Abstract
Our pilot study examined global DNA methylation and telomere length (TL) using DNA from saliva samples provided by 39 participants in the Arkansas Rural Community Health (ARCH) Study. TL was quantified by qPCR, and DNA methylation and DNA methylation age was assessed using the Illumina 850K Epic BeadChip. Ingenuity Pathway Analysis (IPA) was performed to identify biological pathways that were DM between residents of counties with high or low poverty rates and by race [African American descent (AA) versus European American (EA) descent]. Among AA women, hypermethylation was more common in AA residents of counties with low compared to high poverty rates (70% vs. 30%). The top canonical pathways impacted by differential methylation were related to glucocorticoid receptor, p53, and estrogen-dependent breast cancer signaling in AA women. EA women living in low-poverty counties exhibited less hypermethylation of CpGs than those living in high-poverty counties (27% vs. 73%). The top canonical pathways were related to hereditary breast cancer, glucocorticoid receptor, androgen and PI3K/AKT signaling. Several genes involved in telomere maintenance were shown to be DM by county poverty levels. Therefore, the finding of this pilot study suggests county poverty levels may impact DNA methylation patterns in breast cancer-related pathways as well as genes involved in telomere maintenance. Larger studies should confirm our findings.
Keywords: Poverty, residence, genome-wide DNA methylation, breast cancer
Introduction
Living in disadvantaged neighborhoods is a risk factor for adverse health outcomes independent of individual socioeconomic status (1,2). Biological reasons for these geographic disparities are complex, and biomarkers that could signal the potential development of disease conferred by adverse neighborhood conditions are needed. DNA methylation (DNAm) and telomere length (TL) are two possible mechanisms. DNAm is an epigenetic regulator of gene expression that is responsive to environmental stimuli, such as exposures to smoking, arsenic contamination, and alcohol consumption (3,4), but few studies have examined if DNAm is associated with the large geographic variation in life expectancy and disease incidence (5,6). In addition to methylation of specific loci and genes, the concept of DNA methylation age acceleration (DNAmAA) in relation to health and life expectancy is an emerging area of investigation. An age predictor was developed by Horvath using DNAm data from multiple studies and several human tissues, including saliva (7), and DNAm aging has been hypothesized to be a risk factor for aging-related disease and mortality (7,8). In addition, TL, a marker of cellular aging, is a predictor of early mortality independent of biological age (9) as well as risk of cancer and other non-neoplastic diseases (10).
Few studies examined rural populations with aberrant DNAm or TL variability, although persons living in rural area are at increased risk of numerous adverse health outcomes (11-13). In this pilot study, we examined the association of county poverty rates on global DNAm, DNAmAA, salivary TL, and differences in DNAm of telomere-associated genes in Arkansas, a rural state. Since racial differences in genome-wide DNAm exist in healthy women (14), analyses were conducted separately for AA and EA women to assess their methylation status by county poverty levels.
Methods
Study population
The Arkansas Rural Community Health (ARCH) study is a study involving 23,735 Arkansas women recruited at community and cancer awareness events from 2007 to 2012 in both rural and metropolitan centers of Arkansas (15). After informed consent, participants completed questionnaires and provided saliva samples. The questionnaire captured demographic characteristics, breast cancer risk factors, and personal and family history of breast cancer.
Residential location of all participants were geocoded to identify their county using ArcGIS version 10 (Esri, Redlands, CA), as well as percent poverty rate at the census tract level (Figure S1). Ten women of self-reported AA descent each from counties with high poverty rates (>20% of the population) and low poverty rates (<10%) were randomly selected based on the 2008–2012 US Census American Community Survey (16), as well as ten women each of EA descent from high and low poverty rates. This study was approved by the Institutional Review Boards of University of Arkansas for Medical Sciences (IRB# 89071).
Figure S1.
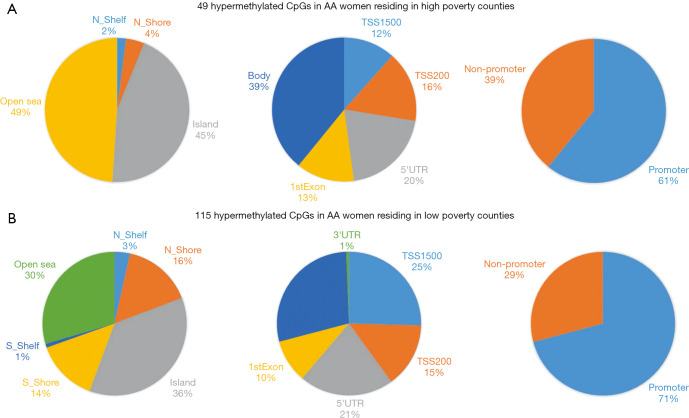
Distribution of differentially methylated CpGs by DNA regions. There were 49 unique hypermethylated CpGs in AA women residing in high poverty counties compared to 115 hypermethylated CpGs unique to low poverty county residence.
Saliva collection and DNA isolation
The saliva samples were collected using the Oragene DNA (OG-500), DNA Genotek, (Ottawa, ON, Canada). DNA was isolated according to the protocol for prepIT-L2P, purchased from the same manufacturer. A 500 µL aliquot of the saliva sample was mixed with 20 µL of prepIT-L2P and ethanol precipitated followed by dilution in Tris-EDTA buffer solution (Sigma-Aldrich, Saint Louis, MO, USA). The DNA samples were quantified on a NanoDropTM 8000 Spectrophotometer (Thermo Fisher Scientific, Watham, MA, USA).
Infinium methylation EPIC BeadChip analysis
Following bisulfite treatment of 1 µg genomic DNA using the EZ DNA Methylation kit (Zymo Research, Irvine, CA), the bisulfite-converted DNA was hybridized onto the Infinium Methylation EPIC BeadChip (Illumina, San Diego, CA) following the Illumina Infinium HD Methylation protocol. The Methylation EPIC BeadChip covers over 850,000 CpG sites with increased genome coverage of regulatory regions and high reproducibility and reliability from the previous versions (17). Whole genome amplification, hybridization, staining and scanning steps for all samples were performed, and the Illumina iScan SQ scanner created images of the single arrays. The intensities of the images were extracted using the Methylation module (v.1.9.0) of the GenomeStudio (v.2011.1) software (Illumina). Raw intensity data as IDAT files were imported into GenomeStudio for the computation of detection P value of the probes. Additional steps, including data import, normalization, filtering and analyses, were performed using the methylation pipeline in Partek Genomics SuiteTM 6.6 (Partek Inc., St. Louis, MO).
Estimation of DNAmAA
DNAm age is a robust measurement derived from the algorithms Horvath developed (7), and has been adapted by studies that used Illumina 450K platform (18,19) as well as the EPIC BeadChip used in this study (20). Briefly, the DNAm age were computed using the R script provided based on the methylation levels at 353 CpG sites (7) without re-training the model on the present data. DNAmAA was defined as the difference between DNAm age and chronological age.
Telomere length in salivary lymphocytes
Average TL was determined using the results of absolute quantitative real time polymerase chain reaction (qPCR) of the repeat copy number to single gene copy number (T/S) ratio (21,22). The mean and standard deviation were calculated using all TL and each sample (T/S) ratio was assigned to a category of high (≥1 standard deviation of the mean value) or low TL (≤1 standard deviation of the mean value) (23).
Data analyses
T-tests and Chi square tests were used to compare variables by county poverty levels and race. Analysis of variance (ANOVA) adjusted for differences on the covariate(s) with Fisher’s Least significant difference contrast method were employed to determine differentially methylated (DM) CpG sites. Hypermethylated CpGs were defined if the average methylation levels were higher than the compared group, and hypomethylated CpGs were defined if the average methylation levels were lower. For pattern recognition in global DNAm profiling, unsupervised hierarchical clustering and Principal Component Analysis (PCA) were used.
Pearson’s correlation was used to calculate the correlation between TL and the DNAmAA. To characterize the methylation patterns, the significant CpGs were divided by functional roles according to their genomic locations such as promoter: within 1,500 bps of a transcription start site (TSS) (TSS1500); within 200 bps of a TSS (TSS200); 5’ untranslated regions (5'UTR); first exon (1stExon); body (non-promoter); 3'UTR (non-promoter); and intergenic regions (24). Genes corresponding to promoter CpGs among the significant DM CpGs were analyzed for their potential biological implications using Ingenuity Pathway Analysis (IPA).
Results
Study population demographics
Biospecimens from 39 women who lived in high or low poverty counties were included. One EA woman was missing the residential info and thus was excluded when comparing poverty levels. The mean (± SD) age at study enrollment was 48.0±12.0 y, mean age at menarche was 12.9±1.5 y, and the mean age at parity was 17.7±9.3 y (Table 1). Most participants were not using birth control pills (84.6%), most had given birth (79.5%), were postmenopausal (56.4%), and did not have a family history of breast cancer (66.7%). In all, 66.7% were overweight, had some college or technical school (46.2%), and drank an alcoholic beverage at least once a month (56.4%). Differences in BMI existed between EA and AAs (P=0.01), in current hormone therapy (P=0.004), and alcohol use between high and low county poverty levels (P=0.03). The mean TL of all women was 0.4±0.02, the mean DNAm age was 79.3±9.9 y, and the mean DNAmAA was 31.3±6.9 (Table 1).
Table 1. Demographic and behavioral characteristics of study participants.
| Characteristics | All | By race | By poverty level*** | |||||
|---|---|---|---|---|---|---|---|---|
| EA (n=20) | AA (n=19) | P* | High (n=18) | Low (n=20) | P | |||
| Age (years), mean ± SD | 48.0±12.0 | 44.9±13.5 | 51.4±9.1 | 0.08 | 48.3±12.4 | 49.1±10.4 | 0.85 | |
| Age at menstrual (years), mean ± SD | 12.9±1.5 | 12.7±1.2 | 13±1.7 | 0.46 | 12.9±1.6 | 12.9±1.4 | 0.82 | |
| Birth control pills, n (%) | 1 | 0.99 | ||||||
| Yes | 4 (10.3) | 2 (10.0) | 2 (10.5) | 2 (11.1) | 2 (10.0) | |||
| No | 33 (84.6) | 17 (85.0) | 16 (84.2) | 15 (83.3) | 17 (85.0) | |||
| Unknown | 2 (5.1) | 1 (5.0) | 1 (5.3) | 1 (5.6) | 1 (5.0) | |||
| Ever given birth, n (%) | 0.95 | 0.43 | ||||||
| Yes | 31 (79.5) | 16 (80.0) | 15 (78.9) | 14 (77.8) | 17 (85.0) | |||
| No | 8 (20.5) | 4 (20.0) | 4 (21.1) | 4 (22.2) | 3 (15.0) | |||
| Age at first child (years), mean ± SD | 17.7±9.3 | 17.6±9.8 | 17.9±8.8 | 0.55 | 18.3±9.3 | 18±9.0 | 0.66 | |
| Number of children | 2.2±1.8 | 2.0±1.8 | 2.6±1.8 | 0.30 | 2.2±1.8 | 2.4±1.9 | 0.88 | |
| Menopausal status, n (%) | 0.92 | 0.99 | ||||||
| Premenopausal | 12 (30.8) | 6 (30.0) | 6 (31.6) | 6 (33.3) | 6 (30.0) | |||
| Postmenopausal | 22 (56.4) | 11 (55.0) | 11 (57.9) | 10 (55.6) | 11 (55.0) | |||
| Unknown | 5 (12.8) | 3 (15.0) | 2 (10.5) | 2 (11.1) | 3 (15.0) | |||
| Current hormone therapy, n (%) | 0.24 | 0.004 | ||||||
| Yes | 5 (12.8) | 4 (20.0) | 1 (5.3) | 0 (0) | 5 (25.0) | |||
| No | 24 (61.5) | 10 (50.0) | 14 (73.7) | 12 (66.7) | 11 (55.0) | |||
| Unknown | 10 (25.6) | 6 (30.0) | 4 (21.1) | 6 (30) | 4 (20.0) | |||
| Family history of breast cancer, n (%) | 0.57 | 0.53 | ||||||
| Yes | 12 (30.8) | 6 (30.0) | 6 (31.6) | 5 (27.8) | 7 (35.0) | |||
| No | 26 (66.7) | 14 (70.0) | 12 (63.2) | 13 (72.2) | 12 (60.0) | |||
| Unsure | 1 (2.6) | 0 (0) | 1 (5.3) | 0 (0) | 1 (5.0) | |||
| BMI (kg/m2) | 31.7±8.6 | 27.4±6.4 | 36.2±8.6 | 0.001 | 31.4±10.5 | 32.5±6.5 | 0.69 | |
| Education, n (%) | 0.34 | 0.34 | ||||||
| Less than high school graduate | 1 (2.6) | 1 (5.0) | 0 (0) | 1 (5.6) | 0 (0) | |||
| High school graduate or GED | 5 (12.8) | 1 (5.0) | 4 (21.1) | 3 (16.7) | 2 (10.0) | |||
| Some college or technical school | 18 (46.2) | 9 (45.0) | 9 (47.4) | 9 (50.0) | 8 (40.0) | |||
| College or post-college graduate | 15 (38.5) | 9 (45.0) | 6 (31.6) | 5 (27.8) | 10 (50.0) | |||
| Alcohol use, n (%) | 0.36 | 0.03 | ||||||
| 2–6 times a week | 2 (5.1) | 0 (0) | 2 (10.5) | 0 (0) | 2 (10.0) | |||
| About once a week | 4 (10.3) | 2 (10.0) | 2 (10.5) | 3 (16.7) | 1 (5.0) | |||
| About once a month | 16 (41.0) | 11 (55.0) | 5 (26.3) | 5 (27.8) | 10 (50.0) | |||
| About once a year | 5 (12.8) | 2 (10.0) | 3 (15.8) | 1 (5.6) | 4 (20.0) | |||
| Never | 11 (28.2) | 5 (25.0) | 6 (31.6) | 9 (50.0) | 2 (10.0) | |||
| Unsure | 1 (2.6) | 0 (0) | 1 (5.3) | 0 (0) | 1 (5.0) | |||
| Telomere length (T/S) | 0.4±0.02 | 0.43±0.03 | 0.43±0.02 | 0.65 | 0.43±0.03 | 0.43±0.02 | 0.64 | |
| DNAmAge | 79.3±9.9 | 76.8±11.0 | 81.9±8.0 | 0.11 | 78.4±11.8 | 81.0±7.1 | 0.41 | |
| Age acceleration | 31.3±6.9 | 32.0±6.4 | 30.5±7.4 | 0.51 | 30.0±8.2 | 31.9±5.1 | 0.39 | |
*, P values represent differences between groups for each characteristic; ***, One EA participant was missing the residential info and was excluded when comparing poverty levels. Continuous variables were evaluated by two-sample t-tests, and chi square (χ2) tests were used to investigate the differences in distributions of categorical variables. EA, European-Americans; AA, African-Americans.
Differences by poverty level on genome-wide methylation profiles among AA women
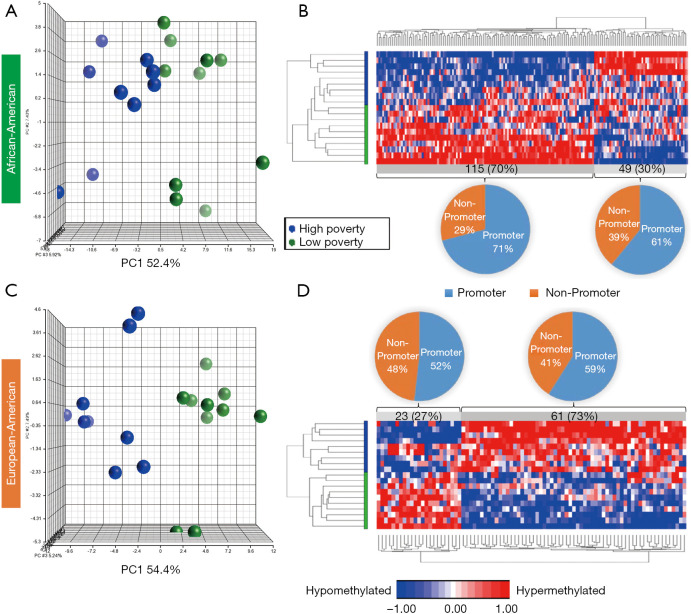
Based on the two-way ANOVA model controlling for alcohol use, 5,489 CpGs (P<0.01) and 164 (P<0.001, absolute fold change ≥1.5) CpGs were DM between high- and low-poverty counties among AA women. Among the 164 DM CpGs (Figure 1), 49 CpGs (29.9%) were hypermethylated in women from high-poverty counties and of which, 45% were within CpG island regions (Figure S1) and 61% were within promoter regions (Figure 1B). On the other hand, 115 CpGs (70.1%) were hypermethylated in women residing in low-poverty counties, and of which, 36% of the sites were within CpG island regions (Figure S1) and 71% were within promoter regions (Figure 1B).
Figure 1.
Differentially methylated (DM) CpGs among AA & EA Women by county poverty level. 3D scatter plots of principal component analysis (PCA) scores on the differentially methylated CpGs among (A) AA and (C) EA by poverty levels. Hierarchical clustering of the top CpG sites (P<0.001, |fold change| ≥1.5) distinguishing high and low poverty levels among (B) AA and (D) EA, as well as pie charts presenting the proportions of DM CpGs in promoter regions.
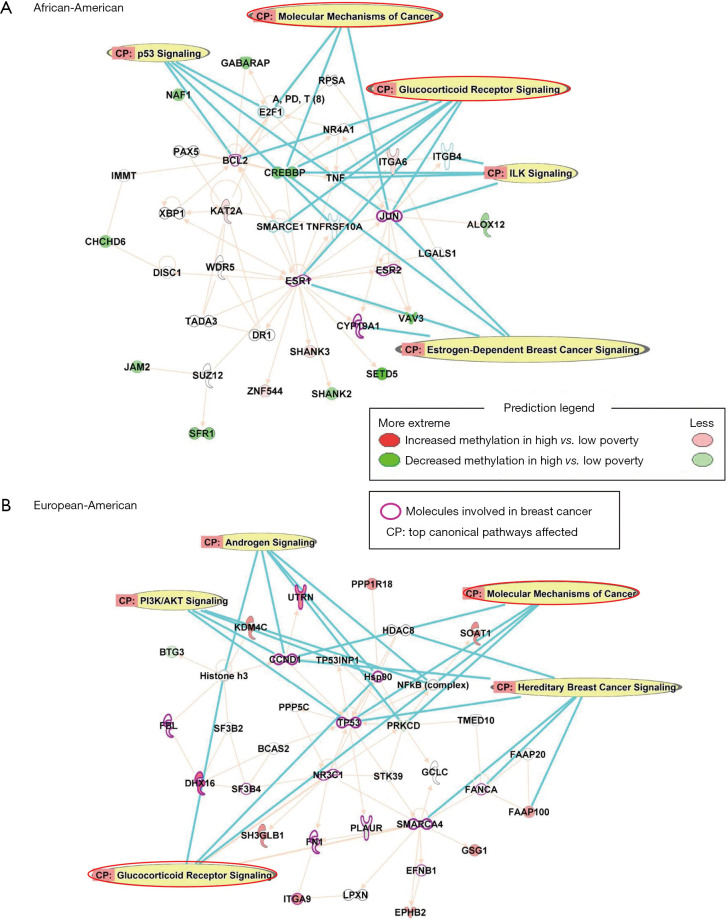
To investigate the potential biological implications, DM CpGs were queried by IPA, and the top networks affected by poverty levels among AA women are associated with Tissue Morphology, Cellular Development, Cellular Growth and Proliferation levels, with five molecules involved in breast cancer (BCL2, JUN, ESR1, ESR2, CYP19A1; Figure 2A). Top canonical pathways included: Glucocorticoid Receptor Signaling, Molecular Mechanisms of Cancer, p53 Signaling, Estrogen-Dependent Breast Cancer Signaling and ILK Signaling.
Figure 2.
Ingenuity pathway analysis revealed networks associated with county poverty levels among (A) AA and (B) EA women. Molecules shown were genes annotated by the DM CpGs with green nodes representing decreased methylation levels for women who were living in high poverty counties when compared to those residing in low poverty counties, and red nodes representing increased in methylation levels from high poverty counties compared to low poverty counties. Genes known as biomarkers for breast cancer were outlined in magenta, and top scoring canonical pathways affected in the network were highlighted in yellow. AA, African-Americans; EA, European-Americans.
Differences by poverty level on genome-wide methylation profiles among EA women
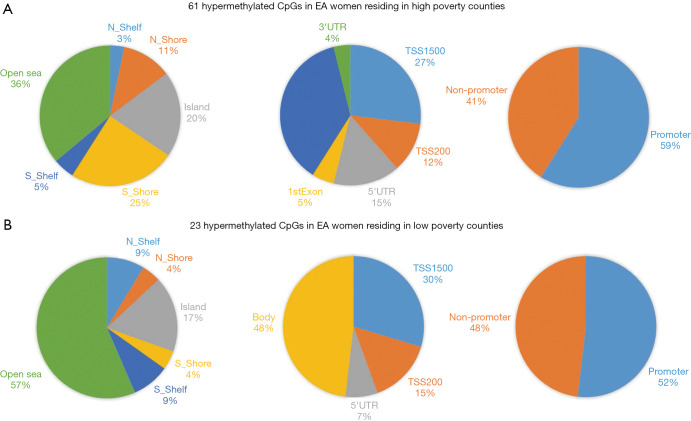
For EA women, based on the two-way ANOVA model controlling for alcohol use, 1,411 CpGs (P<0.01) and 85 (P<0.001, |FC|≥1.5) CpGs were DM between high- and low-county poverty levels. Among the 85 CpGs (Figure 1C,D), 61 CpGs (71.8%) were hypermethylated in women residing in high-poverty counties and of which, 20% of the sites were within CpG island regions (Figure S2) and 59% were within promoter regions (Figure 1D). On the other hand, 23 CpGs (27.1%) were hypermethylated in low-poverty counties (Figure 1D), and of which, 17% of the sites were within CpG island regions (Figure S2) and 52% were within promoter regions (Figure 1D).
Figure S2.
Distribution of differentially methylated CpGs by DNA regions. There were 61 unique hypermethylated CpGs in EA women residing in high poverty counties compared to 23 hypermethylated CpGs unique to low poverty county residence. AA, African-American; EA, European American.
The top networks affected in EA women by poverty levels are associated with Cell Morphology, Cellular Assembly and Organization, Cellular Response to Therapeutics. In all, 14 molecules associated with breast cancer (FBL, CCND1, DHX16, SF3B4, FN1, PLAUR, SMARCA4, FANCA, TP53, Hsp90, UTRN, ITGA9, NR3C1, EFNB1) were also DM (Figure 2B). Top canonical pathways involved in the network are: Hereditary Breast Cancer Signaling, Glucocorticoid Receptor Signaling, Androgen Signaling, PI3K/AKT Signaling, and Molecular Mechanisms of Cancer.
Effects of county poverty levels on DNAmAA
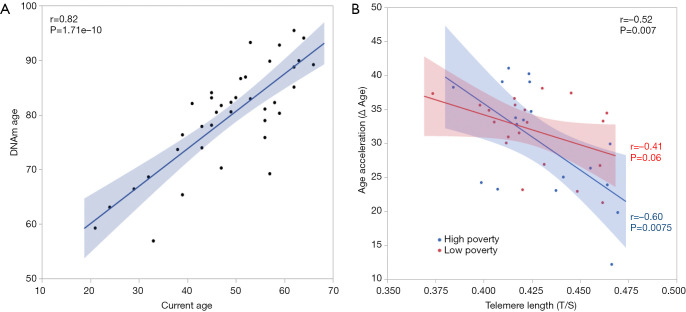
DNAm Age of the 39 women in the study was highly correlated with their chronological age (r=0.82, P=1.71e−10, Figure 3A). No significant differences was found in DNAmAA by race and poverty levels. However, DNAmAA was inverse correlated with the TL (r=−0.52, P=0.0007, Figure 3B) for all women, and the correlation was more significant among women who resided in high poverty (r=−0.6, P=0.0075) than low poverty counties (r=−0.41, P=0.06) (Figure 3B). Although no significant difference in TL by race or by poverty rates (P>0.05) was observed, TL was associated with 100 CpG sites within the above genes for all women (Table S1) when the impact of DNAm in genes reported to be involved in TL (ACYP2, NAF1, OBFC1, RTEL1, TERC, TERT, ZNF208) (25) were examined. Among the 100 CpG sites, four CpGs associated with NAF1, TERC and RTEL1 promoter regions were significantly different by poverty levels among AA women. In EA women, one CpG site in the OBFC1 promoter region (TSS1500) was significantly different according to poverty levels. Likewise, methylated CpG sites in the gene body of NAF1, OBFC1, and ACYP2, RTEL1, and the 3'UTR region of OBFC1 in the 3'UTR region were significantly different by poverty levels among EA women (Table 2).
Figure 3.
Effects of county poverty levels on DNAm age acceleration. (A) Correlation of chronological age versus DNAmAge; and (B) correlation of telomere length and age acceleration by county poverty levels in this study.
Table S1. CpG sites associated with telomere length among all women.
| Probeset ID | UCSC_RefGene_Name | CHR | Relation_to_UCSC_CpG_Island | UCSC_RefGene_Group | r | P | Lower CI | Upper CI |
|---|---|---|---|---|---|---|---|---|
| cg03339910 | RTEL1; RTEL1 | 20 | Island | Body; Body | 0.58 | 0.00013 | 0.32 | 0.76 |
| cg23250191 | TERT; TERT | 5 | Island | TSS200; TSS200 | −0.58 | 0.00015 | −0.76 | −0.32 |
| cg26334826 | RTEL1-TNFRSF6B; RTEL1; RTEL1; RTEL1; RTEL1 | 20 | Island | Body; Body; Body; Body; Body | 0.56 | 0.0003 | 0.29 | 0.74 |
| cg00622799 | RTEL1; RTEL1 | 20 | Island | Body; Body | 0.50 | 0.0014 | 0.21 | 0.71 |
| cg16750953 | TERT; TERT | 5 | Island | Body; Body | −0.48 | 0.002 | −0.69 | −0.19 |
| cg15494117 | TERC | 3 | Island | TSS200 | 0.46 | 0.004 | 0.16 | 0.68 |
| cg12810518 | NAF1; NAF1 | 4 | Island | 1stExon; 1stExon | −0.45 | 0.004 | −0.68 | −0.16 |
| cg01389761 | TERC | 3 | Island | TSS200 | 0.44 | 0.006 | 0.14 | 0.67 |
| cg17249224 | TERT; TERT | 5 | Island | Body; Body | −0.44 | 0.006 | −0.66 | −0.14 |
| cg15974345 | TERT; TERT | 5 | Island | Body; Body | −0.44 | 0.006 | −0.66 | −0.13 |
| cg10973735 | ACYP2; ACYP2 | 2 | Island | 1stExon; 5'UTR | 0.39 | 0.017 | 0.08 | 0.63 |
| cg27236539 | RTEL1; RTEL1 | 20 | Island | TSS200; TSS200 | 0.37 | 0.023 | 0.06 | 0.62 |
| cg02048657 | TERT; TERT | 5 | Island | Body; Body | −0.35 | 0.032 | −0.60 | −0.03 |
| cg23036508 | TERC | 3 | Island | TSS200 | −0.35 | 0.032 | −0.60 | −0.03 |
| g10896616 | TERT; TERT | 5 | Island | TSS200; TSS200 | −0.33 | 0.041 | −0.59 | −0.02 |
| cg17534029 | RTEL1; RTEL1 | 20 | Island | Body; Body | 0.33 | 0.046 | 0.01 | 0.58 |
| cg22989209 | TERT; TERT | 5 | N_Shelf | Body; Body | −0.58 | 0.00012 | −0.76 | −0.33 |
| cg01622668 | NAF1; NAF1 | 4 | N_Shelf | Body; Body | −0.53 | 0.0005 | −0.73 | −0.26 |
| cg07080099 | RTEL1-TNFRSF6B; RTEL1; RTEL1; RTEL1; RTEL1 | 20 | N_Shelf | Body; Body; Body; Body; Body | −0.52 | 0.0008 | −0.72 | −0.24 |
| cg06293931 | OBFC1 | 10 | N_Shelf | Body | −0.52 | 0.0009 | −0.72 | −0.23 |
| cg09218957 | RTEL1-TNFRSF6B; RTEL1; RTEL1; RTEL1; RTEL1 | 20 | N_Shelf | Body; Body; Body; Body; Body | −0.43 | 0.007 | −0.66 | −0.13 |
| cg02601800 | TERT; TERT | 5 | N_Shelf | Body; Body | −0.41 | 0.011 | −0.64 | −0.10 |
| cg13830297 | RTEL1-TNFRSF6B; RTEL1; RTEL1; RTEL1; RTEL1 | 20 | N_Shelf | Body; Body; Body; Body; Body | −0.40 | 0.012 | −0.64 | −0.10 |
| cg12090364 | RTEL1-TNFRSF6B; RTEL1; RTEL1; RTEL1; RTEL1 | 20 | N_Shelf | Body; Body; Body; Body; Body | −0.37 | 0.021 | −0.62 | −0.06 |
| cg26937683 | RTEL1-TNFRSF6B; RTEL1; RTEL1; RTEL1; RTEL1 | 20 | N_Shore | Body; Body; Body; Body; Body | −0.63 | 0.000020 | −0.79 | −0.39 |
| cg05172061 | ACYP2 | 2 | N_Shore | TSS1500 | −0.56 | 0.0003 | −0.74 | −0.29 |
| cg13696431 | TERT; TERT | 5 | N_Shore | Body; Body | −0.54 | 0.0005 | −0.73 | −0.27 |
| cg22738152 | ACYP2 | 2 | N_Shore | TSS1500 | −0.52 | 0.0008 | −0.72 | −0.24 |
| cg04137949 | NAF1; NAF1 | 4 | N_Shore | Body; Body | 0.51 | 0.0010 | 0.23 | 0.72 |
| cg05357717 | RTEL1; RTEL1 | 20 | N_Shore | Body; Body | −0.50 | 0.0013 | −0.71 | −0.22 |
| cg13594182 | TERT; TERT | 5 | N_Shore | Body; Body | −0.49 | 0.002 | −0.70 | −0.20 |
| cg06739590 | TERT; TERT | 5 | N_Shore | Body; Body | −0.48 | 0.002 | −0.69 | −0.19 |
| cg04019076 | TERT; TERT | 5 | N_Shore | Body; Body | −0.48 | 0.003 | −0.69 | −0.18 |
| cg20081540 | RTEL1; RTEL1 | 20 | N_Shore | Body; Body | −0.47 | 0.003 | −0.69 | −0.18 |
| cg16429735 | TERT; TERT | 5 | N_Shore | Body; Body | −0.46 | 0.003 | −0.68 | −0.17 |
| cg17509409 | RTEL1; RTEL1; RTEL1; RTEL1; RTEL1-TNFRSF6B | 20 | N_Shore | TSS1500; TSS1500; TSS1500; TSS1500; TSS1500 | −0.38 | 0.019 | −0.62 | −0.07 |
| cg16336280 | TERT; TERT | 5 | N_Shore | Body; Body | −0.37 | 0.021 | −0.62 | −0.06 |
| cg02538752 | ACYP2 | 2 | N_Shore | TSS1500 | 0.37 | 0.021 | 0.06 | 0.62 |
| cg17173860 | RTEL1; RTEL1; RTEL1; RTEL1; RTEL1-TNFRSF6B; RTEL1-TNFRSF6B; RTEL1; RTEL1; RTEL1; RTEL1 | 20 | N_Shore | ExonBnd; ExonBnd; ExonBnd; ExonBnd; ExonBnd; Body; Body; Body; Body; Body | −0.37 | 0.023 | −0.62 | −0.06 |
| cg01986883 | NAF1; NAF1 | 4 | N_Shore | Body; Body | 0.36 | 0.027 | 0.04 | 0.61 |
| cg04902826 | OBFC1 | 10 | N_Shore | 5'UTR | 0.35 | 0.031 | 0.03 | 0.60 |
| cg15927295 | TERT; TERT | 5 | N_Shore | Body; Body | −0.34 | 0.034 | −0.60 | −0.03 |
| cg08363415 | ACYP2 | 2 | S_Shelf | Body | −0.50 | 0.0013 | −0.71 | −0.22 |
| cg13954681 | ACYP2 | 2 | S_Shelf | Body | 0.33 | 0.045 | 0.01 | 0.59 |
| cg18251019 | OBFC1 | 10 | S_Shore | TSS200 | 0.58 | 0.00012 | 0.32 | 0.76 |
| cg26149131 | ACYP2 | 2 | S_Shore | Body | 0.57 | 0.0002 | 0.31 | 0.75 |
| cg08260673 | ACYP2 | 2 | S_Shore | Body | −0.57 | 0.0002 | −0.75 | −0.30 |
| cg18120808 | NAF1; NAF1 | 4 | S_Shore | TSS1500; TSS1500 | −0.53 | 0.0007 | −0.72 | −0.25 |
| cg25090302 | TERC | 3 | S_Shore | TSS1500 | −0.53 | 0.0007 | −0.72 | −0.25 |
| cg25809480 | RTEL1; RTEL1; RTEL1; RTEL1; RTEL1-TNFRSF6B | 20 | S_Shore | 5'UTR; 5'UTR; 5'UTR; 5'UTR; Body | −0.51 | 0.0012 | −0.71 | −0.22 |
| cg12615982 | TERC | 3 | S_Shore | TSS1500 | −0.49 | 0.002 | −0.70 | −0.20 |
| cg24333189 | TERC | 3 | S_Shore | TSS1500 | −0.49 | 0.002 | −0.70 | −0.20 |
| cg19828863 | OBFC1 | 10 | S_Shore | TSS1500 | −0.48 | 0.002 | −0.69 | −0.19 |
| cg19507224 | OBFC1 | 10 | S_Shore | TSS200 | −0.46 | 0.004 | −0.68 | −0.16 |
| cg07062658 | RTEL1; RTEL1 | 20 | S_Shore | 5'UTR; 5'UTR | 0.44 | 0.006 | 0.13 | 0.66 |
| cg08370839 | OBFC1 | 10 | S_Shore | TSS1500 | −0.42 | 0.009 | −0.65 | −0.11 |
| cg24019832 | OBFC1 | 10 | S_Shore | TSS1500 | 0.41 | 0.010 | 0.11 | 0.65 |
| cg21409704 | NAF1; NAF1 | 4 | S_Shore | TSS1500; TSS1500 | 0.41 | 0.011 | 0.10 | 0.64 |
| cg00352681 | TERT; TERT | 5 | S_Shore | Body; Body | −0.40 | 0.013 | −0.64 | −0.09 |
| cg24309739 | NAF1; NAF1 | 4 | S_Shore | TSS1500; TSS1500 | 0.38 | 0.017 | 0.07 | 0.63 |
| cg24931138 | TERT; TERT | 5 | S_Shore | Body; Body | −0.35 | 0.029 | −0.61 | −0.04 |
| cg20441553 | TERT; TERT | 5 | S_Shore | Body; Body | −0.33 | 0.043 | −0.59 | −0.01 |
| cg14278567 | ACYP2 | 2 | Open sea | Body | −0.72 | 0.0000003 | −0.85 | −0.52 |
| cg01447263 | ACYP2 | 2 | Open sea | Body | −0.62 | 0.00003 | −0.78 | −0.38 |
| cg06749545 | OBFC1 | 10 | Open sea | 3'UTR | −0.61 | 0.00004 | −0.78 | −0.37 |
| cg09031957 | OBFC1 | 10 | Open sea | 3'UTR | −0.61 | 0.00005 | −0.78 | −0.36 |
| cg11319187 | ACYP2 | 2 | Open sea | Body | 0.59 | 0.00009 | 0.34 | 0.77 |
| cg09834789 | OBFC1 | 10 | Open sea | 3'UTR | −0.59 | 0.00011 | −0.76 | −0.33 |
| cg11016558 | OBFC1 | 10 | Open sea | Body | −0.58 | 0.0002 | −0.76 | −0.31 |
| cg21916555 | NAF1; NAF1 | 4 | Open sea | Body; Body | −0.57 | 0.0002 | −0.75 | −0.31 |
| cg13601318 | NAF1; NAF1 | 4 | Open sea | Body; Body | −0.57 | 0.0002 | −0.75 | −0.31 |
| cg07936144 | TERT; TERT; TERT; TERT | 5 | Open sea | ExonBnd; ExonBnd; Body; Body | −0.57 | 0.0002 | −0.75 | −0.31 |
| cg24360131 | ACYP2 | 2 | Open sea | Body | −0.56 | 0.0002 | −0.75 | −0.30 |
| cg03302253 | ACYP2 | 2 | Open sea | Body | −0.55 | 0.0003 | −0.74 | −0.29 |
| cg25656654 | OBFC1 | 10 | Open sea | 3'UTR | −0.54 | 0.0004 | −0.73 | −0.27 |
| cg04920123 | ACYP2 | 2 | Open sea | Body | −0.54 | 0.0005 | −0.73 | −0.27 |
| cg14958080 | TERT; TERT | 5 | Open sea | Body; Body | −0.54 | 0.0005 | −0.73 | −0.26 |
| cg13240013 | ACYP2 | 2 | Open sea | Body | −0.54 | 0.0005 | −0.73 | −0.26 |
| cg16527659 | ACYP2 | 2 | Open sea | Body | −0.53 | 0.0005 | −0.73 | −0.26 |
| cg19128723 | OBFC1 | 10 | Open sea | Body | 0.53 | 0.0006 | 0.26 | 0.73 |
| cg20503346 | ACYP2 | 2 | Open sea | Body | −0.53 | 0.0006 | −0.73 | −0.26 |
| cg10274419 | OBFC1 | 10 | Open sea | 3'UTR | −0.53 | 0.0007 | −0.73 | −0.25 |
| cg19883490 | OBFC1 | 10 | Open sea | Body | −0.52 | 0.0007 | −0.72 | −0.25 |
| cg03725688 | NAF1; NAF1 | 4 | Open sea | Body; Body | −0.51 | 0.0012 | −0.71 | −0.22 |
| cg21640312 | NAF1; NAF1 | 4 | Open sea | Body; Body | −0.50 | 0.0013 | −0.71 | −0.22 |
| cg09058170 | ACYP2 | 2 | Open sea | Body | −0.50 | 0.0014 | −0.71 | −0.21 |
| cg07641791 | OBFC1 | 10 | Open sea | Body | −0.49 | 0.002 | −0.70 | −0.21 |
| cg06511943 | OBFC1 | 10 | Open sea | Body | −0.49 | 0.002 | −0.70 | −0.21 |
| cg17332810 | ACYP2 | 2 | Open sea | Body | −0.49 | 0.002 | −0.70 | −0.20 |
| cg08322053 | ACYP2 | 2 | Open sea | Body | −0.47 | 0.003 | −0.69 | −0.18 |
| cg20382968 | OBFC1 | 10 | Open sea | Body | −0.45 | 0.005 | −0.67 | −0.15 |
| cg16485140 | ZNF208 | 19 | Open sea | Body | −0.42 | 0.008 | −0.65 | −0.12 |
| cg12539618 | RTEL1-TNFRSF6B; RTEL1; RTEL1; RTEL1; RTEL1 | 20 | Open sea | Body; Body; Body; Body; Body | −0.39 | 0.014 | −0.63 | −0.08 |
| cg07072878 | NAF1 | 4 | Open sea | 3'UTR | −0.39 | 0.017 | −0.63 | −0.07 |
| cg06103076 | RTEL1; RTEL1-TNFRSF6B; RTEL1; RTEL1; RTEL1 | 20 | Open sea | 5'UTR; Body; Body; Body; Body | −0.36 | 0.027 | −0.61 | −0.04 |
| cg21939447 | NAF1; NAF1; NAF1; NAF1 | 4 | Open sea | ExonBnd; ExonBnd; Body; Body | −0.34 | 0.036 | −0.60 | −0.02 |
| cg16408679 | ACYP2 | 2 | Open sea | Body | −0.33 | 0.044 | −0.59 | −0.01 |
| cg21651647 | ACYP2 | 2 | Open sea | Body | −0.33 | 0.045 | −0.59 | −0.01 |
| cg08856627 | RTEL1; RTEL1-TNFRSF6B; RTEL1; RTEL1; RTEL1 | 20 | Open sea | 5'UTR; Body; Body; Body; Body | 0.32 | 0.047 | 0.01 | 0.58 |
| cg11005552 | OBFC1 | 10 | Open sea | Body | 0.32 | 0.049 | 0.00 | 0.58 |
Table 2. Telomere length-associated CpG sites significantly different by county poverty levels.
| Gene symbol | Chr | CpG island regions | Gene centric regions | P | Mean beta value | High poverty/low poverty | |||
|---|---|---|---|---|---|---|---|---|---|
| High poverty | Low poverty | Fold-Change | Trend | ||||||
| AA women | |||||||||
| NAF1 | 4 | S_Shore | TSS1500 | 0.002 | 0.46 | 0.58 | −1.6 | Down | |
| NAF1 | 4 | S_Shore | TSS1500 | 0.007 | 0.27 | 0.41 | −1.9 | Down | |
| RTEL1 | 20 | S_Shore | 5'UTR | 0.026 | 0.62 | 0.66 | −1.1 | Down | |
| TERC | 3 | Island | TSS200 | 0.033 | 0.38 | 0.42 | −1.2 | Down | |
| EA women | |||||||||
| NAF1 | 4 | N_Shore | Body | 0.006 | 0.05 | 0.02 | 2.1 | Up | |
| RTEL1 | 20 | Island | Body | 0.014 | 0.28 | 0.20 | 1.6 | Up | |
| NAF1 | 4 | N_Shore | Body | 0.025 | 0.25 | 0.15 | 1.8 | Up | |
| OBFC1 | 10 | Open sea | Body | 0.033 | 0.64 | 0.76 | −1.8 | Down | |
| OBFC1 | 10 | Open sea | 3'UTR | 0.034 | 0.64 | 0.76 | −1.9 | Down | |
| RTEL1 | 20 | N_Shore | Body | 0.035 | 0.85 | 0.87 | −1.2 | Down | |
| OBFC1 | 10 | Open sea | Body | 0.040 | 0.41 | 0.48 | −1.4 | Down | |
| OBFC1 | 10 | S_Shore | TSS1500 | 0.041 | 0.77 | 0.83 | −1.4 | Down | |
| ACYP2 | 2 | Open sea | Body | 0.044 | 0.35 | 0.26 | 1.5 | Up | |
| OBFC1 | 10 | Open sea | 3'UTR | 0.046 | 0.74 | 0.81 | −1.5 | Down | |
| RTEL1 | 20 | Island | Body | 0.046 | 0.40 | 0.30 | 1.6 | Up | |
Discussion
In this pilot study, DNAm patterns differed based on race and county poverty levels. While no significant differences in TL existed, TL was associated with the DNAmAA, and the association was more prominent among women residing in counties with high poverty rates. Genes involved in telomere maintenance were also shown to be DM by county poverty levels.
Contrary to other studies (26,27), the inflammatory pathway was not prominent. Moreover, the pathways identified as DM varied between EA and AA women when county poverty rates were considered. A reversal of hypermethylation patterns between EA and AA women by county poverty existed that could be due to racial differences in single nucleotide polymorphism (SNP) allele frequencies in one-carbon metabolism genes. Significant variations in allele frequencies in eleven genes involved in one-carbon metabolism showed that polygenetic risk scores were significantly associated with breast cancer risk (28). Although the current pilot study was not large enough to include SNP analyses, the differential methylation patterns merit further study.
Both overlap and differences in biological pathways existed that were DM by county poverty and race. In AA women, gene networks involved in estrogen-dependent breast cancer were DM, while hereditary breast cancer networks were DM in EA women. These data suggest gene-environment interactions that are involved in racial differences in breast cancer risk that exists between EA and AA women. Additionally, the number and identity of DM molecules related to breast cancer risk also varied between AA and EA women (5 versus 14). This is consistent with AA women having lower risk of breast cancer compared to EA women. Our results confirm previous findings indicating likely differing etiologic pathways for the development of ER negative breast cancer between AA and EA women (29).
In contrast to other studies (30-32), no significant differences existed in TL by county poverty rate or race, as well as the correlation of TL and age. This could be due to the modest sample size of the current study. However, high correlation of TL and DNAmAA was observed, signifying the potential of epigenetic modifications of life expectancy especially in the rural regions. Smoking status was not available and could have played a role in our findings. Because a high correlation between TL measured in blood compared to saliva exists (33,34), the use of saliva is less likely to be a factor. We did find significant differences in the methylation of several TL-associated genes by county poverty levels, even though TL did not vary significantly. This could be due to laboratory methods for measuring DNAm that are more sensitive than assays for telomere length.
Conclusions
The finding of this pilot study suggests county poverty levels may impact DNAm patterns in breast cancer-related pathways, as well as genes involved in telomere maintenance. Since DNAm is modifiable, identification of methylation patterns impacted by adverse neighborhood conditions could lead to the design of interventions that reduce health disparities experienced by residents in counties with high-poverty rates. Larger studies should confirm our findings.
Acknowledgments
The authors would like to thank all the participants for their contribution to the Arkansas Rural Community Health (ARCH) Study.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Review Boards of University of Arkansas for Medical Sciences (IRB# 89071) and written informed consent was obtained from all participants.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.02.07). The authors have no conflicts of interest to declare.
References
- 1.Merkin SS, Basurto-Davila R, Karlamangla A, et al. Neighborhoods and cumulative biological risk profiles by race/ethnicity in a national sample of U.S. adults: NHANES III. Ann Epidemiol 2009;19:194-201. 10.1016/j.annepidem.2008.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeigler-Johnson CM, Tierney A, Rebbeck TR, et al. Prostate cancer severity associations with neighborhood deprivation. Prostate Cancer 2011;2011:846263. 10.1155/2011/846263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heilig M, Barbier E, Johnstone AL, et al. Reprogramming of mPFC transcriptome and function in alcohol dependence. Genes Brain Behav 2017;16:86-100. 10.1111/gbb.12344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsit CJ. Influence of environmental exposure on human epigenetic regulation. J Exp Biol 2015;218:71-9. 10.1242/jeb.106971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dwyer-Lindgren L, Bertozzi-Villa A, Stubbs RW, et al. Inequalities in Life Expectancy Among US Counties, 1980 to 2014: Temporal Trends and Key Drivers. JAMA Intern Med 2017;177:1003-11. 10.1001/jamainternmed.2017.0918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zolot J. U.S. Life Expectancy Varies Depending on County of Birth. Am J Nurs 2017;117:15. [DOI] [PubMed] [Google Scholar]
- 7.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol 2013;14:R115. 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 2013;49:359-67. 10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mons U, Muezzinler A, Schottker B, et al. Leukocyte Telomere Length and All-Cause, Cardiovascular Disease, and Cancer Mortality: Results From Individual-Participant-Data Meta-Analysis of 2 Large Prospective Cohort Studies. Am J Epidemiol 2017;185:1317-26. 10.1093/aje/kww210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haycock PC, Burgess S, Nounu A, et al. Association Between Telomere Length and Risk of Cancer and Non-Neoplastic Diseases: A Mendelian Randomization Study. JAMA Oncol 2017;3:636-51. 10.1001/jamaoncol.2017.2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zahnd WE, James AS, Jenkins WD, et al. Rural-Urban Differences in Cancer Incidence and Trends in the United States. Cancer Epidemiol Biomarkers Prev 2018;27:1265-74. 10.1158/1055-9965.EPI-17-0430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams F, Thompson E. Disparity in Breast Cancer Late Stage at Diagnosis in Missouri: Does Rural Versus Urban Residence Matter? J Racial Ethn Health Disparities 2016;3:233-9. 10.1007/s40615-015-0132-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H, Liu Y, Zhu Z, et al. Does where you live matter to your health? Investigating factors that influence the self-rated health of urban and rural Chinese residents: evidence drawn from Chinese General Social Survey data. Health Qual Life Outcomes 2017;15:78. 10.1186/s12955-017-0658-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song MA, Brasky TM, Marian C, et al. Racial differences in genome-wide methylation profiling and gene expression in breast tissues from healthy women. Epigenetics 2015;10:1177-87. 10.1080/15592294.2015.1121362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bondurant KL, Harvey S, Klimberg S, et al. Establishment of a southern breast cancer cohort. Breast J 2011;17:281-8. 10.1111/j.1524-4741.2011.01082.x [DOI] [PubMed] [Google Scholar]
- 16.Bureau USC. American Factfinder. 2014.
- 17.Pidsley R, Zotenko E, Peters TJ, et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol 2016;17:208. 10.1186/s13059-016-1066-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horvath S, Gurven M, Levine ME, et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol 2016;17:171. 10.1186/s13059-016-1030-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quach A, Levine ME, Tanaka T, et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY) 2017;9:419-46. 10.18632/aging.101168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis EG, Humphreys KL, McEwen LM, et al. Accelerated DNA methylation age in adolescent girls: associations with elevated diurnal cortisol and reduced hippocampal volume. Transl Psychiatry 2017;7:e1223. 10.1038/tp.2017.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theall KP, Brett ZH, Shirtcliff EA, et al. Neighborhood disorder and telomeres: connecting children's exposure to community level stress and cellular response. Soc Sci Med 2013;85:50-8. 10.1016/j.socscimed.2013.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res 2002;30:e47. 10.1093/nar/30.10.e47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pellatt AJ, Wolff RK, Torres-Mejia G, et al. Telomere length, telomere-related genes, and breast cancer risk: the breast cancer health disparities study. Genes Chromosomes Cancer 2013;52:595-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bibikova M, Barnes B, Tsan C, et al. High density DNA methylation array with single CpG site resolution. Genomics 2011;98:288-95. 10.1016/j.ygeno.2011.07.007 [DOI] [PubMed] [Google Scholar]
- 25.Codd V, Nelson CP, Albrecht E, et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat Genet 2013;45:422-7, 427e1-2. [DOI] [PMC free article] [PubMed]
- 26.Uddin M, Koenen KC, Aiello AE, et al. Epigenetic and inflammatory marker profiles associated with depression in a community-based epidemiologic sample. Psychol Med 2011;41:997-1007. 10.1017/S0033291710001674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith JA, Zhao W, Wang X, et al. Neighborhood characteristics influence DNA methylation of genes involved in stress response and inflammation: The Multi-Ethnic Study of Atherosclerosis. Epigenetics 2017;12:662-73. 10.1080/15592294.2017.1341026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong Z, Yao S, Zirpoli G, et al. Genetic variants in one-carbon metabolism genes and breast cancer risk in European American and African American women. Int J Cancer 2015;137:666-77. 10.1002/ijc.29434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ambrosone CB, Young AC, Sucheston LE, et al. Genome-wide methylation patterns provide insight into differences in breast tumor biology between American women of African and European ancestry. Oncotarget 2014;5:237-48. 10.18632/oncotarget.1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliveira BS, Zunzunegui MV, Quinlan J, et al. Lifecourse Adversity and Telomere Length in Older Women from Northeast Brazil. Rejuvenation Res 2018;21:294-303. 10.1089/rej.2017.1937 [DOI] [PubMed] [Google Scholar]
- 31.Lynch SM, Mitra N, Ravichandran K, et al. Telomere Length and Neighborhood Circumstances: Evaluating Biological Response to Unfavorable Exposures. Cancer Epidemiol Biomarkers Prev 2017;26:553-60. 10.1158/1055-9965.EPI-16-0554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gebreab SY, Riestra P, Gaye A, et al. Perceived neighborhood problems are associated with shorter telomere length in African American women. Psychoneuroendocrinology 2016;69:90-7. 10.1016/j.psyneuen.2016.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell C, Hobcraft J, McLanahan SS, et al. Social disadvantage, genetic sensitivity, and children's telomere length. Proc Natl Acad Sci U S A 2014;111:5944-9. 10.1073/pnas.1404293111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lahnert P. An improved method for determining telomere length and its use in assessing age in blood and saliva. Gerontology 2005;51:352-6. 10.1159/000086374 [DOI] [PubMed] [Google Scholar]