Abstract
Biophysically detailed modeling provides an unmatched method to integrate data from many disparate experimental studies, and manipulate and explore with high precision the resultin brain circuit simulation. We developed a detailed model of the brain motor cortex circuits, simulating over 10,000 biophysically detailed neurons and 30 million synaptic connections. Optimization and evaluation of the cortical model parameters and responses was achieved via parameter exploration using grid search parameter sweeps and evolutionary algorithms. This involves running tens of thousands of simulations requiring significant computational resources. This paper describes our experience in setting up and using Google Compute Platform (GCP) with Slurm to run these large-scale simulations. We describe the best practices and solutions to the issues that arose during the process, and present preliminary results from running simulations on GCP.
Keywords: Brain modeling, Computational neuroscience, Large-scale simulations, Google Cloud Platform
CCS CONCEPTS: Computing methodologies → Modeling and simulation, Applied computing → Life and medical sciences
1. INTRODUCTION
The cerebral cortex is the outermost layer of the brain and is responsible for most high-level functions like vision, language and reasoning, each implemented by circuits in different “bespoke” regions. Biophysically detailed modeling provides an unmatched method to integrate data from many disparate experimental studies, and manipulate and explore with high precision the resulting brain circuit simulation [1–3]. Some examples of brain regions simulated using detailed computational multiscale models include motor cortex (M1), visual (V1) [4], somatosensory (S1), and auditory (A1) cortices, as well as other cortical areas such as hippocampus [3] and cerebellar cortex [5]. We have developed a highly detailed model of M1, simulating a volume of cortical tissue with over 10,000 biophysically detailed neurons and 30 million synaptic connections [6], including 15 neural populations with cell density, distribution and synaptic connectivity patterns derived from experimental data. The model has been used to investigate circuit information pathways, oscillatory coding mechanisms and the role of noradrenaline in modulating output to spinal cord and movement initiation.
Running such simulations and data analysis requires significant computational resources, such as the High Performance Computing (HPC) resources that are available through the National Science Foundation’s Extreme Science and Engineering Discovery Environment (XSEDE) program [7], or cloud computing resources provided by commercial cloud providers such as Google Cloud Platform (GCP) [8]. This is because calculating the electrical currents within each neuron – including synaptic inputs received from thousands of other neurons – requires solving thousands of differential equations every fraction of a millisecond: simulating one second of the full M1 cortical circuit model requires approximately 50 HPC core hours. Distributing cells across compute nodes parallelizes each network simulation, and many of these simulations are typically executed in parallel to explore different parameter combinations. Finding model parameters that reproduce biology and investigating neural coding via in silico experiments involve exploring large parameter spaces [9, 10]. Optimization and evaluation of the cortical model parameters and responses is achieved via parameter exploration methods including grid search parameter sweeps and evolutionary algorithms [9]–[13]. This involves running tens of thousands of simulations, with each simulated second of the full circuit model requiring approximately 96 core hours
We have previously used evolutionary algorithm optimization to train a cortical model to control a virtual arm using XSEDE supercomputers [9]. We were recently awarded the NSF/Internet2 Exploring Clouds for Acceleration of Science (E-CAS) grant [14] to continue our research by simulating large-scale models of brain circuits using GCP. This poster describes our experience in setting up and using GCP with Slurm to run these large-scale simulations, best practices and solutions to the issues that arose during the process, and preliminary results from running simulations on GCP.
2. SETTING UP THE CLOUD ENVIRONMENT
2.1. Requirements
Integration of Slurm [17] as the cluster management and job scheduling system with GCP compute resources is integral for our project computing needs for the following reasons:
Model characteristics:
One mode of running the model requires launching multiple jobs for high throughput computing (HTC), e.g., 1000 jobs in parallel, where each job is an MPI-based HPC job requiring multiple cores. As described in section 1, our study involves performing parameter optimizations, which requires significant HPC resources. For example, grid search parameter optimization or exploration involves searching exhaustively through a specified subset of parameters, evaluating all combinations of parameter values. This requires submitting thousands of jobs simultaneously, one for each parameter combination. Similarly, evolutionary algorithm optimization requires running hundreds or thousands of generations, each evaluating many individuals, where each individual corresponds to a simulation with a parameter combination. Slurm is well suited to handle this task by enabling the user to submit multiple jobs at once to the queue. Slurm then assigns computing resources as they become available.
Elasticity/Walltime:
While the XSEDE resources were free of cost to use, there were limitations, whose specifics varied by system. In general, we were restricted in walltime (hours that a job is allowed to be resident on the systems), number of running jobs at a given time per user, and total number of cores available. GCP supports elastic computing and can auto-scale according to job requirements, and has no specific walltime, number of jobs, or core number restrictions. Using Slurm with GCP enables us to submit multiple such jobs in a batch environment.
Ease of Use:
We had used Slurm for managing our jobs on XSEDE resources, and were familiar with the scripts for usage and management. Additionally, NetPyNE already had built-in support for automatically generating Slurm scripts and submitting Slurm jobs. Setting up GCP with Slurm makes it easier to be used as a central lab resource to run simulations and share results.
2.2. GCP-Slurm Setup
Once the GCP project account was created and credits transferred, user accounts were created for the lab members to access GCP setup. Similar to XSEDE HPC setup, users have to log in to a login node where they can submit the jobs to compute nodes using Slurm scheduler. The Slurm controller node runs the Slurm controller and the database.
A custom image from the CentOS-7 image was created to install and setup our custom software setup: NEURON [15], NetPyNE [16], and Python3. To deploy the cluster, we first downloaded the git repository that contained the Slurm for GCP deployment-manager files. The YAML file, from the git repository, contains the details of the configuration of the deployment, the Slurm version to deploy, and the machine instance types to deploy. The YAML file was modified to include the newly created disk image and the type of instances required for launching the compute nodes. The instructions outlined in the google cloud lab user guide [18] were fairly easy to follow for the initial setup.
Table 1 shows the GCP test instance setup that was needed for running 10 minutes of the M1 model and for performing evolutionary optimization. Once the YAML file was updated we ran the shell scripts that execute the gcloud commands to deploy the cluster, including login, controller and compute nodes.
Table 1:
YAML configuration parameters for running M1 model for 10 minutes and performing evolutionary optimization
| M1 model | Evolutionary Optimization | |
|---|---|---|
|
| ||
| Compute machine type | n1-standard-32 | n1-standard-96 |
| Compute disk size (gb) | 10 | 10 |
| Login machine type | n1-standard-2 | n1-standard-8 |
| Login disk size (gb) | 10 | 10 |
| Controller machine type | n1-standard-32 | n1-highmem-64 |
| Controller disk type (gb) | 2000 | 2000 |
| Compute node memory | 120GB | 360GB |
2.3. Experiences in setting up GCP-Slurm
Some of the initial challenges that we faced while setting up GCP-Slurm integration ranged from disk mounting to latency issues. We were able to resolve most of the issues by communicating with customer support and initiating discussions on the google discussion forum [19]. During the initial setup, /home folder was not automatically mounted via NFS. With helpful discussions from GCP-Slurm forum, the problem was identified as a result of incompatibility between python versions in the disk image and the GCP startup scripts (disk image having Python 3 but the GCP startup scripts required Python 2). We also faced the issue of shutdown of node instances that took over 3 hours. During the shutdown process, Slurm waits for each node to return that it’s shutdown and deleted from the list. An increase in the number of nodes caused an increase in the overall shutdown time. Decreasing the Suspend/Resume timeout settings in the configuration and removing the option to wait for confirmation from the nodes fixed this.
Running simulations on greater than 16 nodes resulted in crashing of the job scheduler. The controller node needed more memory so the issue was resolved when we increased the number of cores/memory for the controller node. The controller instances were initially created with 8 cores and later changed to 32 or 64 cores with high memory to prevent the node from crashing frequently due to processing and memory limitations.
However there are some issues that are still being worked out with the GCP support team. For example, 25% of the simulations crashed with a timeout error from the NEURON simulator that happened on random simulations at random times. After working closely with NEURON developers to debug this issue, and communicating with Google support, we concluded that the timeout error happens due to the internode latency. Our simulations require low-latency network performance necessary for tightly coupled node-to-node communication. We have been whitelisted in the GCP placement groups alpha program that provides clusters with all nodes in the same rack. Placement groups should low internode latency and possibly high-bandwidth that should enable us to run multinode simulations. We explored the use of preemptible VMs that are significantly cheaper (approximately by a factor of 4) than the regular instances, but require the code to be fault tolerant in case of preemption. To use preemptible cores we enabled the option to perform preemptible bursting in the YAML file. Approximately 30–40% of the jobs were preempted when using this option. GCP-Slurm was set up to automatically to resubmit jobs that get preempted.
As part of ECAS grant, we were awarded $100k in GCP credits to demonstrate commercial cloud resources could be used to accelerate science. Most of these credits were spent on 5.2M pre-emptible core hours and 900k non-preemptible core hours, as well as the virtual machines’ RAM memory. Less than 1% was employed on data storage (17k GB month) and networking/disk transfer. This provide us with an estimate of how much we would need to budget if we continue using google cloud beyond the grant period.
3. SIMULATION RESULTS
3.1. Single simulation output
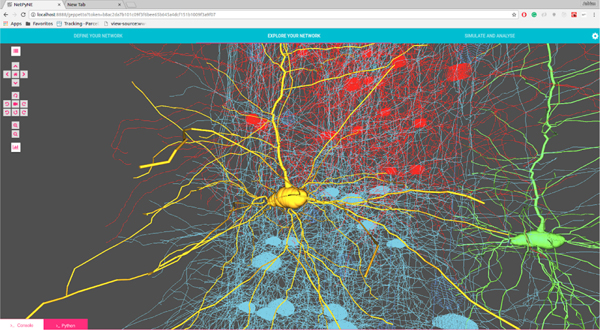
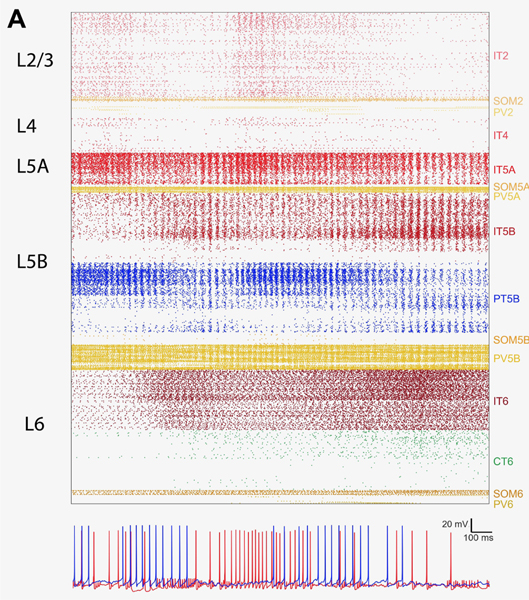
We present here the results from a single simulation of the M1 cortical model, with 10k cells and over 30M synapses. Figure 1 shows a 3D representation of the cortical network, including the detailed morphologies of several pyramidal neurons. Simulating the network required solving the differential equations that describe the electrical properties of each neuron, including their ionic and synaptic currents and membrane voltage (order 10ms), and how they generate action potentials (spikes, order 1 ms), a stiff problem. Membrane voltage traces of two typical network neurons are shown in Figure 2
Figure 1:
3D representation of M1 cortical network, illustrating the detailed morphologies of multiple pyramidal neurons.
Figure 2:
Output of single simulation of the motor cortex (M1) model. Top. Raster plot showing spikes (points) over time (x-axis) for 10k neurons. Bottom. Membrane voltage of two example cells in the network, showing action potentials (brief spikes of increased voltage over baseline).
3.2. Long duration simulation
We then investigated self-organizing criticality and avalanches in the M1 model. Neuronal avalanches are a cascade of bursts of activity in neuronal networks whose size distribution follows a power law. This is a robust and reproducible phenomenon observed experimentally across cortex, but whose physiological mechanisms are not currently known. Published data shows that avalanches exhibited a scale free pattern with a constant power law relation across different time scales. Therefore, investigating this phenomenon required running simulations for long durations, beyond the simulation durations ranging 1–5s we typically employed. The longest simulation we had previously ran was for 60s. Using GCP we were now able to run a simulation of 10 minutes, an order of magnitude higher. The simulation ran on 512 cores (32 nodes x 16 cores/node), took approximately 90 hours, and produced 22,610,549 spikes and 1.2 GB of data. This was possible due to the unconstrained number of hours available on this platform.
3.3. Grid search parameter optimization
We ran simulations on approximately 130K cores simultaneously to perform grid-search parameter optimization in the M1 model. The aim was to find the values for 7 model parameters that produced average population firing rates that approximately matched experimental data. Model parameters included the overall connection strengths between excitatory and inhibitory populations, and the strength of long-range inputs driving the model.
A total of 1152 jobs were run to evaluate different parameter combinations, where each job simulated 2 sec of the M1 model. Each job ran on 48 cores (6 nodes * 8 cores/node) for approximately 2.5 hours, making a total of approximately 130K core hours. Since preemptible nodes were employed, approximately 20% of the jobs were automatically resubmitted after being preempted. The grid search simulations generated a total of 21GB of output data. Exploration of the 1152 output simulations allowed us to find the parameter combination that best reproduced cortical experimental data. The results were presented at Google Next 2018 Cloud talks in San Francisco and London.
4. CONCLUSION AND FUTURE DIRECTIONS
This paper describes our preliminary efforts at using the GCP-Slurm integration to run simulations and optimize the parameters of our large-scale biophysically detailed cortical models. We have provided details of our setup as well as solutions to multiple issues, in the hope that this will be useful to future researchers wishing to use GCP-Slurm for similar projects. Significant effort was required, to setup and maintain Slurm- based instances, especially for researchers not involved in IT-related fields. Nonetheless, we felt that our lab benefited from the GCP-Slurm setup allowing us to flexibly deploy (and remove) clusters with fully customized specifications for tasks with different requirements, and access virtually unlimited resources without queue waiting times.
More rigorous work is still needed to make sure that the setup is fully functional for multiple lab members to use it. Given the access to a large number of simultaneous cores and unrestricted run times through GCP, we are planning to run large scale models of other brain regions, including auditory thalamocortical circuits. Google provided us access to a placement group, which would potentially address the latency issue. We are working with the Google team to be early testers of this feature and provide feedback. We are also planning to do further scaling studies to understand the performance characteristics of NetPyNE/NEURON. This can be done by carrying out detailed profiling and tracing analysis to understand processor and memory level performance characteristics, communication patterns and characteristics, and I/O behavior.
ACKNOWLEDGMENTS
We would like to acknowledge funding support from NSF E-CAS grant 1904444, NIH grants U01EB017695, U24EB028998 and R01DC012947, NYS SCIRB grant DOH01-C32250GG- 3450000, ARO grant W911NF-19–1-0402, and cloud resources from Google.
Footnotes
ACM Reference Format:
Subhashini Sivagnanam, Wyatt Gorman, Donald Doherty, Samuel A Neymotin, Stephan Fang, Hermine Hovhannisyan, William W Lytton, and Salvador Dura-Bernal. 2020. Simulating Large-scale Models of Brain Neuronal Circuits using Google Cloud Platform. In Practice and Experience in Advanced Research Computing (PEARC ‘20), July 26–30, 2020, Portland, OR, USA. ACM, New York, NY, USA, 5 pages. https://doi.org/10.1145/3311790.3399621
Contributor Information
Subhashini Sivagnanam, State University of New York DMC, Brooklyn NY; San Diego Supercomputer Center / University California San Diego, La Jolla CA.
Wyatt Gorman, Google Corporation, Mountain View CA.
Donald Doherty, State University of New York DMC, Brooklyn NY.
Samuel A Neymotin, Nathan Kline Institute for Psychiatric Research, Orangeburg NY USA.
Stephan Fang, Google Corporation, Mountain View CA.
Hermine Hovhannisyan, Google Corporation, Mountain View CA.
William W Lytton, State University of New York DMC, Brooklyn NY; King’s County Hospital, Brooklyn NY.
Salvador Dura-Bernal, State University of New York DMC, Brooklyn NY; Nathan Kline Institute for Psychiatric Research, Orangeburg NY USA.
REFERENCES
- [1].Markram H, Muller E, Ramaswamy S, Reimann MW, Abdellah M, Sanchez CA, et al. Reconstruction and Simulation of Neocortical Microcircuitry. Cell. 2015. pp. 456–492. doi: 10.1016/j.cell.2015.09.029 [DOI] [PubMed] [Google Scholar]
- [2].Hawrylycz M, Anastassiou C, Arkhipov A, Berg J, Buice M, Cain N, et al. Inferring cortical function in the mouse visual system through large-scale systems neuroscience. Proceedings of the National Academy of Sciences. 2016. pp. 7337–7344. doi: 10.1073/pnas.1512901113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bezaire MJ, Raikov I, Burk K, Vyas D, Soltesz I. Interneuronal mechanisms of hippocampal theta oscillations in a full-scale model of the rodent CA1 circuit. eLife. 2016. doi: 10.7554/elife.18566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Arkhipov A, Gouwens NW, Billeh YN, Gratiy S, Iyer R, Wei Z, et al. Visual physiology of the Layer 4 cortical circuit in silico. bioRxiv. 2018. doi: 10.1101/292839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bases Ito M. and implications of learning in the cerebellum — adaptive control and internal model mechanism. Progress in Brain Research. 2005. pp. 95–109. doi: 10.1016/s0079-6123(04)48009-1 [DOI] [PubMed] [Google Scholar]
- [6].Dura-Bernal S, Neymotin SA, Suter BA, Shepherd GMG, Lytton WW. Multiscale dynamics and information flow in a data-driven model of the primary motor cortex microcircuit. doi: 10.1101/201707 [DOI] [Google Scholar]
- [7].Home - XSEDE. [cited 8 Feb 2020]. Available: http://www.xsede.org
- [8].Cloud Computing Services | Google Cloud. In: Google Cloud [Internet]. [cited 1 May 2020]. Available: https://cloud.google.com/
- [9].Dura-Bernal S, Neymotin SA, Kerr CC, Sivagnanam S, Majumdar A, Francis JT, et al. Evolutionary algorithm optimization of biological learning parameters in a biomimetic neuroprosthesis. IBM J Res Dev. 2017;61: 6.1–6.14. doi: 10.1147/JRD.2017.2656758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Neymotin SA, Suter BA, Dura-Bernal S, Shepherd GMG, Migliore M, Lytton WW. Optimizing computer models of corticospinal neurons to replicate in vitro dynamics. J Neurophysiol. 2017;117: 148–162. doi: 10.1152/jn.00570.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Prinz AA, Bucher D, Marder E. Similar network activity from disparate circuit parameters. Nat Neurosci. 2004;7: 1345–1352. Available: https://www.ncbi.nlm.nih.gov/pubmed/15558066 [DOI] [PubMed] [Google Scholar]
- [12].Achard P, De Schutter E. Complex Parameter Landscape for a Complex Neuron Model. PLoS Computational Biology. 2006. p. e94. doi: 10.1371/journal.pcbi.0020094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rumbell TH, Draguljić D, Yadav A, Hof PR, Luebke JI, Weaver CM. Automated evolutionary optimization of ion channel conductances and kinetics in models of young and aged rhesus monkey pyramidal neurons. J Comput Neurosci. 2016;41: 65–90. [DOI] [PubMed] [Google Scholar]
- [14].Exploring Clouds for Acceleration of Science | Internet2. [cited 1 May 2020]. Available: https://www.internet2.edu/vision-initiatives/initiatives/exploring-clouds-acceleration-science/
- [15].NEURON | empirically-based simulations of neurons and networks of neurons. [cited 8 Feb 2020]. Available: www.neuron.yale.edu
- [16].Welcome to NetPyNE’s documentation! — NetPyNE documentation. [cited 1 May 2020]. Available: www.netpyne.org
- [17].Slurm Workload Manager - Documentation. [cited 1 May 2020]. Available: https://slurm.schedmd.com
- [18].Deploy an Auto-Scaling HPC Cluster with Slurm. [cited 1 May 2020]. Available: https://codelabs.developers.google.com/codelabs/hpc-slurm-on-gcp/#0
- [19].Google Groups. [cited 1 May 2020]. Available: https://groups.google.com/forum/#!topic/google-cloud-slurm-discuss/