Abstract
Aspergillus fumigatus causes a range of human and animal diseases collectively known as aspergillosis. A. fumigatus possesses and expresses a range of genetic determinants of virulence, which facilitate colonisation and disease progression, including the secretion of mycotoxins. Gliotoxin (GT) is the best studied A. fumigatus mycotoxin with a wide range of known toxic effects that impair human immune cell function. GT is also highly toxic to A. fumigatus and this fungus has evolved self-protection mechanisms that include (i) the GT efflux pump GliA, (ii) the GT neutralising enzyme GliT, and (iii) the negative regulation of GT biosynthesis by the bis-thiomethyltransferase GtmA. The transcription factor (TF) RglT is the main regulator of GliT and this GT protection mechanism also occurs in the non-GT producing fungus A. nidulans. However, the A. nidulans genome does not encode GtmA and GliA. This work aimed at analysing the transcriptional response to exogenous GT in A. fumigatus and A. nidulans, two distantly related Aspergillus species, and to identify additional components required for GT protection. RNA-sequencing shows a highly different transcriptional response to exogenous GT with the RglT-dependent regulon also significantly differing between A. fumigatus and A. nidulans. However, we were able to observe homologs whose expression pattern was similar in both species (43 RglT-independent and 11 RglT-dependent). Based on this approach, we identified a novel RglT-dependent methyltranferase, MtrA, involved in GT protection. Taking into consideration the occurrence of RglT-independent modulated genes, we screened an A. fumigatus deletion library of 484 transcription factors (TFs) for sensitivity to GT and identified 15 TFs important for GT self-protection. Of these, the TF KojR, which is essential for kojic acid biosynthesis in Aspergillus oryzae, was also essential for virulence and GT biosynthesis in A. fumigatus, and for GT protection in A. fumigatus, A. nidulans, and A. oryzae. KojR regulates rglT, gliT, gliJ expression and sulfur metabolism in Aspergillus species. Together, this study identified conserved components required for GT protection in Aspergillus species.
Author summary
A. fumigatus secretes mycotoxins that are essential for its virulence and pathogenicity. Gliotoxin (GT) is a sulfur-containing mycotoxin, which is known to impair several aspects of the human immune response. GT is also toxic to different fungal species, which have evolved several GT protection strategies. To further decipher these responses, we used transcriptional profiling aiming to compare the response to GT in the GT producer A. fumigatus and the GT non-producer A. nidulans. This analysis allowed us to identify additional genes with a potential role in GT protection. We also identified 15 transcription factors (TFs) encoded in the A. fumigatus genome that are important for conferring resistance to exogenous gliotoxin. One of these TFs, KojR, which is essential for A. oryzae kojic acid production, is also important for virulence in A. fumigatus and GT protection in A. fumigatus, A. nidulans and A. oryzae. KojR regulates the expression of genes important for gliotoxin biosynthesis and protection, and sulfur metabolism. Together, this work identified conserved components required for gliotoxin protection in Aspergillus species.
Introduction
Aspergillus fumigatus is a saprophytic fungus that can cause a variety of human and animal diseases known as aspergillosis [1]. The most severe of these diseases is invasive pulmonary aspergillosis (IPA), a life-threatening infection in immunosuppressed patients [2,3]. A. fumigatus pathogenicity is a multifactorial trait that depends in part on several virulence factors, such as thermotolerance, growth in the presence of hypoxic conditions, evasion and modulation of the human immune system and metabolic flexibility [3–7]. Another important A. fumigatus virulence attribute is the production of secondary metabolites (SMs). The genes that encode SMs are generally organized in biosynthetic gene clusters (BGCs) [8] and A. fumigatus has at least 598 SM-associated genes distributed among 33 BGCs [9,10]. SMs can cause damage to the host immune system, protect the fungus from host immune cells, or can mediate the acquisition of essential nutrients [11–18]. Gliotoxin (GT) is the best studied A. fumigatus SM and has been detected in vivo in murine models of invasive aspergillosis (IA), in human cancer patients [19], and in isolates derived from patients with COVID-19 and aspergillosis secondary co-infections [20].
Numerous modes of action for GT in the mammalian host have been described: (i) GT interferes with macrophage-mediated phagocytosis through prevention of integrin activation and actin dynamics, resulting in macrophage membrane retraction and failure to phagocytose pathogen targets [21]; (ii) GT inhibits the production of pro-inflammatory cytokines secreted by macrophages and the activation of the NFkB regulatory complex [22]; (iii) GT interferes with the correct assembly of NADPH oxidase through preventing p47phox phosphorylation and cytoskeletal incorporation as well as membrane translocation of subunits p47phox, p67phox and p40phox [23]; and (iv) GT inhibits neutrophil chemoattraction by targeting the activity of leukotriene A4 (LTA4) hydrolase, an enzyme that participates in LTA biosynthesis [24].
GT is a sulfur-containing mycotoxin, a member of the epipolythiopiperazines, produced by different fungal species, including Gliocadium fimbriatum (from which it was originally isolated and named accordingly), A. fumigatus and closely related non-pathogenic species [25,26], and also by species of Trichoderma and Penicillium [27–30]. In A. fumigatus, a BGC on chromosome VI contains 13 gli genes responsible for GT biosynthesis and secretion [30]. GT biosynthesis is tightly regulated because it interferes with and depends on several cellular pathways that regulate sulfur metabolism [cysteine (Cys) and methionine (Met)], oxidative stress defenses [glutathione (GSH) and ergothioneine (EGT)], methylation [S-adenosylmethionine (SAM)], and iron metabolism (Fe–S clusters) [31–35]. Regulation of GT biosynthesis involves numerous transcription factors (TFs), protein kinases, transcriptional and developmental regulators, regulators of G-protein signalling as well as chromatin modifying enzymes [30].
Even though the regulation of GT biosynthesis is well characterized, regulation of endogenous protection from GT—which is also highly toxic to the fungus—is less well understood. A. fumigatus self-protection against GT is predicted to be based on the following mechanisms: (i) the major facilitator superfamily transporter GliA, which is part of the GT BGC, catalyses GT efflux; (ii) the reversible enzymatic activity of the oxidoreductase GliT, and (iii) the negative regulation of GT biosynthesis through the off switch mechanism of the S-adenosylmethionine-dependent gliotoxin bis-thiomethyltransferase GtmA. GliA is responsible for transferring GT out of fungal cells and deletion of gliA increases susceptibility to GT [36]. GliT produces the final toxic form of GT by catalysing disulphide bridge closure of the precursor dithiol gliotoxin (dtGT) [30,37]. However, if there is excess GT production, GliT is able to reduce GSH and produce dtGT attenuating GT toxicity [30,37]. GtmA, whose gene is not located in the GT BGC, is able to convert dtGT into bisdethiobis(methylthio)-gliotoxin (bmGT) and to attenuate GT production postbiosynthetically [38–41]. It is thought that the primary role of GtmA is a decrease in GT biosynthesis and not a back up for GliT and toxin neutralisation [30].
Until recently, the TF regulating gliT has remained elusive. It has previously been shown that gliT is not regulated by GliZ, a Zn(II)2Cys6 TF that is part of the gliotoxin BGC and is required for gliotoxin biosynthesis [37]. The TF RglT was shown to regulate not only GliZ but several other gli genes, including gliT [30,36,37,42], through directly binding to the respective promoter regions during GT-producing conditions. Interestingly, RglT and GliT were shown to have similar roles in GT protection in A. nidulans, a fungus that does not produce GT [42]. The genome of A. nidulans lacks homologs for gliA and gtmA. Therefore, the aim of this work is to elucidate additional components required for GT protection in Aspergillus species by comparing the transcriptional response of A. fumigatus and A. nidulans to exogenous GT. By using transcriptional profiling (RNA-seq), we investigate the influence of RglT in GT protection in the GT-producer A. fumigatus and in the GT-non producer A. nidulans. Although they are both members of the genus Aspergillus, the two species are distantly related and the sequence divergence of their genome sequences is on par to that of the human and fish genomes [43,44]. We identified several novel genetic determinants dependent or not on RglT and that could be involved in GT protection, including a novel methyltransferase encoded by mtrA whose deletion confers GT-sensitivity in both A. fumigatus and A. nidulans. We also screened an A. fumigatus deletion library of 484 null mutants and identified, in addition to RglT, 15 TFs that are potentially important for GT self-protection. We found that one of these TFs is a KojR ortholog, previously reported as regulator of the kojic acid production gene cluster in A. oryzae, is important as well for A. nidulans and A. oryzae GT protection and involved in A. fumigatus virulence, GT self-protection and GT and bmGT biosynthesis. KojR regulates the inorganic sulfur assimilation and transssulfuration pathways when A. fumigatus is exposed to GT.
Results
A. fumigatus RglT controls the expression of GT- and other SM-encoding genes when exposed to exogenous GT
To determine which genes are under the transcriptional control of RglT in the presence of exogenous GT, we performed RNA-sequencing (RNA-seq) of the A. fumigatus wild-type (WT) and ΔrglT strains when exposed to 5 μg/ml GT for 3 h. In these conditions, A. fumigatus is protecting itself from the effects of GT, as has previously been shown [45]. Differentially expressed genes (DEGs) were defined as those with a minimum log2 fold change of 2 [log2FC ≥ 1.0 and ≤ -1.0; p-value < 0.05; FDR (false discovery rate) of 0.05]. DEG comparisons were carried out for: (i) the WT strain, comparing the GT condition to the control (GT-free condition) to determine which genes are modulated after 3 h exposure to GT; and (ii) when comparing the rglT deletion strain to the WT strain in the presence of GT to determine which genes are under the regulatory control of RglT in these conditions.
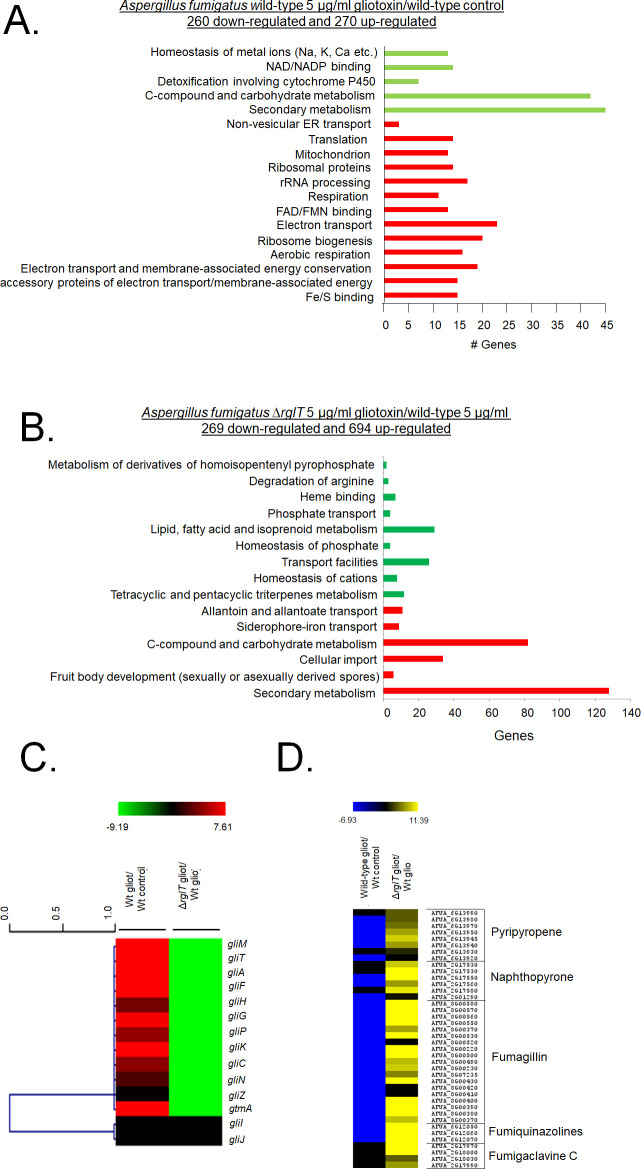
In the WT strain, 260 genes were down-regulated and 270 genes up-regulated when comparing the GT to the control condition (S1 Table). FunCat (Functional Categorisation) (https://elbe.hki-jena.de/fungifun/fungifun.php) analysis for the WT strain comparisons showed a transcriptional up-regulation of genes coding for proteins involved in non-vesicular ER transport, translation, mitochondrion, respiration and electron transport (p-value < 0.01; Fig 1A). FunCat analysis for the down-regulated genes showed enrichment for genes involved in homeostasis of metal ions, detoxification involving cytochrome P450, C-compound and carbohydrate metabolism, and secondary metabolism (p-value < 0.01; Fig 1A). When comparing the WT to the ΔrglT strain in the presence of GT, a total of 269 genes were down-regulated and 694 genes were up-regulated (S2 Table). FunCat enrichment analysis of these DEGs showed a transcriptional up-regulation of genes encoding proteins involved in secondary metabolism, C-compound and carbohydrate metabolism, cellular import, and siderophore-iron transport (p-value < 0.01; Fig 1B). FunCat analysis of down-regulated DEGs showed enrichment of transport facilities, lipid, fatty acid and isoprenoid metabolism, and homeostasis of cations (p-value < 0.01; Fig 1B). These results suggest that A. fumigatus adapts to the prolonged presence of GT through transcriptionally inducing cellular homeostasis and detoxification processes and that RglT is important for these processes.
Fig 1. Functional characterisation (FunCat) of significantly differently expressed genes (DEGs) identified by RNA-sequencing in the A. fumigatus ΔrglT strain.
(A) FunCat analysis of DEGs up-regulated in the A. fumigatus wild-type and in (B) ΔrglT strain in comparison to the wild-type (WT) strains when exposed to 5 μg/ml GT for 3 h. (C) Heat map depicting the log2 fold change (Log2FC) of differentially expressed genes (DEGs), as determined by RNA-sequencing, and encoding enzymes present in the GT BGC required for GT biosynthesis. The gene gtmA was also included in this heat map. (D) Heat map depicting the Log2FC of differentially expressed genes (DEGs), as determined by RNA-sequencing, and encoding enzymes required for secondary metabolite (SM) biosynthesis. In both (C) and (D) log2FC values are based on the wild-type strain exposed to GT in comparison to the wild-type control and the ΔrglT exposed to GT in comparison to the wild-type exposed to GT. Heat map scale and gene identities are indicated. Hierarchical clustering was performed in MeV (http://mev.tm4.org/), using Pearson correlation with complete linkage clustering.
More specifically, RglT is important for the expression of 11 out of the 13 genes present in the GT BGC (including gliT and gliA) as well as gtmA (which is not part of the GT BGC) in the presence of exogenous GT (Fig 1C). Interestingly, gliI and gliJ expression are not altered by the presence of GT in the WT and in the ΔrglT strains (Fig 1C). RNA-seq analysis also revealed transcriptional modulation of other SM-encoding genes. For example, BGC genes encoding components for the biosynthesis of pyripyropene, naphthopyrone, fumagillin, fumiquinazolines, and fumigaclavine were shown to be under the transcriptional control of RglT, with this TF repressing these genes under prolonged GT exposing conditions (Fig 1D).
These results suggest that RglT is important not only for GT biosynthesis and self-protection, but also for the regulation of the expression of genes involved in the production of other SMs.
Comparison of the A. fumigatus and A. nidulans transcriptional responses when exposed to GT
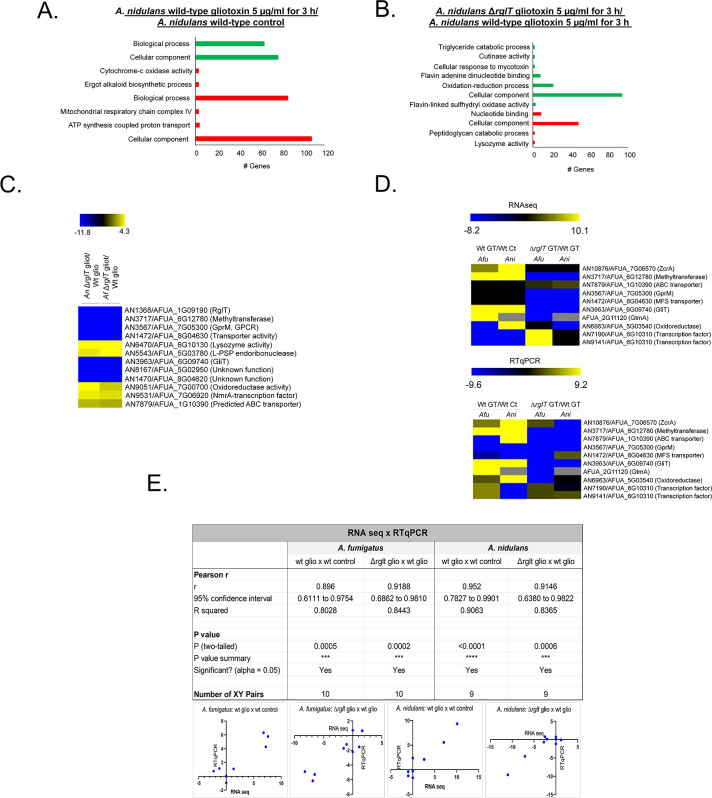
Next, we performed RNA-seq for the A. nidulans WT (the GT MIC for A. nidulans is 30 μg/ml) and ΔrglT strains in the same conditions (5 μg/ml GT for 3 h) as described above and identified DEGs. The same two comparisons were carried out as described above for A. fumigatus (WT GT vs. WT Control and ΔrglT GT vs WT GT). In the A. nidulans WT strain, 678 genes were down-regulated and 675 genes were up-regulated when comparing the GT to the control condition (S3 Table). We were not able to observe any Funcat enrichment for these DEGs and then we employed gene ontology (GO; https://elbe.hki-jena.de/fungifun/fungifun.php) enrichment analyses for the WT strain that showed transcriptional up-regulation of genes coding for proteins involved in mitochondrial function, such as ATP synthesis coupled proton transport and cytochrome-c oxidase activity, as well as ergot alkaloid biosynthetic process and a heterogeneous set of cellular components (p-value < 0.01; Fig 2A). GO analysis for the down-regulated genes showed enrichment for a heterogeneous set of genes involved in biological processes and cellular components (p-value < 0.01; Fig 2A). When comparing the WT to the ΔrglT strain in the presence of GT, 132 genes were down-regulated and 68 genes were up-regulated (S4 Table). GO enrichment analyses of the ΔrglT strain showed a transcriptional up-regulation of genes encoding proteins involved in nucleotide binding and cellular components (p-value < 0.01; Fig 2B). GO analysis of down-regulated DEGs in the ΔrglT strain showed enrichment of flavin adenine dinucleotide binding and oxidation-reduction processes (p-value < 0.01; Fig 2B). In contrast to A. fumigatus, where prolonged exposure to GT resulted in 530 identified DEG in the WT strain (about 5.3% of the A. fumigatus 9,929 protein coding sequences, CDSs), in A. nidulans 1,353 DEG (about 13.5% of the A. nidulans 9,956 CDSs) were identified in the WT strain in these conditions. Similarly, in A. fumigatus, RglT controls the expression of 963 genes (about 9.7% of the CDSs) when exposed to GT for 3 h, whereas the number of genes regulated by RglT in A. nidulans is smaller (200 genes, about 2% of the CDSs). Together, these results suggest very significant differences in the transcriptional response to GT and for the role of RglT in mediating this response between Aspergilus species.
Fig 2. Functional characterisation (FunCat) of significantly differently expressed genes (DEGs) identified by RNA-sequencing in the A. nidulans ΔrglT strain.
(A) FunCat analysis of DEGs up-regulated in the A. nidulans wild-type in the presence of GT when compared to the control condition, and in (B) the ΔrglT strain in comparison to the wild-type (WT) strain when exposed to 5 μg/ml GT for 3 h. (C) Heat map depicting the log2 fold change (Log2FC) of differentially expressed homologous genes (DEHGs), as determined by RNA-sequencing, that have their expression dependent on RglT. (D) Upper panel: heat map depicting the log2 fold change (Log2FC) of 9 DEHGs and gtmA as determined by RNA-sequencing. Lower panel: heat map depicting the log2 fold change (Log2FC) of 9 DEHGs and gtmA as determined by RT-qPCR. Log2FC values are based on the A. fumigatus (Afu) or A. nidulans (Ani) wild-type strains exposed to GT when compared to the Afu or Ani wild-type control and the Afu or Ani ΔrglT strains exposed to GT when compared to the Afu or Ani wild-type exposed to GT. Heat map scale and gene identities are indicated. Hierarchical clustering was performed in MeV (http://mev.tm4.org/), using Pearson correlation with complete linkage clustering. For the RT-qPCR experiments, the values represent the average of three independent biological repetitions (each with 2 technical repetitions). (D) Pearson correlation analysis between the expression of 10 genes as measured by RT-qPCR and RNA-sequencing. A positive correlation was seen for all analysed datasets.
To determine whether a conserved transcriptional response to GT in both A. fumigatus and A. nidulans exists, we searched for homologous genes whose expression pattern was similar (i) between the WT strains and (ii) RglT-dependent in both fungal species in the presence of GT. When A. fumigatus wild-type DEGs were compared to A. nidulans wild-type DEGs, 35 and 11 homologs were up- and down-regulated in both species, respectively (S3 Table). Furthermore, a total of 11 genes were found: six genes were dependent on RglT for expression [GliT (AN3963/AFUA_6G09740), a methyltransferase (AN3717/AFUA_6G12780), GprM (AN3567/AFUA_7G05300), a Major Facilitator Superfamily transporter (AN1472/AFUA_8G04630), AN8167/AFUA_5G02950, and AN1470/AFUA_5G8G04260 with unknown functions]; and the other five genes were dependent on RglT for repression [lysozyme activity (AN6470/AFUA_6G10130), L-PSP endoribonuclease (AN5543/AFUA_5G033780), oxidoreductase activity (AN9051/AFUA_7G00700), NmrA-transcription factor (AN9531/AFUA_7G06920), and an ATP-binding cassette transporter (AN7879/AFUA_1G10390)] (Fig 2C and S4 Table).
We observed 43 homologs that were RglT-independent (S4 Table) and among them, we identified as induced by GT a putative cysteine synthase (AN7113/AFUA_4G03930) and ZrcA, a zinc/cadmium resistance protein (AN10876/AFUA_7G06570) (S4 Table). Another RglT-independent gene is the GliT paralogue AN6963/AFUA_5G03540; interestingly, this gene is upregulated in A. nidulans and downregulated in A. fumigatus upon GT exposure (S4 Table).
RNA-seq results were confirmed by RT-qPCR for the majority of the 9 selected A. fumigatus/A. nidulans homologs and for the A. fumigatus GtmA-encoding gene (A. nidulans lacks a homolog) (Figs 2D and 2E and S1). The expression of these 10 genes showed a high level of correlation with the RNA-seq data (Pearson correlation from 0.896 to 0.952; Fig 2E).
Taken together, our results suggest that the transcriptional responses of A. fumigatus and A. nidulans to GT are very different, although we were able to observe 54 homologs whose transcriptional response was similar in both species. However, we note that we used an equal exposure to GT (5 μg/ml GT for 3 hours) for both species but A. nidulans shows greater susceptibility to GT than A. fumigatus.
A novel methyltransferase (MtrA) is important for GT self-defense in Aspergilli
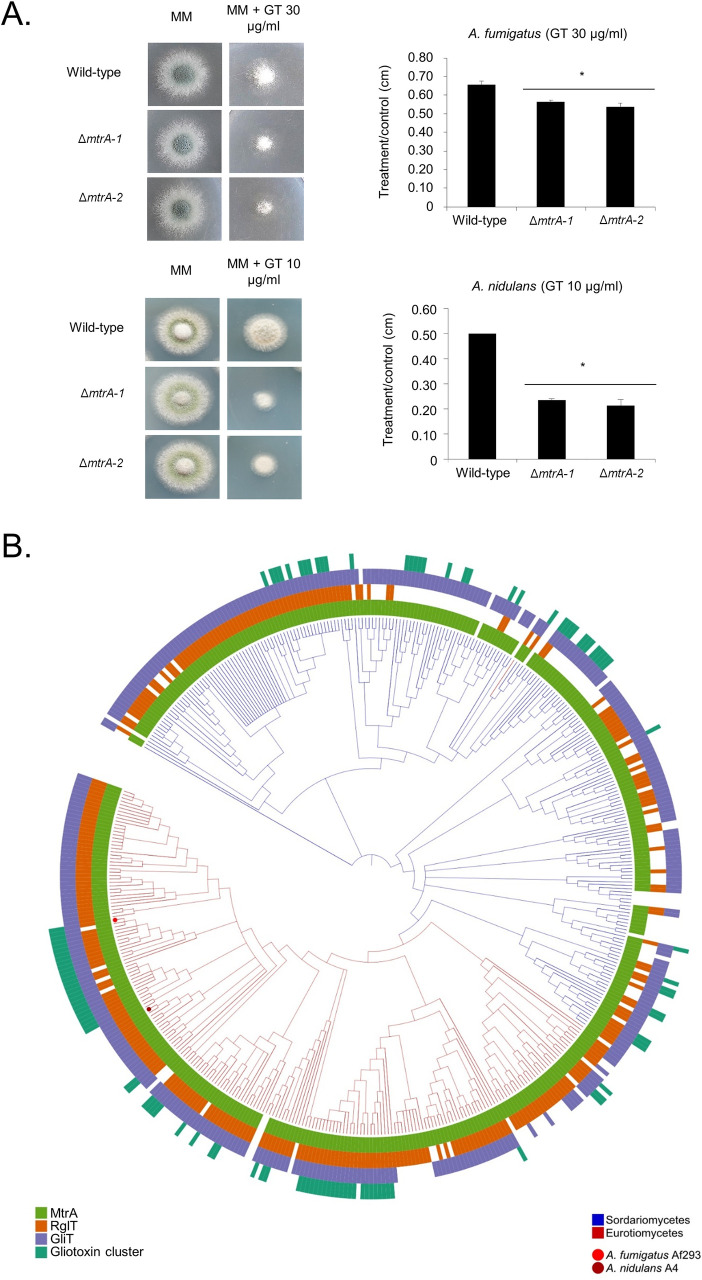
Among the genes dependent on Aspergilli RglT, we identified a methyltransferase (named MtrA, AN3717/AFUA_6G12780). (Fig 2C and S4 Table). We deleted this gene in both A. nidulans and A. fumigatus and tested the growth of two independent transformants for each deletion in the presence of GT (Figs 3A and S3). A. fumigatus ΔmtrA deletion strains showed about 20% growth reduction compared to the corresponding wild-type while A. nidulans ΔmtrA deletion strains presented about 60% growth reduction (Fig 3A). Fig 3B shows the taxonomic distribution of MtrA, RglT, GliT, and GT biosynthetic gene cluster (BGC) homologs across Eurotiomycetes and Sordariomycetes. Phylogenetically informed model testing of the distributions of RglT and MtrA suggests that the pattern of occurrence of RglT is statistically dependent on the distribution of MtrA but not vice-versa. This analysis suggests an evolutionary scenario in which the MtrA-based resistance mechanism is ancestral and the evolutionary recruitment of RglT regulation of GliT occurred subsequently.
Fig 3. The A. fumigatus and A. nidulans ΔmtrA mutants are more sensitive to GT.
(A) The A. fumigatus and A. nidulans wild-type and two independent ΔmtrA strains were grown for 48 hours at 37°C on MM in the absence or presence of 30 or 10 μg/ml of GT, respectively. The results are the average of three repetitions ± standard deviation. Statistical analysis was performed using a one-tailed, paired t-test when compared to the control condition (*, p < 0.01). (B) The phylogenetic distribution of MtrA, RglT, GliT and GT biosynthetic gene cluster homologs across 458 fungal genomes. A 4-gene phylogeny of genomes from Eurotiomycetes (shown by the red branches) and Sordariomycetes (shown by the blue branches). For every tip in the phylogeny, the presence of MtrA, RglT, GliT and gliotoxin BGC homologs is depicted using light green, orange, blue, and dark green, respectively; absences are depicted in white. The dark green bar plots depict how many of the 13 Gli genes are present in the gliotoxin BGC homolog. The tip corresponding to the A. fumigatus Af293 genome is indicated by a red dot, and the tip corresponding to the A. nidulans A4 genome is indicated by a maroon dot.
These results strongly indicate that our approach to compare the transcriptomics of A. fumigatus and A. nidulans ΔrglT mutants was efficient to identify novel genes involved in GT-protection.
Screening of A. fumigatus TF deletion strains for sensitivity to GT
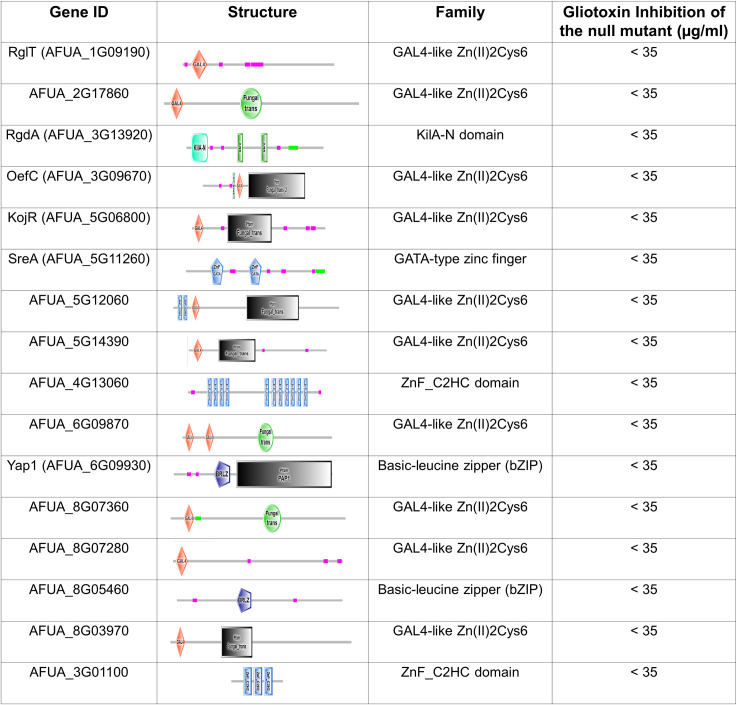
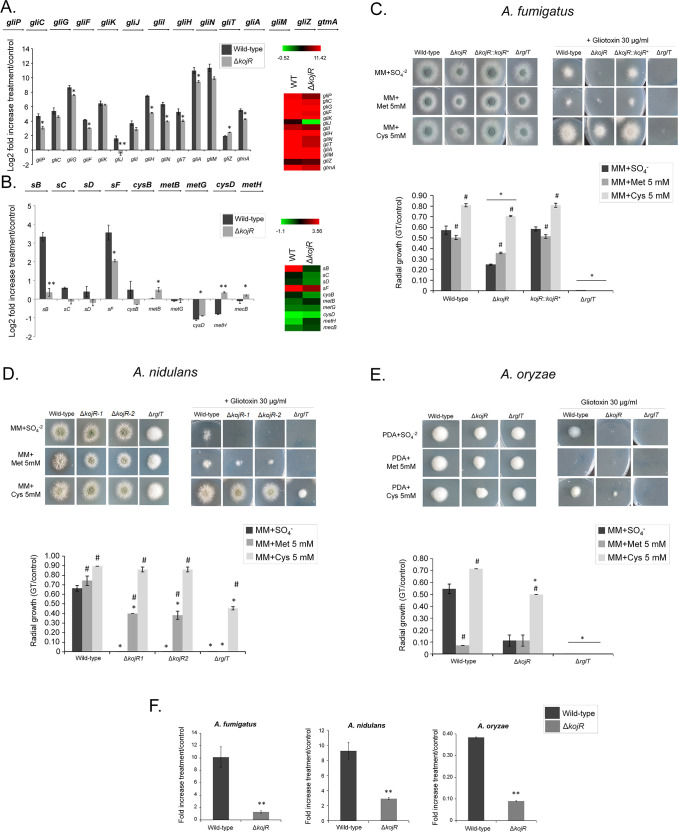
Our RNA-seq results show there are DEGs that are not dependent on RglT and suggest there additional TFs that could be important for GT protection in Aspergilli. To identify these possible TFs involved in the GT self-protection response, we took advantage of the existing A. fumigatus TF null deletion library [46] which we screened for sensitivity to 35 μg/ml of GT (A. fumigatus minimal inhibitory concentration for GT is 70 μg/ml). In addition to ΔrglT, 15 null mutants exhibited increased sensitivity to 35 μg/ml of GT (Figs 4 and 5). Only AFUA_3G09670 (oefA) is up-regulated in the ΔrglT mutant and only AFUA_4G13060 is up-regulated in the wild-type strain in our RNA-seq experiments (S1 and S2 Tables).
Fig 4. List of the transcription factor mutants identified as more sensitive to gliotoxin (the domais organization are based on http://smart.embl-heidelberg.de/).
Fig 5. Identification A. fumigatus TFs important for gliotoxin (GT) self-protection.
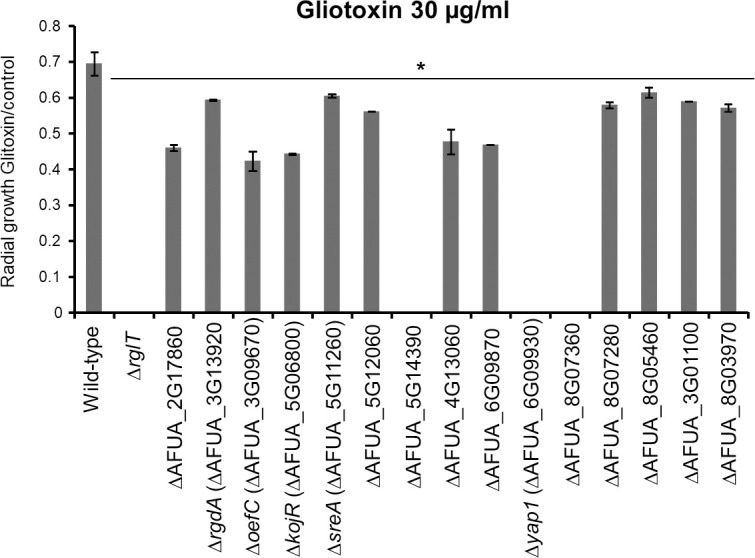
Strains were grown from 104 conidia for 2 days at 37°C on minimal medium (MM) supplemented with 30 μg/ml GT before colony radial diameter was measured. The results are expressed as the radial diameter of the treatment divided by the radial diameter of the growth in the control GT-free condition. The results are the means of three repetitions ± standard deviation. Statistical analysis was performed using a one-tailed, paired t-test when compared to the control condition (*, p < 0.05).
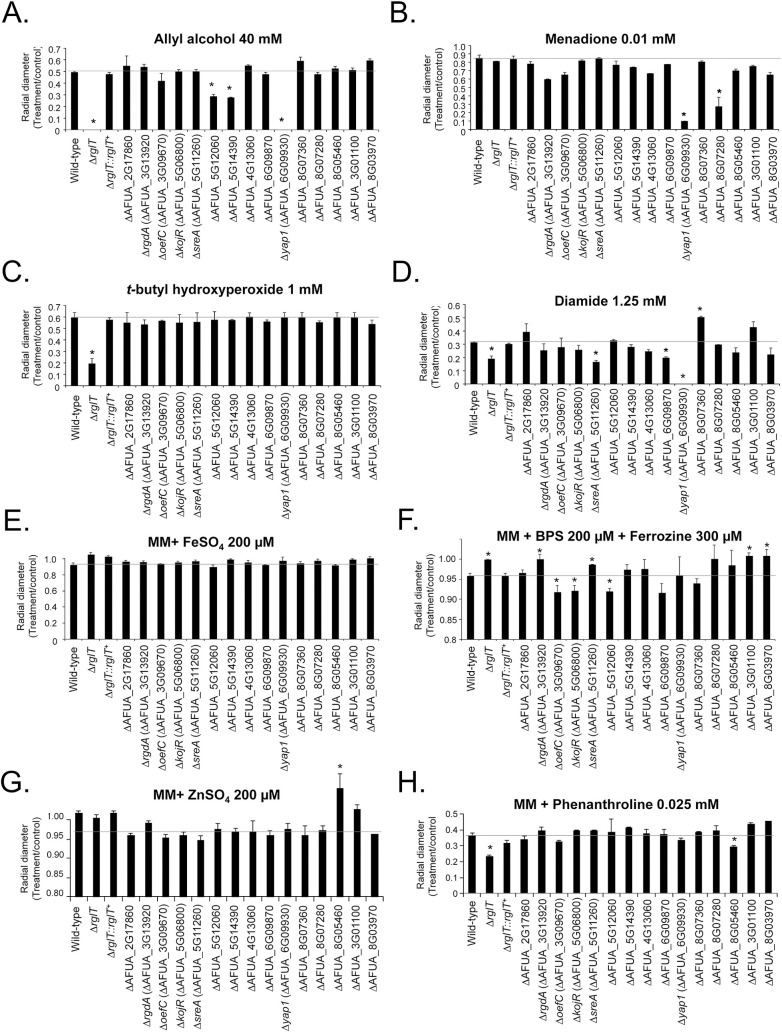
Previous studies have shown that GT biosynthesis and self-protection is intimately linked to sulfur metabolism, oxidative stress resistance, as well as iron and zinc metabolism [47–52]. To identify if any of the TFs identified in the screen are associated with these pathways, the null mutants were screened for growth in the presence of (i) methionine or cysteine as single sulfur sources (S2 Fig); (ii) oxidative stress-inducing agents such as allyl alcohol (converted to the oxidative stress-inducing compound acrolein by alcohol dehydrogenases), t-butyl hydroxyperoxide, menadione, and diamide (a GSH scavenger); and (iii) increased and depleted extracellular iron and zinc concentrations (Fig 6). None of these mutants showed altered growth compared to the WT strain in the presence of methionine or cysteine as single sulfur sources (S2 Fig). Six of the deletion strains [ΔrglT, ΔsreA (ΔAFUA_5G12060), ΔAFUA_5G140390, Δyap1 (ΔAFUA_6G09930), ΔAFUA_6G09870, and ΔAFUA_8G07360] were sensitive to at least one of the four oxidative stress-inducing agents (Fig 6A–6D). The ΔAFUA_8G07360 mutant is resistant to diamide (Fig 6D). Four strains (ΔAFUA_3G13920, ΔAFUA_5G11260, ΔAFUA_3G01100 and ΔAFUA_8G03970) had increased growth, while ΔAFUA_3G09670, ΔAFUA_5G06800, and ΔAFUA_5G12060 presented significantly decreased growth in iron starvation conditions (Fig 6F). Intriguingly, ΔAFUA_8G05460 had reduced growth upon zinc starvation but increased growth upon zinc excess (Fig 6G and 6H). The ΔrglT mutant has reduced growth in zinc starvation conditions but had increased growth in iron starvation conditions (Fig 6F and 6H).
Fig 6. Growth phenotypes of the A. fumigatus GT-sensitive transcription factor deletion strains.
Strains were grown from 104 conidia for 5 days at 37°C on minimal medium supplemented with the indicated compounds. The results are expressed as the radial diameter of the treatment divided by the radial diameter of the growth in the control, drug-free condition. The results are the means of three repetitions ± standard deviation. Statistical analysis was performed using a one-tailed, paired t-test when compared to the control condition (*, p < 0.05). (A) allyl alcohol 40 mM. (B) menadione 0.01 mM. (C) t-butyl hydroxyperoxide 1 mM. (D) diamide 1.25 mM. (E) FeSO4 200 μM. (F) BPS 200 μM+Ferrozine 300 μM. (G) ZnSO4 200 μM. (H) phenanthroline 0.025 mM.
Taken together, these results strongly suggest that some of the mutants identified as more sensitive to GT are also involved in pathways related to oxidative stress and iron and zinc metabolism.
Production of gliotoxin and BmGT in the gliotoxin sensitive mutants
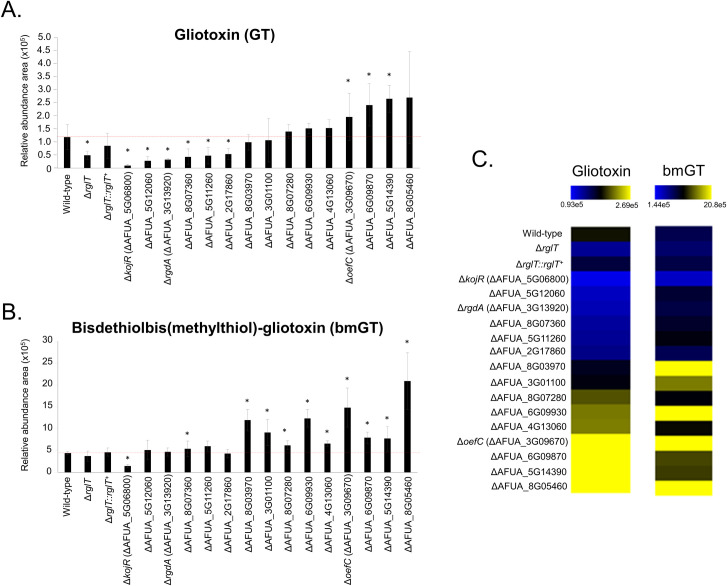
The ΔrglT and ΔgliT strains are not only sensitive to exogenous GT, but they also secrete significantly less GT [37,42]. To determine if the TF deletion strains from our screen are also impaired for GT and bmGT biosynthesis and secretion, we quantified extracellular GT and bmGT in culture supernatants of these strains when grown in GT-inducing conditions. As previously demonstrated [42], high performance liquid chromatography and mass spectrometry experiments showed that ΔrglT has reduced GT and produces a similar amount of bmGT when compared to the WT and rglT complemented strains (S5 Table, Fig 7A–7C). The ΔkojR (ΔAFUA_5G06800), ΔAFUA_5G12060, ΔrgdA (ΔAFUA_3G13920), ΔAFUA_8G07360, ΔsreA (ΔAFUA_5G11260), and ΔAFUA_2G17860 strains secrete significantly less GT when compared to the WT strain (Fig 7A and 7C). In contrast, the ΔoefC (ΔAFUA_3G09670), ΔAFUA_6G09870, and ΔAFUA_5G14390 strains secrete significantly more GT than the WT strain (Fig 7A and 7C). Production of bmGT is decreased in the ΔkojR (ΔAFUA_5G06800) but increased in most of the strains (10 out of 16), with the exception of ΔrglT, ΔAFUA_5G12060, ΔrgdA (ΔAFUA_3G13920), ΔsreA (ΔAFUA_5G11260), and ΔAFUA_2G17860 which produce similar concentrations of bmGT than the WT strain (Fig 7B and 7C). Interestingly, the ΔkojR (ΔAFUA_5G06800) strain is the only TF deletion strain with reduced GT and bmGT levels (Fig 7A–7C), whilst the ΔAFUA_8G03970, ΔAFUA_6G09930, and ΔoefC (ΔAFUA_3G09670) strains have increased levels of GT and bmGT in the supernatants (Fig 7A–7C).
Fig 7. Production of GT and bmGT in the A. fumigatus GT-sensitive TF deletion strains.
(A) Relative abundance of GT and (B) bisdethiobis(methylthio)-gliotoxin (bmGT) as measured by high performance liquid chromatography coupled to mass spectrometry. Results are the average of three repetitions ± standard deviation. Statistical analysis was performed using a one-tailed, paired t-test when comparing quantities to the relative abundance of the WT strain (*, p < 0.005). (C) Heat map of the results from panels (A) and (B). Heat map scale and gene identities are indicated. Hierarchical clustering was performed in MeV (http://mev.tm4.org/), using Pearson correlation with complete linkage clustering.
Our results indicate that several TFs are involved in the induction or repression of GT and bmGT biosynthesis. In contrast, only for the ΔkojR strain are GT and bmGT extracellular levels significantly decreased, and we have chosen this TF for further characterisation.
RglT and KojR are important for GT protection in Aspergilli
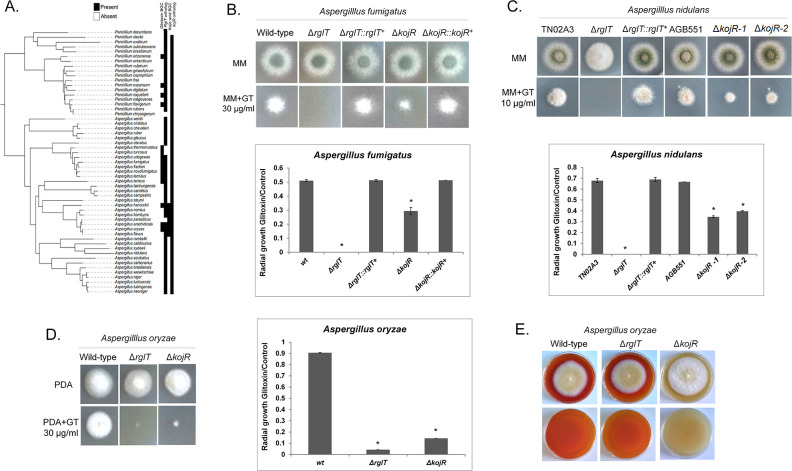
We have previously shown that RglT and GliT in A. fumigatus and A. nidulans have a similar role in protecting each fungus from the deleterious effects of GT [42]. Homologs of RglT and GliT are present in a number of Eurotiomycetes and Sordariomycetes suggesting that this mechanism of GT protection is widespread, including in many species that are not GT producers [42]. Similarly to MtrA, phylogenetically informed model testing led to an evolutionary scenario in which the GliT-based resistance mechanism is ancestral and RglT-mediated regulation of GliT occurred subsequently [42]. To determine whether a phylogenetic relationship exists between RglT and KojR, we screened for the presence of GT and kojic acid BGCs as well as RglT and KojR homologs across Aspergillus and Penicillium species and constructed a phylogenetic tree (Fig 8A). Results suggest that their origins predate the origin of both genera. In contrast, homologs of the kojic acid BGC are present in only A. oryzae and close relatives (Fig 8A). In contrast to RglT and GliT, examination of the phylogenetic distribution patterns of RglT and KojR revealed that they are not significantly correlated (scenario A). The same was true for KojR and the kojic acid BGC, suggesting that KojR was recruited specifically for kojic acid biosynthesis in A. oryzae and close relatives. In contrast, the distribution of the GT biosynthetic gene cluster is correlated with the presence of RglT (scenario B) [42], but not vice-versa; however, scenario A (lack of correlation between their distributions) also fit the observed data well (Table 1).
Fig 8. Aspergilli RglT and KojR homologs are important for gliotoxin production and self-protection.
(A) Distribution of homologs of the gliotoxin and kojic acid biosynthetic gene clusters and of the RglT and KojR homologs amongst Aspergillus and Penicillium species. Homologs of the gliotoxin biosynthetic gene cluster are present across many (but not all) Aspergillus and Penicillium species, suggesting that the cluster was present in their common ancestor. Homologs of the kojic acid biosynthetic gene cluster are present only in Aspergillus oryzae and close relatives, suggesting that the cluster originated much more recently. Orthologs for RglT and KojR are broadly conserved in both genera, suggesting they were present in the Aspergillus–Penicillium common ancestor. Presence or absence of biosynthetic gene cluster homologs and of gene orthologs are depicted in black or white, respectively. (B-D) Pictures and graphs of the A. fumigatus (B), A. nidulans (C) and A. oryzae (D) wild-type, ΔrglT, and ΔkojR strains when grown for 48 h from 104 conidia at 37°C on minimal medium (MM, A. fumigatus, A. nidulans) or PDA (A. oryzae) and MM/PDA+10 or 30 μg of gliotoxin. Graphs correspond to the radial diameter of the colony growth that is depicted in the pictures. Standard deviations represent the average of three biological replicates with *p < 0.05 in a one-tailed, paired t-test. (E) Detection of kojic acid (KA) production in A. oryzae in medium containing 1 mM ferric ion (FeCl3). A red colour indicates the presence of KA chelated with ferric ions. The first and second rows indicate the top and bottom of the plates, respectively.
Table 1. Akaike Information Criterium (AIC) of model fitting for phylogenetic correlation tests.
| Independent | Dependent X | Dependent Y | Interdependent | |
|---|---|---|---|---|
|
X: kojR;
Y: rglT |
0.767 | 0.104 | 0.113 | 0.016 |
|
X: kojR;
Y: kojic acid BGC |
0.704 | 0.101 | 0.170 | 0.025 |
|
X: rglT;
Y: gliotoxin BGC |
0.336 | 0.471 | 0.068 | 0.125 |
BGC = biosynthetic gene cluster
To determine whether KojR is also required for protection from GT in other fungi, the corresponding homologous genes were deleted in A. nidulans (chosen because it does not produce either GT or kojic acid) and A. oryzae (produces kojic acid). Similarly to A. fumigatus, deletion of kojR significantly increased sensitivity to exogenously added GT in both A. nidulans and A. oryzae (Fig 8B–8D). In contrast to deletion of rglT, which made all three species fully sensitive to GT, deletion of kojR resulted in only partial sensitivity to GT (Fig 8B–8D). We cannot explain the increased sensitivity to GT in A. oryzae ΔkojR, but these results suggest that RglT functions downstream of KojR and/or that RglT is regulated by KojR in these conditions. To address this, the A. oryzae WT, the rglT and the kojR deletion strains were first screened for kojic acid production, which can be seen by the intensity of the red color (formed by chelating ferric ions) on plates. As expected, deletion of kojR resulted in the absence of red colour and hence kojic acid production, whereas deletion of rglT did not affect kojic acid production in A. oryzae (Fig 8E). These results suggest that RglT does not regulate kojic acid production in A. oryzae.
GliT and GtmA are partially dependent on Aspergilli KojR
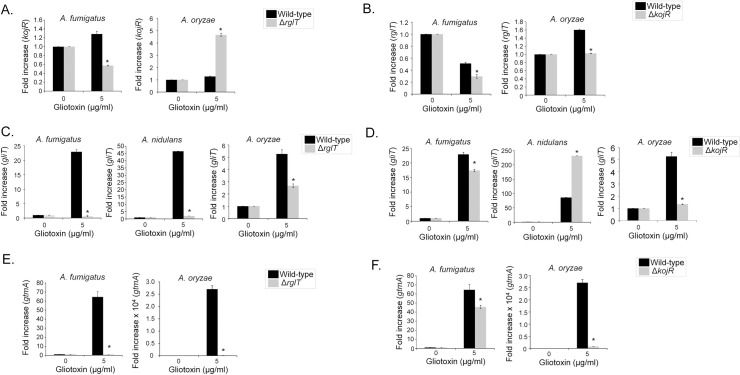
To determine whether KojR is under the transcriptional control of RglT in the presence of GT, transcriptional levels of kojR in the A. fumigatus and A. oryzae rglT deletion strains were measured by RT-qPCR (Fig 9A). In A. fumigatus and A. oryzae, deletion of rglT resulted in significantly decreased and increased kojR transcript levels, respectively, suggesting that RglT regulates kojR in these fungi (Fig 9A). To determine whether rglT is under the transcriptional control of KojR in the presence of GT, rglT transcript levels were quantified by RT-qPCR in the WT and kojR deletion strains (Fig 9B). In the A. fumigatus and A. oryzae ΔkojR strains, rglT transcript levels were significantly reduced, suggesting that KojR regulates rglT expression in the presence of exogenous GT (Fig 9B). We were unable to measure the expression levels of A. nidulans kojR and rglT in the absence or presence of GT because the PCR amplification profiles were similar to the negative control (water), suggesting kojR and rglT have very low levels of expression in this fungus.
Fig 9. GliT and GtmA are dependent on Aspergilli RglT and KojR.
A. fumigatus, A. nidulans, and A. oryzae strains were grown for 21 h at 37°C before 5 μg/ml of GT was added to cultures for 3 hours. RNA was extracted from all fungal strains, reverse transcribed to cDNA and RT-qPCRs were peformed. The expression of gtmA, kojR and rglT in the A. nidulans WT, ΔrglT and ΔkojR strains could not be measured due to either the absence of the gene (gtmA) or the absence of quantifyable mRNA. Results are the average of three biological replicates ± standard deviation. Statistical analysis was performed using a one-tailed, paired t-test when compared to the control condition (*, p < 0.05). (A-D) A. fumigatus, A. nidulans, and A. oryzae wild-type, ΔrglT, and ΔkojR strains were grown for 21 h at 37°C and mycelia was exposed to 5 μg/ml of GT for 3 hours. RT-qPCR analysis for kojR (A), rglT (B), gliT (C and E), and rglT (D and F) genes.
Finally, we further assessed the role of KojR in GT protection in A. fumigatus, A. nidulans, and A. oryzae. We were unable to detect any GT and bmGT production in A. oryzae wild-type, RglT and KojR (S4 Fig). We previously demonstrated that the expression of gliT, encoding an oxidoreductase with a GT neutralising function, is dependent on RglT in both A. fumigatus and A. nidulans when GT was added exogenously to the medium [42] (Figs 2C and 9C). In agreement with these data, the expression of A. oryzae gliT is decreased in the ΔrglT mutant, suggesting that gliT expression is dependent on RglT in all three Aspergillus species (Fig 9C). Furthermore, gliT expression was also dependent on KojR in these fungal species with KojR inducing gliT expression in A. fumigatus (about 20% reduction in the ΔkojR mutant) and A. oryzae (about a three-fold reduction in the ΔkojR mutant) and repressing gliT in A. nidulans (about 2.5-fold increase in the ΔkojR mutant; Fig 9D). Subsequently, gtmA transcript levels were also quantified as gtmA is also important for GT self-protection [38]. The expression of gtmA is also significantly decreased in A. fumigatus and A. oryzae ΔrglT mutants (Fig 9E) and about 20% and 25-fold, respectively, in the A. fumigatus and A. oryzae ΔkojR mutants (Fig 9F). No gtmA homolog is present in A. nidulans.
Together these results suggest: (i) a regulatory interplay exists between RglT and KojR in GT protection that differs between Aspergillus spp; (ii) RglT-dependent GliT protection from exogenous GT is a conserved mechanism in GT-producing and non-producing Aspergillus spp; (iii) the existence of further mechanisms of GT self-protection that significantly differ between GT-producing and non-producing Aspergillus spp; and (iv) A. fumigatus KojR plays a role in the gliT and gtmA regulation in the presence of GT.
A. fumigatus KojR is important for gliJ expression, inorganic sulfur assimilation, and virulence
We decided to investigate the influence of KojR on the expression of other genes that are important for GT production and protection. Fig 10 summarizes the contributions of some of these pathways and the different genes involved in GT biosynthesis and self-defense. We compared the mRNA accumulation of selected genes described in Fig 10 when the wild-type and the ΔkojR mutant were exposed to 5 μg/ml GT for 3 hours (Fig 11A). Thirteen gli genes in the GT BGC, and gtmA, were up-regulated when A. fumigatus wild-type is exposed to GT (Fig 11A). Interestingly, different from the RNA-seq data (Fig 1C), we were able to see increased gliI and gliJ mRNA levels when the wild-type is exposed to GT by using RT-qPCR (Fig 11A). These results emphasize that the gli genes are up-regulated when A. fumigatus is exposed to GT. The gliP, gliG, gliF, gliH, gliN, gliA, gliT, and gtmA (these two last ones previously shown in the Fig 9) have about 20 to 30% less mRNA accumulation in the ΔkojR mutant than in the wild-type while ΔgliJ has a two-fold increase in the wild-type and it is significantly down-regulated in the ΔkojR mutant (Fig 11A). The TF gliZ is about 10% more expressed in the ΔkojR mutant than in the wild-type while gtmA is about 20% less expressed in the ΔkojR mutant (Fig 11A).
Fig 10. Schematic representation of the pathways that are important for GT production and self-defense.
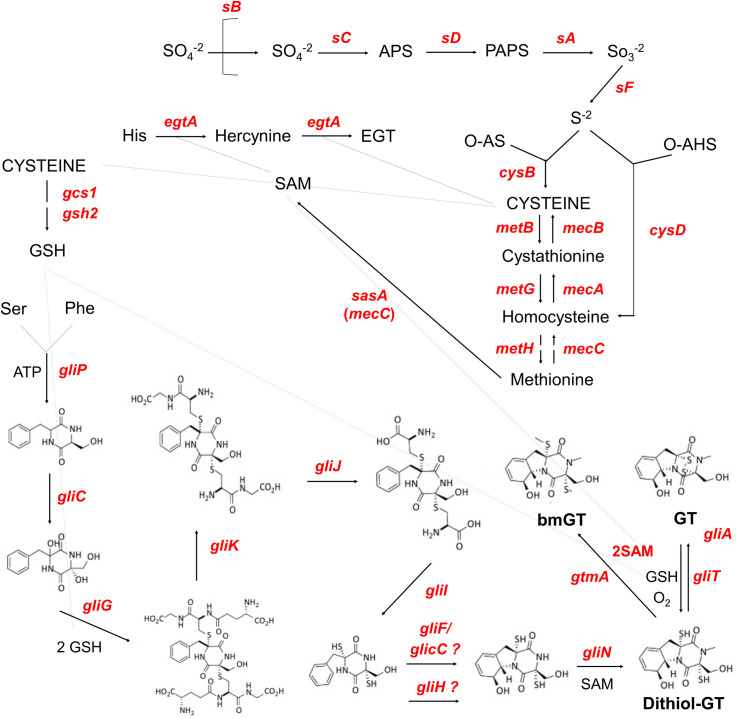
– The pathways of sulfate assimilation, transsulfuration, and GT production are shown in the scheme. The depicted genes are: sB, sulfate transporter; sC, ATP sulfurylase; sD, adenosine 5’-phosphosulfate (APS) kinase; sA, 3’-phosphoadenosine-5’-phosphosulfate (PAPS) reductase; sF, β-subunit of the sulfite reductase; AFUA_6G08920, α-subunit of the sulfite reductase; cysB, cysteine synthase; metB, cystathione-ϒ-synthase; metG, cysthationine-β-lyase; metH, methionine synthase; mecC, 5-adenosylmethionine synthetase; mecA, cystathionine-β-synthase; mecB, cystathionine- ϒ-lyase; cysD, homocysteine synthase; gliP, non-ribosomal peptide synthetase; gliC, cytochrome P450 monooxygenase; gliG, glutathione-S-transferase; gliK, unknown protein; gliJ, membrane dipeptidase; gliI, 1-aminocyclopropane-1-carboxylic acid synthase; gliF, cytochrome P450 monooxygenase; gliC, cytochrome P450 monooxygenase; gliH, acetyl transferase; gliN, methyltransferase; gliT, gliotoxin sulfhydryl oxidase; gliA, major facilitator type glioxin transporter; gcs1, glutamate cysteine-ligase; gsh2, glutathione synthase; egtA, ergothioneine synthase; and gtmA, bis-thiomethyltransferase. Abbreviations: O-AS, O-acetylserine; O-HS, O-acetylhomoserine; GSH, glutathione; SAM, S-adenosyl methionine; EGT, ergothioneine; bmGT, bisdethiobis(methylthio)-gliotoxin; and GT, gliotoxin.
Fig 11. A. fumigatus gliJ and sulfur assimilation genes are dowregulated in the ΔkojR mutant.
– A. fumigatus was grown for 21 h at 37°C before 5 μg/ml of GT was added to cultures for 3 hours. RNA was extracted from the wild-type and ΔkojR strains, reverse transcribed to cDNA and RT-qPCRs were peformed. Results are expressed as log2 fold increase and are the average of three biological replicates ± standard deviation. Statistical analysis was performed using a one-tailed, paired t-test when compared to the control condition (**, p < 0.01 and *, p < 0.005). (A) Genes in the A. fumigatus GT pathway and gtmA. (B) Selected genes in the A. fumigatus sulfur assimilation and transsulfuration pathways. Heat map scale and gene identities are indicated. (C) A. fumigatus, (D) A. nidulans, and (E) A. oryzae wild-type, ΔkojR, ΔkojR::kojR+, and ΔrglT mutants were grown for 48 hours at 37°C on MM+SO4-2, MM+Met 5 mM, and MM+Cys 5 mM in the presence or absence of 10 or 30 μg/ml of GT. The results are the average of three repetitions ± standard deviation. Statistical analysis was performed using a one-tailed, paired t-test when compared to the control condition (*, p < 0.005 when comparing the mutant strains versus the wild-type and and #, p < 0.005 when comparing the growth on MM+Met or MM+Cys versus the growth on MM+SO4-). (F) A. fumigatus, A. nidulans, and A. oryzae were grown for 20 h at 37°C before 5 μg/ml of GT was added to cultures for 3 hours. RNA was extracted from the wild-type and ΔkojR strains, reverse transcribed to cDNA and RT-qPCRs were peformed. Results are the average of three biological replicates ± standard deviation. Statistical analysis was performed using a one-tailed, paired t-test when compared to the control condition (**, p < 0.01 and *, p < 0.005).
Inorganic sulfur assimilation starts by the transport of sulfate by the sulfate transporter sB; through several steps of metabolic transformation, sulfate is subsequently converted into sulfide and then to methionine through the subsequent action of sC, sD, sF, CysB, MetB, CysD, and MetH (Figs 10 and 11B). In the A. fumigatus wild-type strain, there is an increased mRNA accumulation of all the genes of the inorganic sulfur assimilation (sB, sC, sD, sF, and cysB) including its transcriptional regulator metR and a decreased mRNA accumulation of selected genes in the transsulfuration pathway (metB, metG, cysD, metH, and mecB) upon GT exposure (Figs 10 and 11B). In contrast, the sB mRNA accumulation is significantly reduced about 10-fold in the ΔkojR mutant when compared to the wild-type strain (Fig 11B). There is a lower mRNA accumulation of sB, sC, sD, sF, cysB, and cysD when compared to the wild-type strain (Fig 11B). In contrast, metB, metH, and mecB have higher mRNA accumulation than the wild-type and there are no differences between both strains for metG (Fig 11B).
The increased expression of transsulfuration genes (metB, metH, and mecB) when the ΔkojR mutant is exposed to GT suggests that the ΔkojR mutant is trying to synthesize other sulfur metabolic intermediates through the transsulfuration pathway. We reasoned that if ΔkojR mutant has problems in inorganic sulfur assimilation, growth in the presence of organic sulfur sources, such as methionine or cysteine, could suppress the GT-sensitivity observed when ΔkojR is grown on MM+SO4 as a single inorganic sulfur source (Figs 8B and 11C). The GT-sensitivity of the ΔkojR mutant is partially suppressed in MM+cysteine but not in MM+methionine (Fig 11C), strongly emphasizing not only the defects of assimilation of inorganic sulfur by ΔkojR in the presence of GT but also possible defects in methionine assimilation. Interestingly, the A. nidulans ΔkojR GT susceptibility is also suppressed by cysteine and only partially by methionine as a single sulfur source (Fig 11D) while cysteine (but not methionine) partially suppressed A. oryzae GT ΔkojR susceptibility (Fig 11E), strongly suggesting that A. nidulans and A. oryzae KojR homologs are also important for sulfur assimilation during GT protection. Additional evidence for that comes from the KojR-dependency on the expression A. nidulans and A. oryzae of sB sulfate transporter homologs upon GT exposure (Fig 11F). We also observed that GT-sensitivity of both A. fumigatus and A. oryzae ΔrglT mutants could not be suppressed by either methionine or cysteine as a single sulfur souce while A. nidulans ΔrglT GT susceptibility was partially suppressed by cysteine (Fig 11A–11E).
Taken together, these data indicate that although there is a global reduction in the expression of several genes in the GT BGC, including gliT (and also gtmA, not present in the GT BGC), A. fumigatus KojR is essential for the gliJ expression. Furthermore, A. fumigatus KojR is also important for the transcriptional modulation of several genes involved in inorganic sulfur assimilation and transsulfuration in the presence of GT. Cysteine suppression of GT susceptibility in A. nidulans and A. oryzae and trancriptional control of the sA sulfate permease in the presence of GT also indicated that KojR is involved in the regulation of the sulfur pathways in these species upon GT stress.
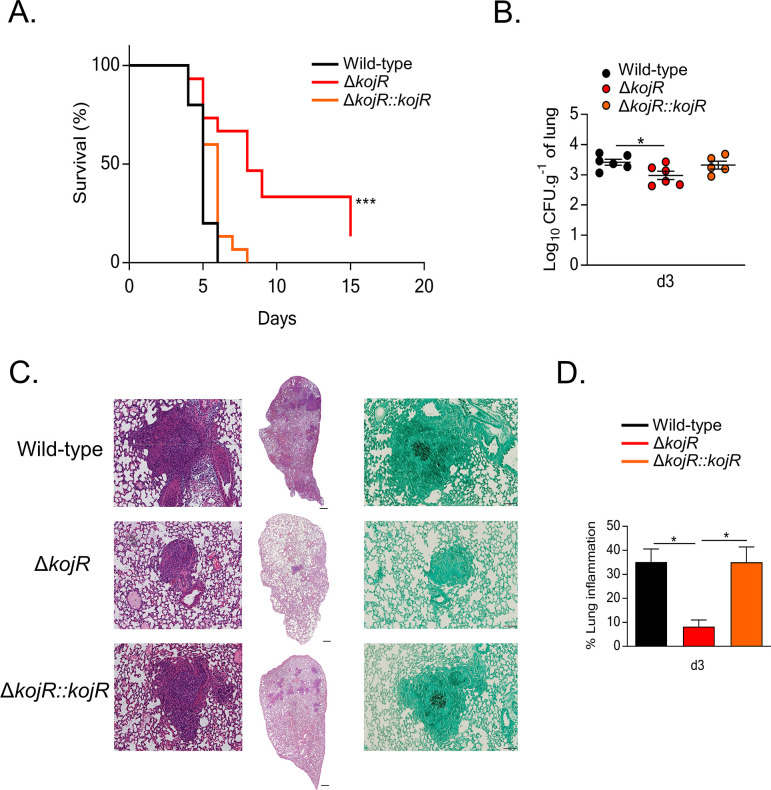
We also investigated if KojR could impact A. fumigatus virulence in a chemotherapeutic murine model of IPA. All mice infected with the WT and ΔkojR::kojR strains died between day 5 and day 7 post-infection (p.i.), whereas 10% of mice infected with the ΔkojR strain survived for the duration of the experiment (Figs 12A and S5). Fungal burden in the lungs was also significantly reduced for the ΔkojR strain after 3 days p.i. when compared to the WT and ΔkojR::kojR strains (Fig 12B). In addition, histopathology of the lung tissue after 3 days p.i. with the ΔkojR strain demonstrated a significantly reduced inflammation score (Fig 12C) and immune cell recruitment to the tissue (Fig 12D) relative to the WT and ΔkojR::kojR strains. These results strongly indicate that KojR is important for A. fumigatus inflammation and virulence.
Fig 12. KojR is required for virulence in a mouse model of invasive aspergillosis.
- (A) Survival curve (n = 10 mice/strain) infected with the indicated A. fumigatus strains. Phosphate buffered saline (PBS) was administered in a negative control group (n = 5). The indicated P values are based on the log-rank, Mantel-Cox, and Gehan-Breslow-Wilcoxon tests comparing the KojR deletion strain to the WT and complemented strains. (B) Fungal burden in murine lungs after 3 days post-infection (p.i.) with the different strains. Murine lungs were excised, ruptured, and resuspended before dilutions were prepared and incubated on plates containing complete medium. Fungal growth was assessed by counting the colony-forming units (CFU) on the plates for each dilution. (C) Histopathology of mice infected with the different strains. Lungs were excised at 3 days post-infection (p.i.) before lung sections were prepared and stained with hematoxylin and eosin (H&E) and/or with Gomori’s methenamine silver (GMS). (D) Inflammation (expressed as %) in murine lungs after 3 days post-infection (p.i.) with the different strains. Murine lungs were excised, and slides of lungs sections were prepared. To quantify lung inflammation of infected animals, inflamed areas on slide images were analyzed using the thresholding tool in ImageJ software. Values are averages ± standard deviations (error bars) of three biological replicates (lungs from different mice). Values that are significantly different in a two-way multiple comparison ANOVA test are indicated as follows: *p-value < 0.05, ***p-value < 0.001.
Discussion
The transcriptional regulation of GT biosynthesis requires many components including TFs, protein kinases, G-protein signalling and chromatin modifying enzymes, which together integrate different signaling pathways [30]. In addition, GT biosynthesis requires GSH, of which the sulfur-containing amino acids methionine and cysteine are biosynthetic precursors; and GT biosynthesis is linked to cellular oxidative stress in mammalian and fungal cells via a yet to be described mechanism [31–35]. In addition, GT self-protection is essential during GT biosynthesis. The three main mechanisms of GT self-protection in A. fumigatus are the GT-neutralising oxidoreductase GliT, the GT efflux pump GliA, and the negative regulation of GT biosynthesis through GtmA [30]. RglT was recently identified as the main TF controlling the expression of gliT and of gtmA in A. fumigatus. The RglT homolog is also important for gliT expression and GT protection in the non-GT producer A. nidulans [42]. Despite this similarity, A. nidulans does not encode homologs of GtmA and GliA, suggesting that (i) these GT protection mechanisms are not required or that (ii) additional, unknown GT protection mechanisms exist in this fungus. It is perhaps not surprising that GtmA and GliA are not present in A. nidulans, because as a non-GT producing fungus, GtmA-mediated attenuation of GT biosynthesis and GT efflux are not required. We currently do not know how the presence of GT is signalled and/or whether this mycotoxin can be taken up by A. nidulans. In this work, the transcriptional response of A. fumigatus and A. nidulans upon prolonged exposure to GT was analysed and found to be significantly different between both fungal species. In agreement, the RglT-dependent regulome in both fungal species is also very different. This may be due to the evolutionary distance between both fungal species which share around 66–67% of amino acid identity, a protein identity comparable to the phylogenetic relationship between mammals and fish [53]. Despite this, our dataset also identified a conserved transcriptional response. These genes encode proteins important for gliotoxin modification, such as the observed RglT-dependent putative methyltransferase MtrA (AN3717/AFUA_6G12780) and one putative oxidoreductase (AN9051/AFUA_7G00700), attenuating directly or indirectly its activity. We have investigated one of these genes, mtrA, and demonstrated that it is important for GT protection in A. fumigatus and A. nidulans. The A. nidulans ΔmtrA is more sensitive to GT than the A. fumigatus ΔmtrA most likely because there are additional mechanisms of GT protection in A. fumigatus, such as GtmA. It remains to be investigated if MtrA is directly methylating GT or other proteins important for GT protection.
To further identify conserved components important for GT self-protection, we screened the A. fumigatus TF deletion library to identify TFs important for this process. We identified 15 TFs (4 have been characterized and 11 have not been characterized), whose deletion caused a significant decrease in growth in the presence of exogenous GT. The 11 uncharacterized TFs can broadly be classed into 4 categories based on their respective GT and bmGT biosynthesis profiles: (i) AFUA_2G17860 and AFUA_5G12060 (whose deletion mutants are GT non-producers and bmGT producers), (ii) AFUA_3G01100, AFUA_5G14390, AFUA_4G13060, AFUA_6G09870, AFUA_8G07280 and AFUA_8G05460 (whose deletion mutants are GT producers and bmGT overproducers), (iii) AFUA_8G03970 and oefC (AFUA_3G09670, whose deletion mutants are GT and bmGT overproducers) and (iv) AFUA_8G07360 (whose deletion mutant is a GT non-producer and bmGT overproducer). They present interesting targets for future mycotoxin-related studies.
In addition, our work identified an additional 4 TFs, which have previously been characterized, as important for GT biosynthesis. Yap1 (AFUA_6G09930, whose deletion mutant is a GT overproducer and bmGT overproducer) is an important regulator of resistance to oxidative stress [54]. SreA (AFUA_5G11260, whose deletion mutant is a GT non-producer and bmGT producer) regulates iron uptake, through repressing iron-regulated genes and siderophore biosynthesis genes in high-iron conditions [55]. The identification of these TFs emphasizes the close connection between GT production and oxidative stress and iron metabolism [30,34,48,49]. RgdA (AFUA_3G13920, whose deletion mutant is a GT non-producer and bmGT producer) has previously been reported as important for hyphal growth, asexual sporulation and virulence [56]. In contrast to our results, these authors observed increased GT production in the ΔrgdA mutant, which may be due to the fact they have used Af293 as a background strain for the rgdA deletion mutant whereas this work used CEA17 as a background strain [56]. Differences in cellular responses between these two A. fumigatus background strains have been reported before and include the role of the calcium-responsive TF CrzA during the caspofungin paradoxical effect [57]. KojR (AFUA_5G06800, whose deletion mutant is a GT non-producer and bmGT non-producer) is the A. oryzae homolog involved in the production of kojic acid [58]. It is perhaps not surprising that our work identified Yap1, SreA and RgdA as involved in GT biosynthesis and self-protection, as oxidative stress, iron metabolism, and fungal growth have all been shown to be important for GT and/or SM biosynthesis [30,34,48,49]. In contrast, the deletion of kojR was the only mutant strain where reduced amounts of GT and bmGT was observed, suggesting that this TF is important for GT metabolism. The role of this TF in GT protection was therefore further characterised in GT-producing A. fumigatus and non-producing (A. nidulans and A. oryzae) Aspergillus species. These results expand significantly the number of TFs involved, and suggest additional complex mechanisms in regulating GT protection and production.
As a preliminary step to understand additional KojR gene targets, we performed extensive RTqPCR experiments with all the gli BGC genes, gtmA, and selected genes involved in the inorganic sulfur assimilation and transsulfurattion pathways. Our results showed a global reduction of 20 to 30% of the expression of the gli genes when the ΔkojR was exposed to GT, except for gliJ, encoding a membrane dipeptidase, that is significantly down-regulated in the mutant. These results strongly indicate that gliJ is a direct or indirect target for A. fumigatus KojR. The blockage of the GT biosynthesis pathway caused by the lack of gliJ expression helps to understand the lack of GT production in this mutant. As previously shown [45], we have also observed that exposure of A. fumigatus to GT increases the expression of genes involved in the inorganic sulfur assimilation and transsulfuration pathways. KojR is also negatively affecting the increased mRNA accumulation of several genes in these pathways upon GT exposure. This could impact the S-adenosyl methione (SAM):S-adenosyl homocysteine (SAH) balance, essential for the methylation of GT by GtmA, helping to explain why there is a decrease in the bmGT production in the A. fumigatus ΔkojR mutant. Interestingly, cysteine supplementation (instead of inorganic sulfur) can suppress the ΔkojR GT-sensitivity, emphasizing the role played by KojR in sulfur assimilation. The main TF for sulfur inorganic assimilation, MetR, is transcriptionally induced in the presence of GT [33] and ΔmetR showed significantly increased sensitivity to GT [59]. It remains to be investigated a possible interaction between KojR and MetR since we observed decreased mRNA accumulation of metR upon GT in the ΔkojR mutant.
We have also shown that KojR is essential for virulence in a chemotherapeutic murine model of IPA. Both ΔkojR and ΔgliP (gliP encodes the nonribosomal peptide synthase of GT BGC) do not produce GT but in contrast to ΔkojR, ΔgliP is still virulent in the chemotherapeutic murine model of IPA [60]. Previously, we observed that ΔrglT is hypovirulent in the same murine model and suggest that these virulence defects could be due to defects in oxidative stress resistance [42]. It is possible the impact of the lack of KojR in the sulfur metabolism affects the growth and virulence of ΔkojR in the murine lungs. Additional experiments are necessary to clarify this and it remains subject to future investigations.
We were able to show a conserved mechanism of protection from GT in these three fungi whereby deletion of kojR and rglT increases sensitivity to exogenous GT. This protection is likely to occur through the RglT-dependent regulation of GliT, where expression of gliT is dependent on RglT in the presence of GT in the three fungal species. Future studies will determine growth of the respective gliT deletion strains in order to confirm this conserved mechanism of GT protection in Aspergillus species. The expression of gliT is also dependent on KojR in all three fungi in the presence of GT; however, KojR is partially required for gliT expression in A. fumigatus and A. oryzae, whereas KojR represses gliT in A. nidulans, which does not produce GT. These results suggest that KojR-dependent regulation of gliT differs between GT-producing and non-producing Aspergillus species and these results are in agreement with our A. fumigatus/A. nidulans RNA-seq dataset. In A. fumigatus and A. oryzae, rglT expression is dependent on KojR and we suggest here that KojR directly regulates rglT and indirectly gliT. This regulatory hierarchy would therefore confer protection from GT in these fungi. In A. fumigatus, kojR expression is dependent on RglT whereas in A. oryzae kojR repression is dependent on RglT. These results suggest that differences in the regulation of both TFs in these fungi.
Interestingly, we observed that both A. fumigatus and A. oryzae ΔrglT and ΔkojR are more sensitive to GT and involved in the positive regulation of the oxidoreductase gliT and gtmA. Although gliT expression is reduced in A. nidulans ΔrglT, it is increased in ΔkojR, suggesting the mechanism of gliT regulation is distinct in this species. Since GliT is essential for A. nidulans GT detoxification, these results indicate that increased expression of gliT in the A. nidulans ΔkojR strain is not enough to confer GT resistance to A. nidulans. These results suggest that KojR is also important for GT detoxification in a GT non-producer and that there are additional mechanisms of GT detoxification regulated by KojR in A. nidulans. Actually, two pieces of evidence suggest that KojR is also affecting the inorganic sulfur assimilation in A. nidulans and A. oryzae: (i) the sB sulfate permease gene is down-regulated in the presence of GT, and (ii) cysteine as a single sulfur source can suppress GT toxicity in these two species. We hypothesize that cysteine can suppress GT susceptibility in all three species because most likely it can bypass the initial steps of inorganic sulfur assimilation, providing directly cysteine for the synthesis of ergothioneine, glutathione, and methionine. Ergothioneine and glutathione are essential for oxiredox balance in the cell and also for GT prodution while methione can provide methyl groups for GT methylation, crucial for bmGT production. Interestingly, cysteine as a single sulfur source can suppress A. nidulans ΔrglT GT sensitivity but not A. fumigatus and A. oryzae ΔrglT GT sensitivity, suggesting diferences in the organic sulfur assimilation in these species during GT detoxification.
Although A. oryzae has the GT BGC, we were unable to detect GT production in the A. oryzae wild-type, ΔrglT and ΔkojR strains (S4 Fig). However, RglT and KojR are still regulating A. oryzae gliT and gtmA. A. oryzae strains have been selected for millenia for the saccharification of starch-rich rice to sugars that are fermentable by Saccharomyces cerevisiae into sake, the high-alcohol rice wine [61]. The A. oryzae-rice mixture (koji-culture) is mixed with additional steamed rice and fermented by S. cerevisiae. This A. oryzae rice starch breakdown and subsequent conversion into alcohol by S. cerevisiae occurs side by side [61]. Extensive genome analysis suggested that, through selection by humans, atoxigenic lineages of A. oryzae evolved into “cell factories” important for the saccharification process [62]. Interestingly, production of the secondary metabolite kojic acid was retained in these strains during the koji-culture [58]. The kojic acid gene cluster is one of the simplest clusters of secondary metabolite production ever reported, composed by three genes: an enzyme containing a FAD-dependent oxidoreductase domain, a transporter, and a kojR encoding a GAL4-like Zn(II)2Cys6 transcription factor [58,63]. As expected, A. oryzae ΔkojR is unable to produce kojic acid [58] but there is no involvement of RglT in kojic acid production.
There is extensive heterogeneity in drug susceptibility, nutritional requirements, and virulence across A. fumigatus clinical and environmental isolates [57,64–66]. It remains to be determined if GT-sensitive mutants in different Aspergillus spp isolates will behave accordingly to what was observed in this manuscript using the available isolates. Our work provides new opportunities to understand GT production and protection, and also highlights significant differences in transcriptional responses to extracellular mycotoxins between fungal species. More importantly, the identification of a conserved transcriptional response to exogenous GT in different Aspergillus species and isolates provides a basis for future studies which will further decipher these protection pathways.
Materials and methods
Ethics statement
The animal procedures were designed as a prospective, randomized, blinded, experimental study, and performed following the guidelines for animal research and the 3R principles of the EU directive. Eight-week-old gender and age-matched C57Bl/6 mice were bred under specific-pathogen-free conditions and kept at the Life and Health Sciences Research Institute (ICVS) Animal Facility. Animal experimentation was performed following biosafety level 2 (BSL-2), and protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Minho. The ethical and regulatory approvals were consented by Ethics Committee for Research in Life and Health Sciences of the University of Minho (SECVS 074/2016). Furthermore, all in vivo procedures followed the EU-adopted regulations (Directive 2010/63/EU) and were conducted according to the guidelines sanctioned by the Portuguese ethics committee for animal experimentation, Direção-Geral de Alimentação e Veterinária (DGAV).
Strains and media
All strains used in this study are listed in S6 Table. Strains were grown at 37°C except for growth on allyl alcohol-containing solid medium, which was carried out at 30°C. Conidia of A. fumigatus and A. nidulans were grown on complete medium (YG) [2% (w/v) glucose, 0.5% (w/v) yeast extract, trace elements] or minimal media (MM) [1% (w/v) glucose, nitrate salts, trace elements, pH 6.5]. Solid YG and MM were the same as described above with the addition of 2% (w/v) agar. Where necessary, uridine and uracil (1.2 g/L) were added. Trace elements, vitamins, and nitrate salt compositions were as described previously [67]. Potato Dextrose Agar (PDA, Difco) was used for A. oryzae. For iron or zinc starvation or excess experiments, strains were growth in solid MM without FeSO₄ or zinc. For phenotypic characterization, plates were inoculated with 104 spores per strain and left to grow for 120 h at 37 or 30°C. All radial growth experiments were expressed as ratios, dividing colony radial diameter of growth in the stress condition by colony radial diameter in the control (no stress) condition.
Phylogenomic inference of Aspergillus–Penicillium species phylogeny
Publicly available gene annotations for Aspergillus and Penicillium species (n = 55) were downloaded from NCBI in February 2021. OrthoFinder, v2.3.8 [68], was used to identify orthologous groups of genes using the protein sequences of the 55 proteomes as input. From the 18,979 orthologous groups of genes, 1,133 were identified to be single-copy and present in all proteomes. All 1,133 genes were aligned using MAFFT, v7.402 [69], with the ‘auto’ option. The corresponding nucleotide sequences were threaded onto the protein alignments using the ‘thread_dna’ function in PhyKIT, v1.1.2 [20]. The resulting nucleotide alignments were trimmed using ClipKIT, v1.1.3 [26], with the ‘smart-gap’ option. All 1,133 genes were concatenated into a single matrix with 2,446,740 sites using the ‘create_concat’ function in PhyKIT [20]. The resulting alignment was used to reconstruct the evolutionary history of the 55 species using IQ-TREE, v2.0.6 [70]. During tree search, the number of trees maintained during maximum likelihood inference was increased from five to ten using the ‘nbest’ option. Bipartition support was assessed using 1,000 ultrafast bootstrap approximations [71].
Comparison of the inferred phylogeny showed that it was nearly identical to the phylogeny inferred in a previous genome-scale examination of evolutionary relationships among Aspergillus and Penicillium species [44]. Minor differences were observed among bipartitions previously determined to harbor high levels of incongruence. These differences concern the placements of Penicillium decumbens and Aspergillus awamori. Differences in gene and taxon sampling may be responsible for these incongruences. All bipartitions received full support.
Identifying homologous biosynthetic gene clusters and orthologs of RglT and kojR
To identify homologs of the biosynthetic gene clusters involved in the production of gliotoxin and kojic acid, the 13 protein sequences of the gliotoxin cluster from Aspergillus fumigatus and the three protein sequences of the kojic acid cluster from Aspergillus oryzae were used as queries during sequence similarity searches to identify homologs in the 55 proteomes. Sequence similarity searches were conducted using NCBI’s blastp function from BLAST+, v 2.3.0 [72], with an expectation value threshold of 1e-4. Across all homologs, the physical proximity of each gene was assessed. We considered biosynthetic gene clusters to be homologous if 7 / 13 genes from the gliotoxin biosynthetic gene cluster were present including the nonribosomal peptide synthase, gliP. For the kojic acid biosynthetic gene cluster, we required that 2 / 3 genes from the kojic acid cluster were present including kojA, the oxidoreductase required for kojic acid production [73]. When examining gene proximity, a gene boundary distance of five genes was used. To identify genes orthologous to RglT and kojR, OrthoFinder results were parsed to identify genes in the same orthologous group as each gene.
Testing the association of the phylogenetic distributions of kojR and RglT
To evaluate if the distributions of kojR, RglT, and their BGCs on the species phylogeny are significantly associated, we examined the correlation of the phylogenetic distribution of the presence and absence patterns of the two genes on the species phylogeny. We used Pagel’s binary character correlation test implemented in phytools, v0.7–70 [74] to examine four scenarios: the distributions of two genes / BGCs are not correlated (scenario A), the distribution of one gene is correlated with the distribution of another gene / BGC but not vice versa (scenarios B and C), and the distributions of two genes / BGCs are correlated (scenario D). The best fitting model was then determined using the weighted Akaike information criterion (AIC).
Mouse model of pulmonary aspergillosis
The A. fumigatus Δku80 pyrG+ CEA17 and ΔkojR strains were grown on malt extract agar for 7 days before infection. To induce immunosuppression, 150 mg/Kg of cyclophosphamide (Sigma) was administrated intraperitoneally on days -4, -1, and +2, while 200 mg/Kg of cortisone 21-acetate (Acros Organics) was administrated on day -3 via the subcutaneous route. On day 0, mice were challenged intranasally with 1×105 live conidia following anesthesia with 75 mg/Kg of ketamine (Ketamidor, Richter Pharma) and 1 mg/Kg of medetomidine (Domtor, Ecuphar). To avoid bacterial infections, animals were treated with 50 μg/mL of chloramphenicol in drinking water ad libitum. For the survival studies, animals (n = 10) were monitored twice daily for 15 days after infection. Animals were weighted and sacrificed in case of 20% loss weight, severe ataxia or hypothermia, and other severe complications. In some experiments, mice (n = 5) were euthanized on day +3 and their lungs were aseptically removed, weighed, and homogenized in 2 mL sterile PBS in a tissue homogenizer (Ultra-Turrax T25 Basic, IKA Works, Inc.). For fungal burden assessment, lung homogenates were plated on Sabouraud 4% dextrose agar (Sigma) supplemented with chloramphenicol at 50 μg/mL (Sigma). For histological analysis, lungs were excised and fixed with 10% buffered formalin solution (Sigma) for at least 48 hours, processed and paraffin embedded. Lung paraffin sections were stained with Hematoxylin and Eosin (H&E) or Gomori’s Methenamine silver (GMS) (Sigma) for pathological examination. Images were acquired using a widefield upright microscope BX61 (Olympus) and a DP70 high-resolution camera (Olympus). Morphometry analyses were carried out on slide images using ImageJ software (v1.50i, NIH, USA).
Generation of deletion mutants for A. oryzae kojR and rglT, A. nidulans kojR and mtrA, and A. fumigatus ΔkojR::kojR+ and mtrA
The DNA fragment for kojR deletion, using the A. oryzae orotidine-5′-decarboxylase gene (pyrG) as a selectable marker, was generated by fusion polymerase chain reaction (PCR) as follows: DNA fragments upstream and downstream of the kojR coding region were amplified by PCR using A. oryzae genomic DNA as a template and the primer sets BR-TF040-L-F + BR-TF040-L-R and BR-TF040-R-F + BR-TF040-R-R, respectively. The pyrG fragment was amplified by PCR using A. oryzae genomic DNA as a template and the primers BR-TF040-P-F and BR-TF040-P-R. Then, these three PCR fragments were mixed, and a second round of PCR with the primers BR-TF040-L-F and BR-TF040-R-R was performed. For rglT deletion, DNA fragments upstream and downstream of the rglT coding region were amplified by PCR using the primer sets BR-TF390-L-F + BR-TF390-L-R and BR-TF390-R-F + BR-TF390-R-R, respectively. The pyrG fragment was amplified by PCR using the primers BR-TF390-P-F and BR-TF390-P-R. These three PCR fragments were mixed, and a second round of PCR without primers was carried out. Then, a third round of PCR with BR-TF390-L-F_Nest and BR-TF390-R-R_Nest was performed. The resultant PCR fragments were introduced into the recipient strain RkuN16ptr1 (Δku70::ptrA, pyrG–) according to [75].
In A. nidulans, the kojR and mtrA genes were deleted using a gene replacement cassettes which were constructed by “in vivo” recombination in S. cerevisiae as previously described by [76]. Thus, approximately 1.0 kb from each 5’-UTR and 3’-UTR flanking region of the targeted ORFs regions were selected for primer design. The primers AN4118 (kojR)/mtrA_pRS426_5UTR_fw and AN4118/mtrA_pRS426_3UTR_rv contained a short homologous sequence to the MCS of the plasmid pRS426. Both the 5- and 3-UTR fragments were PCR-amplified from A. nidulans genomic DNA (TNO2A3 strain). The pyrG gene placed within the cassette as a prototrophic marker was amplified from pCDA21 plasmid using the primers AN4118/mtrA_5UTR_pyrG_rv and AN4118/mtrA_3UTR_pyrG_fw. The cassette was PCR-amplified from these plasmids utilizing TaKaRa Ex Taq DNA Polymerase (Clontech Takara Bio) and used for A. nidulans transformation.
Complementation of the kojR gene in A. fumigatus was achieved via cotransformation. A fragment containing the kojR gene flanked by 1-kb 5′ and 3′ UTR regions was PCR amplified (primers AfukojR complemented 5UTR pRS fw and AfukojR complemented 3UTR pRS rv). This DNA fragment was transformed into the ΔkojR strain along with the plasmid pPTR1 (TaKaRa Bio) containing the pyrithiamine resistance ptrA gene. Mutants were selected on MM supplemented with 1 μg ml−1 pyrithiamine and confirmed with PCR using the external primers (AfukojR 5UTR ext fw and AfukojR 3UTR ext rv). Primer sequences are listed in S7 Table. A. fumigatus mrtA gene was deleted using a gene replacement cassette which was also constructed by “in vivo” recombination in S. cerevisiae. Approximately 1.0 kb from each 5’-UTR and 3’-UTR flanking region of the targeted ORF regions were selected for primer design. The primers Afu6g12780_pRS426_5UTR FW and Afu6g12780_pRS426_3UTR RV contained a short homologous sequence to the MCS of the plasmid pRS426. The pyrG gene placed within the cassette as a prototrophic marker was amplified from pCDA21 plasmid using the primers Afu6g12780_5UTR_pyrG_RV and Afu6g12780_3UTR_pyrG_FW. Southern blot analysis to confirm the mtrA deletion (S5 Fig). Primer sequences are listed in S7 Table.
Screening of A. fumigatus transcription factor deletion strains
A library of 484 A. fumigatus TFKO (transcription factor knock out) strains [46] was screened for sensitivity to gliotoxin. To verify this sensitivity, gliotoxin concentrations lower than the minimum inhibitory concentration (MIC) for the A. fumigatus wild type strain, CEA17, (70μg / mL) [42] were used. Conidia were inoculated in 96-well plates at the final concentration of 104 spores / well, in 200 μl MM supplemented or not with concentrations of gliotoxin ranging from 35 μg/ml (0.5 x MIC of wt) to 70 μg/ml, and incubated at 37°C for 48 h. Three independent experiments were carried out. Validation of this screening was carried out by growing sensitive strains at the concentration of 104 spores in solid MM plates with 30 μg/ml gliotoxin at 37°C, for 48 h.
Gliotoxin and Bis(methylthio)gliotoxin Presence Analysis by ultra-high-performance liquid chromatography (UHPLC) coupled to high-resolution electrospray ionization mass spectrometry (HRESIMS)
All strains were grown on Czapek Dox Agar (CDA) at 37°C in an incubator (VWR International) in the dark over four days. To evaluate the biosynthesis of gliotoxin (GT) and bis(methylthio)gliotoxin (bmGT) in these fungal strains, cultures were extracted with organic solvents and analyzed by mass spectrometry (see below). The agar plates were sprayed with MeOH, chopped with a spatula, and then the contents were transferred into a scintillation vial. Afterwards, acetone (~15 ml) was added to the scintillation vial, and the resulting slurry was vortexed vigorously for approximately 3 min and then let too set at RT for 4 h. Next, the mixture was filtered, and the resulting extract was dried under a stream of nitrogen gas.
HRESIMS experiments utilized a Thermo QExactive Plus mass spectrometer (Thermo Fisher Scientific), equipped with an electrospray ionization source. This was coupled to an Acquity UHPLC system (Waters Corp.), using a flow rate of 0.3 ml/min and a BEH C18 column (2.1 mm x 50 mm, 1.7 μm) that was operated at 40°C. The mobile phase consisted of CH3CN–H2O (Fischer Optima LC-MS grade; both acidified with 0.1% formic acid). The gradient started at 15% CH3CN and increased linearly to 100% CH3CN over 8 min, where it was held for 1.5 min before returning to starting conditions to re-equilibrate.
Extracts were analyzed in biological triplicates and each of these in technical triplicate using selected ion monitoring (SIM) in the positive ion mode with a resolving power of 35,000. For GT, the SIM had an inclusion list containing the mass [M+H]+ = 327.04677, with an isolation window of 1.8 Da and a retention window of 2.5–5.0 min. For bmGT, the SIM had an inclusion list containing the mass [M+H]+ = 357.09373, with an isolation window of 1.8 Da and a retention window of 2.5–4.5 min. The extracts, gliotoxin standard (Cayman Chemical), and bis(methylthio)gliotoxin standard (isolated previously) [26] were each prepared at a concentration of 2.5 mg/ml for the extracts and 0.01 mg/ml for the standards; all extracts were dissolved in MeOH and injected with a volume of 3 μl. To eliminate the possibility for sample carryover, two blanks (MeOH) were injected between every sample, and the standards were analyzed at the end of of the run.
To ascertain the relative abundance of GT and bmGT in these samples, batch process methods were run using Thermo Xcalibur (Thermo Fisher Scientific). For GT, we used a mass range of 327.0437–327.0492 Da at a retention time of 3.24 min with a 5.00 second window. For bmGT, we used a mass range of 357.0928–357.9937 Da at a retention time of 3.30 min with a 5.00 second window.
RNA extraction, RNA-sequencing, cDNA synthesis and RT-qPCR
All experiments were carried out in biological triplicates and conidia (107) were inoculated in liquid MM (A. fumigatus and A. nidulans), or in Potato Dextrose Broth (A. oryzae). Gliotoxin was added for 3 h to the culture medium to a final concentration of 5 μg/ml after strains were grown for 21 h in MM. As GT was dissolved in DMSO, control cultures received the same volume of DMSO for 3 h. For total RNA isolation, mycelia were ground in liquid nitrogen and total RNA was extracted using TRIzol (Invitrogen), treated with RQ1 RNase-free DNase I (Promega), and purified using the RNAeasy kit (Qiagen) according to the manufacturer’s instructions. RNA was quantified using a NanoDrop and Qubit fluorometer, and analyzed using an Agilent 2100 Bioanalyzer system to assess the integrity of the RNA. All RNA had an RNA integrity number (RIN) between 8.0 and 10 (Thermo Scientific) according to the manufacturer’s protocol.
RNA-sequencing was carried out using Illumina sequencing. The cDNA libraries were constructed using the TruSeq Total RNA with Ribo Zero (Illumina, San Diego, CA, USA). From 0.1–1 μg of total RNA, the ribosomal RNA was depleted and the remaining RNA was purified, fragmented and prepared for complementary DNA (cDNA) synthesis, according to manufacturer recommendations. The libraries were validated following the Library Quantitative PCR (qPCR) Quantification Guide (Illumina). Paired-end sequencing was performed on the Illumina NextSeq 500 Sequencing System using NextSeq High Output (2 x 150) kit, according to manufacturer recommendations. The BioProject ID in the NCBI’s BioProject database is PRJNA729661.
For RT-qPCR, the RNA was reverse transcribed to cDNA using the ImProm-II reverse transcription system (Promega) according to manufacturer’s instructions, and the synthesized cDNA was used for real-time analysis using the SYBR green PCR master mix kit (Applied Biosystems) in the ABI 7500 Fast real-time PCR system (Applied Biosystems, Foster City, CA, USA). Primer sequences are listed in S7 Table. RNASeq data was processed to select genes for normalization of the qPCR data, applying the method of [77]. Briefly, genes with a coefficient of variation smaller than 10%, behaving in a normally distributed manner, and with large expression values (FPKM) were selected. After experimental validation, Afu2g02680 (Putative matrix AAA protease) and AN1191 [Small ubiquitin-like modifier (SUMO) protein]. For A. oryzae experiments, tubA (AO090009000281) was used as a normalizer.
Kojic Acid (KA) production by A. oryzae
KA production by the A. oryzae strains was detected by the colorimetric method verifying the intensity of the red color formed by chelating ferric ions [58,78]. The standard medium for KA production [0.25% (w/v) yeast extract, 0.1% (w/v) K2HPO4, 0.05% (w/v) MgSO4–7H2O, and 10% (w/v) glucose, pH 6.0] containing 1 mM ferric ion (FeCl3) was used for detecting KA production by A. oryzae. The color of the medium turned red, indicating the presence of KA chelated with ferric ions. This color change could be observed at around 5 days after cultivation.
Supporting information
(PDF)
(PPTX)
(PDF)
Results show that the A. oryzae WT and deletion strains do not produce (A) gliotoxin or (B) bis(methylthio)gliotoxin. The data are presented as extracted ion chromatograms (XIC) from selected ion monitoring (SIM) using the protonated mass of gliotoxin (C13H15N2O4S2; [M+H]+ = 327.0473) and bis(methylthio)gliotoxin (C15H21N2O4S2; [M+H]+ = 357.0928) with a window of ± 5.0 ppm. GT and bmGT were detected in the A. fumigatus positive control strain Af293.
(PDF)
(PPTX)
(XLS)
(XLSX)
If applicable, the A. fumigatus homologue is also indicated.
(XLSX)
If applicable, the A. fumigatus homologue is also indicated. Also shown are a list of genes that are dependent or independent on RglT for their expression.
(XLSX)
(XLS)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files. The BioProject ID in the NCBI's BioProject database is PRJNA729661. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA729661.
Funding Statement
São Paulo Research Foundation (FAPESP) grant numbers 2016/12948-7 (PAC), 2017/14159-2 (LNAR), and 2018/10962-8 (GHG) and 2016/07870-9 (GHG), and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) 301058/2019-9 and 404735/2018-5 (GHG), both from Brazil. J.L.S. and A.R. are supported by the Howard Hughes Medical Institute through the James H. Gilliam Fellowships for Advanced Study program. AR’s laboratory received additional support from a Discovery grant from Vanderbilt University, the Burroughs Wellcome Fund, the National Science Foundation (DEB-1442113), and the National Institutes of Health/National Institute of Allergy and Infectious Diseases (R56AI146096 and R01AI153356). S.L.K. is supported by the National Institutes of Health via the National Center for Complementary and Integrative Health (F31 AT010558). A.C., R.A.G., and C.D.-O. were supported by the Fundação para a Ciência e a Tecnologia (FCT) (PTDC/MED-GEN/28778/2017, UIDB/50026/2020, and UIDP/50026/2020), the Northern Portugal Regional Operational Program (NORTE 2020), under the Portugal 2020 Partnership Agreement, through the European Regional Development Fund (ERDF) (NORTE-01-0145-FEDER-000039), the ICVS Scientific Microscopy Platform, member of the national infrastructure PPBI - Portuguese Platform of Bioimaging (PPBI-POCI-01-0145-FEDER-022122), the European Union’s Horizon 2020 research and innovation program under grant agreement 847507, and the “la Caixa” Foundation (ID 100010434) and FCT under the agreement LCF/PR/HP17/52190003. Individual support was provided by FCT (SFRH/BD/141127/2018 to C.D.-O.). We thank Universidade do Minho, Portugal and Universidade do São Paulo, Brazil for providing support for the scientific collaboration (G.H.G. and A.C., Edital USP-UMinho 2019. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sugui JA, Kwon-Chung K J, Juvvadi P R, Latgé J P, Steinbach W J. Aspergillus fumigatus and related species. Cold Spring Harb Perspect Med. 2014; Nov 6;5(2):a019786. doi: 10.1101/cshperspect.a019786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van de Veerdonk FL, Gresnigt MS, Romani L, Netea MG, Latgé JP. Aspergillus fumigatus morphology and dynamic host interactions. Nat Rev Microbiol. 2017; Nov;15(11):661–674. doi: 10.1038/nrmicro.2017.90 [DOI] [PubMed] [Google Scholar]
- 3.Latgé JP and Chamilos G. Aspergillus fumigatus and aspergillosis in 2019. Clin. Microbiol. Rev. 2019; 33: e00140–18. doi: 10.1128/CMR.00140-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamei K, and Watanabe A. Aspergillus mycotoxins and their effect on the host. Med. Mycol. 2005; 43: 95–99. doi: 10.1080/13693780500051547 [DOI] [PubMed] [Google Scholar]
- 5.Tekaia F, and Latgé JP. Aspergillus fumigatus: saprophyte or pathogen? Curr. Opin. Microbiol. 2005; 8: 385–392. doi: 10.1016/j.mib.2005.06.017 [DOI] [PubMed] [Google Scholar]
- 6.Abad A, Victoria Fernández-Molina J, Bikandi J, Ramírez A, Margareto J et al. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev. Iberoam. Micol. 2010; 27: 155–182. doi: 10.1016/j.riam.2010.10.003 [DOI] [PubMed] [Google Scholar]
- 7.Grahl N, Shepardson KM, Chung D, and Cramer RA. Hypoxia and fungal pathogenesis: to air or not to air? Eukaryot. Cell. 2012; 11: 560–570. doi: 10.1128/EC.00031-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keller N P. Fungal secondary metabolism: regulation, function and drug discovery. Nat. Rev. Microbiol. 2019; 17: 167–180. doi: 10.1038/s41579-018-0121-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lind AL, Wisecaver JH, Lameiras C, Wiemann P, Palmer JM, Keller NP, et al. Drivers of genetic diversity in secondary metabolic gene clusters within a fungal species. PLoS Biol. 2017; Nov 17;15(11):e2003583. doi: 10.1371/journal.pbio.2003583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mead ME, Knowles SL, Raja HA, Beattie SR, Kowalski CH, Steenwyk JL, et al. Characterizing the Pathogenic, Genomic, and Chemical Traits of Aspergillus fischeri, a Close Relative of the Major Human Fungal Pathogen Aspergillus fumigatus. mSphere. 2019; 4: e00018–19. doi: 10.1128/mSphere.00018-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shwab EK, Bok JW, Tribus M, Galehr J, Graessle S, Keller NP. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 2007; 6: 1656–1664. doi: 10.1128/EC.00186-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Losada L, Ajayi O, Frisvad JC, Yu J, Nierman WC. Effect of competition on the production and activity of secondary metabolites in Aspergillus species. Med. Mycol. 2009; 47: S88–S96. doi: 10.1080/13693780802409542 [DOI] [PubMed] [Google Scholar]
- 13.Yin WB, Baccile JA, Bok JW, Chen Y, Keller NP, Schroeder FC. A nonribosomal peptide synthetase-derived iron (III) complex from the pathogenic fungus Aspergillus fumigatus. J. Am. Chem. Soc. 2013; 135: 2064–2067. doi: 10.1021/ja311145n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiemann P, Lechner BE, Baccile JA, Velk TA, Yin WB, Bok JW, et al. Perturbations in small molecule synthesis uncovers an iron-responsive secondary metabolite network in Aspergillus fumigatus. Front. Microbiol. 2014; 5: 530. doi: 10.3389/fmicb.2014.00530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bignell E, Cairns TC, Throckmorton K, Nierman WC, Keller NP. Secondary metabolite arsenal of an opportunistic pathogenic fungus. Philos. Trans. R. Soc. B Biol. Sci. 2016; 371: 20160023. doi: 10.1098/rstb.2016.0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knox B P, Blachowicz A, Palmer JM, Romsdahl J, Huttenlocher A, Wang CCC, et al. Characterization of Aspergillus fumigatus Isolates from Air and Surfaces of the International Space Station. mSphere. 2016; 1: e00227–16. doi: 10.1128/mSphere.00227-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raffa N, and Keller NP. A call to arms: Mustering secondary metabolites for success and survival of an opportunistic pathogen. PLOS Pathog. 2019; 15: e1007606. doi: 10.1371/journal.ppat.1007606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blachowicz A, Raffa N, Bok JW, Choera T, Knox B, Lim FY, et al. Contributions of spore secondary metabolites to UV-C protection and virulence vary in different Aspergillus fumigatus strains. MBio. 2020; 11: e03415–19. doi: 10.1128/mBio.03415-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis RE, Wiederhold NP, Chi J, Han XY, Komanduri K V., Kontoyiannis DP, et al. Detection of gliotoxin in experimental and human aspergillosis. Infect Immun. 2005; 73: 635–637. doi: 10.1128/IAI.73.1.635-637.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steenwyk JL, Mead ME, de Castro PA, Valero C, Damasio A, Dos Santos RAC, et al. Genomic and Phenotypic Analysis of COVID-19-Associated Pulmonary Aspergillosis Isolates of Aspergillus fumigatus. Microbiol Spectr. 2021; Jun 9:e0001021. doi: 10.1128/Spectrum.00010-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlam D, Canton J, Carreño M, Kopinski H, Freeman SA, Grinstein S. Gliotoxin Suppresses Macrophage Immune Function by Subverting Phosphatidylinositol 3,4,5-Trisphosphate Homeostasis. MBio. 2016; 7: 1–15. doi: 10.1128/mBio.02242-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arias M, Santiago L, Vidal-Garcı´a M, Redrado S, Lanuza P, Comas L, et al. Preparations for invasion: Modulation of host lung immunity during pulmonary aspergillosis by gliotoxin and other fungal secondary metabolites. Front Immunol. 2018; 9: 1–12. doi: 10.3389/fimmu.2018.00001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsunawaki S, Yoshida LS, Nishida S, Kobayashi T, Shimoyama T. Fungal metabolite gliotoxin inhibits assembly of the human respiratory burst NADPH oxidase. Infect Immun. 2004; 72: 3373–3382. doi: 10.1128/IAI.72.6.3373-3382.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.König S, Pace S, Pein H, Heinekamp T, Kramer J, Romp E, et al. Gliotoxin from Aspergillus fumigatus Abrogates Leukotriene B4 Formation through Inhibition of Leukotriene A4 Hydrolase. Cell Chem Biol. 2019; 1–11. doi: 10.1016/j.chembiol.2019.01.001 [DOI] [PubMed] [Google Scholar]
- 25.Knowles SL, Mead ME, Silva LP, Raja HA, Steenwyk JL, Goldman GH, et al. Gliotoxin, a Known Virulence Factor in the Major Human Pathogen Aspergillus fumigatus, Is Also Biosynthesized by Its Nonpathogenic Relative Aspergillus fischeri. mBio. 2020; Feb 11;11(1):e03361–19. doi: 10.1128/mBio.03361-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steenwyk JL, Mead ME, Knowles SL, Raja HA, Roberts CD, Bader O, et al. Variation Among Biosynthetic Gene Clusters, Secondary Metabolite Profiles, and Cards of Virulence Across Aspergillus Species. Genetics. 2020; Oct;216(2):481–497. doi: 10.1534/genetics.120.303549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scharf DH, Heinekamp T, Remme N, Hortschansky P, Brakhage AA, Hertweck C. Biosynthesis and function of gliotoxin in Aspergillus fumigatus. Appl Microbiol Biotechnol. 2012; Jan;93(2):467–72. doi: 10.1007/s00253-011-3689-1 [DOI] [PubMed] [Google Scholar]
- 28.Scharf DH, Brakhage AA, Mukherjee PK. Gliotoxin—bane or boon? Environ Microbiol. 2016; Apr;18(4):1096–109. doi: 10.1111/1462-2920.13080 [DOI] [PubMed] [Google Scholar]
- 29.Welch TR, Williams RM. Epidithiodioxopiperazines. occurrence, synthesis and biogenesis. Nat Prod Rep. 2014; Oct;31(10):1376–404. doi: 10.1039/c3np70097f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dolan SK, O’Keeffe G , Jones GW , Doyle S. Resistance is not futile: Gliotoxin biosynthesis, functionality and utility. Trends Microbiol. 2015; 23: 419–428. doi: 10.1016/j.tim.2015.02.005 . [DOI] [PubMed] [Google Scholar]
- 31.Davis C, Carberry S, Schrettl M, Singh I, Stephens J C, Barry S M, et al. The role of glutathione S-transferase GliG in gliotoxin biosynthesis in Aspergillus fumigatus. Chem. Biol. 2011; 18, 542–552. doi: 10.1016/j.chembiol.2010.12.022 12.022. [DOI] [PubMed] [Google Scholar]
- 32.Struck A W, Thompson M L, Wong L S, Micklefield J.S-adenosyl-methionine-dependent methyltransferases: highly versatileenzymes in biocatalysis, biosynthesis and other biotechnological applications. Chembiochem 2012; 13, 2642–2655. doi: 10.1002/cbic.201200556 [DOI] [PubMed] [Google Scholar]
- 33.Amich J, Schafferer L, Haas H, and Krappmann S. Regulation of sulphur assimilation is essential for virulence and affects iron homeostasis of the human-pathogenic mould Aspergillus fumigatus. PLoS Pathog. 2013; 9:e1003573. doi: 10.1371/journal.ppat.1003573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheridan K J, Lechner B E, Keeffe G O, Keller M A, Werner E R,Lindner H, et al. Ergothioneine biosynthesis and functionality in the opportunistic fungal pathogen, Aspergillus fumigatus. Sci. Rep. 2016; 6:35306. doi: 10.1038/srep35306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Traynor AM, Sheridan KJ, Jones GW, Calera JA, Doyle S. Involvement of Sulfur in the Biosynthesis of Essential Metabolites in Pathogenic Fungi of Animals, Particularly Aspergillus spp.: Molecular and Therapeutic Implications. Front Microbiol. 2019; Dec 13;10:2859. doi: 10.3389/fmicb.2019.02859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang DN, Toyotome T, Muraosa Y, Watanabe A, Wuren T, Bunsupa S, et al. GliA in Aspergillus fumigatus is required for its tolerance to gliotoxin and affects the amount of extracellular and intracellular gliotoxin. Med Mycol. 2014; Jul;52(5):506–18. doi: 10.1093/mmy/myu007 [DOI] [PubMed] [Google Scholar]
- 37.Schrettl M, Carberry S, Kavanagh K, Haas H, Jones GW, O’Brien J, et al. Self-protection against gliotoxin-a component of the gliotoxin biosynthetic cluster, gliT, completely protects Aspergillus fumigatus against exogenous gliotoxin. PLoS Pathog. 2010; 6. doi: 10.1371/journal.ppat.1000952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dolan SK, Owens RA, O’Keeffe G, Hammel S, Fitzpatrick DA, Jones GW, et al. Regulation of nonribosomal peptide synthesis: bis-thiomethylation attenuates gliotoxin biosynthesis in Aspergillus fumigatus. Chem Biol. 2014; Aug 14;21(8):999–1012. doi: 10.1016/j.chembiol.2014.07.006 [DOI] [PubMed] [Google Scholar]
- 39.Dolan SK, Bock T, Hering V, Owens RA, Jones GW, Blankenfeldt W, et al. Structural, mechanistic and functional insight into gliotoxin bis-thiomethylation in Aspergillus fumigatus. Open Biol. 2017; Feb;7(2):160292. doi: 10.1098/rsob.160292 ; PMCID: PMC5356443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith EB, Dolan SK, Fitzpatrick DA, Doyle S, Jones GW. Towards understanding the gliotoxin detoxification mechanism: in vivo thiomethylation protects yeast from gliotoxin cytotoxicity. Microb Cell. 2016; Feb 19;3(3):120–125. doi: 10.15698/mic2016.03.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scharf DH, Habel A, Heinekamp T, Brakhage AA, Hertweck C. Opposed effects of enzymatic gliotoxin N- and S-methylations. J Am Chem Soc. 2014; Aug 20;136(33):11674–9. doi: 10.1021/ja5033106 [DOI] [PubMed] [Google Scholar]
- 42.Ries LNA, Pardeshi L, Dong Z, Tan K, Steenwyk JL, Colabardini AC, et al. The Aspergillus fumigatus transcription factor RglT is important for gliotoxin biosynthesis and self-protection, and virulence. PLoS Pathog. 2020; Jul 15;16(7):e1008645. doi: 10.1371/journal.ppat.1008645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fedorova ND, Khaldi N, Joardar VS, Maiti R, Amedeo P, Anderson MJ, et al. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet. 2008; Apr 11;4(4):e1000046. doi: 10.1371/journal.pgen.1000046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steenwyk JL, Shen XX, Lind AL, Goldman GH, Rokas A. A Robust Phylogenomic Time Tree for Biotechnologically and Medically Important Fungi in the Genera Aspergillus and Penicillium. mBio. 2019; Jul 9;10(4):e00925–19. doi: 10.1128/mBio.00925-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Keeffe G, Hammel S, Owens RA, Keane TM, Fitzpatrick DA, Jones GW et al. RNA-seq reveals the pan-transcriptomic impact of attenuating the gliotoxin self-protection mechanism in Aspergillus fumigatus. BMC Genomics. 2014; 15, 894. doi: 10.1186/1471-2164-15-894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Furukawa T, Van Rhijn N, Fraczek M, Gsaller F, Davies E, Carr P, et al. The negative cofactor 2 complex is a key regulator of drug resistance in Aspergillus fumigatus. Nat Commun. 2020; Jan 22;11(1):427. doi: 10.1038/s41467-019-14191-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mukherjee PK, Hurley JF, Taylor JT, Puckhaber L, Lehner S, Druzhinina I, et al. Ferricrocin, the intracellular siderophore of Trichoderma virens, is involved in growth, conidiation, gliotoxin biosynthesis and induction of systemic resistance in maize. Biochem Biophys Res Commun. 2018; Oct 28;505(2):606–611. doi: 10.1016/j.bbrc.2018.09.170 [DOI] [PubMed] [Google Scholar]
- 48.Winkelströter LK, Dolan SK, Dos Reis TF, Bom VL, de Castro PA, Hagiwara D, et al. Systematic Global Analysis of Genes Encoding Protein Phosphatases in Aspergillus fumigatus. G3 (Bethesda). 2015; May 5;5(7):1525–39. doi: 10.1534/g3.115.016766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jain R, Valiante V, Remme N, Docimo T, Heinekamp T, Hertweck C, et al. The MAP kinase MpkA controls cell wall integrity, oxidative stress response, gliotoxin production and iron adaptation in Aspergillus fumigatus. Mol Microbiol. 2011; Oct;82(1):39–53. doi: 10.1111/j.1365-2958.2011.07778.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seo H, Kang S, Park YS, Yun CW. The Role of Zinc in Gliotoxin Biosynthesis of Aspergillus fumigatus. Int J Mol Sci. 2019; Dec 8;20(24):6192. doi: 10.3390/ijms20246192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vicentefranqueira R, Amich J, Marín L, Sánchez CI, Leal F, Calera JA. The Transcription Factor ZafA Regulates the Homeostatic and Adaptive Response to Zinc Starvation in Aspergillus fumigatus. Genes (Basel). 2018; Jun 26;9(7):318. doi: 10.3390/genes9070318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Traynor AM, Owens RA, Coughlin CM, Holton MC, Jones GW, Calera JA, et al. At the metal-metabolite interface in Aspergillus fumigatus: towards untangling the intersecting roles of zinc and gliotoxin. Microbiology (Reading). 2021; Nov;167(11). doi: 10.1099/mic.0.001106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005; Dec 22;438(7071):1105–15. doi: 10.1038/nature04341 [DOI] [PubMed] [Google Scholar]
- 54.Qiao J, Kontoyiannis DP, Calderone R, Li D, Ma Y, Wan Z, et al. Afyap1, encoding a bZip transcriptional factor of Aspergillus fumigatus, contributes to oxidative stress response but is not essential to the virulence of this pathogen in mice immunosuppressed by cyclophosphamide and triamcinolone. Med Mycol. 2008; Dec;46(8):773–82. doi: 10.1080/13693780802054215 [DOI] [PubMed] [Google Scholar]
- 55.Schrettl M, Kim HS, Eisendle M, Kragl C, Nierman WC, Heinekamp T, et al. SreA-mediated iron regulation in Aspergillus fumigatus. Mol Microbiol. 2008. Oct;70(1):27–43. doi: 10.1111/j.1365-2958.2008.06376.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jun SC, Choi YH, Lee MW, Yu JH, Shin KS. The Putative APSES Transcription Factor RgdA Governs Growth, Development, Toxigenesis, and Virulence in Aspergillus fumigatus. mSphere. 2020; Nov 11;5(6):e00998–20. doi: 10.1128/mSphere.00998-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ries LNA, Rocha MC, de Castro PA, Silva-Rocha R, Silva RN, Freitas FZ, et al. The Aspergillus fumigatus CrzA Transcription Factor Activates Chitin Synthase Gene Expression during the Caspofungin Paradoxical Effect. mBio. 2017; Jun 13;8(3):e00705–17. doi: 10.1128/mBio.00705-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Terabayashi Y, Sano M, Yamane N, Marui J, Tamano K, Sagara J, et al. Identification and characterization of genes responsible for biosynthesis of kojic acid, an industrially important compound from Aspergillus oryzae. Fungal Genet Biol. 2010; Dec;47(12):953–61. doi: 10.1016/j.fgb.2010.08.014 [DOI] [PubMed] [Google Scholar]
- 59.Owens RA, O’Keeffe G, Smith EB, Dolan SK, Hammel S, Sheridan KJ, et al. Interplay between Gliotoxin Resistance, Secretion, and the Methyl/Methionine Cycle in Aspergillus fumigatus. Eukaryot Cell. 2015; Sep;14(9):941–57. doi: 10.1128/EC.00055-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cramer RA Jr, Gamcsik MP, Brooking RM, Najvar LK, Kirkpatrick WR, Patterson TF, et al. Disruption of a nonribosomal peptide synthetase in Aspergillus fumigatus eliminates gliotoxin production. Eukaryot Cell. 2006; Jun;5(6):972–80. doi: 10.1128/EC.00049-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoshizawa K. Sake: Production and flavor. Food Rev. Int. 1999; 15,83–107. [Google Scholar]
- 62.Gibbons JG, Salichos L, Slot JC, Rinker DC, McGary KL, King JG, et al. The evolutionary imprint of domestication on genome variation and function of the filamentous fungus Aspergillus oryzae. Curr Biol. 2012; 22:1403–1409. doi: 10.1016/j.cub.2012.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rokas A, Wisecaver JH, Lind AL. The birth, evolution and death of metabolic gene clusters in fungi. Nat Rev Microbiol. 2018; Dec;16(12):731–744. doi: 10.1038/s41579-018-0075-3 [DOI] [PubMed] [Google Scholar]
- 64.Kowalski CH, Beattie SR, Fuller KK, McGurk EA, Tang YW, Hohl TM, et al. Heterogeneity among Isolates Reveals that Fitness in Low Oxygen Correlates with Aspergillus fumigatus Virulence. mBio. 2016; Sep 20;7(5):e01515–16. doi: 10.1128/mBio.01515-16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Colabardini AC, Wang F, Dong Z, Pardeshi L, Rocha MC, Costa JH, et al. Heterogeneity in the transcriptional response of the human pathogen Aspergillus fumigatus to the antifungal agent caspofungin. Genetics. 2021; Oct 30;220(1):iyab183. doi: 10.1093/genetics/iyab183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ries LNA, Steenwyk JL, de Castro PA, de Lima PBA, Almeida F, de Assis LJ, et al. Nutritional Heterogeneity Among Aspergillus fumigatus Strains Has Consequences for Virulence in a Strain- and Host-Dependent Manner. Front Microbiol. 2019; Apr 24;10:854. doi: 10.3389/fmicb.2019.00854 ; PMCID: PMC6492530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kafer E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv Genet. 1977; 19 33–131. doi: 10.1016/s0065-2660(08)60245-x [DOI] [PubMed] [Google Scholar]
- 68.Emms D M, and Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019; 20, 238. doi: 10.1186/s13059-019-1832-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Katoh K, Standley D M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013; 30, 772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Minh B Q, Schmidt H A, Chernomor O, Schrempf D, Woodhams M D, von Haeseler A, et al. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020; 37, 1530–1534. doi: 10.1093/molbev/msaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoang D T, Chernomor O, von Haeseler A, Minh B Q, Vinh L S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018; 35, 518–522. doi: 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10, 421. doi: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tamano K, Kuninaga M, Kojima N, Umemura M, Machida M, Koike H. Use of the kojA promoter, involved in kojic acid biosynthesis, for polyketide production in Aspergillus oryzae: implications for long-term production. BMC Biotechnol. 2019; Oct 26;19(1):70. doi: 10.1186/s12896-019-0567-x ; PMCID: PMC6814975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Revell L J. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012; 3, 217–223. doi: 10.1111/j.2041-210X.2011.00169.x [DOI] [Google Scholar]
- 75.Jin FJ, Nishida M, Hara S, Koyama Y. Identification and characterization of a putative basic helix-loop-helix transcription factor involved in the early stage of conidiophore development in Aspergillus oryzae. Fungal Genet Biol. 2011; Dec;48(12):1108–15. doi: 10.1016/j.fgb.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 76.Colot HH, Park G, Turner GE, Ringelberg C, Crew CM, Borkovich KA, et al. A highthroughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Nat Acad Sci USA. 2006; 103:10352–10357. doi: 10.1073/pnas.0601456103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yim AK, Wong JW, Ku YS, Qin H, Chan TF, Lam HM. Using RNA-Seq Data to Evaluate Reference Genes Suitable for Gene Expression Studies in Soybean. PLoS One. 2015; Sep 8;10(9):e0136343. doi: 10.1371/journal.pone.0136343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bentley R. Preparation and analysis of kojic acid. Methods Enzymol. 1957; 3, 238–241. [Google Scholar]