Abstract
In a previous study, we proposed to associate spoligotyping and typing with the variable number of tandem DNA repeats (VNTR) as an alternative strategy to IS6110-restriction fragment length polymorphism (RFLP) for molecular epidemiological studies on tuberculosis. The aim of the present study was to further evaluate this PCR-based typing strategy and to describe the population structure of Mycobacterium tuberculosis in another insular setting, Sicily. A collection of 106 DNA samples from M. tuberculosis patient isolates was characterized by spoligotyping and VNTR typing. All isolates were independently genotyped by the standard IS6110-RFLP method, and clustering results between the three methods were compared. The totals for the clustered isolates were, respectively, 15, 60, and 82% by IS6110-RFLP, spoligotyping, and VNTR typing. The most frequent spoligotype included type 42 that missed spacers 21 to 24 and spacers 33 to 36 and derived types 33, 213, and 273 that, together represented as much as 26% of all isolates, whereas the Haarlem clade of strains (types 47 and 50, VNTR allele 32333) accounted for 9% of the total strains. The combination of spoligotyping and VNTR typing results reduced the number of clusters to 43% but remained superior to the level of IS6110-RFLP clustering (ca. 15%). All but one IS6110-defined cluster were identified by the combination of spoligotyping and VNTR clustering results, whereas 9 of 15 spoligotyping-defined clusters could be further subdivided by IS6110-RFLP. Reinterpretation of previous IS6110-RFLP results in the light of spoligotyping-VNTR typing results allowed us to detect an additional cluster that was previously missed. Although less discriminative than IS6110-RFLP, our results suggest that the use of the combination of spoligotyping and VNTR typing is a good screening strategy for detecting epidemiological links for the study of tuberculosis epidemiology at the molecular level.
DNA fingerprinting is an important tool for differentiating the clinical isolates identified as Mycobacterium tuberculosis at the subspecies level and for studying the epidemiology of tuberculosis in a community (11, 32). Although restriction fragment length polymorphism (RFLP) analysis based on the insertion sequence IS6110 is considered a “gold standard” among DNA fingerprinting methods (23, 30), a number of limitations and drawbacks of IS6110 fingerprinting have been demonstrated, both at the theoretical level and at the practical level (18). First, its discriminatory power for isolates with fewer than six IS6110 copies is low and, consequently, secondary typing using another independent genetic marker is often required (3). Second, the existence of IS6110 insertion preferential loci or ipl (4, 17, 34) raises an important issue; since IS6110 typing relies on random IS6110 integration into the genome, the clonality between two isolates by IS6110 typing, even in high-copy-number isolates, may not always indicate recent transmission of tuberculosis (8). Third, IS6110-RFLP is a labor-intensive and time-consuming methodology, and adequate intralaboratory or interlaboratory comparison between profiles is a difficult task requiring intensive input and expertise from investigators. Thus, in order to improve the comparison of RFLP profiles, specialized softwares are needed (22). For these reasons, the worldwide implementation of IS6110-RFLP technique remains difficult and alternative, rapid, simple, and cost-effective genotyping methodologies are required. The advent of “spoligotyping,” an alternative PCR-based technique based on the polymorphism of the direct repeat (DR) locus gave promising results; however, it has been shown to be less discriminative than IS6110-RFLP (15). In this context, the association of two PCR-based techniques was suggested as a potential alternative to IS6110-RFLP (9). Subsequently, the association of spoligotyping and double-repetitive-element PCR (DRE-PCR), a procedure based on the detection of inter-IS6110-PGRS (polymorphic GC-rich sequence) polymorphism (6) was proposed as an alternative to IS6110-RFLP for epidemiological studies of tuberculosis (19, 26). Nevertheless, DRE-PCR reproducibility was recently shown to be suboptimal (16). Consequently, an optimal association of two PCR-based genotyping methods to study tuberculosis epidemiology still remains an open question. Aside from spoligotyping and DRE-PCR (26), ligation-mediated PCR and spoligotyping (1), spoligotyping and typing with the variable number of tandem DNA repeats (VNTR) (5), and recently spoligotyping and PGRS-typing (35) have also been proposed as alternatives to IS6110-RFLP.
The aim of the present investigation was to apply PCR-based genotyping methods on Sicilian isolates previously subjected to IS6110-RFLP typing (20) and to compare their discriminative power to IS6110-RFLP. The discriminative power of each method or each association of methods was calculated by using the Hunter-Gaston Index (HGI [12]). Moreover, some considerations on the genotype population structure of M. tuberculosis in this area were drawn in the light of these new molecular findings. Our results show that the use of a combination of independent PCR-based genotyping methods is a reliable although less-discriminative strategy for performing molecular epidemiologic studies on tuberculosis. We also show that each method produces a very coherent set of data indicative of the clonal structure of M. tuberculosis strain population and that molecular clustering may, in certain epidemiological contexts, further elucidate tuberculosis epidemiology.
MATERIALS AND METHODS
Patients.
The population studied included 106 patients (29 women and 77 men; male/female sex ratio of 2.65). The mean ages among the female group were 45.6 years, excluding two children, and 42.6 years if these two patients are included. The mean age among the male group was 40 years if four children and infants are included among the patients and 42 years if they are excluded. In both populations (males and females), two age groups of 20 to 29 and 50 to 59 years were the most represented. This bimodal distribution may underline most of the reactivation cases that occurred among the age group of 50 to 59 years, whereas “recent” transmission cases were mostly limited to the age group of 20 to 29 years. In Italy, the incidence rate has remained stable between 1995 and 1996 at approximately 9 cases per 100,000 population, underlining the need for a continued surveillance and a more effective control strategy (20).
Clinical isolates and DNAs.
A collection of 106 clinical isolates was collected during a period of approximately 5 years (June 1994 to February 2000). Most of these isolates had been previously characterized by IS6110-RFLP (20). All DNAs were prepared by using the classical cetyltrimethylammonium bromide method (33).
Genotyping methods.
Southern blotting with labeled IS6110 DNA was performed as previously described using an internationally agreed protocol (30). Briefly, M. tuberculosis DNA was digested with PvuII, electrophoresed, and hybridized with a labeled 245-bp PCR-generated probe. Each Southern blot included DNA from M. tuberculosis strain 14323 as an external standard (30). Autoradiographs of the Southern blots were compared by using Gel Manager software (Biosystematica, Tavistock, United Kingdom), and RFLPs were clustered by the unweighted pair group method using arithmetic averages (UPGMA [24]).
Spoligotyping was performed using in-house prepared membranes according to a published protocol (15). A Biodyne C membrane (Pall Biosupport, Portsmouth, England) having 43 covalently bound oligonucleotides was prepared essentially as previously published (15) with slight modifications (31); 6 spacers were derived from M. bovis BCG sequence, and 37 spacers were derived from sequences present in the M. tuberculosis H37Rv DR locus. Briefly, the Biodyne C membrane was incubated for 10 min in 12 ml of a freshly prepared 16% (wt/vol) EDAC (1-ethyl-3-3-dimethyl aminopropyl carbodiimide). It was then rinsed in deionized water for 2 min and placed in a Miniblotter 45 system (Immunetics, Cambridge, Mass.). The first and last slots were filled with 150 μl of diluted black ink. The 43 remaining slots were filled with 150 μl of an amino-linked oligonucleotide solution in 500 mM NaHCO3 (pH 8.4) as reported previously (15). The membrane was removed and inactivated with 100 mM NaOH for 10 min and then rinsed twice in 250 ml of 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])–0.1% sodium dodecyl sulfate (SDS) at 60°C for 5 min and twice in 20 mM EDTA (pH 8) for 15 min at room temperature. It was kept at 4°C in sealed plastic bags until use. A premix was prepared for 44 PCR samples, with each final sample containing PCR reagents (49 μl) and DNA (1 μl). Each tube contained 5 μl of Tth polymerase reaction Mix 10x (Eurobio, Les Ulis, France), 7 μl of 50 mM MgCl2, 0.8 μl of a deoxynucleoside triphosphate mix (dNTPs; 25 mM), 1 μl of each primer (DRa and biotinylated DRb, 20 pmol/μl), 0.5 U of Tth polymerase (Eurobio), and ultrapure water to obtain a final volume of 49 μl. Diluted DNA (from 50 to 300 ng of DNA in 1 μl) was added to the tubes. The samples were subjected after a 3-min denaturation to 96°C to 35 amplification cycles (96°C, 1 min; 54°C, 1 min; 72°C, 30 s) in a first-generation Perkin-Elmer Thermocycler (Perkin-Elmer, Norwalk, Conn.), followed by a final extension step at 72°C for 10 min. The membrane was washed twice for 5 min in 250 ml of 2× SSPE–0.1% SDS and placed in the Miniblotter. Then, 30 μl of each PCR product was diluted into 135 μl of 2× SSPE–SDS 0.1%, heat denatured for 10 min at 100°C, and frozen on ice for 5 min. Each slot was filled with the diluted PCR product. A 1-h hybridization at 60°C in an oven was performed, and the membrane was washed twice for 5 min at 60°C in 250 ml of 2× SSPE–0.5% SDS. Detection was done after a 45-min to 1-h incubation of the membrane in 14 ml of 2× SSPE 0.5% SDS containing 5 μl of streptavidin-POD conjugate (Roche Biochemicals, Meylan, France). It was followed by two 10-min washings at 42°C in 2× SSPE–0.5% SDS, two 5-min washings in 2× SSPE at room temperature, and a final 2-min incubation in 20 ml of ECL Detection Liquids (Enhanced Chemo-Luminescence Detection Kit; Amersham, Buckinghamshire, England). The autoradiograms were developed using standard photochemicals after 10 min to 2 h of exposition on ECL Hyperfilms (Amersham).
VNTR typing was performed as described previously with slight modifications (7). PCR was performed in a total volume of 30 μl containing 3 μl of 10x recombinant Taq buffer (AP-Biotech, Uppsala, Sweden), 2 mM MgCl2, a 30 nM concentration of each primer, a 500 μM concentration of each of the four dNTPs, 3 μl of dimethyl sulfoxide, 0.6 U of recombinant Taq (AP-Biotech), and 50 to 200 ng of DNA sample. Reactions were run in a Perkin-Elmer 9600 Thermocycler. An initial denaturation of 12 min at 95°C was followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 1 min, and extension at 72°C for 2 min, with a final extension step at 72°C for 10 min. An aliquot of the reaction tubes was run on a 3% Metaphor gel (FMC Bioproducts, Rockland, Maine). Molecular weight standards (100-bp ladder or PhiX-HaeIII; AP-Biotech) were run every four to five lanes. The molecular weight determination of PCR fragments was performed using Taxotron software (Taxolab; PAD Grimont, Institut Pasteur, Paris, France) on images digitized using the Video-Copy system (Bioprobe, Montreuil, France). Once the length of the PCR fragments was precisely calculated, the number of copies for each exact tandem repeat (ETR) was deduced according to a previously published scheme (7) and documented as a five-digit number representing allele profiles ETR-A to ETR-E.
Analysis of spoligotyping results.
Spoligotyping results were analyzed as previously described using Excel (Microsoft, Cupertino, Calif.) and Taxotron (27) softwares. The dendrogram was constructed by using UPGMA after pairwise comparison of strains by calculation of the Jaccard Index (13, 24).
Pattern designation.
Each IS-clustered isolate was assigned to its IS6110-defined cluster as previously published (20). Spoligotypes described only once were designated as “orphans.” Spoligotyping-defined shared-types were designated for each clinical isolate after comparison of spoligotyping results to a proprietary database of spoligotyping, which contained 4,500 individual spoligotype patterns of M. tuberculosis representative of more than 50 countries. A limited version of this database was recently described (28). VNTR results are given under the form of a five digit format as reported previously (5).
RESULTS
Spoligotyping of the Sicilian M. tuberculosis clinical isolates.
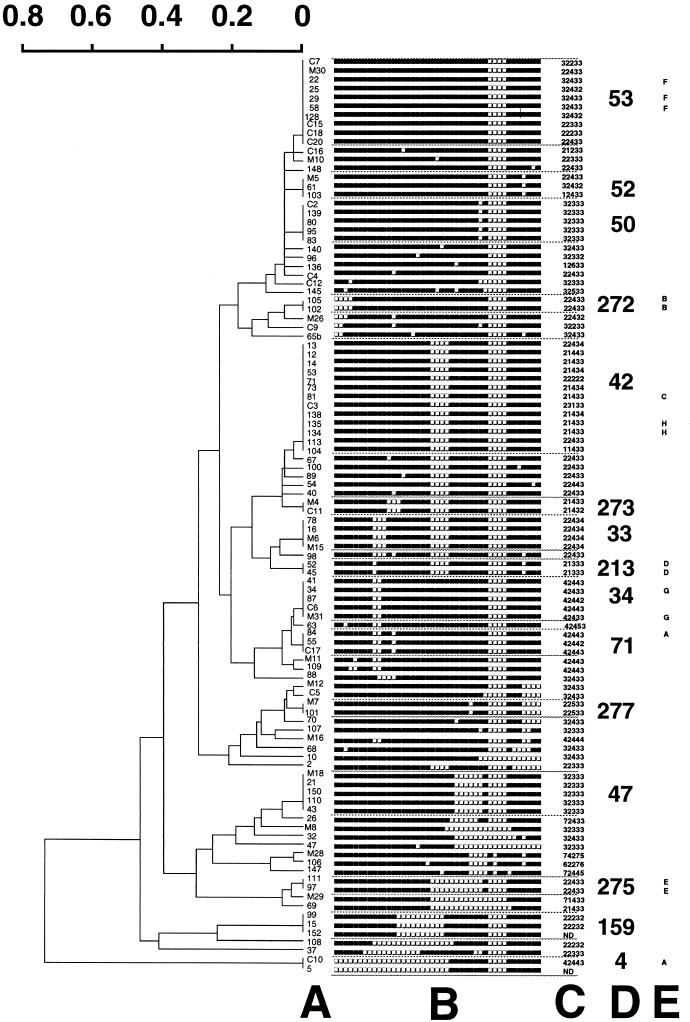
We first characterized the collection of 106 DNAs from Sicilian M. tuberculosis by spoligotyping in a blind study. A total of 104 results were obtained that identified 56 unique patterns. A total of 63 clinical isolates were clustered (60% of total isolates) in 15 clusters, whereas 41 clinical isolates harbored unique profiles. The designation of the spoligotype was attributed by comparison of the results to those contained in a database containing 4,500 spoligotype patterns from 50 countries (Fig. 1 and Table 1). New shared types were created when the patterns were not found into the database (types 272 to 278) either for new observed shared types (types 272, 273, 275, and 277) or when an orphan isolate from this study matched an orphan isolate from our database (types 274, 276, and 278). The distribution of the 63 clustered isolates was as follows: six clusters of 2 isolates (Fig. 1; clusters for types 4, 213, 272, 273, 275, and 277), three clusters of 3 isolates (types 52, 71, and 159), one cluster of 4 isolates (type 33), three clusters of 5 isolates (types 34, 47, and 50), and two large clusters of 13 and 10 isolates, respectively (types 42 and 53). To these 63 clustered isolates, we may also add 16 clinical isolates belonging to a previously identified shared type, according to the database, but being unique in Sicily (Table 1). Thus, the total number of unclustered isolates (isolates for which no matching pattern was detected in the database) dropped to 25 instead of 41 previously (24% instead of 40%). A dendrogram showing the population structure of M. tuberculosis clinical isolates assessed was constructed by pairwise comparison of isolates using the Jaccard Index and the UPGMA algorithm (Fig. 1).
FIG. 1.
Dendrogram and schematic representation of spoligotyping and VNTR typing results obtained with 104 clinical isolates of M. tuberculosis from Sicily. The scale represents the dissimilarity index according to computation of the Jaccard Index and use of the UPGMA algorithm. Columns A, isolate number; B, spoligotype description; C, VNTR typing results; D, shared type identification number; E, IS6110-RFLP cluster.
TABLE 1.
Shared type designations, origins, and references for 16 orphan clinical isolates from Sicily that matched patterns in our databasea
| Strain | Spoligotype designationa
|
Originb | Reference | |
|---|---|---|---|---|
| I | II | |||
| C4 | 37 | S77 | NLD | 15 |
| M2 | 48 | S55 | GBR | 10 |
| 8 | ||||
| M1 | 51 | S229 | FXX | 9 |
| 2 | ||||
| M1 | 86 | NLD | 15 | |
| 0 | ||||
| 40 | 93 | S125 | USA | 25 |
| C16 | 118 | S145 | USA | 25 |
| 96 | 119 | S3 | USA | 25 |
| 136 | 122 | FXX | 28 | |
| 89 | 150 | ITA | 1 | |
| 15 | 159 | ITA | 1 | |
| 100 | 162 | FXX | 9 | |
| C5 | 169 | ITA | 1 | |
| 10 | 237 | S233 | USA | 25 |
| 69 | 274 | RUS | 28 | |
| 26 | 276 | NLD | 29 | |
| 148 | 278 | SEN | 21 | |
Spoligotype designation according to our nomenclature (designation I) or to the American study (designation II) (25).
Origin of publication is according to ISO CODE 3166 available through the United Nations Statistics Division at http://www.un.org./unsd/methods/m49alpha.htm. NLD, The Netherlands; GBR, Great Britain; FXX, Metropolitan France; USA, United States of America; ITA, Italy; RUS, Russia; SEN, Senegal.
VNTR typing of the Sicilian M. tuberculosis clinical isolates.
The same clinical isolates were independently studied by VNTR typing, and the results are shown in column C of Fig. 1. Thirty-three unique types were observed upon VNTR typing with a total of 84 clustered isolates found in 15 clusters (81% of total isolates). The most frequently observed genotype was allele combination 22433 (15 clinical isolates). This is positively correlated to the most prevalent spoligotyping-defined major clade characterized by the absence of spacers 21 to 24 and 33 to 36 (type 42 and derived shared types 33, 213, 273, and 275, plus two orphan isolates (strains 98 and 108), that altogether represent more than 30% of the isolates. The second and third most prevalent genotypes are, respectively, allele combinations 32333 and 32433 (13 and 12 clinical isolates). The first combination is linked to spoligotypes 47 and 50, which belong to the Haarlem family of tubercle bacilli (five isolates each or 4.8%) and which may be tentatively defined by the simultaneous absence of spacer 31 and spacers 33 to 36 and VNTR profile 32333 (16). A third clade totaling 11 isolates (10%) was characterized by the simultaneous absence of spacers 9 and 10 and spacers 33 to 36 (shared types 34 and 71 plus two orphan isolates M11 and 109) and was found to be linked to the first VNTR allele (exact tandem repeat A [ETR-A] equal to four copies). No Beijing type (spoligotype 1, VNTR allele 42435) was observed in Sicily to date, whereas the ubiquitous shared type 53 (VNTR variable) accounted for 10% of the clinical isolates. The description of other less-represented spoligotypes were as follows: spoligotypes types 44 (isolate 140), 150 (isolate 89), 159 (isolates 15, 99, and 152), 162 (isolate 100), and 169 (isolate C5) were identified in an earlier study performed in Verona, Italy (1). Other types were found in the United States or in Cuba, which may be explained by the past history of emigration of Sicilians in the 1950s (Table 1). Finally, three isolates (isolates M28, 106, and 147) were shown to harbor a high exact tandem repeat-A (ETR-A) allele (>4), a characteristic spoligotype (absence of spacers 29-32 and 34), and a low IS6110 copy number (<5). All of these characteristics were recently described in a cluster of clinical isolates from Guinea-Bissau (14). By spoligotyping, these strains were determined to belong to a larger clade of a likely Afro-Asian bovine descent (28). Indeed, a cross-examination of their demographical records confirmed that these cases belonged to patients originating in Asia and/or Africa and may constitute imported tuberculosis cases.
Combination of spoligotyping and VNTR typing.
The IS6110-RFLP results of most of the isolates were described in an earlier study. This study showed that the most prevalent clinical isolates in Sicily harbored 8 to 11 copies of IS6110 (20). Table 2 summarizes the correlation observed between previously defined IS6110-RFLP clusters and clustering results obtained in this study, as well as previous epidemiological findings. In all but one case, the results obtained by the three molecular methods were in total agreement, a finding which confirms the clonality of the isolates studied. These results suggest recent tuberculosis transmission; however, traditional epidemiological investigation did not reveal a direct link between the patients (Table 2). In one case, two IS6110-RFLP-clustered strains were further subdivided by spoligotyping. This result suggests that an unusual mutation event happened in the DR locus and that these two isolates, although very closely related genetically, are not epidemiologically linked. This result concerns a single case of clustering of two pulmonary tuberculosis cases involving patients of 47 and 69 years, respectively, that were not involved in a recent tuberculosis transmission chain. DRE-PCR of these two isolates further confirmed that these two cases were not epidemiologically linked. On the other hand, DRE-PCR confirmed the clonality of the 16 other IS6110-defined clustered strains (results not shown).
TABLE 2.
Correlation between genotyping methods and epidemiologic links
| Isolate no. | IS6110-RFLP pattern | IS copy no. | Spoligotype(s) | VNTR type | DRE-PCR result | Epidemiologic findings |
|---|---|---|---|---|---|---|
| 84, C10 | A | 8 | 4, 71 | 42443 | Different | No apparent linkage |
| 102, 105 | B | 10 | 272 | 22433 | Identical | No apparent linkage |
| 62, 81a | C | 11 | 42 | 21433 | NDb | Living in the same apartment complex |
| 45, 52 | D | 11 | 213 | 21333 | Identical | No apparent linkage |
| 97, 111 | E | 12 | 275 | 22433 | Identical | No apparent linkage |
| 22, 29, 58, 59a | F | 8 | 53 | 32433 | Identical | No apparent linkage |
| M31, 34 | Gc | 9 | 34 | 42433 | Identical | No apparent linkage |
| 134, 135 | H | 8 | 42 | 21433 | Identical | Father and daughter |
DNA not available.
ND, Not done.
This cluster was initially overlooked but was detected by rechecking the RFLP results following indication of clustering by spoligotyping and VNTR typing.
The discriminative power of VNTR typing and spoligotyping was calculated by using the HGI (12). According to this method, this index should be superior to 90% for a typing method to be taken as efficient. In the present study, the HGI was, respectively, 93.9 and 96.75% for VNTR typing and spoligotyping. When they were used in combination, the HGI increased to 99.1%, which is close to the HGI of 99.7% obtained with IS6110-RFLP (Table 3).
TABLE 3.
Comparison of discriminatory power of three different typing systems used individually and in various combinations
| Typing system (no. of isolates) | No. of clustered isolates (%) | Maximum no. of isolates in a cluster | No. of clusters | No. of unique types | HGI (%) |
|---|---|---|---|---|---|
| IS6110-RFLP (102) | 16 (15) | 3 | 7 | 93 | 99.7 |
| Spoligotyping (104) | 63 (60) | 13 | 15 | 56 | 96.75 |
| VNTR (102) | 84 (82) | 15 | 15 | 33 | 93.9 |
| IS6110-RFLP-Spoligotyping (102) | 14 (13) | 3 | 6 | 94 | 99.8 |
| IS6110-RFLP-VNTR (102) | 16 (15) | 3 | 7 | 93 | 99.7 |
| Spoligotyping-VNTR (102) | 44 (43) | 5 | 16 | 74 | 99.1 |
DISCUSSION
In a previous study, we proposed the association of spoligotyping and VNTR typing as an alternative to IS6110-RFLP for molecular epidemiological studies on tuberculosis (5). The aim of the present investigation was to study this typing strategy in another geographical and epidemiological setting, Sicily, and to describe the population structure of M. tuberculosis strains in this region of Italy. Thus, a collection of 106 DNA isolates was independently genotyped by three methods. The totals of the clustered isolates were, respectively, 15, 60, and 82% by IS6110-RFLP typing, spoligotyping, and VNTR typing. The combination of spoligotyping and VNTR typing results reduced the number of clustered isolates to 43%. All but one IS6110-defined cluster were identified by the combination of spoligotyping and VNTR typing, suggesting that the use of a combination of PCR-based, independent genotyping methods is a reliable genotyping strategy for performing molecular epidemiologic studies on tuberculosis. Indeed, the differences observed in clustering level between spoligotyping-VNTR typing versus IS6110-RFLP typing suggest that these loci evolve with different molecular clocks. For example, the IS6110 molecular clock has been shown to be faster than the one of spoligotyping (15, 16).
In Sicily, the molecular epidemiology of M. tuberculosis strains was previously assessed by IS6110-RFLP during a 4-year period (20). Although this study led to the detection of microepidemics in the community, the data suggested that the rate of disease caused by reactivation largely exceeded the rate caused by recent transmission. Our study confirms the former study by validating all but one previously defined IS6110-RFLP cluster. It also extends the description of the genetic diversity of Sicilian M. tuberculosis clinical isolates by including spoligotyping and VNTR typing data, thus allowing us to place the Sicilian M. tuberculosis genotyping data within an Italian historical and geographical framework. As shown in Table 1, similarities between Cuban, American, and Italian spoligotypes were detected, which may be tentatively explained by the past migration history from Sicily toward the Italian peninsula or the American continent. Inversely, similarities between African or Indian and Sicilian strains appear likely to be an expression of more recent population migratory flows to Sicily. In this study, new molecular characteristics of Sicilian M. tuberculosis bacilli were identified which could mirror the influence of the complex nature of Sicilian peopling history on the epidemiology of tuberculosis. For example, a major family of M. tuberculosis isolates (type 42) accounted for 26% of total isolates (Fig. 1) and appears to possess a high biogeographical specificity in our spoligotyping database for Latin America, the Caribbean, and Mediterranean Europe. It is highly significant that type 42 was not present in any of the 1,283 M. tuberculosis isolates recently described in the United States (25). Indeed, this result suggests that the high prevalence of this cluster in Europe may be due to a remnant clone that mirrors a historically prevalent clade in this region. Continuous entry and leaving of M. tuberculosis strains, along with their human hosts, in the Sicilian population contributes to heterogeneity and, consequently, increases the epidemiologic predictive value of finding a molecular cluster by reducing the probability of clustering by chance. Thus, the demographic instability of Sicily (migration into and from the island) and the concentration of the population in large urban centers such as Palermo may contribute to an enhanced reliability of molecular typing compared to a large and stable rural community such as the one found in Arkansas (2). On the other hand, a low prevalence area and an unstable sociodemographic context are conditions where traditional case findings and epidemiological inquiries are more laborious and poorly effective. All of these characteristics fit completely with the epidemiological and demographic situation in the province of Palermo, thus validating clustering via molecular methods.
When we consider methodological issues, a multistep screening strategy combining two highly reproducible but moderately discriminative PCR-based genotyping techniques (spoligotyping and VNTR typing) is an efficient two-step strategy for potentially detecting epidemiologic links in a given community. This multistep screening approach is also interesting because it gradually increases the discriminatory power and consequently retains fewer and fewer strains for subsequent analysis. It also provides a population genetics view of tuberculosis genotype prevalence in a given setting by easily linking local and global epidemiologic issues with the use of spoligotype and VNTR databases. Last, but not least, our results indicate the coherence of molecular typing results using independent genetic markers underlying the clonal structure of tubercle bacillus populations. In the future, the study of various independent molecular clocks of M. tuberculosis genomes through individual as well as combined numerical phylogenetic analysis will help shed light on the monophyletic versus the poly- and/or paraphyletic hypothesis of tuberculosis origin.
ACKNOWLEDGMENTS
This work was supported by the Délégation Générale au Réseau International des Instituts Pasteur et Instituts Associés and the Fondation Française Raoul Follereau, Paris, France.
REFERENCES
- 1.Bonora S, Gutierrez M C, Perri G D, Brunello F, Allegranzi B, Ligozzi M, Fontana R, Concia E, Vincent V. Comparative evaluation of ligation-mediated PCR and spoligotyping as screening methods for genotyping of Mycobacterium tuberculosis strains. J Clin Microbiol. 1999;37:3118–3123. doi: 10.1128/jcm.37.10.3118-3123.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braden C R, Templeton G L, Cave M D, Valway S, Onorato I M, Castro K G, Moers D, Yang Z, Stead W W, Bates J H. Interpretation of restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates from a state with a large rural population. J Infect Dis. 1997;175:1446–1452. doi: 10.1086/516478. [DOI] [PubMed] [Google Scholar]
- 3.Chaves F, Yang Z, El Hajj H, Alonso M, Burman W J, Eisenach K D, Dronda F, Bates J H, Cave D. Usefulness of the secondary probe pTBN12 in DNA fingerprinting of Mycobacterium tuberculosis. J Clin Microbiol. 1996;34:1118–1123. doi: 10.1128/jcm.34.5.1118-1123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang Z, Forbes K J. A Mycobacterium tuberculosis IS6110 preferential locus (ipl) for insertion into the genome. J Clin Microbiol. 1997;35:479–481. doi: 10.1128/jcm.35.2.479-481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filliol I, Ferdinand S, Negroni L, Sola C, Rastogi N. Molecular typing of Mycobacterium tuberculosis based on variable number of tandem DNA repeats used alone, and in association with spoligotyping. J Clin Microbiol. 2000;38:2520–2524. doi: 10.1128/jcm.38.7.2520-2524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman C R, Stoeckle M Y, Johnson W D, Jr, Riley L W. Double-repetitive-element PCR method for subtyping Mycobacterium tuberculosis clinical isolates. J Clin Microbiol. 1995;33:1383–1384. doi: 10.1128/jcm.33.5.1383-1384.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frothingham R, Meeker-O'Connell W A. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998;144:1189–1196. doi: 10.1099/00221287-144-5-1189. [DOI] [PubMed] [Google Scholar]
- 8.Gillespie S H, Dickens A, McHugh T D. False molecular clusters due to nonrandom association of IS6110 with Mycobacterium tuberculosis. J Clin Microbiol. 2000;38:2081–2086. doi: 10.1128/jcm.38.6.2081-2086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goguet de la Salmonière Y-O, Li H M, Torrea G, Bunschoten A, van Embden J D A, Gicquel B. Evaluation of spoligotyping in a study of the transmission of Mycobacterium tuberculosis. J Clin Microbiol. 1997;35:2210–2214. doi: 10.1128/jcm.35.9.2210-2214.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goyal M, Saunders N A, van Embden J D A, Young D B, Shaw R J. Differentiation of Mycobacterium tuberculosis isolates by spoligotyping and IS6110 restriction fragment length polymorphism. J Clin Microbiol. 1997;35:647–651. doi: 10.1128/jcm.35.3.647-651.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heldal E, Döcker H, Caugant D A, Tverdal A. Pulmonary tuberculosis in Norwegian patients: the role of reactivation, re-infection and primary infection assessed by previous mass screening data and restriction fragment length polymorphism. Int J Tuberc Lung Dis. 2000;4:300–307. [PubMed] [Google Scholar]
- 12.Hunter P R, Gaston M A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaccard P. Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat. 1908;44:223–270. [Google Scholar]
- 14.Källenius G, Koivula T, Ghebremichael S, Hoffner S E, Norberg R, Svensson E, Dias F, Marklund B, Svenson S B. Evolution and clonal traits of Mycobacterium tuberculosis in Guinea-Bissau. J Clin Microbiol. 1999;37:3872–3878. doi: 10.1128/jcm.37.12.3872-3878.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J D A. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kremer K, van Soolingen D, Frothingham R, Haas W H, Hermans P W M, Martin C, Palittapongarnpim P, Plikaytis B B, Riley L W, Yakrus M A, Musser J M, van Embden J D A. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol. 1999;37:2607–2618. doi: 10.1128/jcm.37.8.2607-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurepina N E, Sreevatsan S, Plikaytis B B, Bifani P B, Connell N D, Donneelly R J, vanSoolingen D, Musser J M, Kreiswirth B N. Characterization of the phylogenetic distribution and chromosomal insertion sites of five IS6110 elements in Mycobacterium tuberculosis: nonrandom integration in the dnaA-dnaN region. Tubercle Lung Dis. 1998;79:31–42. doi: 10.1054/tuld.1998.0003. [DOI] [PubMed] [Google Scholar]
- 18.McHugh T D, Gillespie S H. Nonrandom association of IS6110 and Mycobacterium tuberculosis: implications for molecular epidemiological studies. J Clin Microbiol. 1998;36:1410–1413. doi: 10.1128/jcm.36.5.1410-1413.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montoro E, Valdivia J, Cardoso-Leao S. Molecular fingerprinting of Mycobacterium tuberculosis isolates obtained in Havana, Cuba, by IS6110 restriction fragment length polymorphism analysis and by the double-repetitive element method. J Clin Microbiol. 1998;36:3099–3102. doi: 10.1128/jcm.36.10.3099-3102.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nastasi A, Mammina C. Epidemiological study of tuberculosis in Palermo, Italy: IS6110 fingerprinting of Mycobacterium tuberculosis strains isolated in the years 1994–1998. Infection. 1999;27:318–322. doi: 10.1007/s150100050036. [DOI] [PubMed] [Google Scholar]
- 21.Niang M N, Goguet de la Salmonière Y O, Samb A, Hane A A, Cisse M F, Gicquel B, Perraut R. Characterization of M. tuberculosis strains from West African patients by spoligotyping. Microbes Infect. 1999;1:1189–1192. doi: 10.1016/s1286-4579(99)00243-9. [DOI] [PubMed] [Google Scholar]
- 22.Salamon H, Segal M R, Ponce de Leon A, Small P. A method accommodating analysis of errors facilitates comparison and clustering of molecular fingerprints. Emerg Infect Dis. 1998;4:159–167. doi: 10.3201/eid0402.980203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Small P M, Hopewell P C, Singh S P, Paz A, Parsonnet J, Ruston D C, Schecter G F, Daley C L, Schoolnik G K. The epidemiology of tuberculosis in San Francisco. N Engl J Med. 1994;330:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- 24.Sneath P H A, Sokal R. Numerical taxonomy: the principles and practices of classification. W. H. San Francisco, Calif: Freeman and Co.; 1973. [Google Scholar]
- 25.Soini H, Pan X, Amin A, Graviss E A, Siddiqui A, Musser J M. Characterization of Mycobacterium tuberculosis isolates from patients in Houston, Texas, by spoligotyping. J Clin Microbiol. 2000;38:669–676. doi: 10.1128/jcm.38.2.669-676.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sola C, Horgen L, Maïsetti J, Devallois A, Goh K S, Rastogi N. Spoligotyping followed by double-repetitive element PCR is a rapid alternative methodology to IS6110-fingerprinting for epidemiological studies of tuberculosis. J Clin Microbiol. 1998;36:1122–1124. doi: 10.1128/jcm.36.4.1122-1124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sola C, Devallois A, Horgen L, Maïsetti J, Filliol I, Legrand E, Rastogi N. Tuberculosis in the Caribbean: using spacer oligonucleotide typing to understand strain origin and transmission. Emerg Infect Dis. 1999;5:404–414. doi: 10.3201/eid0503.990311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sola, C., I. Filliol, C. Guttierez, I. Mokrousov, V. Vincent, and N. Rastogi. An update of a spoligotype database of Mycobacterium tuberculosis: analysis of the biogeographical distribution of shared-types and epidemiological and phylogenetical perspectives. Emerg. Infect. Dis., in press. [DOI] [PMC free article] [PubMed]
- 29.van der Zanden A G, Hoentgen A H, Heilmann F G, Weltvreden E F, Schouls L M, van Embden J D. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis complex in paraffin wax embedded tissues and in stained microscopic preparations. Mol Pathol. 1998;51:209–214. doi: 10.1136/mp.51.4.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Embden J D A, van Gorkom T, Kremer K, Jansen R, van der Zeijst B A M, Schouls L M. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J Bacteriol. 2000;182:2393–2401. doi: 10.1128/jb.182.9.2393-2401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Soolingen D, Borgdorff M W, de Haas P E W, Sebek M M G G, Veen J, Dessens M, Kremer K, van Embden J D A. Molecular epidemiology of Tuberculosis in The Netherlands: a nationwide study from 1993 through 1997. J Infect Dis. 1999;180:726–736. doi: 10.1086/314930. [DOI] [PubMed] [Google Scholar]
- 33.van Soolingen D, Hermans P W M, de Haas P E W, Sool D R, van Embden J D A. The occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warren R M, Simpson S L, Richardson M, van Der Spuy G D, Lombard P, Victor T C, van Helden P D. Mapping of IS6110 flanking regions in clinical isolates of Mycobacterium tuberculosis demonstrates genome plasticity. Mol Microbiol. 2000;37:1405–1416. doi: 10.1046/j.1365-2958.2000.02090.x. [DOI] [PubMed] [Google Scholar]
- 35.Yang Z H, Ijaz K, Bates J H, Eisenach K D, Cave M D. Spoligotyping and polymorphic GC-rich repetitive sequence fingerprinting of Mycobacterium tuberculosis strains having a few copies of IS6110. J Clin Microbiol. 2000;38:3572–3576. doi: 10.1128/jcm.38.10.3572-3576.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]