Abstract
It is well documented that drug responses are related to Absorption, Distribution, Metabolism, and Excretion (ADME) characteristics of individual patients. Several studies have identified genetic variability in pharmacogenes, that are either directly responsible for or are associated with ADME, giving rise to individualized treatments. Our objective was to provide a comprehensive overview of pharmacogenetic variation in the Saudi population. We mined next generation sequencing (NGS) data from 11,889 unrelated Saudi nationals, to determine the presence and frequencies of known functional SNP variants in 8 clinically relevant pharmacogenes (CYP2C9, CYP2C19, CYP3A5, CYP4F2, VKORC1, DPYD, TPMT and NUDT15), recommended by the Clinical Pharmacogenetics Implementation Consortium (CPIC), and collectively identified 82 such star alleles. Functionally significant pharmacogenetic variants were prevalent especially in CYP genes (excluding CYP3A5), with 10–44.4% of variants predicted to be inactive or to have decreased activity. In CYP3A5, inactive alleles (87.5%) were the most common. Only 1.8%, 0.7% and 0.7% of NUDT15, TPMT and DPYD variants respectively, were predicted to affect gene activity. In contrast, VKORC1 was found functionally, to be highly polymorphic with 53.7% of Saudi individuals harboring variants predicted to result in decreased activity and 31.3% having variants leading to increased metabolic activity. Furthermore, among the 8 pharmacogenes studied, we detected six rare variants with an aggregated frequency of 1.1%, that among several other ethnicities, were uniquely found in Saudi population. Similarly, within our cohort, the 8 pharmacogenes yielded forty-six novel variants predicted to be deleterious. Based upon our findings, 99.2% of individuals from the Saudi population carry at least one actionable pharmacogenetic variant.
Introduction
Pharmacogenomics (PGx) studies genetic variations in an individual’s drug metabolizing enzymes, associating these with adverse drug events or the level of drug response [1]. Drug efficacy and toxicity may be predicted from the genetic background of individuals, particularly in respect of the Cytochrome P450 (CYP) family of liver enzymes [2, 3]. These enzymes catalyze the conversion of substances that are metabolized by our bodies, including pharmaceuticals [4]. Overall, the efficacy of a drug is related to its Absorption, Distribution, Metabolism and Excretion (ADME) [3, 5]. The efficacy of a drug may also be associated with drug target polymorphisms. Drug targets can include receptors, enzymes and membrane transporters [2]. CYPs are responsible for deactivation of many drugs through direct metabolic activity or via facilitation of excretion and thus play a central role in ADME related efficacy. CYPs are also important in enzymatic conversion of some drugs from their native to bioactive forms [6].
These differences in drug metabolism highlight the current trend towards individualized pharmacotherapy, such that the right drug is delivered at the right dose to the right patient. A standard dose of a given drug is not always safe, effective or economical in an individual patient [7, 8]. The high incidence of adverse drug events (ADEs) represents a heavy burden for the US health care system. Almost 7 million emergency department visits are related to ADEs each year with an estimated cost of $3.5 billion annually [9]. Large scale genomic studies provide opportunities to associate drug responses with individual pharmacogenetic profiles. Such knowledge may improve drug efficacy, result in better outcomes, and in some instances, prevents life-threatening adverse drug events. Dosing no longer needs to be based on the average drug responses of a patient population but can be personalized, taking into consideration individual pharmacogenomic and environmental variation. There are well-established drug-gene interactions, that include but are not limited to, clopidogrel (CYP2C19), warfarin (CYP2C9, VKORC1 and CYP4F2), thiopurines (TPMT, NUDT15), tacrolimus (CYP3A5) and fluorouracil (DPYD) [10–14]. These medications are commonly used globally with Saudi Arabia being no exception [9, 15]. Protocols targeting use of the right drug at the right dose in the right person, based on genomic data to personalize treatment, have already been clinically implemented successfully in other countries, e.g. the RIGHT protocol in the US and U-PGx project in Austria, Spain, Great Britain, Greece, Italy, Netherlands and Slovenia [16–18]. The allelic frequencies of genes encoding drug-metabolizing enzymes and their phenotypic consequences may vary considerably between ethnic groups. The impact of these allelic variants has been well studied in Caucasians and some other ethnicities, yet poorly in Arabs [5, 19]. This study expands pharmacogenetic knowledge of the Arab population. Description of the allelic spectrum of pharmacogenes, both known and novel variants, their frequency, and phenotypic designation in Saudi nationals, will provide a basis for better clinical management in this population.
During the last decade technological advances have enabled comprehensive mapping of human pharmacogenes [20, 21]. Next Generation Sequencing (NGS) and High Performance Computing (HPC) are two technologies that have enhanced this field [22, 23]. The mining of variants using sequence data from population-based genome programs, provide an opportunity to characterize the pharmacogenomic profiles of each of these groups. Here we describe our findings from the Saudi population.
Results
Mining of NGS data from a total of 11,889 (1,928 PGx gene panels and of 9,961 exomes) unrelated individuals was used to impute allele and haplotype frequencies. We analyzed frequencies of 82 haplotypes distributed across 8 pharmacogenes (Table 1). Nineteen CYP2C9 variants (*2, *3, *5, *6, *7, *8, *9, *11, *12, *14, *24, *32, *33, *36, *39, *43, *44, *45, *60) were identified that jointly accounted for 21.1% of all CYP2C9 alleles in Saudi Arabs, however, only CYP2C9*2 with a minor allele frequency (MAF) of 13.4% and CYP2C9*3 (MAF = 5.3%) were relatively common. Fifteen variant alleles (*2, *3, *4A, *6, *8, *9, *12, *13, *15, *16, *17, *24, *28, *30, *34) were found in CYP2C19 of which *17 and *2 were the most common: 25.9% and 9.6%, respectively. A splice site variant (rs7767746) that is the core allele for CYP3A5*3 was present in 84.7% of the population. Three other alleles (*6, *7 and *8) in CYP3A5 showed MAFs from <0.1% to 2.4%. In CYP4F2, 44.4% of Saudi individuals harbor a *3 allele, the remaining population being wild type. We detected four VKORC1 alleles; the most common was VKORC1*2 (MAF = 53.7%) followed by rs7294, 3730G>A (MAF = 29.2%). Two other VKORC1 variants: 106G>T (rs61742245) and 196G>A (rs72547529) were less commonly observed with MAFs of 2.1% and <0.1%, respectively. Genetic polymorphisms in DPYD and TPMT were rare in the Saudi population. We identified eight variants (rs67376798, rs3918290, rs1801266, rs115232898, rs112766203.1, rs72549304, rs146356975, rs56038477) for DPYD and ten star alleles for TPMT although the overall MAFs for both these were low, 0.7% (DPYD) and 0.9% (TPMT). Two alleles (*3 and *5) were identified in NUDT15. The *3 allele was present with a MAF of 1.8%; *5 being much less common (MAF<0.1%), in the population.
Table 1. Frequencies and functional status of pharmacogenetic alleles in Saudi population.
| Gene | Allele | Core variant | Variant type | Functional Status | Allele Frequency, SA (%) |
|---|---|---|---|---|---|
| CYP2C9 * | |||||
| *1 | None | Normal | 78.9 | ||
| *2 | rs1799853 | Missense (R144C) | Decreased | 13.4 | |
| *3 | rs1057910 | Missense (I359L) | Inactive | 5.3 | |
| *4 | rs56165452 | Missense (I359T) | Decreased | 0 | |
| *5 | rs28371686 | Missense (D360E) | Decreased | 0.2 | |
| *6 | rs9332131 | Frameshift | Inactive | 0.1 | |
| *7 | rs67807361 | Missense (L19I) | Uncertain function | <0.1 | |
| *8 | rs7900194 | Missense (R150H) | Decreased | 0.5 | |
| *9 | rs2256871 | Missense (H251R) | Normal | 0.7 | |
| *11 | rs28371685 | Missense (R335W) | Decreased | 0.6 | |
| *12 | rs9332239 | Missense (P489S) | Decreased | <0.1 | |
| *13 | rs72558187 | Missense (L90P) | Inactive | 0 | |
| *14 | rs72558189 | Missense (R125H) | Decreased | <0.1 | |
| *24 | rs749060448 | Missense (E354K) | Inactive | <0.1 | |
| *32 | rs868182778 | Missense (V490F) | Uncertain function | <0.1 | |
| *33 | rs200183364 | Missense (R132Q) | Inactive | 0.3 | |
| *36 | rs114071557 | Start lost | Uncertain function | <0.1 | |
| *39 | rs762239445 | Missense(R124W) | Inactive | <0.1 | |
| *43 | rs767576260 | Missense (R124W) | Inactive | <0.1 | |
| *44 | rs200965026 | Missense (T130M)) | Decreased | <0.1 | |
| *45 | rs199523631 | Missense (R132W) | Inactive | <0.1 | |
| *60 | rs767284820 | Missense (L467P) | Uncertain function | <0.1 | |
| CYP2C19 * | |||||
| *1 | None | Normal | 63.3 | ||
| *2 | rs4244285 | Splicing defect | Inactive | 9.6 | |
| *3 | rs4986893 | Stop-gain (W212X) | Inactive | 0.1 | |
| *4A | rs28399504 | Start lost | Inactive | <0.1 | |
| *4B | rs28399504, rs12248560 | Start lost, Regulatory | Inactive | 0 | |
| *5 | rs56337013 | Missense (R433W) | Inactive | 0 | |
| *6 | rs72552267 | Missense (R132Q) | Inactive | <0.1 | |
| *7 | rs72558186 | Splicing defect | Inactive | 0 | |
| *8 | rs41291556 | Missense (W120R) | Inactive | 0.1 | |
| *9 | rs17884712 | Missense (R144H) | Decreased | 0.2 | |
| *10 | rs6413438 | Missense (P227L) | Decreased | 0 | |
| *12 | rs55640102 | Stop-lost (X491C) | Uncertain function | <0.1 | |
| *13 | rs17879685 | Missense (R410C) | Normal | 0.4 | |
| *15 | rs17882687 | Missense (I19L) | Normal | 0.4 | |
| *16 | rs192154563 | Missense (R442C) | Decreased | <0.1 | |
| *17 | rs12248560 | Regulatory | Increased | 25.9 | |
| *24 | rs118203757 | Missense (R335Q) | Inactive | <0.1 | |
| *28 | rs113934938 | Missense (V374I) | Normal | <0.1 | |
| *30 | rs145328984 | Missense (R73C) | Uncertain function | 0.1 | |
| *34 | rs367543002, rs367543003 | Missense (P3S, F4L) | Uncertain function | <0.1 | |
| CYP3A5 * | |||||
| *1 | None | Normal | 12.5 | ||
| *2 | rs28365083 | Missense (T398N) | Uncertain function | 0 | |
| *3 | rs776746 | Splicing defect | Inactive | 84.5 | |
| *6 | rs10264272 | Splicing defect | Inactive | 2.4 | |
| *7 | rs41303343 | Frameshift | Inactive | 0.4 | |
| *8 | rs55817950 | Missense (R28C) | Uncertain function | <0.1 | |
| CYP4F2 * | |||||
| *1 | None | Normal | 55.6 | ||
| *3 | rs2108622 | Missense (V433M) | Decreased function | 44.4 | |
| VKORC1 * | |||||
| Wild-type | None | Normal | 15.08 | ||
| 1173C>T (*2) | rs9934438 | Regulatory | Decreased expression | 53.7 | |
| 3730G>A | rs7294 | UTR | Increased | 29.2 | |
| 85G>T | rs104894539 | Missense(V29L) | Increased | 0 | |
| 106G>T | rs61742245 | Missense (D36Y) | Increased | 2.1 | |
| 172A>G | rs104894541 | Missense (R58G) | Increased | 0 | |
| 196G>A | rs72547529 | Missense (V66M) | Increased | <0.1 | |
| 292C>G | rs72547528 | Missense (R98W) | Increased | 0 | |
| 383T>G | rs104894542 | Missense (L128R) | Increased | 0 | |
| DPYD * / ** | |||||
| *1 | None | Normal | 99.82 | ||
| 2846A>T | rs67376798 | Missense (D949V) | Inactive | <0.1 | |
| *2A | rs3918290 | Splicing defect | Inactive | 0.1 | |
| *8 | rs1801266 | Missense(R235W) | Inactive | <0.1 | |
| 557A>G | rs115232898 | Missense (Y186C) | Decreased function | 0.1 | |
| 2279C>T | rs112766203.1 | Missense (T760I) | Decreased function | <0.1 | |
| 1475C>T | rs72549304 | Missense (S492L) | Inactive | <0.1 | |
| 868A>G | rs146356975 | Missense (K290E) | Decreased function | <0.1 | |
| 1236G>A (HapB3) | rs56038477 | Synonymous (E412 =) | Decreased function | 0.5 | |
| TPMT ** | |||||
| *1 | None | Normal | 99.1 | ||
| *2 | rs1800462 | Missense (A80P) | Inactive | <0.1 | |
| *3A | rs1800460, rs1142345 | Missense (A154T, Y240C) | Inactive | 0.3 | |
| *3B | rs1800460 | Missense (A154T) | Inactive | <0.1 | |
| *3C | rs1142345 | Missense (Y240C) | Inactive | 0.4 | |
| *6 | rs75543815 | Missense (Y180F) | Uncertain function | 0 | |
| *8 | rs56161402 | Missense (R215H) | Uncertain function | 0.2 | |
| *12 | rs200220210 | Missense (S125L) | Uncertain function | <0.1 | |
| *24 | rs6921269 | Missense (Q179H) | Uncertain function | 0.1 | |
| *25 | rs377085266 | Missense(C212R) | Uncertain function | <0.1 | |
| *34 | rs111901354 | Missense (R82W) | Uncertain function | <0.1 | |
| NUDT15 * | |||||
| *1 | None | Normal | 98.5 | ||
| *3 | rs116855232 | Missense (R139C) | Inactive | 1.8 | |
| *5 | rs186364861 | Missense (V18I) | Uncertain function | <0.1 |
Functional status of star alleles was defined according to:
* the Pharmacogene Variation Consortium (https://www.pharmvar.org) and the Clinical Pharmacogenetics Implementation Consortium guidelines (https://cpicpgx.org/guidlines/).
**-literature [24].
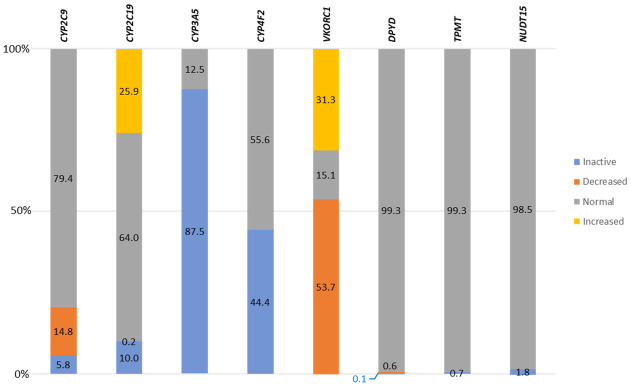
Functional consequences predicted for PGx alleles in the Saudi population were found predominantly in CYP genes. In CYP3A5 we found the highest number (87.5%) of inactive alleles as a result of the frequently observed intronic splice site CYP3A5 *3 variant. CYP4F2 showed decreased function alleles in 44.4% of individuals, whereas, in two other CYP genes (CYP2C9 and CYP2C19) reduced function alleles (inactive or decreased) were less common, being 20.6% and 10.1%, respectively. In other prominent PGx genes, allele function was much more conserved, with only 1.8%, 0.7% and 0.7% of NUDT15, TMPT and DPYD variants predicted to affect activity, respectively. In contrast, functionally, VKORC1 was highly polymorphic with 53.7% of Saudi individuals harboring variants predicted to result in decreased activity, whereas 31.3% carry variants leading to increased metabolic activity (Table 1 and S1 Table, Fig 1).
Fig 1. Combined functional consequences of genetic variations in pharmacogenes, within the Saudi population.
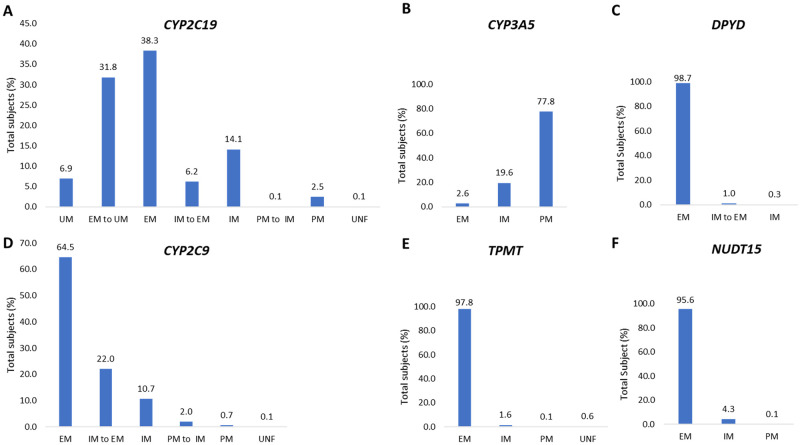
Based on genotypic data and predicted functional consequences of variant alleles we defined genotype-to-phenotype correlations (Fig 2). The phenotyping algorithms were derived from CPIC guidelines which were available only for CYP2C19, CYP2C9, CYP3A5, TPMT, NUDT15, and DPYD. Extensive metabolizer (EM) was the most frequent category for DPYD (98.7%), TPMT (97.8%) and NUDT15 (95.6%). EM status was also the highest (64.5% and 38.3%) for CYP2C9 and CYP2C19 although a significant number of remaining individuals (35.4% and 61.6%) are predicted to carry an altered drug metabolizer status for these two genes. CYP3A5 non-expressers (poor metabolizer, PM) represented 77.8% of the population.
Fig 2. Percentage of the predicted CYP2C19 (A), CYP3A5 (B), DPYD (C), CYP2C9 (D), TPMT (E), NUDT15 (F) metabolizer groups.
EM, extensive metabolizer; IM, intermediate metabolizer; PM, poor metabolizer; UM, ultrarapid metabolizer; UNF, uncertain function.
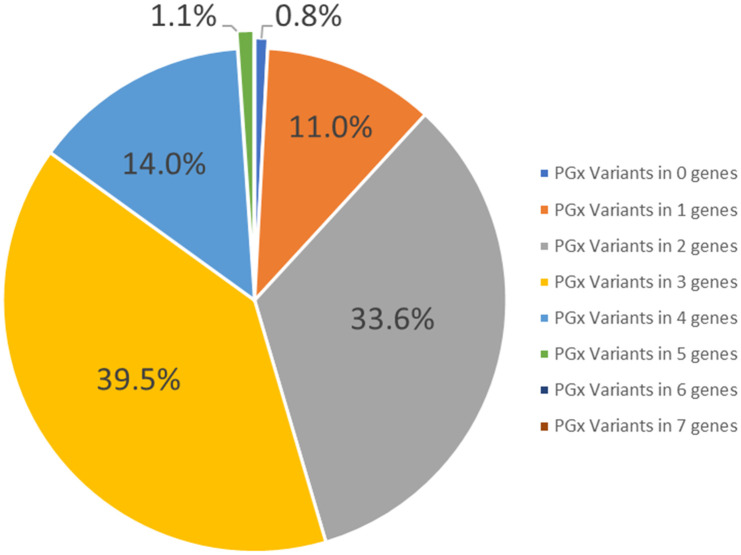
The percentage of Saudi individuals who carry actionable PGx variant(s) is summarized in Fig 3. Of the 1928 Saudi individuals (genotyped using the PGx gene panel), 99.2% carry at least one actionable PGx allele, with a maximum of 5 detected in 1.1% of the population.
Fig 3. Percentage of Saudi subjects carrying actionable variants in zero to five pharmacogenomics genes.
Of all 62 previously reported, predicted to be pathogenic (based on a two-fold scoring) rare variants (MAF<1%), four (1 stop-gain, 2 frameshift and 2 missense variants) with an aggregated frequency of 0.67% were uniquely observed in Saudi individuals when compared with other populations (European, Finish, Hispanic, African, South Asian, East Asian, Ashkenazi Jews and Arabs). Two missense variants were only present in Arabs (Table 2 and S2 Table). Next, we identified 46 novel sequence alterations in seven of the eight PGx genes studied. They included 5 stop-gain, 5 splice site, 1 frameshift, and 35 missense variants with an ADME score of ≥84%. DPYD revealed the largest number (n = 19) of novel alterations, the most frequent being DPYD:p.Ile971Thr, having a MAF of 0.00055 (Table 3 and S3 Table).
Table 2. Rare pharmacogenetic variants present in Saudi population in comparison to other populations.
| Minor allele frequency (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | RSID | Variant type | SA | AFR | AMR | ASJ | EAS | FIN | NFE | SAS | GME | QARB | Kaviar |
| CYP2C9 | rs771127798 | Frameshift | 0.076 | 0 | 0 | 0 | 0 | 0 | <0.01 | 0 | 0 | 0 | <0.01 |
| CYP2C9 | rs200985348 | Stop-gained | 0.017 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | <0.01 |
| CYP3A5 | rs1267703650 | Missense (I442T) | 0.135 | 0 | <0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CYP4F2 | rs780094643 | Missense(G417V) | 0.130 | 0 | 0 | 0 | 0 | 0 | <0.01 | 0 | 0.1 | 0 | <0.01 |
| CYP4F2 | rs763539865 | Frameshift | 0.421 | 0 | 0 | 0 | 0 | 0 | 0 | <0.01 | 0 | 0 | <0.01 |
| DPYD | rs568132506 | Missense(P86L) | 0.324 | 0 | 0.009 | 0 | 0 | 0 | 0.006 | <0.01 | 0.05 | 0.3 | 0.007 |
A complete summary of rare variants in Saudi population is presented in S2 Table. SA, Saudi Arabia; AFR, Africans; AMR, Latin/Admixed Americans; ASJ, Ashkenazi Jews; EAS, East Asians; FIN, European Finish; NFE, Non-Finish European; SAS, South Asians; GME, Greater Middle East Variome; QARB, Qatari Arabs; RSID, reference SNP cluster ID.
Table 3. Novel pharmacogenomics variants in Saudi population.
| Gene | Variant | Variant type | Minor allele frequency, SA (%) |
|---|---|---|---|
| CYP2C19 | NM_000769.4:c.914C>A:p.Thr305Asn | missense | 0.00420557 |
| CYP2C19 | NM_000769.4:c.332-1G>A | splice acceptor | 0.00841114 |
| CYP2C19 | NM_000769.4:c.482-2A>G | splice acceptor | 0.00841114 |
| CYP2C19 | NM_000769.4:c.1034T>C:p.Met345Thr | missense | 0.01682227 |
| CYP2C19 | NM_000769.4:c.1071A>C:p.Arg357Ser | missense | 0.00841114 |
| CYP2C9 | NM_000771.4:c.1023C>G:p.Asp341Glu | missense | 0.00420557 |
| CYP2C9 | NM_000771.4:c.893G>C:p.Gly298Ala | missense | 0.01261670 |
| CYP2C9 | NM_000771.4:c.961+1G>A | splice donor | 0.00841114 |
| CYP2C9 | NM_000771.4:c.1061A>G:p.Glu354Gly | missense | 0.00420557 |
| CYP2C9 | NM_000771.4:c.1198G>T:p.Glu400Ter | stop gained | 0.01682227 |
| CYP2C9 | NM_000771.4:c.1243G>T:p.Glu415Ter | stop gained | 0.00420557 |
| CYP3A5 | NM_000777.5:c.1205A>G:p.His402Arg | missense | 0.00420557 |
| CYP3A5 | NM_000777.5:c.1120G>A:p.Glu374Lys | missense | 0.00420557 |
| CYP3A5 | NM_000777.5:c.1067T>C:p.Leu356Pro | missense | 0.00420557 |
| CYP3A5 | NM_000777.5:c.1063T>C:p.Tyr355His | missense | 0.00420557 |
| CYP3A5 | NM_000777.5:c.957T>A:p.Tyr319Ter | stop gained | 0.01682227 |
| CYP3A5 | NM_000777.5:c.931A>G:p.Ser311Gly | missense | 0.00420557 |
| CYP3A5 | NM_000777.5:c.409T>C:p.Phe137Leu | missense | 0.00420557 |
| CYP3A5 | NM_000777.5:c.219-2A>G | splice acceptor | 0.00420557 |
| CYP4F2 | NM_001082.5:c.1288A>T:p.Asn430Tyr | missense | 0.02102784 |
| CYP4F2 | NM_001082.5:c.1231G>C:p.Gly411Arg | missense | 0.00420557 |
| CYP4F2 | NM_001082.5:c.985G>A:p.Gly329Ser | missense | 0.00420557 |
| CYP4F2 | NM_001082.5:c.889G>T:p.Asp297Tyr | missense | 0.00841114 |
| DPYD | NM_000110.4:c.2836delG:p.Ala946LeufsTer2 | frameshift | 0.00841114 |
| DPYD | NM_000110.4:c.1526C>G:p.Ser509Ter | stop gained | 0.00420557 |
| DPYD | NM_000110.4:c.958+1G>A | splice donor | 0.00841114 |
| DPYD | NM_000110.4:c.390T>A:p.Cys130Ter | stop gained | 0.00420557 |
| DPYD | NM_000110.4:c.2912T>C:p.Ile971Thr | missense | 0.05467239 |
| DPYD | NM_000110.4:c.2310C>G:p.Ile770Met | missense | 0.00420557 |
| DPYD | NM_000110.4:c.2137A>C:p.Asn713His | missense | 0.00420557 |
| DPYD | NM_000110.4:c.2083T>G:p.Cys695Gly | missense | 0.00420557 |
| DPYD | NM_000110.4:c.1804C>A:p.Pro602Thr | missense | 0.00420557 |
| DPYD | NM_000110.4:c.1679T>C:p.Ile560Thr | missense | 0.00420557 |
| DPYD | NM_000110.4:c.1657C>T:p.Pro553Ser | missense | 0.00420557 |
| DPYD | NM_000110.4:c.1591G>A:p.Val531Met | missense | 0.00420557 |
| DPYD | NM_000110.4:c.1405A>G:p.Met469Val | missense | 0.00420557 |
| DPYD | NM_000110.4:c.1309G>A:p.Ala437Thr | missense | 0.00841114 |
| DPYD | NM_000110.4:c.1076T>C:p.Val359Ala | missense | 0.00420557 |
| DPYD | NM_000110.4:c.574C>T:p.Leu192Phe | missense | 0.00420557 |
| DPYD | NM_000110.4:c.431C>T:p.Ala144Val | missense | 0.01682227 |
| DPYD | NM_000110.4:c.217C>T:p.Leu73Phe | missense | 0.00420557 |
| DPYD | NM_000110.4:c.194C>A:p.Thr65Lys | missense | 0.00420557 |
| TPMT | NM_000367.5:c.581G>A:p.Gly194Asp | missense | 0.00841114 |
| TPMT | NM_000367.5:c.454A>G:p.Arg152Gly | missense | 0.01682227 |
| TPMT | NM_000367.5:c.202C>A:p.Pro68Thr | missense | 0.00420557 |
| VKORC1 | NM_024006.6:c.404G>A:p.Cys135Tyr | missense | 0.01261670 |
Discussion
Inter-individual differences in drug efficacy drive current trends towards personalized pharmacotherapy targeting delivery of the right drug, at the right dose to the right patient. A standard dose of a given drug is not always safe, effective or economical in an individual patient [7, 8]. Mining of large-scale NGS data is a very powerful tool for cataloging the range and frequency of genetic variation in populations [25]. We used whole exome and PGx gene panel NGS data to estimate pharmacogenetic diversity in the Saudi population, thus far poorly recorded in current databases, compared to many other ethnic groups.
Our analysis provides the most comprehensive overview of PGx variability (predicted to be clinically relevant), of 8 phase I and phase II enzymes, in the Saudi population, published to date. We found that 61.6% of the Saudi cohort carry actionable CYP2C19 alleles, which may be associated with an increased risk of major adverse cardiovascular events during antiplatelet therapy with clopidogrel. In this instance ADEs range from stent thrombosis in poor and intermediate metabolizers, to bleeding risk in rapid and ultrarapid metabolizers. This drug was prescribed to several thousand patients who were treated at King Faisal Specialist Hospital and Research Centre, Riyadh, Saudi Arabia (KFSHR&RC) last year alone. Similar to European, African and Ashkenazi populations, CYP2C19*17 was the most frequent allele. CYP2C19*30 was unique to Arabs, with CYP2C19*13 and CYP2C19*15 detected in Saudi individuals, observed only in Africans (*13 and *15) and Jews (*15). Actionable CYP2C9 alleles associated with metabolism of warfarin were identified in 35.4% of Saudis.
Furthermore, the CYP4F2*3 and VKORC1*2 variants responsible for increased [26] and decreased warfarin activity [12] respectively, were strongly represented in our study population. CYP4F2 acts as an important counterpart to VKORC1 in limiting excessive accumulation of vitamin K [27, 28]. Inappropriate warfarin dosing underlies one of the most frequently reported adverse events, acute haemorrhages being one of the most common emergency visits in the US [29]. At KFSH&RC alone, warfarin is prescribed for several thousand patients every year. According to updated CPIC guidelines, genotypes of CYP2C9, VKORC1 and CYP4F2, should be considered together to estimate therapeutic warfarin dosing. One of the key factors strongly considered in dosing algorithms include ethnicity and population related genetic information. The majority of PGx data underpinning these guidelines arises from European, African American and East Asian ancestry [12]. Very little is known about pharmacogenetics in Arabs. Our study shows that the frequency of CYP2C9*2, *3, and VKORC1*2 in the Saudi population is similar to that of Europeans[25]. Other CYP2C9 variants common in Africans and present in Europeans (e.g. CYP2C9*5, *6, *8, and *11), that should be considered in warfarin dosing algorithms due to associated bleeding risk, show low occurrence in the Saudi population. Based on our findings, and subject to clinical validation, dosing recommendations for warfarin in Saudi patients should follow those for non-African ancestry, as recommended in CPIC guidelines [22]. However, studying the impact of a significantly higher frequency, of the functionally inactive CYP2C9*33 allele, on warfarin dosing in the Saudi population is strongly indicated. The vast majority of the Saudi population carries the CYP3A5*3 variant that results in a truncated mRNA with loss of protein expression [30]. Frequency of the *3 allele varies extremely across human populations and is correlated with distance from the equator. Equatorial populations may experience shortage of water and a sodium retaining phenotype in hot climates [31]. Our findings show frequency of this allele in the Saudi population, to be similar to that in six other populations (Ashkenazi, European, American, Finish and both Asians) [24, 25]. This gene catalyzes the metabolism of tacrolimus, a mainstay immunosuppressant. Patients with the CYP3A5*3 allele require the standard dose of this medication [13]. At KFSH&RC alone ~4000 patients received tacrolimus last year, and 22.2% of these may be normal metabolizers (2.6%) or intermediate metabolizers (19.6.%), requiring an increased tacrolimus dose to achieve a successful outcome. Clinical validation of this would be required, particularly given relatedness of donors and recipients in a consanguineous population, where histoincompatibility may be less than observed elsewhere.
Genetic variation in TPMT and NUDT15 are strongly linked to the risk for adverse reactions, to thiopurines commonly used for treatment of malignant and non-malignant conditions [11]. The “normal” starting doses are generally high based on clinical trials which are enriched in wild-type individuals. Full doses are tailored for normal metabolizers and may cause acute toxicity in intermediate and poor metabolizers [32]. Thiopurine tolerance is highly correlated with genetic ancestry [33]. The functionally inactive TPMT*3A allele is much less common in Saudi individuals relative to American, European and Ashkenazi populations [24, 25]. CPIC guidelines recommend a customized dose of thiopurines in compound intermediate metabolizers (intermediate metabolizers in both TMPT and NUDT15 [11]. We identified 0.03% (n = 3) compound intermediate metabolizers in Saudi population. Genetic variation in DPYD is a strong predictor of adverse risk related to use of the chemotherapeutic agent fluorouracil, commonly used in the treatment of various malignancies. Many cases have been reported of severe toxicities or even lethal outcome due to the DPYD poor or null metabolizer phenotype [34]. In our study we identified 1.3% of Saudi individuals who carry either a functionally normal allele plus one null or one functionally decreased allele, and would be predicted to be intermediate metabolizers. Reduced doses of fluorouracil may be indicated for these individuals [10]. More importantly our study detected in the Saudi population, the DPYD rare pathogenic mutation (c.257C>T) that may be responsible for severe toxicity in heterozygous patients or lethality in homozygous cancer patients treated with fluoropyrimidines [35]. We found this variant to be significantly enriched in the Saudi population with approximately 1 in every 333 individuals heterozygous for this allele. This DPYD allele is also present in the Qatari population (0.3%) whereas it is very rare in other populations, with frequencies (relative to the Saudi population) <36-fold in Americans, <52-fold in Europeans, <99-fold in South Asians, and was absent in other compared populations (Table 2 and S2 Table). Given the high rate of consanguinity (~60%) in Saudi Arabia, we can expect relative to outbred populations, a higher incidence of homozygotes for the DPYD (c.257C>T) mutation. Consanguinity increases the probability of a mate to be a carrier of the same recessive allele [36]. Thus, genotyping DPYD in the Saudi population may have greater clinical relevance. In most of the pharmacogenes screened we observed alleles shared with other Arabs [19, 24, 37], and some unique to the Saudi population. Amongst those shared with other Arabs, some were observed at significantly (p<0.05) different frequencies (S1 and S2 Tables).
Large-scale NGS data mining enables discovery of novel and rare pharmacogenetic alterations [3]. They are often population specific alleles and are not incorporated within current pharmacogenomic assays. Our study shows that such variants are present in the Saudi population, with computational algorithms predicting their functional significance in multiple instances. They may significantly add to knowledge of potentially actionable variants in ADME genes within the Saudi population and should be further investigated. Novel variants require experimental validation to test their functional effects in drug response [38]. Our study highlights the value of mining large NGS databases as a powerful tool, to improve knowledge of genomic variation within ADME genes, and stimulate their further investigation and eventual implementation in clinical practice. The data we present from one of the larger Middle Eastern countries, provides the most comprehensive overview of pharmacogenetic variants in Arabs, who to date are underrepresented in international genomic databases. We believe it will have both immediate and near-term clinical implications, expanding the application of pharmacogenetics and the practice of precision or individualized medicine in Arab patients.
Study limitations
The clinical impact of variants identified by this study remain in question as information from relevant clinical trials are limited. While PGx variants are predicted to be actionable in other populations, one cannot assume that these variants will ultimately have the same impact in the Saudi population without clinical verification. Another limitation of our study is the technical constraints of exome sequencing; non-coding regions and loci with high genomic complexity are poorly, or not covered at all. Structural changes and copy number variations which may be relevant are not reliably identified by whole exome or gene panel sequencing. Thus, we were not able to call star alleles with whole gene deletions, duplications or hybrids that are common in the assignment of CYP2D6 alleles. Accordingly, we did not include CYP2D6 in our analysis.
Furthermore, actionable variants located in non-coding regions CYP2C19 rs12248560, CYP3A5 rs776746, VKORC1 rs9934438, VKORC1 rs7294, DPYD rs67376798 were not covered by whole exome sequencing, our data for these being exclusively obtained from the PGx custom gene panel only.
Methods
Manuscript was based on access to fully anonymized data from Saudi Human Genome Project for which waiver of consents was granted by the IRB of King Faisal Specialist Hospital and Research Center. The dataset used for mining of pharmacogenomic variants comprised 9,961 exomes and 1,928 PGx custom gene panels (genes are listed in S4 Table), from unrelated Arab individuals sequenced by the Saudi Human Genome Program (SHGP) between 2015 and 2019, as part of a comprehensive investigation of rare diseases in the Saudi population [39, 40]. We studied eight genes for which the Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines are curated (https://cpicpgx.org/guidelines/) and present on FDA labels (https://www.fda.gov.Drugs/ScienceReseach/ucm572698). CYP star allele assignment and their clinical function was derived from Pharmacogene Variation Consortium (https://www.pharmvar.org/genes/) and CPIC allele functional tables. Metabolizer types were inferred based on CPIC guidelines and the Pharmacogenomics Knowledgebase (PharmGKB) https://www.pharmgkb.org/ and they were defined as follows: ultrarapid metabolizer (UM), intermediate metabolizer (IM), extensive/normal metabolizer (EM), poor metabolizer (PM), rapid metabolizer (RM), IM to EM and PM to EM. Our method for Star allele calling was based upon using the Stargazer algorithm (v.1.0.8). This algorithm performs statistical haplotype phasing using Beagle [41] with reference samples from the 1000 Genomes Project [42]. The Beagle method is based on localized haplotype-cluster model, which is an empirical linkage disequilibrium model that can take the local structure in the data into consideration. The Beagle algorithm is accurate and runs fast due to the use of an EM-based algorithm that literately fits the best model to the data. Afterwards, the phased haplotypes computed by Beagle are then matched to publicly available star allele information, mostly in the PharmVar database (https://www.pharmvar.org) and PharmGKB (https://www.pharmgkb.org/). Finally, Stargazer reports the star allele findings in a tabular format along with prediction of the related metabolizer information. Frequencies of intronic and UTR variants were covered only by the PGx panel and their frequency was calculated based on the cohort of 1928 individuals. Variants with MAF <1% were defined as rare and genetic alterations with frequencies that exceeded the observed frequencies in other populations (European, Finish, Hispanic, African, South Asian, East Asian, Ashkenazi Jews and Arabs) by >20-fold were considered as being “Saudi-specific”. A Chi-square test was used to determine the statistical difference for allele frequencies between different populations. A p-value less than 0.05 was considered significant. Next, we classified alleles as novel if they were not observed in: 1000 Genomes (phase3), gnomAD (v.3.1.1), Exac (v.0.3) and Kaviar (v.160204). Functional consequence of PGx rare Saudi-specific and novel variants was predicted using a two-fold approach. Any variants with a high IMPACT rating, such as frameshift indels or stop loss variants were considered to be deleterious [43, 44]. We then applied the ADME-optimized framework that is an ensemble of deleteriousness prediction methods for predicting deleteriousness in pharmacogenes. We used 18 prediction algorithms to compute the ADME scores including CADD, SIFT, PolyPhen, LRT (likelihood ratio test), MutationAssessor, FATHMM, FATHMM-MKL, PROVEAN, VEST3, DANN, MetSVM, MetaLR, GERP++, SiPhy, PhyloP-vertebrate, PhyloP-mammalian, PhastCons-vertebrate, and PhastCons-mammalian. ADME scores larger than 84% were considered to affect pharmacogene functionality [45]. We used phenotypes generated from Stargazer for CYP2C19, CYP2C9, CYP3A5, DPYD, NUDT15 and TPMT to determine the percentage of individuals predicted to have actionable PGx variants. For VKORC1 (rs9934438) and CYP4F2*3 (rs2108622), individuals carrying heterozygous (CT) or homozygous (TT) and heterozygous (GA) or homozygous (AA), respectively were considered to have an actionable variant in those genes.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We acknowledge the support of the Saudi Human Genome Program, King Abdulaziz City for Science and Technology for supporting the work described in this manuscript.
Data Availability
Data cannot be shared publicly because of National policy. Data are available from the King Faisal Specialist Hospital and Research Centre Institutional Data Access / Ethics Committee (contact via ora@kfshrc.edu.sa) for researchers who meet the criteria for access to confidential data.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Schildcrout JS, Denny JC, Bowton E, Gregg W, Pulley JM, Basford MA, et al. Optimizing drug outcomes through pharmacogenetics: a case for preemptive genotyping. Clin Pharmacol Ther. 2012;92(2):235–42. Epub 2012/06/29. doi: 10.1038/clpt.2012.66 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson JA. Drug target pharmacogenomics: an overview. Am J Pharmacogenomics. 2001;1(4):271–81. Epub 2002/06/27. doi: 10.2165/00129785-200101040-00004 . [DOI] [PubMed] [Google Scholar]
- 3.Fujikura K, Ingelman-Sundberg M, Lauschke VM. Genetic variation in the human cytochrome P450 supergene family. Pharmacogenet Genomics. 2015;25(12):584–94. Epub 2015/09/05. doi: 10.1097/FPC.0000000000000172 . [DOI] [PubMed] [Google Scholar]
- 4.Kozyra M, Ingelman-Sundberg M, Lauschke VM. Rare genetic variants in cellular transporters, metabolic enzymes, and nuclear receptors can be important determinants of interindividual differences in drug response. Genet Med. 2017;19(1):20–9. Epub 2016/04/22. doi: 10.1038/gim.2016.33 . [DOI] [PubMed] [Google Scholar]
- 5.Zhou Y, Ingelman-Sundberg M, Lauschke VM. Worldwide Distribution of Cytochrome P450 Alleles: A Meta-analysis of Population-scale Sequencing Projects. Clin Pharmacol Ther. 2017;102(4):688–700. Epub 2017/04/06. doi: 10.1002/cpt.690 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caudle KE, Dunnenberger HM, Freimuth RR, Peterson JF, Burlison JD, Whirl-Carrillo M, et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med. 2017;19(2):215–23. Epub 2016/07/22. doi: 10.1038/gim.2016.87 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramos E, Doumatey A, Elkahloun AG, Shriner D, Huang H, Chen G, et al. Pharmacogenomics, ancestry and clinical decision making for global populations. Pharmacogenomics J. 2014;14(3):217–22. Epub 2013/07/10. doi: 10.1038/tpj.2013.24 . [DOI] [PubMed] [Google Scholar]
- 8.Johnson JA, Burkley BM, Langaee TY, Clare-Salzler MJ, Klein TE, Altman RB. Implementing personalized medicine: development of a cost-effective customized pharmacogenetics genotyping array. Clin Pharmacol Ther. 2012;92(4):437–9. Epub 2012/08/23. doi: 10.1038/clpt.2012.125 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji Y, Skierka JM, Blommel JH, Moore BE, VanCuyk DL, Bruflat JK, et al. Preemptive Pharmacogenomic Testing for Precision Medicine: A Comprehensive Analysis of Five Actionable Pharmacogenomic Genes Using Next-Generation DNA Sequencing and a Customized CYP2D6 Genotyping Cascade. J Mol Diagn. 2016;18(3):438–45. Epub 2016/03/08. doi: 10.1016/j.jmoldx.2016.01.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amstutz U, Henricks LM, Offer SM, Barbarino J, Schellens JHM, Swen JJ, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Dihydropyrimidine Dehydrogenase Genotype and Fluoropyrimidine Dosing: 2017 Update. Clin Pharmacol Ther. 2018;103(2):210–6. Epub 2017/11/21. doi: 10.1002/cpt.911 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Relling MV, Schwab M, Whirl-Carrillo M, Suarez-Kurtz G, Pui CH, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for Thiopurine Dosing Based on TPMT and NUDT15 Genotypes: 2018 Update. Clin Pharmacol Ther. 2019;105(5):1095–105. Epub 2018/11/18. doi: 10.1002/cpt.1304 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson JA, Caudle KE, Gong L, Whirl-Carrillo M, Stein CM, Scott SA, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics-Guided Warfarin Dosing: 2017 Update. Clin Pharmacol Ther. 2017;102(3):397–404. Epub 2017/02/16. doi: 10.1002/cpt.668 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birdwell KA, Decker B, Barbarino JM, Peterson JF, Stein CM, Sadee W, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP3A5 Genotype and Tacrolimus Dosing. Clin Pharmacol Ther. 2015;98(1):19–24. Epub 2015/03/25. doi: 10.1002/cpt.113 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott SA, Sangkuhl K, Stein CM, Hulot JS, Mega JL, Roden DM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94(3):317–23. Epub 2013/05/24. doi: 10.1038/clpt.2013.105 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Driest SL, Shi Y, Bowton EA, Schildcrout JS, Peterson JF, Pulley J, et al. Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin Pharmacol Ther. 2014;95(4):423–31. Epub 2013/11/21. doi: 10.1038/clpt.2013.229 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bielinski SJ, Olson JE, Pathak J, Weinshilboum RM, Wang L, Lyke KJ, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc. 2014;89(1):25–33. Epub 2014/01/07. doi: 10.1016/j.mayocp.2013.10.021 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89(3):464–7. Epub 2011/01/29. doi: 10.1038/clpt.2010.279 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blagec K, Koopmann R, Crommentuijn-van Rhenen M, Holsappel I, van der Wouden CH, Konta L, et al. Implementing pharmacogenomics decision support across seven European countries: The Ubiquitous Pharmacogenomics (U-PGx) project. J Am Med Inform Assoc. 2018;25(7):893–8. Epub 2018/02/15. doi: 10.1093/jamia/ocy005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sivadas A, Scaria V. Pharmacogenomic survey of Qatari populations using whole-genome and exome sequences. Pharmacogenomics J. 2018;18(4):590–600. Epub 2018/05/04. doi: 10.1038/s41397-018-0022-8 . [DOI] [PubMed] [Google Scholar]
- 20.Wright GEB, Carleton B, Hayden MR, Ross CJD. The global spectrum of protein-coding pharmacogenomic diversity. Pharmacogenomics J. 2018;18(1):187–95. Epub 2016/10/26. doi: 10.1038/tpj.2016.77 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bush WS, Crosslin DR, Owusu-Obeng A, Wallace J, Almoguera B, Basford MA, et al. Genetic variation among 82 pharmacogenes: The PGRNseq data from the eMERGE network. Clin Pharmacol Ther. 2016;100(2):160–9. Epub 2016/02/10. doi: 10.1002/cpt.350 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohn I, Paton TA, Marshall CR, Basran R, Stavropoulos DJ, Ray PN, et al. Genome sequencing as a platform for pharmacogenetic genotyping: a pediatric cohort study. NPJ Genom Med. 2017;2:19. Epub 2017/12/22. doi: 10.1038/s41525-017-0021-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chua EW, Cree SL, Ton KN, Lehnert K, Shepherd P, Helsby N, et al. Cross-Comparison of Exome Analysis, Next-Generation Sequencing of Amplicons, and the iPLEX((R)) ADME PGx Panel for Pharmacogenomic Profiling. Front Pharmacol. 2016;7:1. Epub 2016/02/10. doi: 10.3389/fphar.2016.00001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Lauschke VM. Comprehensive overview of the pharmacogenetic diversity in Ashkenazi Jews. J Med Genet. 2018;55(9):617–27. Epub 2018/07/05. doi: 10.1136/jmedgenet-2018-105429 . [DOI] [PubMed] [Google Scholar]
- 25.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91. Epub 2016/08/19. doi: 10.1038/nature19057 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caldwell MD, Awad T, Johnson JA, Gage BF, Falkowski M, Gardina P, et al. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008;111(8):4106–12. Epub 2008/02/06. doi: 10.1182/blood-2007-11-122010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danese E, Montagnana M, Johnson JA, Rettie AE, Zambon CF, Lubitz SA, et al. Impact of the CYP4F2 p.V433M polymorphism on coumarin dose requirement: systematic review and meta-analysis. Clin Pharmacol Ther. 2012;92(6):746–56. Epub 2012/11/08. doi: 10.1038/clpt.2012.184 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang JE, Jorgensen AL, Alfirevic A, Williamson PR, Toh CH, Park BK, et al. Effects of CYP4F2 genetic polymorphisms and haplotypes on clinical outcomes in patients initiated on warfarin therapy. Pharmacogenet Genomics. 2009;19(10):781–9. Epub 2009/09/11. doi: 10.1097/FPC.0b013e3283311347 . [DOI] [PubMed] [Google Scholar]
- 29.Shehab N, Sperling LS, Kegler SR, Budnitz DS. National estimates of emergency department visits for hemorrhage-related adverse events from clopidogrel plus aspirin and from warfarin. Arch Intern Med. 2010;170(21):1926–33. Epub 2010/11/26. doi: 10.1001/archinternmed.2010.407 . [DOI] [PubMed] [Google Scholar]
- 30.Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27(4):383–91. Epub 2001/03/30. doi: 10.1038/86882 . [DOI] [PubMed] [Google Scholar]
- 31.Thompson EE, Kuttab-Boulos H, Witonsky D, Yang L, Roe BA, Di Rienzo A. CYP3A variation and the evolution of salt-sensitivity variants. Am J Hum Genet. 2004;75(6):1059–69. Epub 2004/10/20. doi: 10.1086/426406 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans WE, Horner M, Chu YQ, Kalwinsky D, Roberts WM. Altered mercaptopurine metabolism, toxic effects, and dosage requirement in a thiopurine methyltransferase-deficient child with acute lymphocytic leukemia. J Pediatr. 1991;119(6):985–9. Epub 1991/12/01. doi: 10.1016/s0022-3476(05)83063-x . [DOI] [PubMed] [Google Scholar]
- 33.Yang JJ, Landier W, Yang W, Liu C, Hageman L, Cheng C, et al. Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J Clin Oncol. 2015;33(11):1235–42. Epub 2015/01/28. doi: 10.1200/JCO.2014.59.4671 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoff PM, Ansari R, Batist G, Cox J, Kocha W, Kuperminc M, et al. Comparison of oral capecitabine versus intravenous fluorouracil plus leucovorin as first-line treatment in 605 patients with metastatic colorectal cancer: results of a randomized phase III study. J Clin Oncol. 2001;19(8):2282–92. Epub 2001/04/17. doi: 10.1200/JCO.2001.19.8.2282 . [DOI] [PubMed] [Google Scholar]
- 35.Del Re M, Quaquarini E, Sottotetti F, Michelucci A, Palumbo R, Simi P, et al. Uncommon dihydropyrimidine dehydrogenase mutations and toxicity by fluoropyrimidines: a lethal case with a new variant. Pharmacogenomics. 2016;17(1):5–9. Epub 2015/12/15. doi: 10.2217/pgs.15.146 . [DOI] [PubMed] [Google Scholar]
- 36.Alkuraya FS. Autozygome decoded. Genet Med. 2010;12(12):765–71. Epub 2010/12/30. doi: 10.1097/GIM.0b013e3181fbfcc4 . [DOI] [PubMed] [Google Scholar]
- 37.Al-Eitan LN. Pharmacogenomic landscape of VIP genetic variants in Jordanian Arabs and comparison with worldwide populations. Gene. 2020;737:144408. Epub 2020/02/03. doi: 10.1016/j.gene.2020.144408 . [DOI] [PubMed] [Google Scholar]
- 38.Ingelman-Sundberg M, Mkrtchian S, Zhou Y, Lauschke VM. Integrating rare genetic variants into pharmacogenetic drug response predictions. Hum Genomics. 2018;12(1):26. Epub 2018/05/26. doi: 10.1186/s40246-018-0157-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abouelhoda M, Sobahy T, El-Kalioby M, Patel N, Shamseldin H, Monies D, et al. Clinical genomics can facilitate countrywide estimation of autosomal recessive disease burden. Genet Med. 2016;18(12):1244–9. Epub 2016/04/29. doi: 10.1038/gim.2016.37 . [DOI] [PubMed] [Google Scholar]
- 40.Saudi Mendeliome G. Comprehensive gene panels provide advantages over clinical exome sequencing for Mendelian diseases. Genome Biol. 2015;16:134. Epub 2015/06/27. doi: 10.1186/s13059-015-0693-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Browning BL, Zhou Y, Browning SR. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am J Hum Genet. 2018;103(3):338–48. Epub 2018/08/14. doi: 10.1016/j.ajhg.2018.07.015 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81(5):1084–97. Epub 2007/10/10. doi: 10.1086/521987 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. Epub 2015/03/06. doi: 10.1038/gim.2015.30 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howe KL, Achuthan P, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, et al. Ensembl 2021. Nucleic Acids Res. 2021;49(D1):D884–D91. Epub 2020/11/03. doi: 10.1093/nar/gkaa942 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Y, Mkrtchian S, Kumondai M, Hiratsuka M, Lauschke VM. An optimized prediction framework to assess the functional impact of pharmacogenetic variants. Pharmacogenomics J. 2019;19(2):115–26. Epub 2018/09/13. doi: 10.1038/s41397-018-0044-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
Data cannot be shared publicly because of National policy. Data are available from the King Faisal Specialist Hospital and Research Centre Institutional Data Access / Ethics Committee (contact via ora@kfshrc.edu.sa) for researchers who meet the criteria for access to confidential data.