Abstract
The high incidence of glioblastoma recurrence necessitates additional therapeutic strategies. Heterogeneous populations of cells, including glioma stem cells (GSC) have been implicated in disease recurrence. GSCs are able to survive irradiation and temozolomide (TMZ) treatment due to upregulation of DNA damage pathways. One potential strategy to target treatment-resistant tumor populations may be via the integrated stress response (ISR). Modulation of the ISR pathway also allows for sensitization of treatment-resistant cells to TRAIL. We generated a novel cell-based death receptor assay to identify potent inducers of ISR-dependent DR5 expression. We used this assay to screen compounds from three commercially available libraries, and identified 1-benzyl-3-cetyl-2-methylimidazolium iodide (NH125) as a potent inducer of DR5 expression. NH125 engages the EIF2α–ATF4–CHOP axis culminating in DR5 expression at low micromolar doses. Expression of CHOP plays a critical role in NH125-mediated TRAIL synergy. Treatment of GSC with NH125 produces a marked reduction in viability when compared with other cell lines. NH125-treated GSC also synergize with lower doses of TRAIL when compared with all other cell lines tested. Transcriptional analysis of NH125-treated GSC uncovers a unique profile that involves activation of ISR and GADD45 pathways. Treatment of GSC xenografts with encapsulated PEG–PCL–NH125 leads to a sustained decrease in tumor volume.
Introduction
Glioblastoma multiforme (GBM) is a highly aggressive brain tumor that is accompanied by a poor prognosis. Management typically involves maximally achievable surgical resection followed by adjuvant chemotherapy and radiotherapy. This tri-modality treatment results in a median patient survival of about 14.6 months, with a 5-year survival rate of 9.8% (1, 2). Despite the availability of aggressive treatment options, recurrence is nearly universal (3). Heterogeneous populations of cells, including glioma stem cells (GSC), are thought to confer treatment resistance. Several studies have demonstrated the presence of stem cells in both pediatric and adult brain tumors (4–6). GSC are able to survive irradiation and temozolomide (TMZ) treatment due to upregulation of DNA damage pathways and other unique properties (7, 8). Intracranial implantation of GSC in rodents leads to the development of tumors that mimic the features of malignant gliomas (4–6). These findings suggest that GSC that survive adjuvant treatment may contribute to the recurrence of GBM.
Identifying therapeutic strategies that complement standard of care treatment may prove effective in addressing GSC. TRAIL has shown promise in targeting a variety of malignant cells while leaving normal cells unharmed (9, 10). TRAIL induces cell death through activation of the extrinsic apoptotic pathway following binding of death receptors DR4 and DR5 (11, 12). Treatment with TRAIL bypasses many of issues leading to GBM recurrence, and may effectively complement standard of care treatment. However, implementation of TRAIL in the clinical setting has proven challenging. Phase I and II clinical trials have demonstrated that TRAIL is well tolerated, but does not produce significant antitumor activity (13, 14). Lack of efficacy may be due to a variety of mechanisms, such as upregulation of cellular FLICE inhibitory proteins (cFLIP) and inhibitors of apoptosis (IAP; refs. 13, 14). TRAIL sensitizers may prove advantageous in overcoming some of these resistance mechanisms.
Increased expression of death receptors is an established method of TRAIL sensitization. Signaling through the integrated stress response (ISR) pathway bypasses p53-dependent DNA damage pathways leading to the expression of C/EBP Homologous Protein (CHOP) and DR5 (15–17). Melanoma cells treated with Tunicamydn, a known ER stress agent, demonstrate upregulation of ISR intermediates leading to enhanced DR5 expression and TRAIL sensitization (17). Similarly, Bortezomib was shown to upregulate key ISR intermediates leading to TRAIL sensitization in glioma and pancreatic cancer cells (18, 19). Previously, we have shown that Nelfinavir leads to DR5-dependent TRAIL synergy via expression of CHOP (20). These data affirm that ISR-mediated DR5 expression is an effective means of TRAIL sensitization. However, exploration of the ISR pathway in GSCs has been limited. We propose that ISR inducers will uncover novel therapeutic strategies in GSCs, which may help in eradicating these treatment-resistant cells.
Materials and Methods
Cell lines and reagents
U251, A549, H1299, H1703, and Ovcar3 were obtained from the ATCC. GL261 was obtained from the NCI Division of Cancer Treatment and Diagnosis database. U87 was gift from Dr. Laura Johnson (UPenn, Philadelphia, PA). Normal human astrocytes (NHA) were a gift from Timothy Chan, MD PhD (MSKCC, New York, NY). T4213 GSC was a gift from Dr. Yi Fan (UPenn), NS039 and HK296 GSC were gifts from Dr. Harley Kornblum (UCLA, Los Angeles, CA).
U251, NHA, and GL261 were cultured in DMEM (4.5 g/L glucose; Invitrogen) in the presence of 10% FBS. H1299, H1703, and Ovcar3 were cultured in RPMI1640, supplemented with 25 mmol/L Hepes and 10% FBS. A549 were cultured in Ham’s F12K in the presence of 10% FBS. U87 were cultured in improved MEM, supplemented with 10 mmol/L Hepes and 10% FBS. T4213 were cultured in Neurobasal A supplemented with 0.5X B-27 without vitamin A (Invitrogen), 1 mmol/L sodium pyruvate, 1 × Glutamax, 20 ng/mL EGF, and 20 ng/mL FGF (Peprotech). NS039 and HK296 were cultured in DMEM/F12 supplemented with 1 × B-27, 1.0 mmol/L pyruvate, 50 ng/mL EGF, 20 ng/mL FGF, and 5 μg/mL Heparin Sulfate (Sigma Aldrich). All media were supplemented with 100 units/mL pen-icillin and 100 mg/mL streptomycin (Invitrogen). All cells were cultured in a humidified incubator at 37° C and 5% CO2. Cell lines were certified mycoplasma free by the MycoAlert Assay (Cambrex) on a regular basis.
1-Benzyl-3-cetyl-2-methylimidazolium iodide (NH125) and Tunicamycin were purchased from Abcam. Temozolomide, Salubrinal, and Nelfinavir were purchased from Sigma-Aldrich. Stock solutions of all compounds were generated using DMSO and stored at −20°C.
Creation of reporter cell line
The pGL3-DR5 plasmid was a gift from Dr. Hong-Gang Wang (Penn State, Hershey, PA; ref. 16). This plasmid contained a DR5 5’ flanking sequence (−552 to −7) with a CHOP binding region and mutant NF-kB region (Supplementary Fig. S1). The DR5 5’ flanking sequence was subcloned into the pGL4.21 plasmid (Promega) upstream of the luciferase reporter using a Rapid DNA Ligation Kit (Roche). DH5α containing the DR5-Luc plasmid were expanded under ampicillin selection followed by DNA extraction using QIAprep Miniprep Kit (Qiagen). Insertion of the DR5 sequence was confirmed using an E-Gel following restriction digestion with Sac1 and Xho1. Selected clones were expanded in LB Broth under ampicillin selection followed by DNA extraction using a Qiagen Maxi Prep Kit. U251 cells were transfected with the DR5-Luc plasmid using Lipofectamine 2000 (Invitrogen), and grown under puromycin (2 μg/mL) selection. Single clones were selected and expanded using DMEM supplemented with Puromycin (1 μg/mL). U251 DR5-Luc clones were probed for luciferase expression following Nelfinavir treatment, leading to identification of a single positive clone. Dose-dependent expression of DR5 and Luciferase in this clone was confirmed via Western Blot analysis and bio-luminescent imaging (IVIS Spectrum; PerkinElmer).
Cell-based death receptor screen
U251 DR5-Luc cells were plated at 1.0 × 104 cells/well in 96-well plates. Plates were then incubated in a humidified environment at 37°C and 5% CO2 for 24 hours. Compound from three libraries, the InhibitorSelect Kinase Library III (Calbiochem), and the ScreenWell Redox and Protease Inhibitor Libraries (Enzo Clinical Labs) were added to each plate at 2 or 10 μmol/L. Tunicamycin (1 μg/mL) was used as a positive control, whereas DMSO (1%, v/v) served as a negative control. Steady-Glo reagent was added to plates 24 hours after addition of compound, and plates were imaged on an IVIS Lumina II. The mean (μ) and standard deviation (σ) for each plate was used to transform total flux for each compound (xi) into z-scores using the formula z = (xi - μ)/σ. Calculation of Z-prime for this screening assay was based upon the formula set forth by Zhang and colleagues utilizing Tunicamycin and DMSO as a positive and negative controls, respectively (21).
Western blot analysis
Cells were lysed in the presence RIPA buffer (Thermo Scientific) supplemented with 1 mmol/L PMSF, 1 mmol/L sodium orthovanadate, 1 × Complete MINI (Roche). Protein concentration was determined using a BSA standard curve and Pierce 660 nmol/L reagent. Denatured protein was then loaded on a 4% to 12% Bis-Tris gel (Invitrogen), and subsequently transferred onto a PVDF membrane. Membranes were blocked in non-fat milk followed by incubation in primary antibody overnight at 4°C. Secondary antibody incubation was for 1 hour at room temperature. All antibodies were obtained from Cell Signaling Technology with the exception of ATF3 (Santa Cruz Biotechnology) and Ran (BD Biosciences). Membranes were incubated in ECL Prime (GE Healthcare) and exposed to autoradiography film. Films were then developed using Konica Minolta SRX-101A x-ray developer. Membranes were re-probed following incubation with Western Blot Restore Buffer (Thermo Scientific). Films were scanned at high resolution (600 dpi) and densitometry was performed using Image Studio Lite (Version 5.2; Li-Cor).
Generation of CHOP knockout cell line
U251 CHOP Knockout cells were generated using the CRISPR-Cas9 system (GeneCopoeia). Plasmids containing single sgRNAs along with the Cas9 endonuclease were stably introduced into U251 using Endofectin (GeneCopeiea). Single colonies were isolated following selection with 700 μg/mL of G418 and capture of mCherry positive cells using the Kuiqpick system (NeuroInDx). Efficacy of different sgRNA/Cas9 combinations was assessed using the T7 Endonuclease Assay (New England Biolabs). Of note, U251 clones containing two sgRNA demonstrated the most efficacious targeting of CHOP. Promising clones were treated with 2.5 and 5.0 μmol/L NH125 and 1.0 μg/mL Tunicamycin. Protein was collected and analyzed for expression of ATF4, CHOP, and DR5 via Western blot analysis. Absence of CHOP protein and abrogated DR5 expression signified successful knockout of CHOP.
CHOP knockdown in GSC
T4213 were transfected with either nontargeting pooled siRNA or pooled siRNA against CHOP (GE Dharmacon) for 48 hours using Lipofectamine RNAiMAX (Invitrogen). Cells were then incubated with either 0.1% DMSO or 2.5 μmol/L NH125 during the final 24 hours of transfection followed by cell lysis. Lysate was then probed for key ISR signaling intermediates to assess the impact of CHOP knockdown.
Compound dilution studies, cell viability, and caspase activation
Cells were plated at density of 1.0 × 104 cells/well in 96-well plates using their respective media. Plates were then incubated in a humidified environment at 37°C and 5% CO2 for 24 hours. An 11-point dilution series of NH125 from a high dose of 40 μmol/L down to 0.039 μmol/L was added to each plate. Cells treated with 0 μmol/LNH125 (0.1% DMSO) served as a vehicle control. Plates were incubated with NH125 for 24 hours at 37°C and 5% CO2, followed by addition of Cell Titer Glo (Promega) or Caspase 3/7-Glo (Promega). Bio-luminescent readings were obtained using a Synergy HT luminometer (BioTek). Raw bio-luminescent values were normalized to their vehicle-treated controls, and then fit to a modified Hill Equation using OriginPro 8 (OriginLab) in order to calculate IC50 values. All dilution series experiments were performed as biological triplicates, and repeated at least three times.
To study the effects of serum on GSC, cells were plated in the presence of 10% FBS for 24 hours, and then treated in the presence of 10% FBS for an additional 24 hours, followed by addition of Cell Titer Glo.
In separate experiments, T4213 cells were co-incubated with NH125 and 10 μmol/L Salubrinal for 24 hours at 37°C and 5% CO2, followed by addition of Cell Titer Glo.
Statistical significance was determined in OriginPro8 using unpaired t tests, with P values less than 0.05 considered significant.
Sphere formation assay
T4213 cells were plated at 2,000 cells/well in 6-well plates, and then allowed to incubate overnight at 37°C and 5% CO2. Cells were treated with 0.1% DMSO, 2.5 μmol/L NH125, or 250 μmol/L TMZ in biological triplicate for 7 days. On day 7, individual wells were imaged on a Lionheart Fx microscope at 4 × magnification following addition of NucBlue (Hoechst 33342; Invitrogen). Circular blue objects greater than 50 μm in diameter were counted as neurospheres. Neurosphere counts were then divided by the total number of circular blue objects greater than 10 μm to obtain a percentage of sphere formation. Phase contrast images of neurospheres were taken under 10× magnification 24 hours after treatment with NH125 or Temozolomide.
TRAIL synergy
NH125-treated U251 and U251 CHOP knockout cells were co-incubated with 25 ng/mL of TRAIL (PeproTech) for 4 hours. NH125-treated U251, T4213, and NS039 were also co-incubated with 5.0 ng/mL TRAIL for 4 hours. In separate experiments U251, CHOP knockout, T4213, and NS039 cells were treated with a TRAIL only dilution series for 4 hours. Cell viability was determined after 24 hours using Cell Titer Glo and the Synergy HT plate reader. The combination index method was used to assess NH125 and TRAIL synergy (22). Combination indices were calculated using CompuSyn with values less than <1.0 being indicative of TRAIL synergy.
Immunofluorescence
Individual GSCs were cystospun onto Thermo Shandon coated slides using Rotofix 32a (Hettich) centrifuge at 800 RPM for 5 minutes. Cells were fixed with 4% paraformaldehyde and incubated in blocking buffer for 1 hour at room temperature, followed by incubation in biotin conjugated anti-CD133 (Miltenyi Biotec) overnight at 4° C. Secondary antibody dilution was for 1 hour at room temperature using a Strepavidin conjugated Alexa Fluor antibody (Thermo Fisher Scientific). Slides were mounted using Vectashield with DAPI (Vector Labs), and imaged under 63X (oil immersion) using a Zeiss Axio Observer inverted fluorescent microscope.
T4213 neurospheres were plated in single wells of a poly-D-lysine culture slide. Culture slides were then incubated overnight in a humidified environment at 37°C and 5% CO2. Slides were fixed with 4% paraformaldehyde, and then incubated overnight at 4°C with primary antibodies against SOX2 (Cell Signaling Technology) and Nestin (Abcam).
Whole transcriptome analysis
U251, NHA, and T4213 were treated with 0.1% DMSO or 2.5 μmol/L NH125 in biological triplicate. Total RNA was isolated using the NucleoSpin RNA Kit (Macherey-Nagel) after 24 hours of treatment. RNA concentration and quality was assessed using a NanoDrop ND-1000 before determining the integrity of each sample on an Agilent 2100 Bioanalyzer. Sequencing libraries were assembled from 500 ng of RNA using the QuantSeq 3’ mRNA-Seq Library Prep Kit FWD (Lexogen). Quality control of the libraries was performed on an Agilent 2100 Bioanalyzer followed by quantification with the KAPA Library Quantification Kit. Libraries were sequenced on an Illumina NextSeq 500 with the help of the Wistar Genomics Core (University of Pennsylvania), generating single-end 75-bp reads. These reads were trimmed and aligned to the reference genome GRCh38 using the Bluebee data analysis pipeline. The resulting read counts were assembled in count matrices in R, followed by normalization and differential gene expression using the DESeq2 package. Heatmaps were generated using R, and pathway analysis was performed using ingenuity pathway analysis (IPA) software (Qiagen). Raw and processed read counts were uploaded to NCBI Gene Expression Omnibus under accession number GSE102505.
Generation of PEG-PCL encapsulated NH125
Encapsulation of NH125 in polyethylene glycol-polycaprolactone (PEG-PCL) was performed by the Chemical and Nanoparticle Synthesis Core at the University of Pennsylvania. NH125 was dissolved in chloroform at 20 mg/mL. Separately, PEG (4000)-PCL (3000) copolymer was dissolved in chloroform at a concentration of 50 mg/mL. A solution (200 μL) of PEG-PCL (4 mg) and NH125 (1 mg) was added directly to a glass vial containing 4 mL of Millipore water, and the resulting mixture was sonicated for approximately 5 minutes. The emulsion was then allowed to stand overnight to evaporate the chloroform. The mixture was purified by dialysis (molecular weight cutoff of 3.5 kDa) against water for 24 hours to remove free NH125. Drug loading efficiency (DLE%) and drug loading content (DLC%) were calculated according to the following formulas: DLE (wt%) = (weight of loaded drug/weight of drug in feed) × 100%, DLC (wt%) = (weight of loaded drug/weight of drug loaded micelles) × 100%. The size and size distributions of the NH125 micelles were measured with dynamic light scattering (DLS) in water at 25°C.
Tumor implantation in mice and NH125 treatment
All animal work was approved by the Institute for Animal Care and Use Committee at the University of Pennsylvania. NS039 cells were resuspended in PBS, and then 1.5 × 106 cells were injected into the right flank of 8-week-old female athymic nude mice (Charles River Laboratories). Tumor dimensions were measured on a weekly basis using digital calipers following 2 weeks of growth. Tumor volume was calculated using the formula V = 0.5 × (L × W2). Once tumors reached a volume of approximately 50 mm3, mice were randomly divided into two groups, and treated with either 3 mg/kg empty PEG-PCL (vehicle) or PEG-PCL containing NH125 (PEG-PCL-NH125). All treatments were administered via an intratumoral route in anesthetized mice. A total of two treatments were administered, with each treatment separated by 24 hours. Mice were weighed at least twice weekly throughout all experiments. Statistical comparison was performed on tumor volumes from day 45 using an unpaired t test assuming unequal variance. P values less than 0.05 were considered significant.
Results
NH125 is a potent inducer of DR5 expression
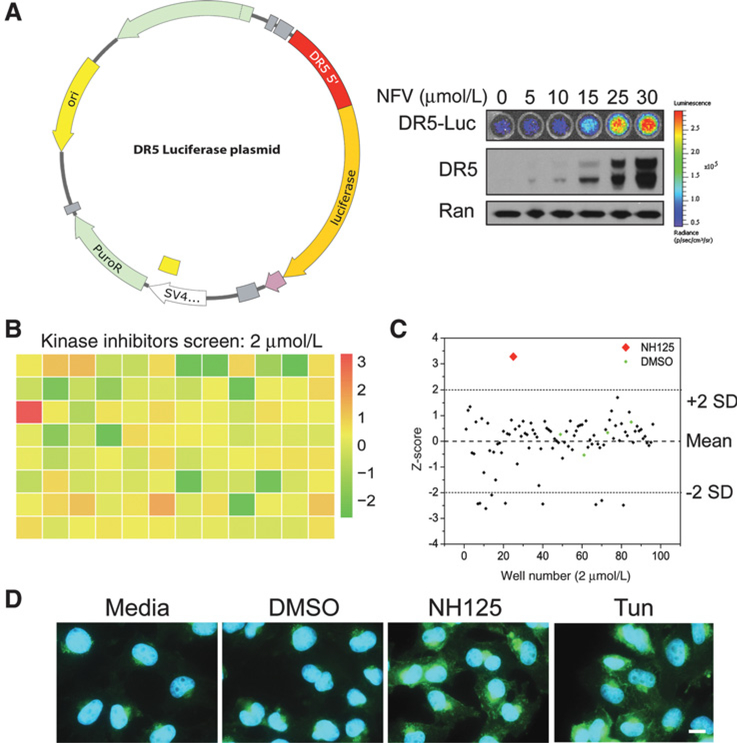
We developed a reporter system to identify CHOP-mediated inducers of DR5 (Fig. 1A). We validated reporter function using Nelfinavir, which has been shown induce DR5 expression through modulation of CHOP (20). Both immunoblot and bio-luminescent assays demonstrated dose-dependent increases in DR5 expression in response to Nelfinavir (Fig. 1A). Following validation of our reporter cell line, we sought to identify potent inducers of DR5 expression by screening libraries of known kinase, redox, and protease inhibitors. A total of 223 compounds were screened at two concentrations (2 and 10 μmol/L), generating a single promising hit (Fig. 1B; Supplementary Fig. S1). Of note, our screening assay had a Z’-factor of 0.64 using Tunicamycin and DMSO as positive and negative controls, respectively (21). The single hit came from a plate containing kinase inhibitors screened at 2 μmol/L, and was identified as NH125. NH125 produced a bio-luminescent signal that was greater than two SDs above the plate mean (Fig. 1C). NH125 was included on the kinase inhibitor plate due to its characterization as an eEF2 kinase inhibitor (23). However, recent studies suggest that this classification may not capture its true mechanism (24, 25). Bio-luminescent imaging of an NH125 dilution series revealed peak activity at 2.5 μmol/L in U251 (Supplementary Fig. S2). Decreasing bio-luminescence and cell viability were seen at higher doses (Supplementary Fig. S2). Increases in DR5 protein following NH125 treatment were confirmed using immunofluorescence (Fig. 1D). Increased DR5 expression was also observed in cells treated with 1.0 μg/mL Tunicamycin, but absent in DMSO- and mock-treated cells (Fig. 1D).
Figure 1.
NH125 is identified as a potent inducer of death receptor 5 (DR5) expression. A, The 5’ flanking sequence of DR5 (red region) was subcloned into the PGL4.21 plasmid upstream of the luciferase reporter. This plasmid construct was then stably introduced into U251 cells. Bioluminescence and immunoblots were performed 24 hours following incubation with increasing doses of Nelfinavir. A dose-dependent increase in bioluminescence that closely matched DR5 expression was observed following Nelfinavir treatment. B, Raw bioluminescence values from U251 DR5 Luciferase cells incubated with kinase inhibitors (2 μmol/L) were normalized using z-scores. Normalized values were then displayed using a red-green-yellow heatmap to emphasize a single positive hit (red square). C, A scatter plot of z-scores demonstrates that a single compound produces a z-score of greater than two SDs above the plate mean. This compound would later be identified as NH125. D, Representative immunofluorescent images of U251 cells treated with media, 0.1% DMSO, 2.5 μmol/L NH125, or 1 μg/mL Tunicamycin (Tun) taken at 63× magnification (scale bar = 15 μm). Images are presented as an overlay of DAPI (blue) and DR5 (green), with both NH125 and Tun leading to increased DR5 expression when compared with their vehicle-treated counterparts.
Increased DR5 expression is due to upregulation of key ISR intermediates
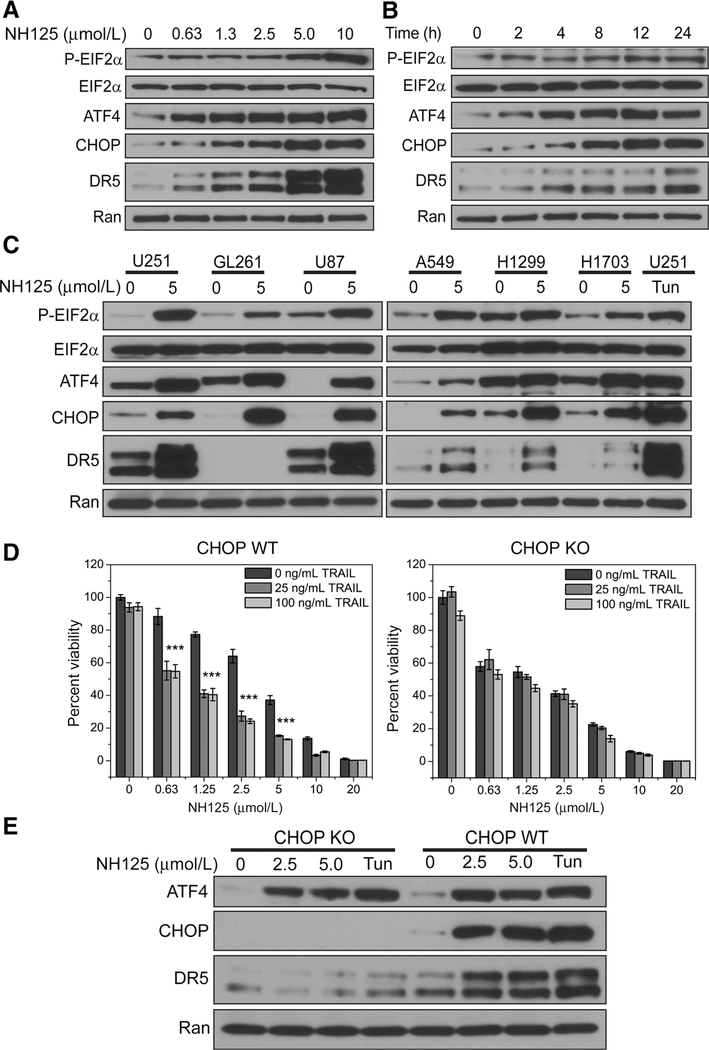
We chose to probe for upstream regulators of DR5 by examining key signaling intermediates EIF2α, ATF4, and CHOP. Treatment of U251 cells with increasing concentrations of NH125 lead to dose-dependent increases in EIF2α phosphorylation, ATF4, and CHOP expression at 24 hours (Fig. 2A). These dose-dependent increases in ATF4 and CHOP expression were accompanied by an increase in DR5 expression (Fig. 2A). Activation of the EIF2α-ATF4-CHOP pathway was noted as earlier as 2 hours, and continued to increase up to 24 hours (Fig. 2B). The EIF2α-ATF4- CHOP pathway was also activated in NH125-treated U87-MG and mouse-derived GL261 (Fig. 2C). Concomitant upregulation of DR5 was also seen in NH125-treated U87-MG (Fig. 2C). DR5 expression was not detected in GL261 due to poor specificity of antibodies against the mouse death receptor (Fig. 2C). Increases in EIF2α phosphorylation along with ATF4-CHOP-DR5 expression were also observed in A549, H1299, and H1703 at 24 hours (Fig. 2C). Tunicamycin-treated U251 served as a positive control demonstrating activation EIF2a-ATF4-CHOP signaling axis followed by DR5 expression (Fig. 2C). These findings demonstrate that NH125 induces phosphorylation of EIF2α followed by ATF4, CHOP, and DR5 expression in a broad range of cancer cell lines.
Figure 2.
NH125 treatment leads to key ISR signaling events culminating in CHOP-mediated DR5 TRAIL synergy. A, U251 cells were treated with increasing concentrations of NH125 for 24 hours and probed for ISR signaling events. Incubating U251 cells with increasing concentrations of NH125 produces an increase in EIF2α phosphorylation accompanied by an increase in ATF4, CHOP, and DR5 expression. B, In a parallel set of experiments, lysate from U251 incubated with 2.5 μmol/L NH125 for 0,2,4,8,12, and 24 hours was probed for ISR signaling events. EIF2a phosphorylation, ATF4, CHOP, and DR5 expression occur as early as 2 hours. C, EIF2a phosphorylation, ATF4, CHOP, and DR5 expression are observed across a panel of glioma and non-small cell lung cancer cells treated with 5 mmol/L NH125 for 24 hours. This pattern is also observed in U251 cells treated with 1 μg/mL of Tunicamycin for24 hours. D, U251 cells were treated with increasing concentrations of NH125 for 24, and then co-incubated with either 25 ng/mL or 100 ng/mL of TRAIL for 4 hours. Raw viability data were normalized to 0.1% DMSO-treated U251, and means and SDs from biological triplicates were plotted. Addition of 25 or 100 ng/mL of TRAIL produces a noticeable decrease in cell viability (*** indicates combination index <0.5, see Supplementary Table S1). Following successful knockout of CHOP, U251 cells were treated with increasing concentrations of NH125 for 20 hours, and then co-incubated with either 25 or 100 ng/mL of TRAIL for 4 hours. Raw viability data were normalized to 0.1% DMSO-treated CHOP knockout cells, and means and SDs from biological triplicates were plotted. Knockout of CHOP led to an abrogation of TRAIL-mediated cell death at both 25 and 100 ng/mL. E, Knockout of CHOP leads a dramatic reduction in DR5 expression following treatment with 2.5 and 5.0 μmol/L NH125 for 24 hours. Similarly, CHOP knockout cells treated with 1 μg/mL Tunicamycin for 24 hours also display decreased DR5 expression.
CHOP has a critical a role in DR5-dependent TRAIL synergy
Addition of 25 ng/mL of TRAIL to NH125-treated U251 cells leads to a decrease in cell viability across a range of NH125 concentrations (Fig. 2D). Increasing the concentration of TRAIL to 100 ng/mL did not lead to a further decrease in cell viability, suggesting that 25 ng/mL of TRAIL is sufficient to produce a synergistic interaction (CI < 0.5; Fig. 2D; Supplementary Table S1). Knockout of CHOP led to abrogation of TRAIL synergy following addition of both 25 and 100 ng/mL of TRAIL (CI > 1.0; Fig. 2D; Supplementary Table S1). Lysate of NH125-treated CHOP knockout cells revealed absent CHOP protein and diminished DR5 expression (Fig. 2E). U251 CHOP knockout cells also demonstrated increased sensitivity to NH125 treatment when compared with their wild-type counterpart (Fig. 2D).
GSC proliferation is greatly diminished following in vitro NH125 treatment
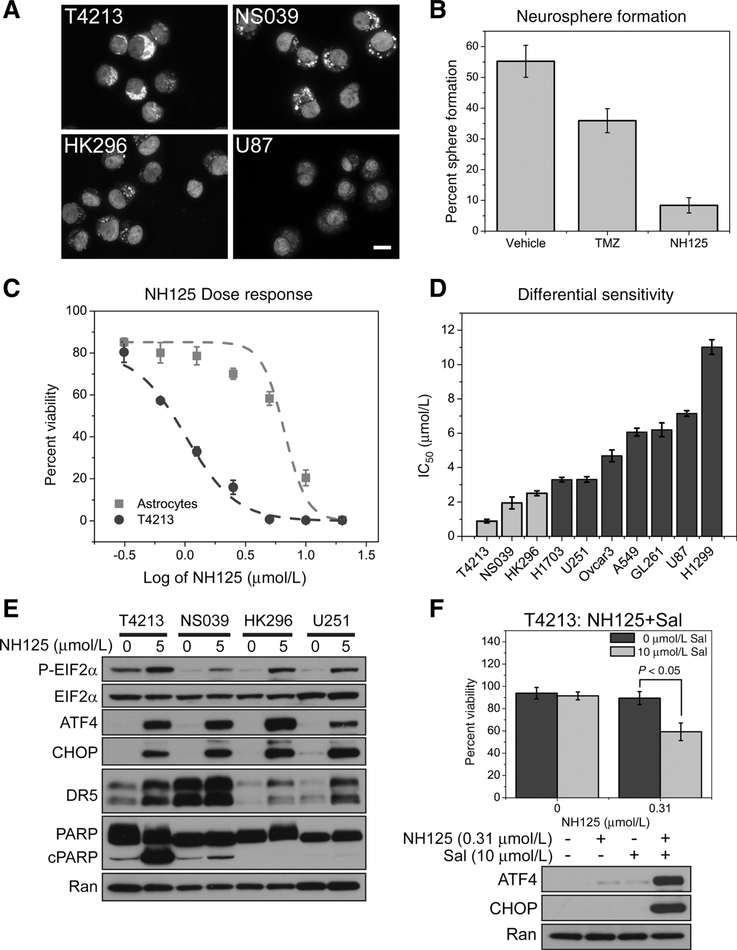
We explored the effect of NH125 treatment in a panel of GSC. GSC are glioma-derived tumor cells that express stem cell markers, demonstrate a capacity for self-renewal, and recapitulate tumor formation in animal models (26–29). GSC present a therapeutic challenge due to their resistance to DNA-damaging agents, and the ability initiate tumor formation (7, 8). Our panel of GSC exhibit varying degrees of CD133 expression, and form neurospheres in culture (Fig. 3A; Supplementary Fig. S3). T4213 also demonstrate SOX2 and Nestin expression (Supplementary Fig. S3). T4213 cells exhibited clonal proliferation into neurospheres following both vehicle (0.1% DMSO) and Temozolomide treatment. However, sphere formation was dramatically reduced from 55% following vehicle treatment down to 8% following NH125 treatment (Fig. 3B). Phase contrast images reveal largely intact neurospheres following Temozolomide treatment (Supplementary Fig. S4). However, NH125 treatment resulted in a striking loss of neurosphere integrity, with cells acquiring a shrunken and fragmented morphology (Supplementary Fig. S4).
Figure 3.
GSC that enrich for CD133 experience a reduction in viability following NH125-mediated ISR signaling. A, Representative immunofluorescence images of untreated GSCs taken under 63× magnification (scale bar = 15 μm). Images are presented as an overlay of CD133 (cytoplasmic staining) and DAPI (nuclear staining), showing that each stem cell line has varying degrees of CD133 expression when compared with U87. B, Assaying for sphere formation following a 7-day treatment with either vehicle, TMZ or NH125 reveals a decrease in neurospheres following NH125 treatment. Means and SDs are calculated from biological triplicates. C, An identical NH125 dilution series is applied to both NHAs and T4213 GSC, revealing a six-fold decrease in the IC50 for T4213. Data points represent means and SDs of normalized viability calculated from biological triplicates. D, IC50 values from a panel of NH125-treated cell lines highlight GSC (light gray) sensitivity to NH125. Means and SDs are calculated from three independent experiments. E, GSC and U251 were incubated with either 0 μmol/L NH125 (0.1% DMSO) or 5 μmol/L NH125 for 24 hours. Lysate was then probed for ATF4, CHOP, DR5, and PARP cleavage. All cell lines demonstrate an increase in ATF4, CHOP, and DR5 expression. PARP cleavage is observed in NH125- treated T4213 and NS039 at the time and concentration tested. F, DMSO and NH125 T4213 were co-incubated with 10 μmol/L Salubrinal (Sal) for 24 hours. Means and SDs are calculated from three independent experiments revealing a decrease in viability following the addition of Sal (P < 0.05). Only combination treatment of T4213 with NH125 and Sal produces an increase in ATF4 and CHOP expression.
GSC are more sensitive to in vitro NH125 treatment
NH125 treatment produced an approximately six-fold shift in the IC50 in T4213 when compared with NHAs, with only 33% viable cells at a concentration of 1.25 μmol/L (log10 = 0.1 μmol/ L; Fig. 3C). Treatment of T4213, NS039, and HK296 with an NH125 dilution series generated IC50’s of 0.89, 1.90, and 2.50 μmol/L, respectively (Fig. 3D; Supplementary Table S2). NH125 treatment also produced an increase in EIF2α phosphorylation accompanied byincreases in ATF4, CHOP, and DR5 in our panel of GSC (Fig. 3E). PARP cleavage was observed in NH125- treated T4213 and NS039, but absent in HK296 at the time and concentration tested (Fig. 3E). However, increases in Caspase 3/7 activity were observed in all NH125-treated GSC when compared with their vehicle-treated counterparts (Supplementary Fig. S5). By comparison, PARP cleavage and Caspase 3/7 activity were detected in U251 cells at 10 μmol/L (Supplementary Fig. S6). Salubrinal was utilized to establish the role of ISR signaling in GSC viability. Salubrinal has been shown to block EIF2aα dephosphorylation by selectively inhibiting protein phosphatase 1 (PP1; ref. 30). Co-incubation of T4213 cells with low-dose NH125 and 10 μmol/L Salubrinal led to an appreciable decrease in cell viability (P < 0.05, Fig. 3F). As expected, Salubrinal potentiated the expression of ATF4 and CHOP in T4213-treated with nanomolar concentrations of NH125 (Fig. 3F). These findings further implicate NH125 activation of the ISR pathway in GSC viability. We considered that growth in serum-free media may enhance GSC sensitivity to ISR inducing agents. Addition of serum caused T4213 cells to acquire a differentiated morphology accompanied by a three-fold increase in the IC50 from 0.89 to 2.71 micromolar (P < 0.05, Supplementary Fig. S7). However, removal of serum from U251 cells prior to treatment with NH125 had a negligible impact on the IC50 (Supplementary Fig. S7), suggesting that differentiation status is involved in GSC sensitivity to NH125.
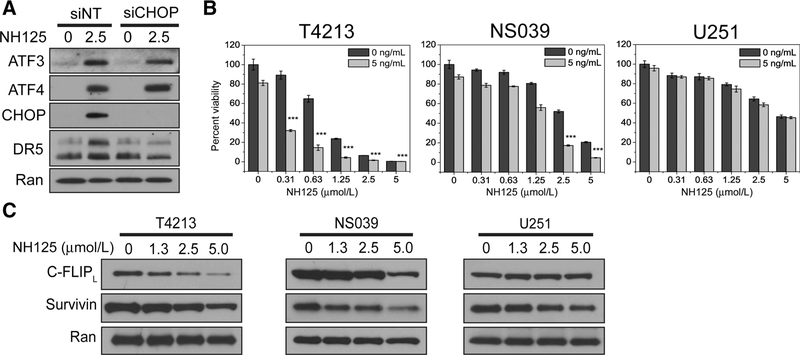
NH125 induces CHOP-mediated DR5 expression and TRAIL synergy in GSC
Pooled siRNAs were utilized to explore the role of CHOP expression in NH125-treated T4213. Incubation of T4213 with NH125 leads to an increase in DR5 expression at 24 hours. However, knockdown of CHOP in T4213 results in a decrease in DR5 expression following NH125 incubation (Fig. 4A). These findings further support a CHOP-mediated mechanism for DR5 expression in NH125-treated GSC. NH125-treated GSCwere also found to be exquisitely sensitive to TRAIL. Addition of 5 ng/mL TRAILwas sufficient to produce a synergistic interaction inT4213 and NS039 (Fig. 4A; Supplementary Table S3). However, this synergistic interaction was absent in NH125-treated U251 incubated with 5 ng/mL of TRAIL (Fig. 4A; Supplementary Table S3). This discrepancy in TRAIL sensitivity may be due to preferential degradation of IAP in NH125-treated GSC. Incubation of T4213 and NS039 with an NH125 dilution series produces a dose- dependent decrease in cFLIPL and Survivin (Fig. 4B). By comparison this dose-dependent effect is less prominent in NH125-treated U251 (Fig. 4B).
Figure 4.
NH125 treatment leads to CHOP-mediated synergy with low-dose TRAIL, and a decrease In C-FLIPL and Survlvln In GSC. A, T4213 transfected with either nontargeting or CHOP-specific pooled siRNAs were treated with either 0.1% DMSO or 2.5 μmol/L NH125 for 24 hours. An absence of CHOP leads to diminished DR5 expression following NH125 treatment despite intact ATF4 and ATF3. B, T4213, NS039, and U251 cells were treated with increasing concentrations of NH125 for 20 hours, and then co-incubated with 5 ng/mL of TRAIL for 4 hours. Raw viability data were normalized to cells treated with 0.1% DMSO, and means and SDs from biological triplicates are presented. Addition of 5 ng/mL of TRAIL produces a synergistic decrease in viability at nanomolar and low micromolar concentrations of NH125 in T4213 and NS039 (*** indicates combination index <0.5, see Supplementary Table S3). This synergistic decrease in viability is not observed in U251. C, A dose-dependent decrease in the long isoform of C-FLIP and Survivin is seen in NH125-treated T4213 and NS039 after 24 hours. A marginal decrease in C-FLIP and Survivin is observed in NH125-treated U251 after 24 hours.
Transcriptome analysis uncovers a unique profile in GSC
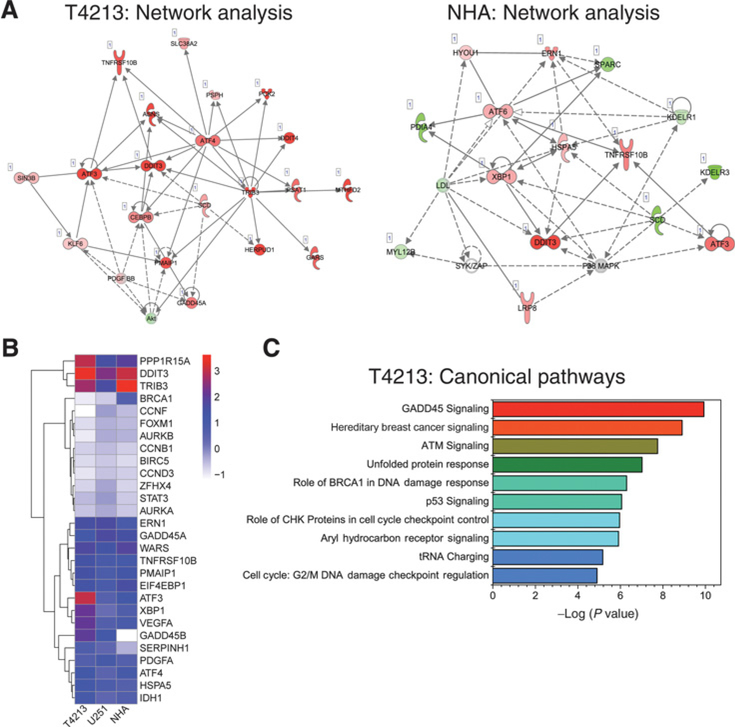
We performed whole transcriptome analysis to identify changes in gene expression following NH125 treatment. We utilized the QuantSeq 3’mRNA Library Kit from Lexogen due to its ability to accurately detect gene expression values. Analysis of differential gene expression using the IPA software package revealed a high score for the cellular compromise network in NH125-treated T4213 (score of 36). Key ISR transcripts, such as ATF4, TRIB3, DDIT3 (CHOP), and ATF3 were strongly upregulated in this network (Fig. 5A). Additionally, ATF4, TRIB3, ATF3, and CEBPB were identified as nodes that exhibited high levels of connectivity to other members of the network (Fig. 5A). For instance, ATF4 is linked to ATF3, and the pro-apoptotic molecules, DDIT3, TNFRSF10B (DR5), TRIB3, and PMAIP1. ATF4 is also indirectly linked to the prosurvival molecule GADD45A (Fig. 5A). In NH125-treated NHA, ATF6, XBP1, and HSPA5 (ER chaperone BIP) acted as the strongest nodes (Fig. 5A). This implicates involvement of the IRE1α-ATF6 and XBP1 pathways of the ER stress response in NH125-treated NHA. Increased expression of ATF3, DDIT3, and TNFRSF10B was common between both networks, with ATF3 expression highest in NH125-treated T4213 (Fig. 5A). Direct comparison of log2 fold change in a subset of genes with low FDR (<0.1) revealed an increase in DDIT3 expression inNH125 treated T4213, U251, and NHA (Fig. 5A). However, expression of ATF3, XBP1, PPP1R15A (GADD34), and GADD45B was highest in T4213, with ATF3 showing the largest change in expression (Fig. 5B). Interestingly, the top three canonical pathways identified in NH125-treated T4213 involved growth arrest and response to DNA damage (Fig. 5C). By comparison, EIF2a signaling is identified as one of the top canonical pathways in both NH125-treated U251 and NHA (Supplementary Fig. S8).
Figure 5.
Transcriptional profiling of NH125-treated GSC reveals unique connections between ISR intermediates and GADD45 signaling. A, A subset of genes with a low P-adjusted value (<0.1) underwent Core Analysis using IPA software. Analysis of the cellular compromise network in T4213 revealed increased expression (red shading), and strong connectivity (solid lines) of ATF4, TRIB3, ATF3, and DDIT3. Increased connectivity was seen with ATF6, HSPA5,XBP1, and ATF3 in NH125-treated NHA. B, A heatmap representation of differential gene expression from T4213, U251, and NHA treated with 2.5 μmol/L NH125. Genes for comparison were chosen from known ER and ISR mediators, cell-cycle regulators, and stem cell markers. Only genes that shared low P-adjusted values (<0.1) across all three cell lines were used to generate the heatmap. Each square is color coded based upon the log2fold change in gene expression between NH125-treated and vehicle-treated controls. Red coloring indicates the strongest expression, whereas blue and white coloring indicates weaker levels of expression. Rows are clustered using an unsupervised hierarchal clustering method based upon minimization of the Euclidean distance. All three cell lines showed similar increases in DDIT3 and TRIB3 expression. However, T4213 demonstrated increased expression in ATF3, PPP1R15A, XBPT, GADD45B, and VEGFA when compared with U251 and T4213. C, The 10 most significant canonical pathways from analysis of the transcriptional data of NH125-treated T4213. Significance is calculated using the Fischer exact test to determine gene sets that closely match pathways from Ingenuity’s Knowledge Base. A red-green-blue heatmap is used to emphasize the most significant canonical pathways.
PEG-PCL-NH125 micelle characterization
PEG-PCL encapsulation was performed to enhance delivery of NH125 to xenograft tumors and minimize delivery to healthy tissue. Stable PEG-PCL micelles containing NH125 were generated with an average hydrodynamic diameter of 25 ± 5 nm, and an average polydispersity index (PDI) of <0.2. The drug loading efficiency of PEG-PCL-NH125 micelles was between 60% and 65%. The drug loading content of PEG-PCL-NH125 micelles was between 10% and 13%. In vitro treatment of U251 with 2.5 μg/mL PEG-PCL-NH125 produced an increase in CHOP and DR5 when compared with PEG-PCL-treated U251 (Supplementary Fig. S9).
Injection of encapsulated NH125 decreases GSC tumor volume
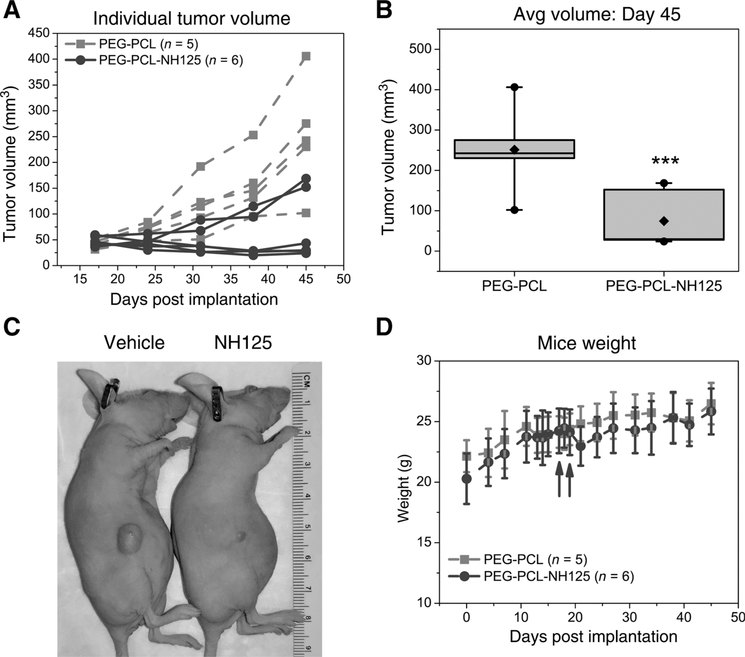
We performed intratumoral delivery of encapsulated NH125 to maximize its therapeutic window. Localized delivery of chemotherapeutic agents has been clinically validated through the use of carmustine loaded wafers (Gliadel) in glioma. Direct injection of 3 mg/kg PEG-PCL-NH125 to NS039 flank tumors produced a sustained decrease in tumor volume in four of six treated tumors over the course of 4 weeks (Figs 6A). Tumor volume was significantly decreased by an average of 177 mm3 in PEG-PCL-NH125 treated tumors when compared to PEG-PCL on day 45 (P < 0.05, Fig. 6B). Visual inspection revealed a dramatic reduction in gross tumor volume in four out of six mice 5 weeks following PEG-PCL-NH125 treatment (Fig. 6C). Both vehicle and PEG-PCL-NH125 treated mice continued to gain weight throughout the entire study (Fig. 6D).
Figure 6.
NS039 flank tumors dramatically shrink following treatment with PEG-PCL encapsulated NH125. A, Tumor volumes from individual NS039 flank tumors following intratumoral treatment with either 3 mg/kg PEG-PCL (n = 5 mice) or 3 mg/kg PEG-PCL-NH125 (n = 6 mice). Four of six PEG-PCL-NH125-treated flank tumors had tumor volumes less than 50 mm3 in the 4 to 5 weeks that tumor volume was tracked. B, Box plot representation of tumor volume on day 45 post-implantation. Boxes are bounded by the 25th and 75th percentile, and include the median tumor volume. Whiskers represent the 5th and 95th percentile of tumor volume. Black diamonds and circles represent the mean, minimum, and maximum tumor volumes, respectively. Comparison of average tumor volume on day 45 between PEG-PCL (n = 5) and PEG-PCL-NH125 treated tumors (n = 6), demonstrates an approximately 177 mm3 difference in tumor volume (*** indicates P < 0.05). C, A representative image of mice bearing a NS039 flank tumor following treatment with 3 mg/kg PEG-PCL or PEG-PCL-NH125. A dramatic reduction in tumor volume is observed 5 weeks (day 52 post-implantation) following treatment. D, Average weight of mice prior, during, and after treatment with 3 mg/kg intratumoral PEG-PCL and PEG-PCL-NH125. Dark gray arrows indicate when both treatments were delivered on day 17 and 19, respectively. Mice weight does not differ between the two treatment groups throughout the course of the study.
Discussion
We utilized our novel cell based death receptor assay to identify NH125 as a potent inducer of DR5 expression. We then demonstrate that NH125 treatment leads to activation of the integrated stress signaling pathway, which is required for DR5 expression. These findings represent a new mechanism of action for NH125 that is reproducible throughout a panel of cancer cells. Prior to our study, NH125 was identified as an eEF2 kinase inhibitor (23). Additional work demonstrated that the eEF2 mechanism may be responsible for TRAIL sensitization in glioma cell lines (31). However, subsequent studies found that NH125 treatment leads to an increase in eEF2 phosphorylation (24, 25). Furthermore, activation of ISR and ER stress signal pathways has been shown to lead to an increase in eEF2 phosphorylation. For instance, cells exposed to hypoxic stress signal through ATF4 leading to eEF2 phosphorylation (32, 33). Because ATF4 signaling halts global translation during cellular stress, an increase in eEF2 phosphorylation is expected in this setting (32, 33). Thus, increased eEF2 phosphorylation is expected if NH125 is truly behaving as an ISR inducing agent.
The ISR pathway also plays a critical role in NH125-mediated DR5 expression in a variety of cells regardless of p53 status. These findings support the conclusion that NH125 engages p53 independent pathways leading to DR5 expression and cell death. We found that this p53-independent mode of action was useful in addressing difficult to treat tumor populations, such as GSC. CD133 positive GSC have been shown to survive irradiation through increased expression of ATM and CHK1 (7). Furthermore, CD133 positive GSC are largely resistant to Temozolomide treatment (8, 34). Temozolomide treatment of T4213 GSC had a limited impact on clonal proliferation and neurosphere integrity in the in vitro setting. However, exposure to NH125 produced a dramatic loss in both clonal proliferation and neurosphere integrity. These suggest a role for NH125 and similar ISR inducing agents in targeting tumor populations that are resistant to DNA damaging agents.
NH125 modulation of ISR signaling allows for TRAIL synergy in a wide range of cell types. The mechanism behind this effect is multilayered. NH125 leads to an increase in DR5 expression primarily through modulation of CHOP (Fig. 7). In GSC, NH125 treatment also leads to a decrease in Survivin and cFLIPL (Fig. 7). Increased expression of DR5 is a proven strategy to sensitize tumor cells to TRAIL. Bortezomib has been shown to increase DR5 expression in NSCLC, and sensitize primary human astrocytomas to TRAIL (18,35). Similarly, ONC201 has been shown to upregulate ATF4, DR5, and sensitize a broad range of malignant cells to TRAIL (36). Thus, NH125 fits into the existing paradigm of modulating ISR and ER stress intermediates to drive DR5 expression. Modulation of DR5 expression may not be sufficient in overcoming TRAIL resistance in certain cells. Increased expression of cFLIPs and IAPs, such as Survivin, have been shown to confer resistance to TRAIL (13, 14, 37). Survivin and cFLIP disrupt signaling events during the caspase cascade by blunting the pro-apoptotic activity of effector caspases following activation of DR5 (13, 14). Targeting cFLIP and Survivin has shown promise in sensitizing TRAIL resistant melanoma and glioma cell lines (37, 38). Because NH125 produces a dose-dependent decrease in cFLIPL and Survivin in GSC, it may prove useful in targeting tumor cells that are resistant to TRAIL.
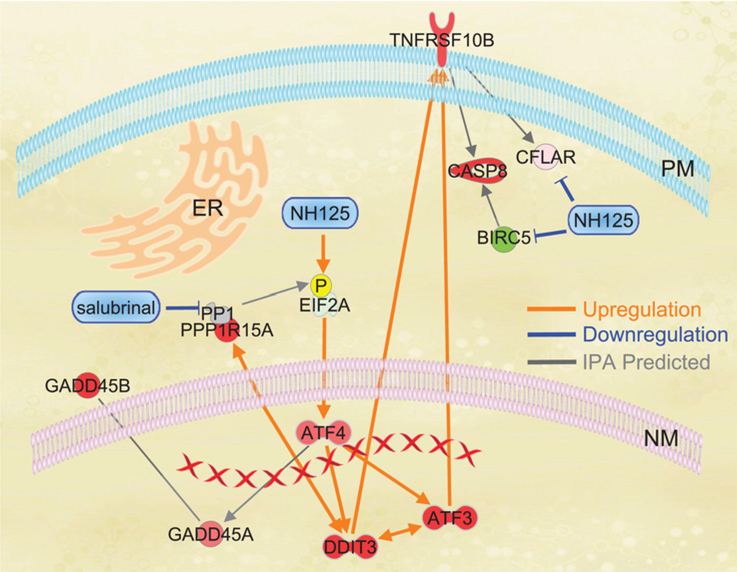
Figure 7.
Overlay of IPA expression data and integrated stress signaling events in NH125-treated GSC. Red and green shading are reflective of increased or decreased expression respectively as predicted by IPA. Orange lines are experimentally observed interactions that lead to an increase in expression. Blue lines are experimentally validated interactions that lead to a decrease in expression or degradation/inhibition of a signaling molecule. Gray lines are interactions predicted by IPA algorithms. Experimental data demonstrate that incubation with NH125 leads to phosphorylation of EIF2α, increased translation of ATF4, expression of DDIT3 (CHOP), and ATF3. Interconnection of ATF3 and DDIT3 was predicted by IPA with both signaling events contributing to an increase in TNFRSF10B (DR5) expression. Translation of ATF4 was found to be indirectly linked to increased expression of GADD45A and GADD45B through IPA. NH125 leads a decrease in BIRC5 (Survivin) and CLFAR (C-FLIP) protein. BIRC5 (Survivin) interacts with CASP8 (Caspase 8), and CLFAR (C-FLIP) interacts directly with TNFRSF10B. Whole transcriptome analysis and co-incubation studies with Salubrinal provide evidence that expression of DDIT3 and PPP1R15A (GADD34)/PP1 are interconnected. Co-incubation with Salubrinal inhibits PPP1R15A blocking de-phosphorylation of EIF2α.
Although NH125 treatment leads to classical ISR signaling events, prolonged ISR activation in the presence of Salubrinal leads to an appreciable decrease in GSC viability. This stands in contrast to the established findings that Salubrinal protects against ISR/ER stress by halting translation and allowing cells to recover from the stressor (30). However, Salubrinal has also been shown to enhance proteasome inhibitor mediated ER stress in multiple myeloma cell lines (39). The proposed mechanism for this effect involves restoring EIF2α phosphorylation thereby preventing cells from entering a quiescent state (39). We speculate that a similar mechanism maybe activated following NH125 treatment of GSC. NH125 treatment may lead to quiescence through ATF4-mediated GADD45 signaling (Fig. 7; 40). Transcriptional profiling of NH125-treated T4213 demonstrates strong nodal involvement of ATF4 in ISR signaling, including an indirect connection between ATF4 and GADD45A (Fig. 5A and Fig. 7). Furthermore, GADD45B is strongly upregulated in T4213 when compared with both U251 and NHA (Fig. 5A and Fig. 7). Both GADD45A and GADD45B have proven roles in protecting cells against the DNA damaging effects of UV irradiation, and controlling the G2-M checkpoint (41, 42). Our data would suggest that NH125 treatment may in fact lead to GADD45-mediated cell-cycle changes, and co-incubation with Salubrinal prolongs EIF2α phosphorylation shifting cells away from GADD45 induced quiescence towards apoptosis (Fig. 7). We can speculate that overcoming GADD45 signaling may allow for further reductions in GSC viability.
Changes in the ATF3 expression were also identified through transcriptional analysis. ATF3 has a broad range of activity in cancer cells. It has been shown to have an integral role in the stress response following PERK mediated phosphorylation of EIF2α (43). Stress-induced expression of ATF3 also leads to increased expression of pro-apoptotic molecules, such as TNFRSF10B (Fig. 7; refs. 44, 45). However, overexpression of ATF3 has been shown to have pro-survival advantage, even leading to a pro-metastatic phenotype in breast cancer cells (45, 46). In glioblastoma, ATF3 has been linked to TGF-β through BMI1, and stress-induced expression of ATF3 results in upregulation of NOXA (47, 48). We found that NH125 treatment leads to increased expression of ATF3 (Fig. 4A and Fig. 7). However, it is unclear whether ATF3 knockdown confers a survival advantage or sensitizes T4213 to NH125 treatment. Either scenario is possible given ATF3’s complex role in stress response signaling. Further exploration of ATF3 signaling in NH125-treated GSC remains a future area of study.
The mechanistic strategy of targeting cancer stem cells through manipulation of the ISR has been explored in several studies. Recently, inducers of ER stress have been shown to cause intestinal cancer stem cells to differentiate making them more susceptible to chemotherapy (49). We were able to re-characterize NH125 as an ISR inducing agent that exhibits unique behavior in GSC. We then demonstrate that this mechanism has exciting therapeutic implications for treatment resistant GSC. Direct application of PEG-PCL encapsulated NH125 to GSC flank tumors leads to a sustained reduction in tumor volume in four out of six mice. Although flank tumor models do not fully recapitulate the tumor microenvironment and blood brain barrier of brain tumors, our pre-clinical findings highlight the potential of NH125 against GSC in vivo. One could speculate that encapsulated NH125 could be applied to a resection cavity followed by systemic delivery of an immunomodulatory agent to treat residual disease. Direct injection of encapsulated NH125 to a distant disease site may be coupled with systemic therapy to elicit an abscopal response. This strategy has been explored in preclinical studies, where the delivery of immunomodulatory agents to the tumor microenvironment, along with the delivery of systemic immunostimulatory agents leads to the eradication of distant disease (50).
Nanoparticle encapsulation of NH125 may be necessary to accommodate its elongated hydrophobic tail, and improve its uptake in target tissues. Localized injection of encapsulated NH125 may be the best approach to achieve a meaningful therapeutic window in a clinical setting. In nude mice, direct injection of encapsulated NH125 did not produce any appreciable adverse health effects. Evaluation of blood and organ toxicity will be essential to determine the presence of systemic toxicity. Additional studies may also include absorption, distribution, metabolism, and excretion testing. However, we felt that these studies were beyond the scope of our present work. Regardless, our findings point to a promising and understudied therapeutic strategy of modulating the ISR to treat GSC, a resistant population of cells that have been implicated in glioblastoma recurrence.
Supplementary Material
Implications.
Taken together, these data suggest that engaging the ISR pathway represents a promising strategy to target treatment refractory GSC that have been implicated in glioblastoma recurrence.
Acknowledgments
The authors would like to acknowledge Harley Kornblum, MD, PhD, for generously sharing his patient-derived GSC lines; Gary D. Kao, MD, PhD, for his thoughtful insight and discussions; Rodrigo Gier for his assistance with Quantseq Library Prep and differential gene expression analysis; and Xiangsheng Xu and Bryan Manning for their help with immunofluorescence staining. This work was supported by: NIH/NINDS (K08 NS076548, to J.F. Dorsey), NIH/NCI (R01 CA201071, to J.F. Dorsey), NIH/NCI (R01 CA181429, to J.F. Dorsey and S. Sheikh), and the Burroughs Wellcome Career Award for Medical Scientists (1006792, to J.F. Dorsey), University of Pennsylvania NeuroOncology Innovation Pilot Grant (to J.F. Dorsey), University of Pennsylvania Department of Radiation Oncology HTS Pilot Grant (to J.F. Dorsey and S. Sheikh).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoom MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987–96. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapywith concomitant and adjuvanttemozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 2009;10:459–66. [DOI] [PubMed] [Google Scholar]
- 3.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med 2008;359: 492–507. [DOI] [PubMed] [Google Scholar]
- 4.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res 2003;63:5821–8. [PubMed] [Google Scholar]
- 5.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, DeVitis S, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 2004;64:7011–21. [DOI] [PubMed] [Google Scholar]
- 6.Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad SciUSA 2003;100:15178–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006;444:756–60. [DOI] [PubMed] [Google Scholar]
- 8.Liu G, Yuan X, Zeng Z, Tunici P, NgH, Abdulkadir IR, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer 2006;5:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M, et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med 1999;5:157–63. [DOI] [PubMed] [Google Scholar]
- 10.Kelley SK, Harris LA, Xie D, Deforge L, Totpal K, Bussiere J, et al. Preclinical studies to predict the disposition of Apo2L/tumor necrosis factor-related apoptosis-inducing ligand in humans: characterization of in vivo efficacy, pharmacokinetics, and safety. J Pharmacol Exp Ther 2001;299:31–8. [PubMed] [Google Scholar]
- 11.Allen JE, El-Deiry WS. Regulation of the human TRAIL gene. Cancer Biol Ther 2012;13:1143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, El-Deiry WS. TRAIL and apoptosis induction by TNF-family death receptors. Oncogene 2003;22:8628–33. [DOI] [PubMed] [Google Scholar]
- 13.Dimberg LY, Anderson CK, Camidge R, Behbakht K, Thorburn A, Ford HL. On the TRAIL to successful cancer therapy? Predicting and counteracting resistance against TRAIL-based therapeutics. Oncogene 2013;32:1341–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemke J, von Karstedt S, Zinngrebe J, Walczak H. Getting TRAIL back on track for cancer therapy. Cell Death Differ 2014;21:1350–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheikh MS, Burns TF, Huang Y, Wu GS, Amundson S, Brooks KS, et al. p53-dependent and -independent regulation of the death receptor KILLER/DR5 gene expression in response to genotoxic stress and tumor necrosis factor alpha. CancerRes 1998;58:1593–8. [PubMed] [Google Scholar]
- 16.Yamaguchi H, Wang HG. CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J Biol Chem 2004;279:45495–502. [DOI] [PubMed] [Google Scholar]
- 17.Jiang CC, Chen LH, Gillespie S, Kiejda KA, Mhaidat N, Wang YF, et al. Tunicamycin sensitizes human melanoma cells to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by up-regulation of TRAIL-R2 viathe unfolded protein response. Cancer Res 2007;67:5880–8. [DOI] [PubMed] [Google Scholar]
- 18.Koschny R, Holland H, Sykora J, Haas TL, Sprick MR, Ganten TM, et al. Bortezomib sensitizes primary human astrocytoma cells of WHO grades I to IV for tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Clin Cancer Res 2007;13:3403–12. [DOI] [PubMed] [Google Scholar]
- 19.Nawrocki ST, Carew JS, Pino MS, Highshaw RA, Dunner K Jr, Huang P, et al. Bortezomib sensitizes pancreatic cancer cells to endoplasmic reticulum stress-mediated apoptosis. Cancer Res 2005;65:11658–66. [DOI] [PubMed] [Google Scholar]
- 20.Tian X, Ye J, Alonso-Basanta M, Hahn SM, Koumenis C, Dorsey JF. Modulation of CCAAT/enhancer binding protein homologous protein (CHOP)-dependent DR5 expression by nelfinavir sensitizes glioblastoma multiforme cells to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). J Biol Chem 2011;286:29408–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang JH, Chung TD, Oldenburg KR. Asimplestatistical parameterforuse in evaluation and validation ofhigh throughput screening assays. J Biomol Screen 1999;4:67–73. [DOI] [PubMed] [Google Scholar]
- 22.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 1984;22:27–55. [DOI] [PubMed] [Google Scholar]
- 23.Arora S, Yang JM, Kinzy TG, Utsumi R, Okamoto T, Kitayama T, et al. Identification and characterization of an inhibitor of eukaryotic elongation factor 2 kinase against human cancer cell lines. Cancer Res 2003;63: 6894–9. [PubMed] [Google Scholar]
- 24.Chen Z, Gopalakrishnan SM, Bui MH, Soni NB, Warrior U, Johnson EF, et al. 1-Benzyl-3-cetyl-2-methylimidazolium iodide (NH125) induces phosphorylation of eukaryotic elongation factor-2 (eEF2): a cautionary note on the anticancer mechanism of an eEF2 kinase inhibitor. J Biol Chem 2011;286:43951–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devkota AK, Tavares CD, Warthaka M, Abramczyk O, Marshall KD, Kaoud TS, et al. Investigating the kinetic mechanism of inhibition of elongation factor 2 kinase by NH125: evidence of a common in vitro artifact. Biochemistry 2012;51:2100–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alan Mitteer R, Wang Y, Shah J, Gordon S, Fager M, Butter PP, et al. Proton beam radiation induces DNA damage and cell apoptosis in glioma stem cells through reactive oxygen species. Sci Rep 2015;5:13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarkaria JN, Carlson BL, Schroeder MA, Grogan P, Brown PD, Giannini C, et al. Use of an orthotopic xenograft model for assessing the effect of epidermal growth factor receptor amplification on glioblastoma radiation response. Clin Cancer Res 2006;12:2264–71. [DOI] [PubMed] [Google Scholar]
- 28.Laks DR, Crisman TJ, Shih MY, Mottahedeh J, Gao F, Sperry J, et al. Large- scale assessment of the gliomasphere model system. Neuro Concol 2016; 18:1367–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Develop 2015;29:1203–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science 2005;307:935–9. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Cheng Y, Zhang L, Ren X, Huber-Keener KJ, Lee S, et al. Inhibition ofeEF-2 kinase sensitizes human glioma cells to TRAIL and down-regulates Bcl-xL expression. Biochem Biophys Res Commun 2011;414:129–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. Hypoxia- induced energy stress regulates mRNAtranslation and cell growth. Mol Cell 2006;21:521–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L, Wise DR, Diehl JA, Simon MC. Hypoxic reactive oxygen species regulate the integrated stress response and cell survival. J Biol Chem 2008; 283:31153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beier D, Rohrl S, Pillai DR, Schwarz S, Kunz-Schughart LA, Leukel P, et al. Temozolomide preferentially depletes cancer stem cells in glioblastoma. Cancer Res 2008;68:5706–15. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Yue P, Chen S, Hu L, Lonial S, Khuri FR, et al. The proteasome inhibitor PS-341 (bortezomib) up-regulates DR5 expression leading to induction of apoptosis and enhancement of TRAIL-induced apoptosis despite up-regulation of c-FLIP and survivin expression in human NSCLC cells. Cancer Res 2007;67:4981–8. [DOI] [PubMed] [Google Scholar]
- 36.Kline CL, Van den Heuvel AP, Allen JE, Prabhu VV, Dicker DT, El-Deiry WS. ONC201 kills solid tumor cells by triggering an integrated stress response dependent on ATF4 activation by specific eIF2alpha kinases. Sci Signal 2016;9:ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chawla-Sarkar M, Bae SI, Reu FJ, Jacobs BS, Lindner DJ, Borden EC. Downregulation of Bcl-2, FLIP or IAPs (XIAP and survivin) by siRNAs sensitizes resistant melanoma cells to Apo2L/TRAIL-induced apoptosis. Cell Death Differ 2004;11:915–23. [DOI] [PubMed] [Google Scholar]
- 38.Son YG, Kim EH, Kim JY, Kim SU, Kwon TK, Yoon AR, et al. Silibinin sensitizes human glioma cells to TRAIL-mediated apoptosis via DR5 up-regulation and down-regulation of c-FLIP and survivin. Cancer Res 2007; 67:8274–84. [DOI] [PubMed] [Google Scholar]
- 39.Schewe DM, Aguirre-Ghiso JA. Inhibition of eIF2alpha dephosphorylation maximizes bortezomib efficiency and eliminates quiescent multiple myeloma cells surviving proteasome inhibitor therapy. Cancer Res 2009;69: 1545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang HY, Jiang L, Wek RC. The eukaryotic initiation factor-2 kinase pathway facilitates differential GADD45a expression in response to environmental stress. J Biol Chem 2007;282:3755–65. [DOI] [PubMed] [Google Scholar]
- 41.Gupta M, Gupta SK, Hoffman B, Liebermann DA. Gadd45a and Gadd45b protect hematopoietic cells from UV-induced apoptosis via distinct signaling pathways, including p38 activation and JNK inhibition. J Biol Chem 2006;281:17552–8. [DOI] [PubMed] [Google Scholar]
- 42.Jin S, Tong T, Fan W, Fan F, Antinore MJ, Zhu X, et al. GADD45-induced cell cycle G2-M arrest associates with altered subcellular distribution ofcyclin B1 and is independent of p38 kinase activity. Oncogene 2002;21: 8696–704. [DOI] [PubMed] [Google Scholar]
- 43.Jiang HY, Wek SA, McGrath BC, Lu D, Hai T, Harding HP, et al. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol Cell Biol 2004;24:1365–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edagawa M, Kawauchi J, Hirata M, Goshima H, Inoue M, Okamoto T, et al. Role of activating transcription factor 3 (ATF3) in endoplasmic reticulum (ER) stress-induced sensitization of p53-deficient human colon cancer cells to tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis through up-regulation of death receptor 5 (DR5) by zerumbone and celecoxib. J Biol Chem 2014; 289:21544–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka Y, Nakamura A, Morioka MS, Inoue S, Tamamori-Adachi M, Yamada K, et al. Systems analysis of ATF3 in stress response and cancer reveals opposing effects on pro-apoptotic genes in p53 pathway. PLoS One 2011;6:e26848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolford CC, McConoughey SJ, Jalgaonkar SP, Leon M, Merchant AS, Dominick JL, et al. Transcription factor ATF3 links host adaptive response to breast cancer metastasis. J Clin Invest 2013;123:2893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gargiulo G, Cesaroni M, Serresi M, de Vries N, Hulsman D, Bruggeman SW, et al. In vivo RNAi screen for BMI1 targets identifies TGF-beta/BMP-ER stress pathways as key regulators of neural- and malignant glioma-stem cell homeostasis. Cancer Cell 2013;23:660–76. [DOI] [PubMed] [Google Scholar]
- 48.Yoo YD, Lee DH, Cha-Molstad H, Kim H, Mun SR, Ji C, et al. Glioma-derived cancer stem cells are hypersensitive to proteasomal inhibition. EMBO Rep 2017;18:150–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wielenga MC, Colak S, Heijmans J, van Lidth de Jeude JF, Rodermond HM, Paton JC, et al. ER-stress-induced differentiation sensitizes colon cancer stem cells to chemotherapy. Cell Rep 2015;13:490–4. [DOI] [PubMed] [Google Scholar]
- 50.Kwong B, Gai SA, Elkhader J, Wittrup KD, Irvine DJ. Localized immunotherapy via liposome-anchored Anti-CD137 + IL-2 prevents lethal toxicity and elicits local and systemic antitumor immunity. Cancer Res 2013;73: 1547–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.