Abstract
We characterized 22 human clinical strains of Streptococcus bovis by genotypic (16S rRNA gene sequence analysis [MicroSeq]; Applied Biosystems, Foster City, Calif.) and phenotypic (API 20 Strep and Rapid ID32 Strep systems (bioMerieux Vitek, Hazelton, Mo.) methods. The strains, isolated from blood, cerebrospinal fluid (CSF), and urine, formed two distinct 16S ribosomal DNA sequence clusters. Three strains which were associated with endocarditis urinary tract infection (UTI), and sepsis clustered with the S. bovis type strain ATCC 33317 (cluster 1); other closely related type strains were S. equinus and S. infantarius. Nineteen strains clustered at a distance of about 2.5% dissimilarity to the S. bovis type strain (cluster 2) and were associated with central nervous system (CNS) disease in addition to endocarditis, UTI, and sepsis. All strains were distinct from S. gallolyticus. Within cluster 2, a single strain grouped with ATCC strain 43143 (cluster 2a) and may be phenotypically distinct. All the other strains formed a second subgroup (cluster 2b) that was biochemically similar to S. bovis biotype II/2 (mannitol negative and beta galactosidase, alpha galactosidase, beta glucuronidase, and trehalose positive). The API 20 Strep system identified isolates of cluster 2b as S. bovis biotype II/2, those of cluster 1 as S. bovis biotype II/1, and that of cluster 2a as S. bovis biotype I. There was an excellent correlation of biotype and genotype: S. bovis biotype II/2 isolates form a separate genospecies distinct from the S. bovis, S. gallolyticus, and S. infantarius type strains and are the most common isolates in adult males.
S. bovis is a human pathogen associated with endocarditis, sepsis, and meningitis (2, 3, 4, 6, 7, 8, 10). Since the early 1980s genetic and biochemical diversity among S. bovis has been noted (4, 7, 8). Several groups studied this diversity and devised schemes to distinguish strains by biotype. S. bovis strains from humans are said to be biotype I (or typical) if, among other traits, they ferment mannitol and produce glucan and biotype II (or variant) if they cannot ferment mannitol or produce glucan. The S. bovis biotype II strains are further divided into type II/1 and type II/2 by the ability of the later group to produce beta-galactosidase and beta-glucuronidase and ferment trehalose but not glycogen (4, 8).
Recently the S. bovis group has been further defined based on 16S rRNA gene sequence, ribotyping, and whole-cell protein electrophoresis patterns. S. infantarius, with a 1.8% difference by 16S rRNA gene sequence from the S. bovis type strain, is phenotypically like S. bovis biotype II/1 (1, 9). Ribotyping patterns were used to differentiate all related species from S. infantarius and distinguish two subspecifics within the group (9). S. gallolyticus, originally isolated from Koala dung, is phenotypically that of biotype I (5, 9). Devriese et al. examined strains previously identified as S. bovis by both phenotypic methods and whole-cell protein electrophoresis and reported that many of the human strains associated with endocarditis and animal (goat, pigeon, and cow) strains were S. gallolyticus (5). By comparing our strains with the more recently described species, we wished to determine the distribution of the various S. bovis groups within our population. Although there have been some previous reports of frequency of occurrence of the S. bovis groups (7), none have been based on 16S rRNA gene sequence analysis of strains from a single population. We characterized by genotypic and phenotypic methods 21 human strains of S. bovis that were isolated in a single hospital from adult males and one referred strain from a child. The correlation of phenotype with genotypic cluster is presented here.
MATERIALS AND METHODS
The clinically significant strains of S. bovis were collected over a period of 10 years, with no two isolates collected in the same month. From 1998 to 2000, the isolates were sequential. They were not a single genetically related cluster. After initial isolation, strains were frozen at −70°C until further study. Presumptive phenotypic identification was performed by Gram stain, colony morphology, sheep blood hemolysis, and catalase reaction. For biochemical testing, streptococcal strains were grown at 37°C on Columbia agar plates (Remel, Lenexa, Kans., and BBL, Becton Dickinson, Cockeysville, Md.) in anaerobic conditions. Biochemical testing was performed using the API 20 Strep system (version 6) and the Rapid ID32 Strep system (bioMerieux Vitek). The interpretation of all these tests was done according to manufacturers' instructions. 16S rRNA gene sequence identification was performed at MIDI Labs (Newark, Del.) and Houston VA laboratories using the MicroSeq 500 Gene kit (Applied Biosystems) according to the manufacturer's specifications. Test strain sequences were compared against the MicroSeq 16S rRNA gene sequence database. The database contains sequences from 1,297 different species (1,187 type strains), including 36 type strains from the genus Streptococcus, 18 type strains from the genus Enterococcus, and 2 type strains from the genus Aerococcus. Within the S. bovis group, S. bovis type strain ATCC 33317 was sequenced. Sequence data obtained from GenBank on non-type strains was also included in the analysis. These strains and their accession numbers are as follows: S. bovis, ATCC 43143; S. bovis, AF104114.1; S. infantarius, AF177729; and five S. bovis sequences—AF082730.1, AF202263.1, AF135453.1, AF104109.1, and AB002481.1.
RESULTS
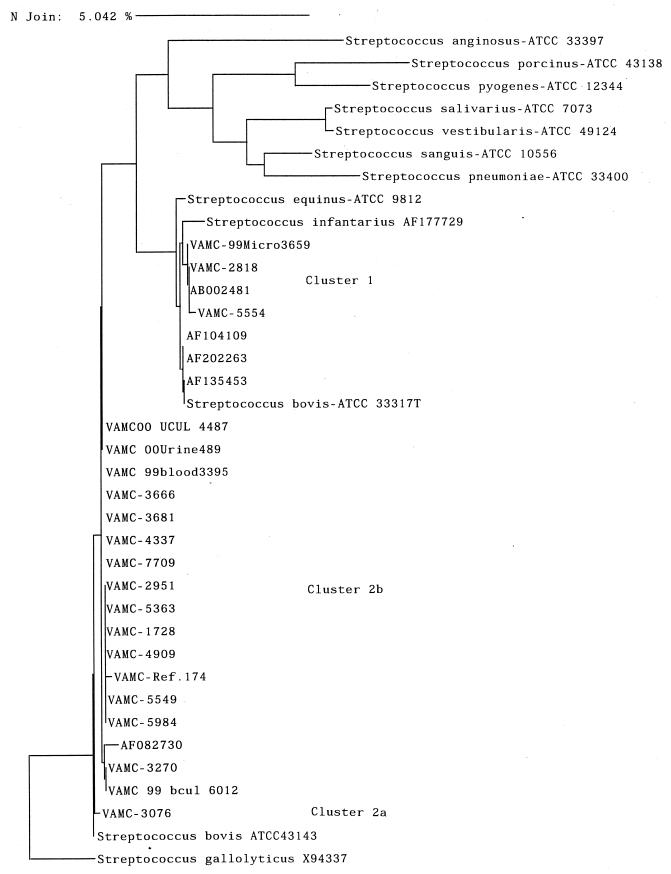
The relatedness of strains as determined by 16S rRNA gene sequence analysis is shown in a dendrogram (Fig. 1). Although sequence comparisons were performed on all 22 of our isolates, all strains in the MicroSeq database (including 36 streptococcal type strains), and all sequences in GenBank, for convenience, only the closely related or better-known type strains, close sequences from GenBank, and 20 of our strains are shown on the dendrogram. The strains from our study are prefaced with “VAMC”, and others are as listed as in Materials and Methods. Details of all clinical strains are given in Table 1. There are two major genogroups into which the strains cluster. Cluster 1 includes three clinical strains, three type strains (S. bovis ATCC 33317, S. equinus ATCC 9812, and S. infantarius AF177729), and four sequences from GenBank. Cluster 2 includes 19 clinical strains, S. bovis ATCC 43143, and a sequence from GenBank. These two clusters differ by about 2.5%. Within cluster 2, a single strain grouped with ATCC 43143 (cluster 2a), while all the other strains (18) formed a second subgroup (cluster 2b), for which there was only one similar sequence in the databases, belonging to an undescribed organism isolated from a human. Strain 4337 and all strains in the identical group show 2.43% difference in sequence from S. bovis type strain ATCC 33317. Cluster 2a differs from 2b by about 0.8%. The S. gallolyticus sequence (Fig. 1) did not cluster within the S. bovis-S. infantarius-S. equinus node.
FIG. 1.
Dendrogram of phylogenetic relationships among Streptococcus bovis isolates and ATCC type strains. Distances were calculated by the neighbor-joining method. The bar at the top indicates a 5% difference measured horizontally.
TABLE 1.
Biochemical characteristics by strain and genotypic cluster of strains isolated in this studya
| Cluster | Strain designation | Source | API biotype no. | API 32S biotype no. | Clinical association |
|---|---|---|---|---|---|
| 1 | 99B3659 | Blood | 5240443 | 22013001110 | Sepsis, colon cancer |
| 1 | 2818 | Cardiac tissue | 5240441 | 22013001110 | Endocarditis |
| 1 | 5554 | Urine | 5240440 | 22013001110 | UTI |
| 2b | 00U4487 | Urine | NDb | UTI | |
| 2b | 00U489 | Urine | 5450451 | UTI | |
| 2b | 99B3395 | Blood | 5650450 | 63077203150 | Sepsis |
| 2b | 96B3666 | Blood | 5650450 | 63077001150 | Sepsis, GI bleed |
| 2b | 96B3681 | Blood and CSF | 5650450 | 63077201150 | Meningitis |
| 2b | 4337 | Brain tissue | 5650450 | Subdural empyema | |
| 2b | 99U7709 | Urine | 5640450 | UTI | |
| 2b | 2951 | Blood | 5650450 | Sepsis, GI bleed | |
| 2b | 5363 | Blood | 5650450 | 63077001150 | Endocarditis |
| 2b | 97B1728 | Blood | 5650450 | 63077201150 | Spontaneous peritonitis |
| 2b | 98B4909 | Blood | 5650450 | 63077001150 | Sepsis |
| 2b | 5538 | Blood | 5650450 | 23077201150 | Sepsis, GI bleed |
| 2b | ref174 | CSF | 5650450 | 63077001150 | Encephalitis and meningitis |
| 2b | 5549 | Urine | 5650450 | 23077201150 | UTI |
| 2b | 5984 | Urine | 5650450 | 23077201150 | UTI |
| 2b | 7057 | Urine | 5650450 | 63077003150 | Unknown |
| 2b | 3270 | Blood | 5650450 | 63077001150 | Endocarditis |
| 2b | 99B6012 | Blood | 5650450 | 63077201150 | Endocarditis |
| 2a | 3076 | Blood | 5250553 | 20237061110 | Endocarditis, meningitis |
The right-hand portion of the strain designation is the same as in Fig. 1. As stated in text, there were isolates associated with UTI endocarditis and sepsis in both larger groups.
ND, not determined.
Table 1 shows the source, API20 Strep system biotype, and Rapid ID32 Strep biotype of all the tested clinical strains. The three strains that clustered with the S. bovis type strain ATCC 33317 were isolated from blood, cardiac tissue, and urine, while strains in cluster 2 were isolated from the central nervous system (CNS) in addition to blood and urine. All organisms were positive for hydrolysis of esculin, production of acetoin and leucine aminopeptidase, and production of acid from lactose, maltose, raffinose, and sucrose. The significant tests that were negative were hydrolysis of hippurate and urea and production of acid from ribose, l-arabinose, and sorbitol. Our data for tests that were variable or important in distinguishing related strains are shown in Table 2 in the first three data columns. The other seven columns show comparative data from sources as indicated. We found that strains that are different genetically also show consistent biochemical differences. For example, all the S. bovis strains in cluster 1 are trehalose negative and those in cluster 2 are trehalose and beta-galactosidase positive. Within this cluster, one strain, 3076 (cluster 2a), is biochemically distinct and genetically closer to S. bovis strain ATCC 43143. The rest of the strains are in cluster 2b. The API 20 Strep system identified isolates of cluster 2b as S. bovis biotype II/2, those of cluster 2a as S. bovis biotype I (although the test results are not as described by others [7] for S. bovis biotype I), and those of cluster 1 as S. bovis biotype II/1. Both the API 20 Strep system and the Rapid ID32 Strep system identified all strains as S. bovis.
TABLE 2.
Summary of differential characteristics of the strainsa
| Characteristic | Our data (% positive) for cluster:
|
Data from references 5 and 9
|
Data from API 20 Strep chart (% positive)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 (n = 3) | 2a (n = 1) | 2b (n = 17) | S. Infantarius | S. bovis biotype II/1 | S. bovis biotype II/2 | S. gallolyticus | Typical biotype I | S. bovis biotype II/1 | S. bovis biotype II/2 | |
| Production of: | ||||||||||
| Beta-glucosidase | 100 | 100 | 100b | v/+ | + | + | + | NDe | ND | ND |
| Beta-glucuronidase | 0 | 0 | 100 | − | − | + | − | 4 | 1 | 88 |
| Alpha-galactosidase | 100 | 0 | 100 | + | + | v | + | 71 | 86 | 85 |
| Beta-galactosidase | 0 | 100 | 100 | − | − | + | − | 14 | 17 | 94 |
| Beta-mannosidase | 0 | 0 | 100b | − | − | + | v | ND | ND | ND |
| Acid from: | ||||||||||
| Trehalose | 0 | 100 | 100 | − | − | + | + | 90 | 30 | 99 |
| Raffinose | 100d | 100d | 100 | v | + | v | + | 90 | 99 | 72 |
| Inulin | 0 | 0 | 0 | − | + | − | + | 63 | 61 | 13 |
| Starchc | fast | fast | slow | +/v | + | − | + | 100 | 73 | 40 |
| Glycogen | 33 | 100 | 0 | +/− | + | − | + | 90 | 65 | 18 |
| Pullulans | 33 | 100 | 0b | v/− | − | − | + | ND | ND | ND |
| Melibiose | 0 | 0 | 20b | v/− | + | − | − | ND | ND | ND |
| Mannitol | 0 | 0 | 0b | − | − | − | + | 86 | 0 | 0 |
Our data, which were obtained by testing our strains on both the API 20 Strep and API 32 Strep systems, is shown in the first group of columns. It is contrasted with data in subsequent columns from references 5 and 9 combined and the API 20 Strep chart.
Only 11 strains tested.
With extended incubation (1 week), all strips became positive: fast, positive in 24 to 48 h; slow, positive in >48 h.
Positive on API 20 Strep but negative on the rapid ID 32 Strep.
ND, not in API 20 Strep database.
DISCUSSION
Recently two new species closely related to the S. bovis-S. equinus group have been described. S. infantarius, so named because it had been isolated from infant's stools, was distinguished based on 16S rRNA gene sequence and ribotyping. S. gallolyticus was reported as isolated from both human and animal specimens. We began this study because the genetic and biochemical diversity of the S. bovis group had not clearly distinguished whether there would be separate biotype or genotype groups for isolates from different clinical settings, e.g., adults and infants, humans and animals, urine and CNS, or isolates associated with colon cancer and isolates not associated with colon cancer. A previous excellent study indicated that biotype I was more commonly associated with underlying cancer or gastrointestinal (GI) disease and that biotype II/2 occurred less frequently (7). Confirming this and clarifying the occurrence of genetically defined strains could lead to a better understanding of the pathogenic role of S. bovis.
In fact, the 22 human strains of S. bovis that we characterized by both 16S rRNA gene sequence analysis and biochemical methods formed two distinct 16S rRNA sequence clusters. However, the genotypic clusters did not clearly segregate the isolates according to source, age of patient, or type of infection. Both clusters 1 and 2 contained isolates that were obtained from blood and urine and associated with sepsis, endocarditis, and urinary tract disease. The single strain from a child that we examined clustered with the majority of our strains from adults, not with the S. infantarius strain as we thought it might. One difference that might be explored further is that the three isolates that were associated with CNS disease were in cluster 2. Although we did not purposefully compare human and animal strains, it is interesting that our analysis of a serendipitous strain isolated from a septic dog placed it in cluster 1 (data not shown) with the S. bovis type strain ATCC 33317, also isolated from an animal.
The genotypic clusters did, however, segregate the isolates into biochemically homogeneous biotypes. The three strains that clustered with S. bovis type strain ATCC 33317 (originally isolated from cow dung) were trehalose negative and compatible with biotype II/1, as is the type strain (4), and also similar to the published description of S. infantarius (see Table 2). All the other strains were similar to S. bovis biotype II/2 in that they produced beta-galactosidase and beta-glucuronidase and fermented trehalose but not glycogen. A single strain that was closest to ATCC strain 43143 (originally isolated from human blood) was also most similar biochemically to S. bovis biotype II/2; the slightly different biochemical pattern may not be significant. In contrast to the results of Devriese (5), we found no strains phenotypically similar to S. bovis biotype I by most descriptions (i.e., none were mannitol positive, even though the Strep API 20 code book called one isolate S. bovis biotype I), and none of our S. bovis strains clustered with the S. gallolyticus sequence.
Thus, in contrast to studies on other populations, S. bovis biotype II/2 is the most frequently occurring of the various S. bovis group biotypes within our adult, predominantly male population. The phenotypic and genotypic characteristics demonstrate that they are clearly distinct from the S. bovis, S. gallolyticus, and S. infantarius type strains and represent a hitherto unnamed species.
Nucleotide sequence accession numbers.
Sequences for strains VAMC blood3395 and VAMC 3076 have been deposited in GenBank under accession numbers AF313406 and AF313408, respectively.
REFERENCES
- 1.Bovet A, Grimont F, Collins M D, Benaoudia F, Devine C, Regnault B, Grimont P A. Streptococcus infantarius sp. nov. related to Streptococcus bovis and Streptococcus equinus. Adv Exp Med Biol. 1997;418:393–395. doi: 10.1007/978-1-4899-1825-3_94. [DOI] [PubMed] [Google Scholar]
- 2.Clarridge J E, III, Osting C, Jalali M, Osborne J, Waddington M. Genotypic and phenotypic characterization of “Streptococcus milleri” group isolates from a veterans administration hospital population. J Clin Microbiol. 1999;37:3681–3687. doi: 10.1128/jcm.37.11.3681-3687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen L, Dunbar S, Clarridge J E., III Streptococcus bovis infection of the central nervous system: report of two cases and review. Clin Infect Disease. 1997;25:819–823. doi: 10.1086/515537. [DOI] [PubMed] [Google Scholar]
- 4.Coykendall A. Classification and identification of the viridans streptococci. Clin Microbiol Rev. 1989;2:315–328. doi: 10.1128/cmr.2.3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devriese L A, Vandamme P, Pot B, Vanrobaeys M, Kersters K, Haesebrouck F. Differentiation between Streptococcus gallolyticus strains of human clinical and veterinary origins and Streptococcus bovis strains from the intestinal tracts of ruminants. J Clin Microbiol. 1998;36:3520–3523. doi: 10.1128/jcm.36.12.3520-3523.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moellering R C., Jr . Enterococcus species, Streptococcus bovis and Leuconostoc species. In: Mandell G, Bennett J, Dolin R, editors. Principles and practice of infectious diseases. New York, N.Y: Churchill Livingstone; 2000. pp. 2147–2156. [Google Scholar]
- 7.Ruoff K, Miller S I, Garner C V, Ferraro M J, Calderwood S B. Bacteremia with Streptococcus bovis and Streptococcus salivarius: clinical correlates of more accurate identification of isolates. J Clin Microbiol. 1989;27:305–308. doi: 10.1128/jcm.27.2.305-308.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruoff K, Whiley R A, Beighton D. Streptococcus. In: Murray P R, editor. Manual of clinical microbiology. 7th ed. Washington, D.C.: ASM Press; 1999. pp. 283–296. [Google Scholar]
- 9.Schlegel L, Grimont F, Collins M D, Regnault B, Grimont P A, Bouvet A. Streptococcus infantarius sp. nov., Streptococcus infantarius subsp. infantarius subsp. nov. and Streptococcus infantarius subsp. coli subsp. nov., isolated from humans and food. Int J Syst Evol Microbiol. 2000;50:1425–1434. doi: 10.1099/00207713-50-4-1425. [DOI] [PubMed] [Google Scholar]
- 10.Tabibian N, Clarridge J E. Streptococcus bovis septicemia and large bowel neoplasia: four new cases and review of the literature. Am Fam Physician. 1989;39:227–229. [PubMed] [Google Scholar]