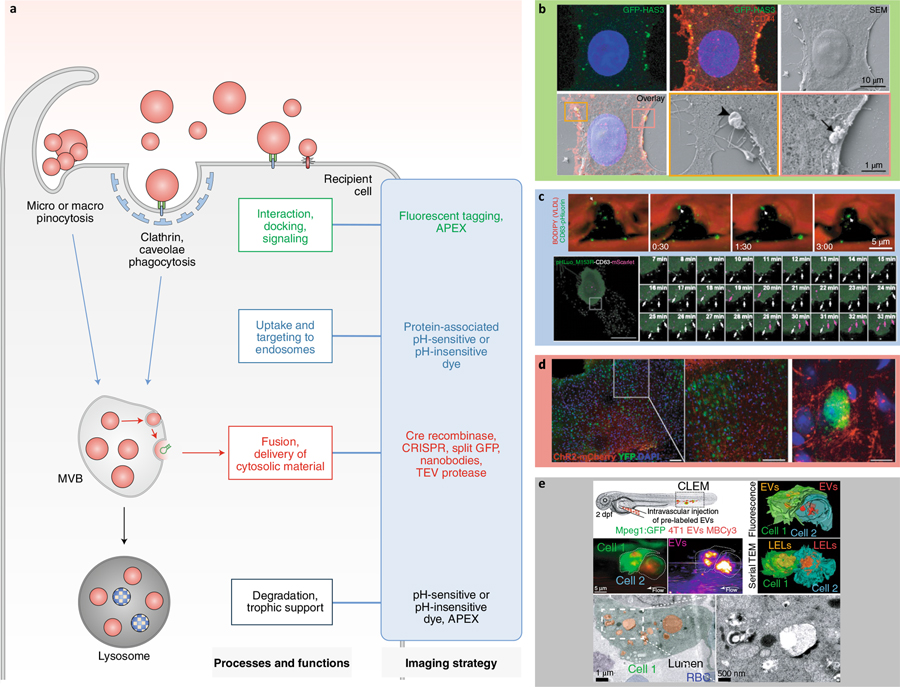
a, Different tagging strategies (blue box) reveal distinct aspects of EV–cell interactions. TEV, tobacco etch virus. b–e, Imaging strategies to track the fate and functions of EVs. b, Correlative light and scanning electron microscopy shows GFP-HAS3-labeled EVs interacting with the PM of receiving cells115. Arrowhead, large EV; arrow, cluster and individual EVs of different sizes. c, Tracking uptake of endogenous CD63-pHluorin-labeled EVs in the D. rerio vasculature17 (top); and tracking double-labeled pHluorin-CD63-mScarlet EVs inside and outside HT1080 cells48 (bottom). VLDL, very low density lipoprotein. Arrows, internalized EV (top). White arrows, external EVs; purple arrows, internal EVs (bottom). d, Ex vivo mapping of EV mRNA using a Cre recombinase strategy in the mouse brain118. e, Correlative light and electron microscopy shows Membright Cy3 lipid dye-labeled EVs accumulating in endolysosomes in D. rerio patrolling macrophages.19 dpf, days post-fertilization; LEL, late endo-lysosome; RBC, red blood cell.
Imaging interaction of EVs with recipient cells.
EVs can function by engaging PM-localized receptors at the target cell membrane, such as in antigen presentation, as super-cytokines, or as carriers of morphogens and ligands for pattern recognition receptors42,54,63,108,111–113. Whereas uptake of EVs has been amply demonstrated by (live) imaging, visualization of EV interaction with the PM has been reported on just a few occasions60, and only recently with live imaging in vitro59,114 and in vivo17,19. Direct observation115 might currently be limited due to a lack of suitable reporters. Indeed, whereas most studies adding labeled EVs to target cells show intracellular accumulation rather than PM labeling, this does not preclude previous EV–PM interaction, especially as functional cargo delivery appears to be a rare event from the ‘bulk EV flow’ perspective. For certain EVs, uptake might indeed be a prerequisite to function, but for other EVs, uptake followed by degradation could instead reflect an end-of-life event after signaling through PM receptors. To date, most reporter systems for EV function have focused on cytoplasmic cargo delivery rather than signal induction. Understanding fusion-independent EV functions thus requires combined microscopy approaches, such as CLEM (Fig. 4b), in vitro35,60,113 and in vivo17,19,61, to cover the full range from whole organism to subcellular at sufficient resolution with light-microscopy or EM ultrastructural resolution.
Imaging cellular uptake of EVs.
EVs are widely reported to deliver contents into the cytoplasm of recipient cells such as signaling proteins, RNA binding proteins, genetic material, metabolites, and enzymes. However, we know little about the fusion events or transporter systems necessary for such delivery. Often, studies follow uptake in bulk, and lack the resolution to study the fate of single EVs. Recently, EM has been used to examine EV uptake in vivo17,19. Live imaging approaches can reveal other details of EV fate, such as acidification of EV-containing compartments after uptake in vitro48 and in vivo17, distinguishing ‘storage’ from degradation (Fig. 4c). V-ATPase (Box 1) inhibitors might be required if uptake and degradation are highly efficient in target cells or to facilitate detection of rare events. Note that the choice of dye (for example, lipid or genetic protein labeling) determines what is being followed after EV uptake. Over time, labels might no longer represent intact EVs, but rather the trafficking of the label itself or of lipid or protein fragments.
Imaging EV function in recipient cells.
EVs elicit phenotypic responses in proximally and distally located cells. Reporter systems have been developed to visualize transfer of mRNAs21,22,100, miRNAs39,116, shRNAs23, and proteins117. Cytoplasmic delivery presupposes endosomal escape by EV-endosome fusion to avoid lysosomal degradation of EV cargos. So far, detection of cargo transfer by live imaging is limited to induction of a global signal at the cellular scale (Fig. 4d). Further resolution is needed to locate and elucidate endosomal escape, demanding new technological developments for single-molecule cargo tracking and to observe potential fusion (Box 1) of endocytosed EVs with the host membrane. Interestingly, in vivo mouse studies indicate that cargo transfer occurs at low ‘efficiency’ in the absence of a specific stimulus21,100. However, in certain pathological models, the functional uptake of EVs can be higher118, highlighting the need to study pathological situations in model organisms.
Several reports indicate a trophic support function of EVs via lysosomal degradation17,119. Lysosomal targeting can be studied by EM17,19 (Fig. 4e) or by live imaging using EV reporters with different acid sensitivity20,120. Live imaging in vivo revealed rapid internalization and degradation of injected or endogenous EVs by professional phagocytes (for example, macrophages and monocytes) and especially pinocytes (for example, scavenger endothelial cells). Therefore, some EVs might function without message delivery17,19. Although trophic function is not strictly incompatible with ‘message transfer’, a yet-unresolved question is whether EV-mediated communication is stochastic or deterministic from a donor cell perspective. Do cells release a large amount of EVs agnostically, letting the recipient cell determine whether to respond via an ‘activation status’ that determines cytoplasmic cargo delivery118? Or do cells release a limited number of ‘magic bullet’ EVs that are tailored for specific communication? The latter is currently supported in the immunological synapse setting35,53,60, but is perhaps less evident beyond this close cell–cell contact setting. These ‘magic bullets’ might be present within the main flow but possess molecular traits that promote capture, facilitate back-fusion (Box 1), or prevent degradation. Thus, tracking bulk EV flow may divert our attention from the rare EV-target cell interactions, the ‘magic bullets’ that do not follow bulk flow fate.
Technological strategies are important to monitor events in the transfer process121, but perhaps the most pressing need is to develop more fundamental knowledge of rare, ‘magic-bullet’ events. When we know the players, we can image the co-packaging of cognate molecules and targeting molecules into ILVs and EVs to follow EV lifespan events in real time, from biogenesis to target cell interactions.
Conclusion
Imaging technology has matured such that we can study most details of the EV lifespan in vivo using diverse tags and microscopy approaches, especially in optically transparent organisms. What is at stake is profound. Imaging biogenesis will distinguish EV subpopulations perhaps associated with distinct functions, and enable a firm nomenclature. By following the biodistribution of EVs in vivo, we will not only assess their capacity to cross biological barriers but also gain insight into their range of actions and their efficiency in reaching target cells previously identified in vitro. In vitro technologies can then be used to dissect mechanisms in more detail, lifting the veil around the important events that in vivo imaging has started to reveal73,106,122.
How EVs act as mediators of intercellular signaling is poorly understood. By following the fate of EVs in vivo, we will gain insight into their in vivo targets and functions. Direct imaging of the release of EV contents into recipient cells is needed to identify whether cargo transfer or signaling interaction (or both), is responsible for the effects of EVs. Although most studies focus on EV functions requiring EV uptake and cargo transfer into recipient cells, mounting evidence points towards extracellular roles for EVs involving neither uptake nor cargo delivery42,63,108,111,112,123. It is unclear how common extracellular roles are in vivo compared to intracellular functions, and whether EVs mainly act systemically rather than locally. The rapid clearance of the majority of injected EVs by the liver and spleen might indicate that many EVs function in waste disposal or trophic support. Therefore, it is important to determine the route taken by endogenous EVs in vivo and the amount of EVs necessary to impact target tissues. Following specific subclasses of EVs in vivo will aid in addressing these key questions, and reveal whether EV communication is stochastic and inefficient or rather relies on specialized EVs to transfer messages.
Knowing the in vivo characteristics of EVs, such as their half-life, biodistribution and targeting mechanisms, also supports their clinical application as biomarkers, drug carriers, or intrinsic modulators of pathological and physiological processes124–126. In vivo imaging approaches reveal the time and location of EV-subtype release and the biological fluids in which they are distributed or accumulate. This ‘hot spot’ mapping could optimize strategies to timely harvest the most relevant EVs for diagnosis or disease monitoring. High-resolution imaging of injected EVs purposed for drug delivery can likewise reveal EV pharmacokinetics (half-life, biodistribution, clearance), fate, and effects on recipient cells in real-time. This supports the development of engineering and administration protocols for efficient biodistribution and targeting, minimal clearance, and improved drug delivery efficiency in clinical practice. Monitoring EV dynamics in vivo will also identify drug targets for modulating EV release, uptake and degradation, influencing pharmacokinetics and EV-intrinsic functions. Thus, in vivo imaging approaches will not only provide crucial insight into fundamental aspects of the EV lifespan but will also benefit clinical development of EV-based drug delivery systems127.
The future of the field critically depends on a systematic approach comparing the pros and cons of each EV labeling and imaging strategy, in vitro and in vivo, to establish their relevance and determine good practices. We foresee development of important synergies between imaging methods and other techniques such as synthetic biology tools to investigate EV biology in vivo; for instance, by controlling and validating EV secretion and fate in vivo and to facilitate downstream analysis of specific EV subpopulations ex vivo. Imaging is now part of the toolbox of scientists studying EVs, who will work with other nanoscientists to further elucidate the biology and therapeutic applications of EVs.
Acknowledgements
The authors acknowledge financial support from the International Society for Extracellular Vesicles, the French Society of Extracellular vesicles, the Société Française des Microscopies, the ITMO BCDE for their support for the organization of the ISEV workshop ‘EV imaging in vivo’ that provided the basis for this review. We thank the workshop organizing committee members G.D.’A., V.H., E.-M.K.-A., X.L., and K.W. for their help. We thank P. Stahl (Washington University School of Medicine, USA) for stimulating discussions and insight. F.J.V. is supported by INCa 2019–125, E.B.C. thanks M. Dustin for support through ERC AdG 670930. D.R.F.C. is supported by the BBSRC (BB/P006205/1) and Cancer Research UK (A28052). V.H. and J.G.G. are funded by La Ligue contre le Cancer, by INCa (PLBIO19–291), by Plan Cancer (Nanotumor) and through institutional funds from University of Strasbourg and INSERM. K.R. is supported by the UEF Cell and Tissue Imaging Unit, Biocenter Kuopio and Biocenter Finland. We apologize to colleagues for any relevant work that could not be cited due to space restrictions.
Footnotes
Competing interests
D.R.F.C. is employed by Evox Therapeutics. S.E.A. serves on the Scientific Advisory Board of EVOX Therapeutics. All other authors have no competing interests.
Peer review information Rita Strack was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Van Niel G, D’Angelo G & Raposo G Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol 19, 213–228 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Jiang D et al. Migrasomes provide regional cues for organ morphogenesis during zebrafish gastrulation. Nat. Cell Biol 21, 966–977 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Huang Y et al. Migrasome formation is mediated by assembly of micron-scale tetraspanin macrodomains. Nat. Cell Biol 21, 991–1002 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Nicolás-Ávila JA et al. A network of macrophages supports mitochondrial homeostasis in the heart. Cell 183, 94–109.e23 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Melentijevic I et al. C. elegans neurons jettison protein aggregates and mitochondria under neurotoxic stress. Nature 542, 367–371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol 20, 332–343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bálint et al. Supramolecular attack particles are autonomous killing entities released from cytotoxic T cells. Science 368, 897–901 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marki A et al. Elongated neutrophil-derived structures are blood-borne microparticles formed by rolling neutrophils during sepsis. J. Exp. Med 218, e20200551 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schubert D A brief history of adherons: the discovery of brain exosomes. Int. J. Mol. Sci 21, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harding C, Heuser J & Stahl P Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol 97, 329–339 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raposo G et al. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med 183, 1161–1172 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yáñez-Mó M et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 4, 27066 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Budnik V, Ruiz-Cañada C & Wendler F Extracellular vesicles round off communication in the nervous system. Nat. Rev. Neurosci 17, 160–172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stahl PD & Raposo G Extracellular vesicles: exosomes and microvesicles, integrators of homeostasis. Physiology 34, 169–177 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Boulanger CM, Loyer X, Rautou P-E & Amabile N Extracellular vesicles in coronary artery disease. Nat. Rev. Cardiol 14, 259–272 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Jeppesen DK et al. Reassessment of exosome composition. Cell 177, 428–445.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verweij FJ et al. Live tracking of inter-organ communication by endogenous exosomes in vivo. Dev. Cell 48, 573–589.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 18.van der Vos KE et al. Directly visualized glioblastoma-derived extracellular vesicles transfer RNA to microglia/macrophages in the brain. Neuro. Oncol 18, 58–69 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyenne V et al. Studying the fate of tumor extracellular vesicles at high spatiotemporal resolution using the zebrafish embryo. Dev. Cell 48, 554–572.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Fazeli G, Trinkwalder M, Irmisch L & Wehman AMC elegans midbodies are released, phagocytosed and undergo LC3-dependent degradation independent of macroautophagy. J. Cell Sci 129, 3721–3731 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ridder K et al. Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLoS Biol. 12, e1001874 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zomer A et al. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 161, 1046–1057 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Jong OG et al. A CRISPR-Cas9-based reporter system for single-cell detection of extracellular vesicle-mediated functional transfer of RNA. Nat. Commun 11, 1113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonçalves MST Fluorescent labeling of biomolecules with organic probes. Chem. Rev 109, 190–212 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Pužar Dominkuš P et al. PKH26 labeling of extracellular vesicles: characterization and cellular internalization of contaminating PKH26 nanoparticles. Biochim. Biophys. Acta Biomembr 1860, 1350–1361 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Corso G et al. Systematic characterization of extracellular vesicles sorting domains and quantification at the single molecule–single vesicle level by fluorescence correlation spectroscopy and single particle imaging. J. Extracell. Vesicles 8, 1663043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collot M et al. MemBright: a family of fluorescent membrane probes for advanced cellular imaging and neuroscience. Cell Chem. Biol 26, 600–614.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Dehghani M, Gulvin SM, Flax J & Gaborski TR Exosome labeling by lipophilic dye PKH26 results in significant increase in vesicle size. Preprint at bioRxiv 10.1101/532028 (2019). [DOI] [Google Scholar]
- 29.Kuffler DP Long-term survival and sprouting in culture by motoneurons isolated from the spinal cord of adult frogs. J. Comp. Neurol 302, 729–738 (1990). [DOI] [PubMed] [Google Scholar]
- 30.Gray WD, Mitchell AJ & Searies CD An accurate, precise method for general labeling of extracellular vesicles. MethodsX 2, 488–496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chuo ST-Y, Chien JC-Y & Lai CP-K Imaging extracellular vesicles: current and emerging methods. J. Biomed. Sci 25, 91 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mckinnon KM et al. Labeling extracellular vesicles for nanoscale flow cytometry. Sci. Rep 7, 1878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao Z et al. Acetylcholinesterase is not a generic marker of extracellular vesicles. J. Extracell. Vesicles 8, 1628592 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai CP et al. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat. Commun 6, 7029 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mittelbrunn M et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun 2, 282 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathieu M, Martin-Jaular L, Lavieu G & Théry C Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol 21, 9–17 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Badr CE & Tannous BA Bioluminescence imaging: progress and applications. Trends Biotechnol. 29, 624–633 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi Y et al. Visualization and in vivo tracking of the exosomes of murine melanoma B16-BL6 cells in mice after intravenous injection. J. Biotechnol 165, 77–84 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Lai CP et al. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano. 8, 483 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu AYT et al. Multiresolution imaging using bioluminescence resonance energy transfer identifies distinct biodistribution profiles of extracellular vesicles and exomeres with redirected tropism. Adv. Sci 7, 2001467 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoshino A et al. Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen G et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 560, 382–386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghoroghi S et al. Ral GTPases promote breast cancer metastasis by controlling biogenesis and organ targeting of exosomes. eLife 10, 61539 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaborowski MP et al. Membrane-bound Gaussia luciferase as a tool to track shedding of membrane proteins from the surface of extracellular vesicles. Sci. Rep 9, 17387 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shinoda H, Shannon M & Nagai T Fluorescent proteins for investigating biological events in acidic environments. Int. J. Mol. Sci 19, 1548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan S et al. Glutamine deprivation alters the origin and function of cancer cell exosomes. EMBO J. 39, e103009 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sung BH, Ketova T, Hoshino D, Zijlstra A & Weaver AM Directional cell movement through tissues is controlled by exosome secretion. Nat. Commun 6, 7164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sung BH et al. A live cell reporter of exosome secretion and uptake reveals pathfinding behavior of migrating cells. Nat. Commun 11, 2092 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verweij FJ et al. Quantifying exosome secretion from single cells reveals a modulatory role for GPCR signaling. J. Cell Biol 217, 1129–1142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bebelman MP et al. Real-time imaging of multivesicular body–plasma membrane fusion to quantify exosome release from single cells. Nat. Protoc 15, 102–121 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Beer KB et al. Degron-tagged reporters probe membrane topology and enable the specific labelling of membrane-wrapped structures. Nat. Commun 10, 3490 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mustonen AM et al. First in vivo detection and characterization of hyaluronan-coated extracellular vesicles in human synovial fluid. J. Orthop. Res 34, 1960–1968 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Choudhuri K et al. Polarized release of T-cell-receptor-enriched microvesicles at the immunological synapse. Nature 507, 118–123 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saliba DG et al. Composition and structure of synaptic ectosomes exporting antigen receptor linked to functional CD40 ligand from helper T cells. eLife 8, e47528 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ambrose AR, Hazime KS, Worboys JD, Niembro-Vivanco O & Davis DM Synaptic secretion from human natural killer cells is diverse and includes supramolecular attack particles. Proc. Natl Acad. Sci. USA 117, 23717–23720 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanwar SS, Dunlay CJ, Simeone DM & Nagrath S Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip 14, 1891–1900 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Icha J, Weber M, Waters JC & Norden C Phototoxicity in live fluorescence microscopy, and how to avoid it. BioEssays 39, 1700003 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Spikes JD Photosensitization in mammalian cells. in Photoimmunology (eds Parrish JA et al.) 23–49 (Springer, 1983). [Google Scholar]
- 59.Elgamal S, Colombo F, Cottini F, Byrd JC & Cocucci E Imaging intercellular interaction and extracellular vesicle exchange in a co-culture model of chronic lymphocytic leukemia and stromal cells by lattice light-sheet fluorescence microscopy. Methods Enzymol. 645, 79–107 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Buschow SI et al. MHC II in dendritic cells is targeted to lysosomes or T cell-induced exosomes via distinct multivesicular body pathways. Traffic 10, 1528–1542 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Hurbain I et al. Microvilli-derived extracellular vesicles govern morphogenesis in Drosophila wing epithelium. Preprint at bioRxiv 10.1101/2020.11.01.363697 (2020). [DOI] [Google Scholar]
- 62.González-Méndez L et al. Polarized sorting of Patched enables cytoneme-mediated Hedgehog reception in the Drosophila wing disc. EMBO J. 39, e103629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gross JC, Chaudhary V, Bartscherer K & Boutros M Active Wnt proteins are secreted on exosomes. Nat. Cell Biol 14, 1036–1045 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Matusek T et al. The ESCRT machinery regulates the secretion and long-range activity of Hedgehog. Nature 516, 99–103 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Gradilla AC et al. Exosomes as Hedgehog carriers in cytoneme-mediated transport and secretion. Nat. Commun 5, 5649 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Tassetto M, Kunitomi M & Andino R Circulating immune cells mediate a systemic rnai-based adaptive antiviral response in drosophila. Cell 169, 314–325.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J et al. C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr. Biol 24, 519–525 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wehman AM, Poggioli C, Schweinsberg P, Grant BD & Nance J The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr. Biol 21, 1951–1959 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hyenne V et al. hRAL-1 controls multivesicular body biogenesis and exosome secretion. J. Cell Biol 211, 27–37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Härkönen K et al. CD44s Assembles hyaluronan coat on filopodia and extracellular vesicles and induces tumorigenicity of MKN74 gastric carcinoma. Cells Cells 8, 276 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verweij FJ, Hyenne V, Van Niel G & Goetz JG Extracellular vesicles: catching the light in zebrafish. Trends Cell Biol. 29, 770–776 (2019). [DOI] [PubMed] [Google Scholar]
- 72.Abels ER et al. Glioblastoma-associated microglia reprogramming is mediated by functional transfer of extracellular miR-21. Cell Rep. 28, 3105–3119.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gupta D et al. Quantification of extracellular vesicles in vitro and in vivo using sensitive bioluminescence imaging. J. Extracell. vesicles 9, 1800222 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Men Y et al. Exosome reporter mice reveal the involvement of exosomes in mediating neuron to astroglia communication in the CNS. Nat. Commun 10, 4136 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baumeister R & Ge L The worm in us - Caenorhabditis elegans as a model of human disease. Trends Biotechnol. 20, 147–148 (2002). [DOI] [PubMed] [Google Scholar]
- 76.Howe K et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fortini ME, Skupski MP, Boguski MS & Hariharan IK A survey of human disease gene counterparts in the Drosophila genome. J. Cell Biol 150, F23–F30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Santoriello C & Zon LI Hooked! Modeling human disease in zebrafish. J. Clin. Invest 122, 2337–2343 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caygill EE & Brand AH The GAL4 system: a versatile system for the manipulation and analysis of gene expression. in Drosophila: Methods and Protocols 2nd edn (ed. Dahmann C) 33–52 (Humana Press, 2016). [DOI] [PubMed] [Google Scholar]
- 80.Port F et al. A large-scale resource for tissue-specific CRISPR mutagenesis in Drosophila. eLife 9, e53865 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Albadri S, De Santis F, Di Donato V & Del Bene F CRISPR/ Cas9-mediated knockin and knockout in zebrafish. in Genome Editing in Neurosciences. Research and Perspectives in Neurosciences (eds. Jaenisch R, Zhang F & Gage F) 41–49 (2017). [PubMed] [Google Scholar]
- 82.Muntasell A, Berger AC & Roche PA T cell-induced secretion of MHC class II-peptide complexes on B cell exosomes. EMBO J. 26, 4263–4272 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lachenal G et al. Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol. Cell. Neurosci 46, 409–418 (2011). [DOI] [PubMed] [Google Scholar]
- 84.Li J et al. Serum-free culture alters the quantity and protein composition of neuroblastoma-derived extracellular vesicles. J. Extracell. vesicles 4, 26883 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rocha S et al. 3D cellular architecture affects microRNA and protein cargo of extracellular vesicles. Adv. Sci 6, 1800948 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thippabhotla S, Zhong C & He M 3D cell culture stimulates the secretion of in vivo like extracellular vesicles. Sci. Rep 9, 13012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cao J et al. Three-dimensional culture of MSCs produces exosomes with improved yield and enhanced therapeutic efficacy for cisplatin-induced acute kidney injury. Stem Cell Res. Ther 11, 206 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim M, Yun H-W, Park DY, Choi BH & Min B-H Three-dimensional spheroid culture increases exosome secretion from mesenchymal stem cells. Tissue Eng. Regen. Med 15, 427–436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lehrich BM, Liang Y & Fiandaca MS Foetal bovine serum influence on in vitro extracellular vesicle analyses. J. Extracell. Vesicles 10, e12061 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen L, Ma L & Yu L WGA is a probe for migrasomes. Cell Discov. 5, 13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ma L et al. Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Res. 25, 24–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Addi C et al. The Flemmingsome reveals an ESCRT-to-membrane coupling via ALIX/syntenin/syndecan-4 required for completion of cytokinesis. Nat. Commun 11, 1941 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Miesenbock G, De Angelis DA & Rothman JE Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394, 192–195 (1998). [DOI] [PubMed] [Google Scholar]
- 94.Cashikar AG & Hanson PI A cell-based assay for CD63-containing extracellular vesicles. PLoS One 14, e0220007 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wegner CS et al. Ultrastructural characterization of giant endosomes induced by GTPase-deficient Rab5. Histochem. Cell Biol 133, 41–55 (2010). [DOI] [PubMed] [Google Scholar]
- 96.Mathieu M et al. Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nat. Commun 10.1038/s41467-021-24384-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lenzini S, Bargi R, Chung G & Shin JW Matrix mechanics and water permeation regulate extracellular vesicle transport. Nat. Nanotechnol 15, 217–223 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mu W, Rana S & Zöller M Host matrix modulation by tumor exosomes promotes motility and invasiveness. Neoplasia 15, 875–IN4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wiklander OPB et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J. Extracell. Vesicles 4, 26316 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ridder K et al. Extracellular vesicle-mediated transfer of functional RNA in the tumor microenvironment. Oncoimmunology 4, e1008371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Riau AK, Ong HS, Yam GHF & Mehta JS Sustained delivery system for stem cell-derived exosomes. Front. Pharmacol 10, 1368 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rilla K et al. Extracellular vesicles are integral and functional components of the extracellular matrix. Matrix Biol. s 75–76, 201–219 (2019). [DOI] [PubMed] [Google Scholar]
- 103.Pastuzyn ED et al. The neuronal gene arc encodes a repurposed retrotransposon gag protein that mediates intercellular RNA transfer. Cell 172, 275–288.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ashley J et al. Retrovirus-like Gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell 172, 262–274.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Edgar JR, Manna PT, Nishimura S, Banting G & Robinson MS Tetherin is an exosomal tether. eLife 5, e17180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Morad G et al. Tumor-derived extracellular vesicles breach the intact blood-brain barrier via transcytosis. ACS Nano. 13, 13853 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Alvarez-Erviti L et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol 29, 341–345 (2011). [DOI] [PubMed] [Google Scholar]
- 108.Denzer K et al. Follicular dendritic cells carry MHC class II-expressing microvesicles at their surface. J. Immunol 165, 1259–1265 (2000). [DOI] [PubMed] [Google Scholar]
- 109.Gao L et al. Tumor-derived exosomes antagonize innate antiviral immunity. Nat. Immunol 19, 233–245 (2018). [DOI] [PubMed] [Google Scholar]
- 110.Vilcaes AA, Chanaday NL & Kavalali ET Interneuronal exchange and functional integration of synaptobrevin via extracellular vesicles. Neuron 109, 971–983.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ko SY et al. Cancer-derived small extracellular vesicles promote angiogenesis by heparin-bound, bevacizumab-insensitive VEGF, independent of vesicle uptake. Commun. Biol 2, 386 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Neumann CJ & Cohen SM Long-range action of Wingless organizes the dorsal-ventral axis of the Drosophila wing. Development 124, 871–880 (1997). [DOI] [PubMed] [Google Scholar]
- 113.Tkach M et al. Qualitative differences in T-cell activation by dendritic cell-derived extracellular vesicle subtypes. EMBO J. 36, 3012–3028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Heusermann W et al. Exosomes surf on filopodia to enter cells at endocytic hot spots and shuttle within endosomes to scan the ER. J. Cell Biol 213, 173–184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Arasu UT, Härkönen K, Koistinen A & Rilla K Correlative light and electron microscopy is a powerful tool to study interactions of extracellular vesicles with recipient cells. Exp. Cell. Res 376, 149–158 (2019). [DOI] [PubMed] [Google Scholar]
- 116.Thomou T et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 542, 450–455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sterzenbach U et al. Engineered exosomes as vehicles for biologically active proteins. Mol. Ther 25, 1269–1278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kur I-M et al. Neuronal activity triggers uptake of hematopoietic extracellular vesicles in vivo. PLoS Biol. 18, e3000643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Frühbeis C et al. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 11, e1001604 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Khmelinskii A et al. Incomplete proteasomal degradation of green fluorescent proteins in the context of tandem fluorescent protein timers. Mol. Biol. Cell 27, 360–370 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Joshi BS, De Beer MA, Giepmans BNG & Zuhorn IS Endocytosis of extracellular vesicles and release of their cargo from endosomes. ACS Nano. 14, 32 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cao H et al. In vivo real-time imaging of extracellular vesicles in liver regeneration via aggregation-induced emission luminogens. ACS Nano. 13, 3522–3533 (2019). [DOI] [PubMed] [Google Scholar]
- 123.Webber JP et al. Differentiation of tumour-promoting stromal myofibroblasts by cancer exosomes. Oncogene 34, 290–302 (2015). [DOI] [PubMed] [Google Scholar]
- 124.Lener T et al. Applying extracellular vesicles based therapeutics in clinical trials - an ISEV position paper. J. Extracell. vesicles 4, 30087 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fais S et al. Evidence-Based clinical use of nanoscale extracellular vesicles in nanomedicine. ACS Nano. 10, 3886–3899 (2016). [DOI] [PubMed] [Google Scholar]
- 126.Kalluri R & LeBleu VS The biology, function, and biomedical applications of exosomes. Science 367, eaau6977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Androuin A, Verweij FJ & Van Niel G Zebrafish as a preclinical model for extracellular vesicle-based therapeutic development. Adv. Drug Deliv. Rev 10.1016/j.addr.2021.05.025 (2021). [DOI] [PubMed] [Google Scholar]
- 128.Liégeois S, Benedetto A, Garnier J-M, Schwab Y & Labouesse M The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J. Cell Biol 173, 949–961 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Koles K et al. Mechanism of evenness interrupted (Evi)-exosome release at synaptic boutons. J. Biol. Chem 287, 16820–16834 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Corrigan L et al. BMP-regulated exosomes from male reproductive glands reprogram female behavior. J. Cell Biol. 206, 671–688 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wolf P The nature and significance of platelet products in human plasma. Br. J. Haematol 13, 269–288 (1967). [DOI] [PubMed] [Google Scholar]
- 132.Nunez EA, Wallis J & Gershon MD Secretory processes in follicular cells of the bat thyroid. III. The occurrence of extracellular vesicles and colloid droplets during arousal from hibernation. Am. J. Anat 141, 179–201 (1974). [DOI] [PubMed] [Google Scholar]
- 133.Trams EG, Lauter CJ, Norman Salem J & Heine U Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim. Biophys. Acta - Biomembr 645, 63–70 (1981). [DOI] [PubMed] [Google Scholar]
- 134.Johnstone RM, Bianchini A & Teng K Reticulocyte maturation and exosome release: transferrin receptor containing exosomes shows multiple plasma membrane functions. Blood 74, 1844–1851 (1989). [PubMed] [Google Scholar]
- 135.Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ & Sixma JJ Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood 94, 3791–3799 (1999). [PubMed] [Google Scholar]
- 136.Yang T et al. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm. Res 32, 2003–2014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]


