Abstract
EMRSA-15 is one of the most important strains of epidemic methicillin-resistant Staphylococcus aureus (EMRSA) found in the United Kingdom. It was originally characterized by weak lysis with phage 75 and production of enterotoxin C but not urease. Two variant strains of EMRSA-15 which show a broader phage pattern than the progenitor strain have emerged. A total of 153 recent clinical isolates representing classical EMRSA-15 (55 isolates) or these phage variants (98 isolates) were compared by SmaI macrorestriction profiles in pulsed-field gel electrophoresis (PFGE) as well as by urease and enterotoxin C production. Eight of the 98 isolates were shown to be other unrelated strains by both PFGE and their production of urease, a misidentification rate of 8% by phage typing. Seventy-one EMRSA-15 isolates were enterotoxin C negative, and the majority of these were sensitive to phage 81. Examination of PFGE profiles and Southern blotting studies suggest that the enterotoxin C gene locus is encoded on a potentially mobile DNA segment of ca. 15 kb. After elimination of the eight non-EMRSA-15 isolates, the remaining 145 were characterized by PFGE, yielding 22 profiles. All profiles were within five band differences of at least one other profile. Classical EMRSA-15 isolates showed nine PFGE profiles, with the majority of isolates (68%) in profile B1. Six of these nine PFGE profiles were unique to the classical EMRSA-15 isolates. Among the phage variants of EMRSA-15, 16 profiles were seen, but the majority of isolates (83%) fell into 1 of 4 profiles (B2, B3, B4, and B7) which correlated well with phage patterns. The most divergent PFGE profiles among the EMRSA-15 isolates had as many as 12 band differences from one another, suggesting that in examining isolates belonging to such a temporally and geographically disseminated epidemic strain, the range of PFGE profiles must be regarded as a continuum and analyzed by relating the profiles back to the most common or progenitor profile.
Staphylococcus aureus is the leading cause of surgical wound infections and the second most frequent cause of bacteremia in the hospital setting (7). Methicillin-resistant S. aureus (MRSA) first appeared in the early 1960s but virtually disappeared during the 1970s in the United Kingdom. It reappeared in the early 1980s (18), and over the last two decades, strains with resistance to an extended range of antibiotics have emerged to pose a major threat to public health. MRSA isolates may comprise as much as 45% of the total number of S. aureus isolated from patients with bacteremia (2). Thousands of isolates are sent each year to the S. aureus Reference Service (SaRS) at the Central Public Health Laboratory (CPHL) for typing; there they are subdivided primarily on the basis of their susceptibilities to 27 phages (19, 21). Where additional discrimination between strains is required, SmaI digests of chromosomal DNA are subjected to pulsed-field gel electrophoresis (PFGE) (24).
Epidemic strains of MRSA are defined as those which have been identified in two or more patients in two or more hospitals (13). The first epidemic MRSA (EMRSA) strain, designated EMRSA-1, was recognized in 1981 (17) and continued to cause outbreaks in hospitals until the late 1980s. A second EMRSA strain, EMRSA-2, emerged in the late 1980s (22) and was followed closely by 12 other EMRSA strains described-during a survey carried out in 1987 and 1988 (13). EMRSA-15 emerged during 1991 and rapidly displaced most of the other EMRSA strains (1). It has now spread to, and is endemic in, hundreds of hospitals across the United Kingdom; it has also recently been identified as causing outbreaks in Australia, New Zealand, Germany, Sweden, and Finland. The strain classically is characterized by weak lysis with phage (φ) 75 of the international set, production of enterotoxin C, and nonproduction of urease (24).
In recent years two “variant” strains of EMRSA-15 have been recognized by SaRS. They were identified as EMRSA-15 variants despite producing additional phage reactions, notably with phages 42E, 81, 83C, and 90, because they were phenotypically similar to classical EMRSA-15 isolates (i.e., in colonial morphology, toxin production, and lack of urease production) and arose in hospitals with large circulating EMRSA-15 populations. Subsequent investigation of some of these isolates by PFGE revealed that they gave SmaI digestion patterns identical or closely related to those of the classical EMRSA-15 isolates, and they were coded as phage variants “42E” and “83C.” Subsequently, some of these variants have lost the reaction with phage 75 which was initially a defining characteristic of EMRSA-15.
This study was undertaken to determine, firstly, whether all isolates classified as EMRSA-15 phage variants are genotypically EMRSA-15; secondly, whether variation in phage pattern is related to variation in PFGE profile; and finally, whether any other characteristics of the strain vary with particular changes in phage pattern and/or PFGE profile.
MATERIALS AND METHODS
Bacterial strains.
In total, 153 clinical isolates of EMRSA-15 and the EMRSA-15 phage variants “42E” and “83C” were examined. Isolates were selected for inclusion in the following manner. Phage typing records of isolates from 1998 were used to identify all 55 hospitals from disparate geographical locations in England, Wales, and the Republic of Ireland which had sent phage variants of EMRSA-15 to SaRS. In some hospitals, multiple different phage variants (i.e., patterns showing reaction differences from one another) were found, so a single representative of each phage variant pattern was included from each hospital. In total, 98 isolates representing the phage variants of EMRSA-15 were selected for further study. In addition, classical EMRSA-15 isolates (based on a weak reaction with phage 75 only) were selected as controls from 49 of the 55 hospitals (no classical EMRSA-15 isolates were present in 6 of the hospitals). Upon subsequent enterotoxin testing, the “classical” isolates from six hospitals were found to be enterotoxin C negative. Due to this finding, an additional classical isolate from each of these hospitals, which was enterotoxin C positive, was included. The EMRSA-15 type strain (NCTC 13142) was included as a control strain for enterotoxin C production, PFGE, and phage typing. Additional controls were NCTC 8325, ATCC 29213 (susceptibility testing), and the enterotoxin-producing strains NCTC 10652 (enterotoxin A; gene sea), NCTC 10654 (enterotoxin B; seb), NCTC 10655 (enterotoxin C; sec), NCTC 10656 (enterotoxin D; sed), and NCTC 11963 (toxic shock syndrome toxin [TSST-1], tst). Isolates were stored on nutrient agar (NA) slopes at room temperature and recovered by subculture on NA plates followed by overnight incubation at 37°C. All isolates were maintained on Microbank beads (Prolab, UK) at −70°C.
Phenotypic characterization of strains.
Strains were phage typed in triplicate using the international phage set (21) and local experimental phages 88A, 90, 83C, and 932 (19). Phage typing of the isolates was performed at 100 times the routine test dilution (100× RTD) because most United Kingdom MRSA isolates are nontypeable at RTD (23). Isolates were tested for coagulase, urease production, and antimicrobial susceptibility by standard methods as previously described (13).
The following agents were tested by the agar dilution method on Isosensitest agar (Oxoid, Ltd., Basingstoke, United Kingdom) with 2% horse blood and interpreted using the following resistance breakpoints: ciprofloxacin, >1 mg/liter; erythromycin, ≥0.5 mg/liter; fusidic acid, ≥1 mg/liter; gentamicin, >1 mg/liter; kanamycin, >4 mg/liter; neomycin, >4 mg/liter; methicillin, ≥4 mg/liter; mupirocin, ≥8 mg/liter; streptomycin, >4 mg/liter; rifampin, >0.12 mg/liter; teicoplanin, ≥4 mg/liter; tetracycline, ≥1 mg/liter; and vancomycin, ≥4 mg/liter.
mecA PCR.
Isolates which were sensitive to methicillin were examined for carriage of the mecA gene by PCR according to the method of Bignardi et al. (6).
Enterotoxin detection.
All isolates were screened for the presence of enterotoxin genes A through D (sea through sed) and the TSST-1 gene (tst) by the method of Johnson et al. (10), but DNA was extracted by boiling in 5% Chelex 100 (Bio-Rad Laboratories, Hercules, Calif.) and lysates were centrifuged to remove cell debris. The supernatant was transferred to a fresh tube, and 5 μl was used as a template in the PCRs (20). Selected isolates were also tested for the production of enterotoxins A through D using a reverse passive latex agglutination kit (SET-RPLA; Oxoid Ltd.) according to the manufacturer's instructions.
PFGE.
PFGE was performed by the method of Kaufmann (11). Briefly, DNA was extracted from overnight cultures grown at 37°C on NA and restriction digested with SmaI (Boehringer GmbH, Mannheim, Germany) overnight at 30°C according to the manufacturer's instructions. Digested DNA was electrophoresed in 1.2% agarose gels for 30 h with a ramped pulse time of 1 to 80 s using a CHEF DRII or CHEF Mapper (Bio-Rad Laboratories). DNA fragments were visualized by staining with 0.5 μg of ethidium bromide/ml. Gels were photographed under UV illumination, and the data were saved to a floppy disk prior to analysis. Gel data were analyzed with GelCompar software (Applied Maths, Kortrijk, Belgium).
Probe generation, Southern blotting, and hybridization.
A subset of isolates were examined for enterotoxin C gene (sec) carriage by Southern blotting. A 699-bp biotin-labeled sec probe was generated by PCR using the following primers: TGT ATC AGC AAC TAA AGT TAA GTC and AAA GGCAAG CAC CGA AG. PCR was performed in a total volume of 100 μl containing 2.5 U of Taq polymerase, 200 μM deoxynucleoside triphosphates, 68 μM biotin-16-dUTP, 2 mM MgCl2, 20 mM Tris-HCl (pH 8.4), 50 mM KCl, and 5 μl of template DNA (prepared as described above). Cycling conditions consisted of 1 cycle of denaturation at 96°C for 2 min followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 52°C for 1 min, and extension at 72°C for 1 min, with a final extension at 72°C for 5 min. The PCR product was purified using a QIAquick PCR purification kit (Qiagen Ltd., Crawley, United Kingdom), and the probe concentration was estimated spectrophotometrically at 260 nm. DNA fragments from PFGE gels were transferred to nylon membranes (Hybond N; Amersham, Little Chalfont, Buckinghamshire, United Kingdom) by vacuum blotting according to the method of Kaufmann et al. (12) with the following modifications: the fragmentation solution (0.25 M HCl) was applied for 25 min, the denaturation solution (0.5 M NaOH–1.5 M NaCl) was applied for 1 h, the neutralization solution (1.5 M NaCl–0.5 M Tris) was applied for 30 min, and then blotting was undertaken using 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 2 h at 5 × 103 Pa. Hybridization was performed by the method of Kaufmann et al. (12) using the sec probe at a concentration of 1 μg/ml. Commercially available biotinylated λ DNA digested with HindIII at a concentration of 1 μg/ml was used as a probe to detect the λ concatamer size standards on the PFGE gels. Hybridization was detected with the BlueGene Nonradioactive Detection System (Gibco BRL, Life Technologies, Paisley, United Kingdom).
RESULTS
Coagulase production and methicillin resistance.
All isolates were coagulase positive, and 149 of 153 isolates were resistant to methicillin. Of the four methicillin-sensitive isolates, one was positive for the mecA gene by PCR. The other three isolates were included in the study despite being methicillin sensitive because their phage patterns were as expected and loss of the mecA gene upon storage is known to occur (14).
Susceptibility to other antimicrobial agents.
The sensitivity patterns for the classical and variant EMRSA-15 isolates were similar, with all isolates susceptible to gentamicin, neomycin, teicoplanin, and vancomycin and the majority susceptible to fusidic acid, kanamycin, mupirocin, rifampin, streptomycin, and tetracycline (Table 1). However, almost all classical EMRSA-15 isolates (94%) were resistant to ciprofloxacin, and the majority were resistant to erythromycin (80%). Resistance to these two antibiotics was less frequent among the variant isolates (76 and 62%, respectively).
TABLE 1.
Antimicrobial susceptibilities of EMRSA-15 isolates
| Antimicrobial agents (breakpoint MIC, mg/liter) | No. (%) of isolates resistant
|
||
|---|---|---|---|
| Classical isolates (n = 55) | Variant isolates (n = 90) | Total (n = 145) | |
| Ciprofloxacin (>1) | 52 (94.4) | 68 (75.6) | 120 (83) |
| Erythromycin (≥0.5) | 44 (80) | 56 (62.2) | 100 (69) |
| Fusidic acid (≥1) | 2 (3.6) | 1 (1.1) | 3 (2) |
| Gentamicin (>1) | 0 (0) | 0 (0) | 0 (0) |
| Kanamycin (>4) | 1 (1.8) | 0 (0) | 1 (0.7) |
| Methicillin (≥4) | 54 (98.2) | 89 (98.9) | 143 (99) |
| Mupirocin (≥8) | 0 (0) | 1 (1.1) | 1 (0.7) |
| Neomycin (>4) | 0 (0) | 0 (0) | 0 (0) |
| Rifampin (≥0.12) | 1 (1.8) | 0 (0) | 1 (0.7) |
| Streptomycin (>4) | 1 (1.8) | 0 (0) | 1 (0.7) |
| Teicoplanin (≥4) | 0 (0) | 0 (0) | 0 (0) |
| Tetracycline (>1) | 0 (0) | 3 (3.3) | 3 (2) |
| Vancomycin (≥4) | 0 (0) | 0 (0) | 0 (0) |
Phage typing.
All 55 clinical isolates included as classical EMRSA-15 isolates gave the expected phage reaction of 75wk (coded as phage pattern 1). Isolates of the variant strains reacted with a combination of the expected phages: 42E, 75, 81, 83C, and 90 (Table 2). Certain weak reactions were variable when strains were typed in triplicate, but there was always a core of conserved reactions for each isolate. Ten major phage patterns could be delineated. Isolates belonging to phage variant “83C” inevitably had a strong reaction with this phage and reacted variably with phages 42E, 81, and 90 (coded as phage pattern 2). The phage patterns defined among this group were 83C/90 (with or without 42E—phage pattern 2a) and 83C (phage pattern 2b). Isolates belonging to phage variant “42E” always reacted strongly with phage 42E and reacted variably with phages 75, 81, 83C, and 90 (coded as phage pattern 3). The phage patterns defined among this group were 42E/75/81/90 (3a), 42E/81/83C/90 (3b), 42E/75/81/83C/90 (3c), 42E/81/83C (3d), 42E/75/90 (3e), and 42E/75/81/83C (3f). In addition, one isolate gave the phage pattern 42E/79/81(phage pattern 4). Patterns were further subdivided on the basis of reaction strength (Table 2).
TABLE 2.
Characteristics of phage patterns and PFGE profiles of isolates
| Lytic phage reactions at 100× RTDa (phage pattern code) | PFGE profile | No. of isolates | No. of band differences from B1 | SEC | Urease |
|---|---|---|---|---|---|
| Classical EMRSA-15 isolates | |||||
| 75wk (1) | B1 | 37 | 0 | + | − |
| 75wk (1) | B3 | 5 | 1 | − | − |
| 75wk (1) | B5 | 7 | 2 | + | − |
| 75wk (1) | B7 | 1 | 3 | − | − |
| 75wk (1) | B12 | 1 | 3 | + | − |
| 75wk (1) | B13 | 1 | 2 | + | − |
| 75wk (1) | B17 | 1 | 2 | + | − |
| 75wk (1) | B19 | 1 | 3 | + | − |
| 75wk (1) | B24 | 1 | 2 | + | − |
| Variant EMRSA-15 isolates | |||||
| 83C/90wk ± 42E (2a) | B2 | 19 | 1 | + | − |
| 83C/90wk (2a) | B1 | 1 | 0 | + | − |
| 83C/90wk (2a) | B18 | 2 | 5 | + | − |
| 83C (2b) | B6 | 1 | 3 | + | − |
| 42E/75wk/81/90wk (3a) | B3 | 28 | 1 | − | − |
| 42E/81/83C ± 90wk (3b or 3d) | B4 | 21 | 2 | − | − |
| 42E/75/81/83C/90wk (3c) | B7 | 7 | 3 | − | − |
| 42E/81wk/83C (3d) | B9 | 1 | 4 | − | − |
| 42E/81wk/83Cwk (3d) | B10 | 1 | 4 | − | − |
| 42E/81/83C (3d) | B11 | 1 | 6 | − | − |
| 42E/81/83C (3d) | B15 | 2 | 3 | − | − |
| 42E/81/83C/90wk (3b) | B16 | 2 | 5 | − | − |
| 42E/75wk/81wk/83C/90wk (3c) | B8 | 1 | 4 | − | − |
| 42E/75/81/83C/90 (3c) | B14 | 1 | 5 | − | − |
| 42E/75wk/90wk (3e) | B1 | 1 | 0 | + | − |
| 42E/75wk/90wk (3e) | B21 | 1 | 3 | − | − |
| 42E/75wk/81/83C (3f) | B23 | 1 | 5 | + | − |
| Non-EMRSA-15 isolates among the variant isolates | |||||
| 42E/81 ± 83C (3d) | C (1–5) | 5 | 16–17 | − | + |
| 42Ewk/81/83C (3d) | Unique | 1 | 14 | − | + |
| 42Ewk/79wk/81wk (4) | Unique | 1 | 15 | − | + |
| 83Cwk (2b) | Unique | 1 | 16 | − | + |
Reactions in boldface are strong reactions in the phage type; underlined reactions varied in strength between different phage typing runs. wk, weak reaction.
PFGE and urease results.
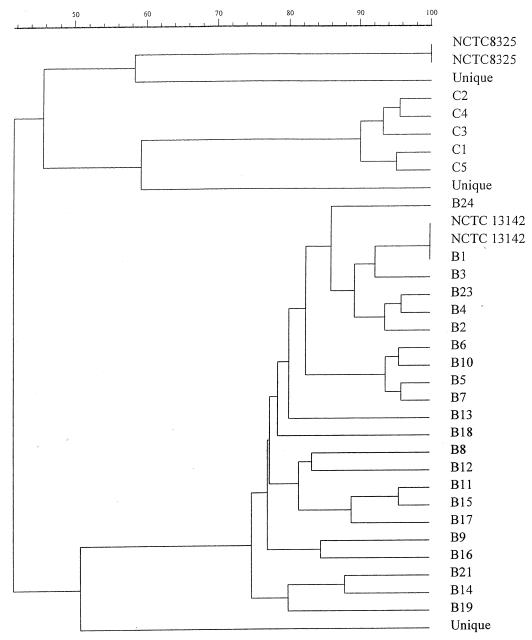
Initially PFGE data were analyzed by comparing band differences between PFGE profiles. The data were used to generate a dendrogram of percent relatedness calculated by the Dice coefficient and represented by UPGMA (Fig. 1), and five clearly different PFGE “clusters” were identified. The clusters with multiple isolates were coded B (145 isolates) and C (five isolates), and the others with only a single representative were considered unique. All isolates within cluster B showed more than 75% relatedness, with only 40% relatedness to the other four clusters and to NCTC 8325. The other clusters in turn showed no more than 65% relatedness to each other. Each of the clusters differed from all other clusters by at least 14 bands. Cluster B accounted for 145 of 153 (95%) isolates, including all 55 classical EMRSA-15 isolates. All cluster B isolates were urease negative. In contrast, the cluster C isolates and the three unique isolates were urease positive. These results suggest that these isolates had been misclassified by phage typing, i.e., an error rate of 8% for phage identification of EMRSA-15 variants. One of the methicillin-sensitive isolates belonged to cluster B (giving the classical phage pattern of 75wk), while the other two had unique PFGE profiles. PFGE variants occurred within clusters B and C, and these are described in more detail below. Representative profiles are shown in Fig. 2.
FIG. 1.
Dendrogram of percent relatedness of PFGE profiles from putative EMRSA-15 isolates calculated using the Dice coefficient and represented by UPGMA. Band tolerances were set at 1.0%.
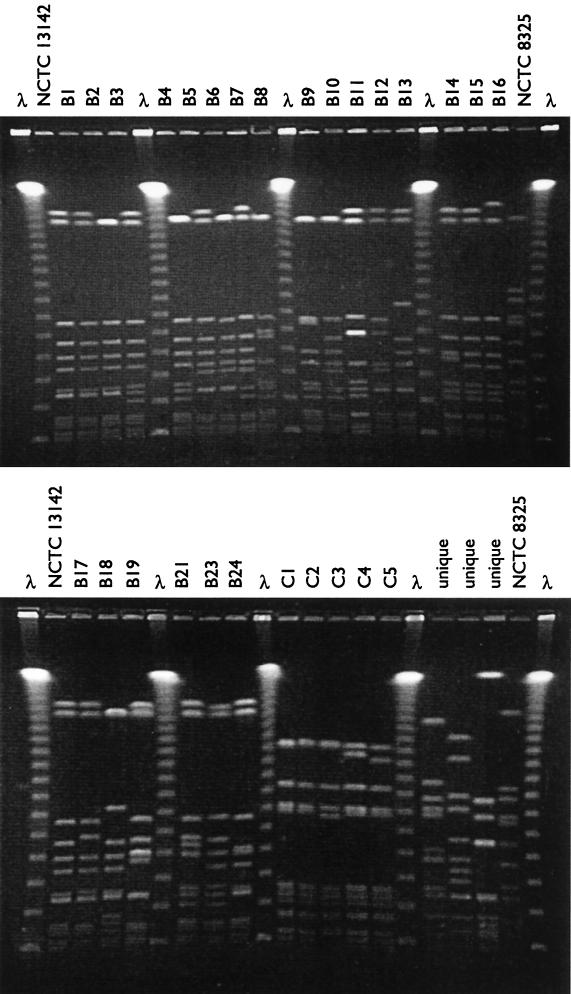
FIG. 2.
SmaI restriction profiles of putative EMRSA-15 isolates. (Top): PFGE profiles B1 through B16; (bottom) PFGE profiles B17 through B24 and the non-EMRSA-15 profiles (C1 through C5 and unique profiles). A commercial molecular weight marker consisting of concatemers of lambda DNA, the EMRSA-15 control strain NCTC 13142, and S. aureus strain NCTC 8325 were included as controls on all gels.
Subtyping isolates within the B cluster.
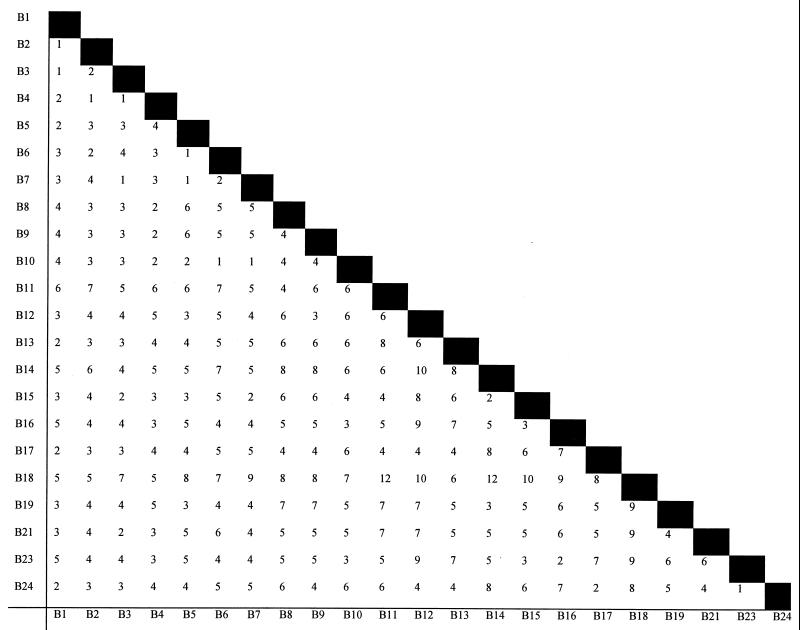
Among the 145 isolates within the EMRSA-15 cluster (B), 22 PFGE profiles, differing by at least one band, were seen (B1 through 19, B21, B23, and 24). A band difference matrix was generated using these data (Fig. 3). The EMRSA-15 type strain was designated B1, as were 36 other classical EMRSA-15 isolates (67% of classical isolates). The remaining 18 classical EMRSA-15 isolates produced eight PFGE profiles with 1 to 3 band differences from the progenitor pattern, B1 (Table 2). One classical EMRSA-15 isolate was methicillin sensitive, and its profile, B24, differed from B1 by a single band shift (a ca. 225-kb band was replaced by a ca. 190-kb band). Variant EMRSA-15 isolates showed 16 different PFGE profiles including B1, although this profile accounted for only two isolates. Four profiles, B2, B3, B4, and B7, accounted for 75 isolates (83% of phage variants), with the rest of the profiles represented by 1 or 2 isolates. The four main profiles were associated with phage patterns 2a, 3a, 3b or 3d, and 3c, respectively (Table 2). However, the correlation between these four phage patterns and PFGE profiles was not absolute. Four isolates exhibited one of the phage patterns listed above but produced different PFGE profiles (B8, B14, and B16). Further, five classical EMRSA-15 isolates had profile B3 and one had profile B7.
FIG. 3.
Band difference matrix of PFGE profiles of EMRSA-15 isolates. Numbers represent the band differences between PFGE profiles listed on the x and y axes.
Correlation of phage and PFGE patterns with geographical location and hospital.
There was no obvious link between geographical location and the EMRSA-15 phage variants or PFGE subtypes (data not shown). Hospitals represented by more than one isolate usually showed multiple PFGE and phage variants, and there was no correlation between the PFGE profiles of the classical and variant isolates in the 49 hospitals represented by both.
Enterotoxin gene carriage and production.
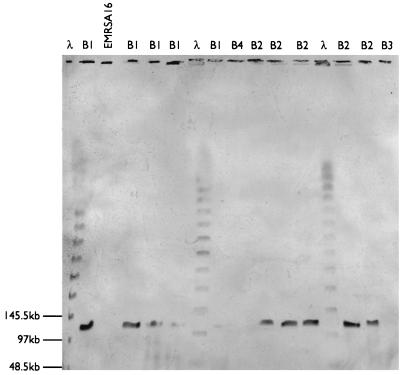
Enterotoxin genes sea through sed and tst were not detected in isolates from PFGE cluster C or among those with unique PFGE profiles (i.e., the eight non-EMRSA-15 isolates). All isolates from PFGE cluster B were negative for genes sea, seb, sed, and tst, although 74 carried the sec gene. These included all isolates belonging to subtypes B1, B2, B5, B6, B18, and B23, which were sec positive by PCR and/or Southern blotting. All isolates belonging to the remaining B subtypes were negative for sec (by PCR and Southern blotting) and did not produce enterotoxin C. The sec gene was localized to a ca. 110-kb fragment on PFGE gels (Fig. 4). Isolates carrying the enterotoxin C gene appeared to have a band doublet at this position in their PFGE profiles, whereas isolates without the gene appeared to have a single band at this position and an extra band at ca. 95 kb (Fig. 5). There was a clear association between absence of the enterotoxin C gene and sensitivity to φ81 (Table 3).
FIG. 4.
Hybridization of a probe for sec to SmaI restriction profiles of enterotoxin C-positive and -negative EMRSA-15 isolates. Size standards were visualized by probing with biotin-labeled λ digested with HindIII. Isolates belonging to PFGE profiles B1 and B2 were sec positive by PCR, and those belonging to profiles B3 and B4 were negative. EMRSA-16 was included as a negative control.
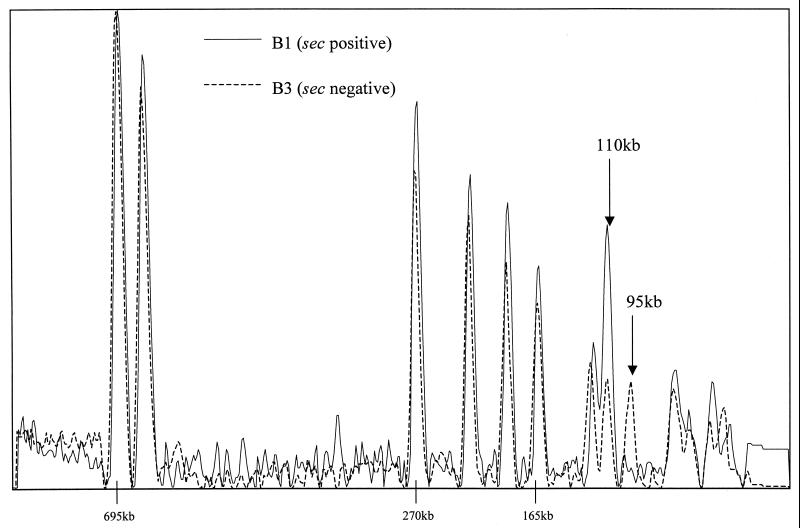
FIG. 5.
Overlaid densitometric curves of the PFGE profiles of variants B1 and B3 illustrating the difference in peak heights of the band at 110 kb in the profile. Band sizes quoted are approximate.
TABLE 3.
Relationship between sensitivity to phage 81 and carriage of the sec gene among EMRSA-15 isolates
| Isolates | No. φ81 sensitive and:
|
No. φ81 resistant and:
|
||
|---|---|---|---|---|
| sec positive | sec negative | sec positive | sec negative | |
| Classical EMRSA-15 | 49 | 6 | 0 | 0 |
| Phage variant EMRSA-15 | 24 | 1 | 1 | 64 |
| Total | 73 | 7 | 1 | 64 |
DISCUSSION
This study was undertaken to extend our knowledge of the characteristics of one of the two major EMRSA strains in the United Kingdom. Previously, EMRSA-15 had been defined by a weak reaction with φ75 of the international set, enterotoxin C production, and nonproduction of urease. Over the past decade, phage variants of this strain have arisen across the United Kingdom, but given that this strain has been circulating for at least 9 years, it is not suprising that some genetic change has taken place. A similar broadening of the phage typing pattern was observed with the epidemic penicillinase-producing 80/81 strain of the 1950s (21). This highly transmissable and virulent strain was shown to have spread across, and persisted in, several continents. The original phage pattern of 80/81 widened to 52/52A/80/81 over time and in different geographic locations. A defective prophage was responsible for the resistance of the original 80/81 strain to φ52 and φ52A, and replacement of this prophage by other phages resulted in the typing pattern difference (25). Presumably, a similar mechanism is responsible for the alteration of the EMRSA-15 phage pattern, although we have not yet attempted to identify the phages responsible for this.
Identification of EMRSA-15 phage variants.
More than 90% of isolates classified as phage variants of EMRSA-15 by phage typing were shown to be bona fide EMRSA-15 by PFGE, validating the usefulness of this quick and cost-effective method for typing. In addition, the fact that all the strains misidentified as EMRSA-15 were urease producers confirms that this simple test would eliminate the majority of false identifications. Furthermore, the phage patterns of the variant strains were generally stable and a core of strong reactions was identified. Although a well-established rule exists for interpreting phage patterns at RTD (two or more strong reaction differences define unrelated strains), this has not yet been validated at 100× RTD, and in any case it was intended for use only with geographically and temporally related isolates. This study suggests that these reaction difference rules may need to be modified for strains that require typing at 100× RTD.
Discrimination between EMRSA-15 isolates by PFGE.
The demonstration of different PFGE profiles among, and unique to, the classical EMRSA-15 isolates illustrates the role that genetic events such as point mutations, insertions, and deletions may play in altering the PFGE patterns of closely related isolates. The profile (B24) of one classical EMRSA-15 isolate, which was methicillin sensitive, differed from the progenitor pattern B1 by a single band shift from ca. 225 kb to ca. 190 kb. The size of the shift in molecular weight equates well with that published for the mec locus in United Kingdom MRSA isolates (9) and suggests that it is the loss of this genetic element which has generated subtype B24 from B1. The concordance between PFGE profile and phage pattern for variant isolates is indicative of the link between loss or gain of prophages from the genome (resulting in widening of the phage typing pattern) and changes in either SmaI restriction sites or fragment sizes responsible for the PFGE banding patterns. Interestingly, three PFGE profiles, B1, B3, and B7, were represented by both variant and classical isolates. Most B1 isolates were classical EMRSA-15 exhibiting the 75wk pattern, but two isolates reacted with phages 83C/90 and 42E/75/90 respectively. Conversely, although the majority of B3 and B7 isolates were EMRSA-15 variants, five B3 isolates and one B7 isolate gave the classical pattern 75wk. These findings suggest that replacement of prophages by other circulating phages may cause changes in the PFGE profile which are not reflected by changes in the phage pattern and vice versa. This is supported by data from Arbeit (3), who showed that different phages could be obtained from isolates with identical PFGE patterns and also that the same phage could insert into different fragments, creating strains which had the same genetic composition but PFGE patterns which differed by as many as 4 bands. These findings illustrate that no typing system is absolute and that sometimes even a simple system such as phage typing can give as much, if not more, information than a complex DNA-based typing system such as PFGE.
Criteria for interpretation of PFGE data.
For epidemiological typing, knowledge of both strain identity and variability within the strain allows us to make judgments on whether direct cross-infection or independent acquisition has taken place. However, interpretation of PFGE data obtained from a widespread strain such as EMRSA-15 can be problematic with regard to both strain identity and variability.
For local epidemiological studies, criteria of strain relatedness such as those described by Tenover et al. (26) with the modifications of Goering (8) are often applied. These criteria suggest that isolates showing 1 to 3 band differences from the outbreak (or progenitor) strain are probably related and part of the outbreak and that those showing 4 to 6 band differences may be related. However, these criteria are validated only for use within epidemiologically and temporally defined (<6 months) outbreaks. Isolates in this study were chosen from geographically diverse locations and were temporally spaced. Despite this, the majority of isolates within the B cluster fell within 1 to 3 band differences from the progenitor pattern, B1. These findings suggest that most isolates belonging to the B cluster differ by one genetic event from B1 and that even the most divergent member of the B cluster, B11, may differ from B1 by only two genetic events (giving a 6-band difference in the PFGE profile). How then, do we distinguish between cross-infection and independent acquisition of a strain such as EMRSA-15 when closely related PFGE profiles are obtained from isolates recovered during a suspected outbreak? A recent study by MacFarlane et al. (16) retrospectively analyzed two putative outbreaks of EMRSA-15 within a United Kingdom hospital. In one outbreak they found several different PFGE profiles differing by 1 to 4 bands, whereas in the second a single PFGE profile was identified. They suggested that in a highly clonal organism such as EMRSA-15, perhaps even single band differences between isolates may be of epidemiological significance. Prospective epidemiological studies involving hospitals with a large circulating EMRSA-15 population and known phage and/or PFGE subtypes may be helpful in attempting to answer this question.
Although most PFGE profiles obtained from isolates within the B cluster were closely related to the progenitor profile B1, higher numbers of band differences were seen when the more-divergent PFGE variant profiles were compared with each other. For example, B11 and B18 differ by 12 bands. Comparing these two profiles in isolation and interpreting the results using the Tenover criteria would lead to their classification as different strains. Although two isolates with such widely differing PFGE profiles are unlikely to represent an incident of cross-infection, classifying them as belonging to different genetic lineages is erroneous. This suggests that in dealing with a widely disseminated strain such as EMRSA-15, the range of PFGE profiles must be regarded as a continuum and cutoff points such as 4 to 6 band differences (which are appropriate for isolates from putative temporally and geographically related outbreaks) are too stringent for determining strain identity. It also emphasizes the importance of relating profiles back to the most common (or progenitor) profile for analysis.
Carriage of the sec gene.
Of particular interest was the appearance of enterotoxin C-negative variants of EMRSA-15. These had previously been reported for some isolates with the classical EMRSA-15 phage pattern (24) but had not been correlated with a change in PFGE profile. Examination of sec-negative isolates showed that loss of this gene was associated with a specific change in PFGE profile, namely, a marked decrease in the intensity of the ca. 110-kb band hybridizing to the sec gene probe in sec-positive isolates, concomitant with the appearance of an extra band at ca. 95 kb. Furthermore, the sec gene probe failed to hybridize with the remaining ca. 110-kb band in the profile of sec-negative variants. Examination of peak heights in the densitometric traces of the sec-positive and -negative isolates supports the view that sec-positive isolates are characterized by a doublet band at ca. 110 kb, whereas in sec-negative strains only a single band is present (Fig. 5). These findings suggest that the sec gene is carried on a piece of DNA of ca. 15 kb, which appears to have been excised from the genome in sec-negative variants.
It is known that enterotoxin genes are often located on mobile elements. Enterotoxin A is phage encoded (5), enterotoxin D is plasmid borne (4), and TSST-1 is encoded on a pathogenicity island (15). The relatively small size of the fragment change suggests that enterotoxin C is not phage encoded, since the average size of the phage genome is ca. 45 to 50 kb. Studies on the TSST-1 pathogenicity islands SAPI1 and SAPI2 have shown that the tst gene is carried on a 17-kb segment of DNA that can be mobilized by certain phages (15). It may be, therefore, that enterotoxin C is encoded on a similar pathogenicity island, since the size of the element appears to be similar and there is a phage association in that the majority of the isolates which were sec negative were sensitive to φ81. Further studies to test this hypothesis are under way.
Overall, this study has shown the usefulness of both phage typing and PFGE for monitoring the evolution of a prevalent strain of MRSA within the United Kingdom and has suggested further avenues of exploration concerning transmission of virulence factors within S. aureus.
ACKNOWLEDGMENTS
We thank Tyrone Pitt and Barry Cookson for critical comments on the manuscript and Maria Mena, Mark Ganner, and Marina Warner for technical assistance.
A. Gil-Setas was supported by a grant from the Spanish Society of Infectious Diseases and Clinical Microbiology. S. Murchan was funded by the EU DG-XII HARMONY project and participated in this project to facilitate method development for HARMONY.
REFERENCES
- 1.Anonymous. Methicillin-resistant Staphylococcus aureus (MRSA) Communicable Dis Rep. 1992;2:21. [PubMed] [Google Scholar]
- 2.Anonymous. Methicillin resistance in Staphylococcus aureus isolated from blood in England and Wales: 1994 to 1998. Communicable Dis Rep. 1999;9:65. , 68. [PubMed] [Google Scholar]
- 3.Arbeit R D. Laboratory procedures for epidemiologic analysis. In: Crossley K B, Archer G L, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 253–286. [Google Scholar]
- 4.Bayles K W, Iandolo J J. Genetic and molecular analyses of the gene encoding staphylococcal enterotoxin D. J Bacteriol. 1989;171:4799–4806. doi: 10.1128/jb.171.9.4799-4806.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betley M J, Mekalanos J J. Staphylococcal enterotoxin A is encoded by phage. Science. 1985;229:185–187. doi: 10.1126/science.3160112. [DOI] [PubMed] [Google Scholar]
- 6.Bignardi G E, Woodford N, Chapman A, Johnson A P, Speller D C E. Detection of the mecA gene and phenotypic detection of resistance in Staphylococcus aureus isolates with borderline or low-level methicillin resistance. J Antimicrob Chemother. 1996;37:53–63. doi: 10.1093/jac/37.1.53. [DOI] [PubMed] [Google Scholar]
- 7.Boyce J M. Epidemiology and prevention of nosocomial infections. In: Crossley K B, Archer G L, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 309–329. [Google Scholar]
- 8.Goering R V. The molecular epidemiology of nosocomial infection: an overview of principles, application and interpretation. In: Specter S, et al., editors. Rapid detection of infectious agents. New York, N.Y: Plenum Press; 1998. pp. 131–157. [Google Scholar]
- 9.Hiramatsu K, Ito T, Hanaki H. Mechanisms of methicillin and vancomycin resistance in Staphylococcus aureus. Clin Infect Dis. 1999;5:221–239. [Google Scholar]
- 10.Johnson W M, Tyler S D, Ewan E P, Ashton F E, Pollard D R, Rozee K R. Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. J Clin Microbiol. 1991;29:426–430. doi: 10.1128/jcm.29.3.426-430.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaufmann M E. Pulsed-field gel electrophoresis. In: Woodford N, Johnson A P, editors. Molecular bacteriology: protocols and clinical applications. Totawa, N.J: Humana Press; 1998. pp. 33–50. [Google Scholar]
- 12.Kaufmann M E, Pitcher D G, Pitt T L. Ribotyping of the bacterial genome. In: Chart H, editor. Methods in practical laboratory bacteriology. Boca Raton, Fla: CRC Press Inc.; 1994. pp. 123–138. [Google Scholar]
- 13.Kerr S, Kerr G E, Makintosh C A, Marples R R. A survey of methicillin-resistant Staphylococcus aureus affecting patients in England and Wales. J Hosp Infect. 1990;16:35–48. doi: 10.1016/0195-6701(90)90047-r. [DOI] [PubMed] [Google Scholar]
- 14.Lacey R W, Stokes A. Studies on recently isolated cultures of methicillin-resistant Staphylococcus aureus. J Gen Microbiol. 1979;114:329–339. doi: 10.1099/00221287-114-2-329. [DOI] [PubMed] [Google Scholar]
- 15.Lindsay J A, Ruzin A, Ross H F, Kurepina N, Novick R P. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol Microbiol. 1998;29:527–543. doi: 10.1046/j.1365-2958.1998.00947.x. [DOI] [PubMed] [Google Scholar]
- 16.MacFarlane L, Walker J, Borrow R, Oppenheim B A, Fox A J. Improved recognition of MRSA case clusters by the application of molecular subtyping using pulsed-field gel electrophoresis. J Hosp Infect. 1999;41:29–37. doi: 10.1016/s0195-6701(99)90034-8. [DOI] [PubMed] [Google Scholar]
- 17.Marples R R, Cooke E M. Workshop on methicillin-resistant Staphylococcus aureus. J Hosp Infect. 1985;6:342–348. [PubMed] [Google Scholar]
- 18.Marples R R, Reith S. Methicillin-resistant Staphylococcus aureus in England and Wales. Communicable Dis Rep Rev. 1992;2:R25–R29. [PubMed] [Google Scholar]
- 19.Marples R R, Richardson J F, de Saxe M J. Bacteriological characters of strains of Staphylococcus aureus submitted to a reference laboratory related to methicillin resistance. J Hyg Camb. 1986;96:217–223. doi: 10.1017/s0022172400065980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Neill G L, Ogunsola F T, Brazier J S, Duerden B I. Modification of a PCR ribotyping method for application as a routine typing scheme for Clostridium difficile. Anaerobe. 1996;2:205–209. [Google Scholar]
- 21.Parker M T. The significance of phage-typing patterns in Staphylococcus aureus. In: Easmon C S F, Adlam C, editors. Staphylococci and staphylococcal infections. London, England: Academic Press Inc.; 1983. pp. 33–62. [Google Scholar]
- 22.Richardson J F, Marples R R. Another strain of methicillin-resistant Staphylococcus aureus epidemic in London. Lancet. 1988;ii:748. doi: 10.1016/s0140-6736(88)90224-3. [DOI] [PubMed] [Google Scholar]
- 23.Richardson J F, Quoraishi A H M, Francis B J, Marples R R. Beta-lactamase-negative, methicillin-resistant Staphylococcus aureus in a newborn nursery: report of an outbreak and laboratory investigations. J Hosp Infect. 1990;16:109–121. doi: 10.1016/0195-6701(90)90055-s. [DOI] [PubMed] [Google Scholar]
- 24.Richardson J F, Reith S. Characterization of a strain of methicillin-resistant Staphylococcus aureus (EMRSA-15) by conventional and molecular methods. J Hosp Infect. 1993;25:45–52. doi: 10.1016/0195-6701(93)90007-m. [DOI] [PubMed] [Google Scholar]
- 25.Rountree P M, Asheshov E H. Further observations on changes in the phage-typing pattern of phage type 80/81 staphylococci. J Gen Microbiol. 1961;26:111–122. doi: 10.1099/00221287-26-1-111. [DOI] [PubMed] [Google Scholar]
- 26.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]