Abstract
Background
Acute myeloid leukemia (AML) is characterized by genetic and epigenetic mutations that lead to a block in differentiation as well as unrestrained proliferation. Many epigenetic regulators have been shown to be important for AML initiation and development. Among these, bromodomain-containing protein 9 (BRD9), an epigenetic regulator, was recently identified as a critical factor required for AML development. Hence targeting BRD9 may provide a new therapeutic strategy. Thus, we investigated the role of BRD9 inhibitor I-BRD9 in AML cells and its potential mechanisms.
Methods
Cell Counting Kit-8 (CCK-8) assays were performed to explore the growth inhibitory effects of I-BRD9 on AML cells. Flow cytometry was used to examine the effects of I-BRD9 on apoptosis, Edu incorporation, and cell differentiation. Apoptotic pathway activation was confirmed by western blot. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was employed to analyze cell death and cell cycle-related gene expression.
Results
I-BRD9 significantly reduced AML cells growth. This is accompanied by decreased Edu incorporation and dramatic cell death. Mechanistically, cell death induced by I-BRD9 was largely blocked by the pan-caspase inhibitor Z-VAD-FMK and, to a lesser extent, by Ferrostatin-1.Furthermore, apoptotic markers including the cleavage of PARP, Capase9, and Capsese3, were induced by I-BRD9, which were rescued by pretreatment with Z-VAD-FMK. In addition, I-BRD9 treatment increased IRE3, CDKN1A, and CDKN2B expression in AML cells, possibly leading to the observed decrease in Edu incorporation. Together, these data strongly suggested that I-BRD9 induced growth inhibition in AML cells was dependent on apoptosis and cell cycle inhibition.
Conclusions
Our data support the important role of BRD9 in AML cells; moreover, the BRD9 inhibitor I-BRD9 could be potentially useful in the treatment of AML .
Keywords: Acute myeloid leukemia (AML), epigenetics, I-BRD9, BRD9, Cell growth
Introduction
Acute myeloid leukemia (AML) is a hematological malignancy characterized by neoplastic clones that are unable to differentiate and have unrestrained proliferation (1). This condition eventually leads to the disruption of normal hematopoiesis, bone marrow failure, and mortality if left untreated (1,2). Despite rapid advances in the knowledge of leukemogenesis, the therapeutic options remain limited, and the mainstream therapies for AML have remained largely unchanged in recent decades. Genetic mutations or translocations in key genes including but not limited to FLT3, KIT, RAS, RUNX1, MLL, CEBPA, and RARA have been shown to play important roles in leukemogenesis (3-5). However, compared to other tumors, AML has a relatively low rate of recurrent somatic mutations. Besides genetic mutations, epigenetic dysregulation is another type of important regulator of leukemogenesis and AML development, with associated epigenetic factors including DOT1L, LSD1, HDACs, PRMT1, PRMT5, and BET bromodomain-containing proteins (6-10). Indeed, many inhibitors or drugs targeting epigenetic factors have entered clinical trials for the treatment of AML (11,12). Thus, targeting epigenetic factors that required for leukemogenesis and leukemia maintenance may represent a promising future therapeutic method for the treatment of AML.
BRD9 is a bromodomain-containing protein that was recently identified as a subunit of SWI/SNF chromatin remodeling complexes (13). It functions mainly by binding to acetylated lysine on post-transcriptionally modified histone proteins to regulate transcription (13). In addition to regulating transcription, BRD9 also binds to acetylated RAD54 and is involved in the homologous recombination pathway, which is important for cancer cell survival (14). BRD9 is overexpressed in AML and preferably binds to enhancer regions in a cell type-specific manner (15). Depletion of BRD9 in AML cells leads to a strong proliferation defect accompanied by the induction of apoptosis (15). These data suggest that BRD9 is a potential therapeutic target in cancer, especially in AML cells.
Although BRD9 has been shown to be important in several malignancies including AML (15,16), whether targeting BRD9 with small-molecule probes or inhibitors could be beneficial in AML remains unsolved. I-BRD9 is a selective cellular chemical probe for BRD9 that may potently inhibit BRD9 (17). Here we showed that I-BRD9 treatment of AML cells resulted in decreased cell proliferation and increased apoptosis and ferroptosis without inducing differentiation. We further demonstrated that I-BRD9 induced several known targets of BRD9 in a cell-specific manner. Altogether, our data suggest that the targeting of BRD9 by I-BRD9 could be of potential use in treating AML. We present the following article in accordance with the MDAR checklist (available at https://dx.doi.org/10.21037/tcr-21-42).
Methods
Cell culture
The NB4 and MV4-11 AML cell lines were purchased from Cellcook (www.cellcook.com). Briefly, the cells were maintained in RPMI1640 and IMDM medium supplemented with 10% fetal bovine serum. The cells were kept in a humidified incubator at 37 °C with 5% CO2.
Cell viability assay
Cell viability was assayed using the Cell Counting Kit-8 (CCK-8) (#BA00208, Bioss, China). Briefly, 1×104cells were seeded into 96-well plates and treated with different concentrations of I-BRD9 (#HY-18975, MCE, USA) for the indicated time. At the end point, 10 µL of CCK-8 was added to each well and incubated for 2–3 h at 37 °C before the absorbance was detected at 450nM on a microplate reader (Varioskan Flash, Thermo Fisher Scientific, USA).
Regarding the inhibitor rescue assay, cells plated in 96-well plates were first treated with 20 µM Z-VAD-FMK (#HY-16658B, MCE, USA), 1 µM Ferrostatin-1 (#HY-100579, MCE, USA), 100 µM a-Tocopherol (#HY-N068, MCE, USA), 10 µM Necrostatin-2 (#HY-14622, MCE, USA), 1 µM (E)-Necrosulfonamide (MLKLi) (#HY-100573, MCE, USA), 5 µM Chloroquine diphosphate salt (#C6628, Sigma, USA), 5 nM Bafilomycin A1 (#A8510, Solarbio, China) for 2 h before I-BRD9 treatment. These inhibitors plus I-BRD9 were further incubated for 72 h and assayed for viability by the CCK-8 assay.
Flow cytometry
Edu assay
Edu incorporation was assessed after planting cells in 12-well plates using the Click-iT™ Plus EdU Alexa Fluor™ 647 Flow Cytometry Assay Kit (#C10635, Thermo Fisher Scientific, USA). Edu was added to subconfluent NB4 and MV4-11 to a final concentration of 10 µM, and the cells were incubated for 2 h before being collected, washed twice with phosphate buffer saline (PBS), and stained according to the manufacturer’s instructions.
Apoptosis assay
Cell death flow cytometry was performed using an Annexin V/ propidium iodide (PI) kit (#A211-01, Vazyme, China). In brief, cells were treated accordingly, collected, washed twice with PBS, and stained with Annexin V in binding buffer for 20 min at room temperature kept from light. After washing with PBS, PI was added to each sample. The samples were then analyzed using a BD FACSCANTO II system (BD Biosciences, USA).
Cell differentiation
The cells were stained with CD11b (#101217, Biolegend, USA, 1:200 dilution), CD14 (#301812, Biolegend, USA, 1:200 dilution), and CD38 (#303506, Biolegend, USA, 1:400 dilution) for 20min at room temperature kept from light. The cells were then washed with PBS and analyzed on a BD FACSCANTO II system (BD Biosciences, USA). All flow cytometry data were analyzed with FlowJo V10.
Western blot
To analyze protein expression, cells were directly lysed with loading buffer (#NP0008, Thermo Fisher Scientific, USA) with dithiothreitol (DTT;#R0861, Thermo Fisher Scientific, USA), separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and blotted with PARP (#9542, Cell Signaling Technology, USA,1:1000 dilution), caspase 9 (#52873, Cell Signaling Technology, USA,1:1000 dilution), caspase 3 (#14220, Cell Signaling Technology, USA,1:1000 dilution) and Lamin B (#AF1408, Beyotime, China, 1:1,000 dilution). The blots were developed on a Gel Imager instrument (CHEMIDOCTMXRS+ System, Bio-Rad, USA). The intensity of bands was quantified using ImageJ.
Real-time quantitative polymerase chain reaction (qRT-PCR)
Total RNA was extracted using TRIzol solution (#15596018, Thermo Fisher Scientific, USA). 1µg of total RNA was then reverse transcribed using the PrimeScript RT reagent Kit with gDNA Eraser (#RR047A, Takara, Japan) to produce cDNA. qRT-PCR was performed with SYBR Green Master Mix (#RR820A, Takara, Japan) and analyzed on a RT-PCR system (CFX96 Touch, Bio-Rad, USA). The primers used are listed below:
Human BRD9: Fwd-5': GCAATGACATACAATAGGCCAGA, Rev-5': GAGCTGCCTGTTTGCTCATCA;
Human DDIT3: Fwd-5': GGAAACAGAGTGGTCATTCCC, Rev-5': CTGCTTGAGCCGTTCATTCTC;
Human IER3: Fwd-5': CAGCCGCAGGGTTCTCTAC, Rev-5': GATCTGGCAGAAGACGATGGT;
Human SOCS3: Fwd-5': CCTGCGCCTCAAGACCTTC, Rev-5': GTCACTGCGCTCCAGTAGAA;
Human CDKN1A: Fwd-5': TGTCCGTCAGAACCCATGC, Rev-5': AAAGTCGAAGTTCCATCGCTC;
Human CDKN2B: Fwd-5': TGTCCGTCAGAACCCATGC, Rev-5': AAAGTCGAAGTTCCATCGCTC;
Human GAPDH: Fwd-5': GGAGCGAGATCCCTCCAAAAT, Rev-5': GGCTGTTGTCATACTTCTCATGG.
Statistical analysis
Data are presented as means with error bars representing standard deviation (SD). Statistical significance was determined by two-tailed Student's t-test comparisons if not stated otherwise. Significant differences are indicated by asterisks (*P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001).
Results
I-BRD9 inhibits AML cell proliferation
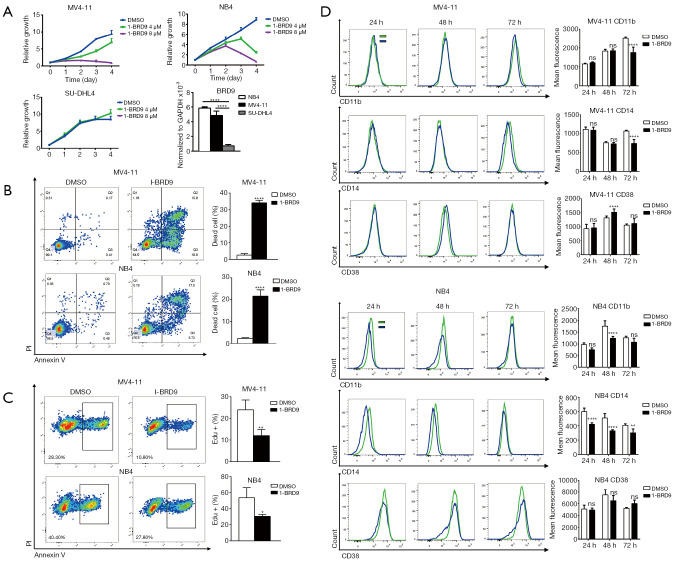
To explore the potential beneficial effect of targeting BRD9 by I-BRD9, we used two AML cell lines, NB4 and MV4-11. NB4 cells reportedly over-express BRD9 (15). And our expression analysis showed that MV4-11 cells expressed a similar level of BRD9 compared to NB4 cells (Figure 1A). These cells were treated with 4 and 8 µM of I-BRD9 for 24, 48, 72, and 96 h. According to the CCK8 assay, I-BRD9 dose-dependently inhibited NB4 and MV4-11 cell growth; however, MV4-11 cells appeared to be more sensitive to I-BRD9 (Figure 1A). To test the specificity of I-BRD9, we further used the SU-DHL-4 cell line, which shows low BRD9 expression (Figure 1A). Treatment of I-BRD9 in SU-DHL-4 cells showed a marginal effect on cell growth. Together these data suggest the on-target effect of I-BRD9 on BRD9 over-expression cells lines. The growth inhibitory effect of I-BRD9 in AML cells was accompanied by increased cell death as shown by Annexin V/PI staining (Figure 1B), and cell cycle arrest, as shown by decreased Edu incorporation (Figure 1C). Since cell death and cell cycle arrest could be a consequence of AML cell differentiation, we further checked whether I-BRD9 treatment led to cell differentiation. We did not observe any indication of cell differentiation induced by I-BRD9, as there was no increase in the expression of several myeloid differentiation markers, including CD11b, CD14, and CD38 (Figure 1D). These data suggested that the targeting of BRD9 by I-BRD9 in AML cells induced cell cycle arrest and cell death independent of triggering cell differentiation.
Figure 1.
I-BRD9 inhibits NB4 and MV4-11 cell growth. (A) CCK-8 viability assay for NB4, MV4-11, and SU-DHL4 cells treated with 4 and 8 µM I-BRD9 for 96 h (n=3, mean ± SD, two-tailed t-test); qPCR analysis of BRD9 in NB4, MV4-11, and SU-DHL4 cells (n=3, mean ± SD, two-tailed t-test). (B) Flow cytometry Annexin V/PI analysis of NB4 and MV4-11 cell death induced by 8 µM I-BRD9 treatment for 48 h (n=3, mean ± SD, two-tailed t-test). (C) Flow cytometry Edu assay analysis of NB4 and MV4-11 cell proliferation following treatment with 8 µM I-BRD9 for 48 h (n=3, mean ± SD, two-tailed t-test). (D) Flow cytometry analysis of CD11b, CD14 and CD38 expression in NB4 and MV4-11 cells treated with 8 µM I-BRD9 for 24, 48 and 72 h (n=3, mean ± SD, two-tailed t-test). *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001. CCK-8, Cell Counting Kit-8; qPCR, quantitative polymerase chain reaction; SD, standard deviation.
Z-VAD rescues I-BRD9 induced cell death
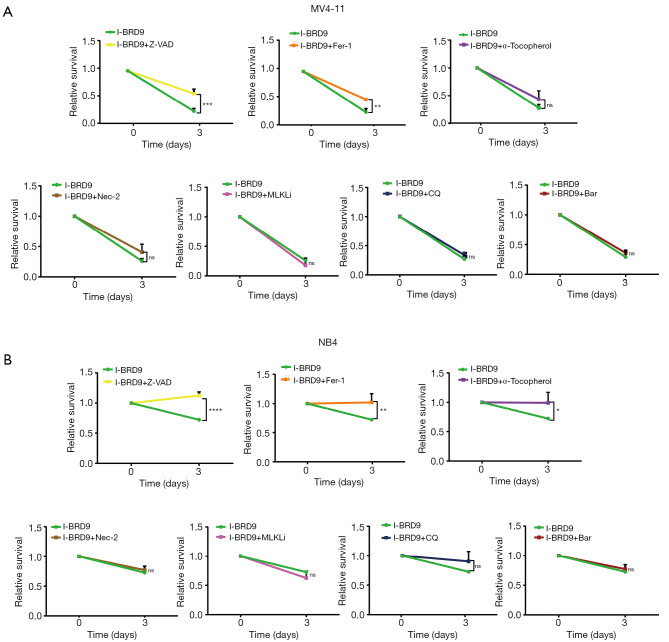
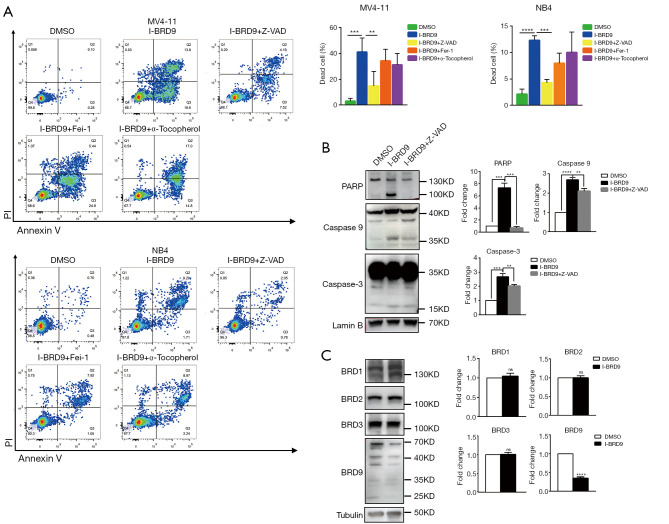
To further understand the mechanism by which I-BRD9-induced cell death, we tested several cell death pathway inhibitors to determine the type of cell death induced by I-BRD9 treatment. NB4 and MV4-11 cells were treated with Z-VAD-FMK (Z-VAD), Ferrostatin-1 (Fer-1), a-Tocopherol, Necrostatin-2 (Nec-2), (E)-Necrosulfonamide (MLKLi), Chloroquine (CQ), and Bafilomycin A1 (Baf) 2 h before I-BRD9 treatment, and then co-incubated with I-BRD9 for 72h. According to the CCK8 assay, apoptosis inhibitor Z-VAD-FMK and ferroptosis inhibitor Fer-1 significantly rescued I-BRD9-induced cell death in both cell lines (Figure 2A,B). In NB4 cells, a-Tocopherol, another inhibitor of ferroptosis, also significantly rescued cell death (Figure 2A,B). Furthermore, in both cell lines, Z-VAD-FMK resulted in a stronger rescue than Fer-1. These observations were further confirmed by Annexin V/PI staining, which showed a stronger rescue of I-BRD9 induced cell death by Z-VAD-FMK than Fer-1 and a-Tocopherol (Figure 3A). These data suggested that I-BRD9 mainly induced apoptosis and, to a lesser extent, ferroptosis in AML cells. This was further supported by the observation that I-BRD9 treatment resulted in an increase in apoptosis markers, such as the cleaved forms of PARP, caspase 3, and caspase 9 in MV4-11 cells, which were rescued by Z-VAD-FMK (Figure 3B). I-BRD9 also decreased the expression of the BRD9 protein, without significantly affecting other BRDs, such as BRD1, BRD2, and BRD3, indicating that cell death was specifically induced by the inhibition of BRD9 (Figure 3C).
Figure 2.
Z-VAD-FMK rescues I-BRD9-induced cell death. (A) CCK-8 viability assay in MV4-11 cells treated with the indicated inhibitors and 8 µM I-BRD9 for 72 h (n=3, mean ± SD, two-tailed t-test,). (B) CCK-8 viability assay in NB4 cells treated with the indicated inhibitors and 8 µM I-BRD9 for 72h (n=3, mean ± SD, two-tailed t-test). *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001. CCK-8, Cell Counting Kit-8; SD, standard deviation.
Figure 3.
I-BRD9 induces apoptosis in MV4-11 and NB4 cells. (A) Flow cytometry Annexin V/PI analysis of cell death in NB4 and MV4-11 cells treated with the indicated inhibitors and 8 µM I-BRD9 for 48 h (n=4, mean ± SD, two-tailed t-test). (B) Western blots and quantifications of PARP, Caspase 9, Caspase 3, and loading control Lamin B in MV4-11 cells pretreated with 20 µM Z-VAD-FMK and then co-incubated with 8 µM I-BRD9 for 48 h (n=3, mean ± SD, two-tailed t-test). (C) Western blots and quantifications of BRD1, BRD2, BRD3, BRD9,and loading control Tubulin in MV4-11 cells treated with 8 µM I-BRD9 for 48 h (n=3, mean ± SD, two-tailed t-test). *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001. SD, standard deviation.
I-BRD9 induces common and cell type-specific target genes
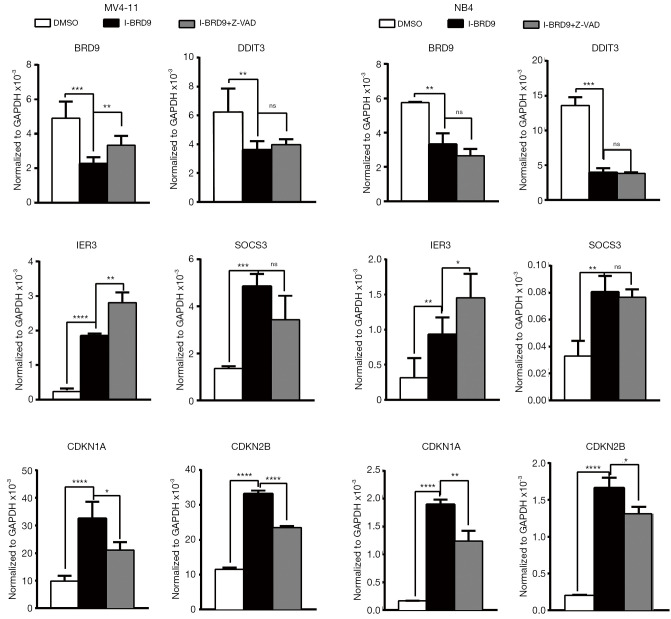
As previously reported, BRD9 can bind to cell type-specific target genes and regulate their expression (15). Among these genes, several are important factors that regulate cell cycle progression and apoptosis. Thus, we evaluated the expression of BRD9, DDIT3, IER3, SOCS3, CDKN1A, and CDKN2B after 24 h I-BRD9 treatment in NB4 and MV4-11 cells. Consistent with the previous report, I-BRD9 induced distinct gene expression in these two AML cell lines; for instance, IER3 expression was increased in MV4-11 cells by I-BRD9 treatment (Figure 4), while SOCS3 expression was much lower in NB4 cells than in MV4-11 cells based on their normalized values. However, CDKN1A and CDKN2B expression were significantly increased in both cell line (Figure 4) and were partially rescued by Z-VAD-FMK, suggesting an important role of these two cell cycle inhibitors in regulating the phenotype induced by I-BRD9. Interestingly, BRD9 expression was also downregulated by I-BRD9, suggesting that BRD9 could regulate its own expression. Taken together, the induction of cell cycle inhibitors and cell death promoter genes might contribute to I-BRD9-induced growth inhibition.
Figure 4.
I-BRD9 induces common and cell type-specific target genes. qRT-PCR analysis of BRD9, DDIT3, IER3, SOCS3, CDKN1A, and CDKN2B in MV4-11 and NB4 cells pretreated with 20µM Z-VAD-FMK and co-incubated with 8 µM I-BRD9 for 24 h (n=3, mean ± SD, two-tailed t-test). *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001. qRT-PCR, real-time quantitative polymerase chain reaction; SD, standard deviation.
Discussion
The targeting of epigenetic factors is emerging as a promising method for the treatment of hematological malignancies. Several epigenetic inhibitors for AML are currently in clinical trials, including type I PRMTs inhibitor GSK3368715, PRMT5 inhibitor GSK3326595, HDACs inhibitors, and Dot1l inhibitor Pinometostat (11,12,18,19). More recently the importance of bromodomain-containing proteins in hematological malignancies has been recognized, such as BRD4, which led to the development of BRD4 inhibitors that have entered clinical trials for the treatment of AML (20). The present study focused on another important bromodomain-containing protein BRD9, which was recently shown to be crucial for AML development in the animal model and was also overexpressed in AML patients (15). We found that targeting the bromodomain-containing protein BRD9 with the small molecule inhibitor I-BRD9 significantly decreased the growth of two AML cell lines. This was accompanied by increased cell death and decreased cell proliferation. Furthermore, we observed that MV4-11 was more sensitive to I-BRD9 compared to NB4. This difference observed is probably becauseI-BRD9 induced higher expression levels of genes involved in promoting cell death in MV4-11 cells compared to NB4 cells. This also fits the reported function ofBRD9 binding to cell-specific genes in different cell types, suggesting that certain AML subtypes may be more responsive to I-BRD9.
In addition to inducing apoptosis, I-BRD9 treatment also increased ferroptosis, although this was not its main effect. Ferroptosis is a novel type of regulated cell death that may be a complementary method to tackle cancers, especially in tumors in which the apoptosis induction pathway is mutated or blocked (21). Thus, it would be interesting to determine whether I-BRD9 could act in synergy with ferroptosis- inducing drugs, such as RSL3 (22), to strengthen its tumor-killing effects. In summary, I-BRD9 showed a strong inhibitory role in AML cells, which may have applications in treating AML alone or in combination with other drugs.
Acknowledgments
We thank Editage for Language editing assistance.
Funding: This work was supported by grants from the National Key R&D Program of China (2017YFC1001903 to JPC) and the Military Logistics Project (AWS17J007).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Institutional ethical approval was waived.
Footnotes
Reporting Checklist: The authors have completed the MDAR checklist. Available at https://dx.doi.org/10.21037/tcr-21-42
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tcr-21-42
Peer Review File: Available at https://dx.doi.org/10.21037/tcr-21-42
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-42). The authors have no conflicts of interest to declare.
References
- 1.Döhner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med 2015;373:1136-52. 10.1056/NEJMra1406184 [DOI] [PubMed] [Google Scholar]
- 2.Estey EH. Acute myeloid leukemia: 2019 update on risk-stratification and management. Am J Hematol 2018;93:1267-91. 10.1002/ajh.25214 [DOI] [PubMed] [Google Scholar]
- 3.Madan V, Shyamsunder P, Han L, et al. Comprehensive mutational analysis of primary and relapse acute promyelocytic leukemia. Leukemia 2016;30:2430. 10.1038/leu.2016.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med 2016;374:2209-21. 10.1056/NEJMoa1516192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tyner JW, Tognon CE, Bottomly D, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562:526-31. 10.1038/s41586-018-0623-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung N, Fung TK, Zeisig BB, et al. Targeting Aberrant Epigenetic Networks Mediated by PRMT1 and KDM4C in Acute Myeloid Leukemia. Cancer Cell 2016;29:32-48. 10.1016/j.ccell.2015.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magliulo D, Bernardi R, Messina S. Lysine-Specific Demethylase 1A as a Promising Target in Acute Myeloid Leukemia. Front Oncol 2018;8:255. 10.3389/fonc.2018.00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.San José-Enériz E, Gimenez-Camino N, Agirre X, et al. HDAC Inhibitors in Acute Myeloid Leukemia. Cancers (Basel) 2019;11:1794. 10.3390/cancers11111794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarighat SS, Santhanam R, Frankhouser D, et al. The dual epigenetic role of PRMT5 in acute myeloid leukemia: gene activation and repression via histone arginine methylation. Leukemia 2016;30:789-99. 10.1038/leu.2015.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuber J, Shi J, Wang E, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011;478:524-8. 10.1038/nature10334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gambacorta V, Gnani D, Vago L, et al. Epigenetic Therapies for Acute Myeloid Leukemia and Their Immune-Related Effects. Front Cell Dev Biol 2019;7:207. 10.3389/fcell.2019.00207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wingelhofer B, Somervaille TCP. Emerging Epigenetic Therapeutic Targets in Acute Myeloid Leukemia. Front Oncol 2019;9:850. 10.3389/fonc.2019.00850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Wang S, Troisi EC, et al. BRD9 defines a SWI/SNF sub-complex and constitutes a specific vulnerability in malignant rhabdoid tumors. Nat Commun 2019;10:1881. 10.1038/s41467-019-09891-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Q, Huang J, Zhang C, et al. The bromodomain containing protein BRD-9 orchestrates RAD51-RAD54 complex formation and regulates homologous recombination-mediated repair. Nat Commun 2020;11:2639. 10.1038/s41467-020-16443-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Gaudio N, Di Costanzo A, Liu NQ, et al. BRD9 binds cell type-specific chromatin regions regulating leukemic cell survival via STAT5 inhibition. Cell Death Dis 2019;10:338. 10.1038/s41419-019-1570-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brien GL, Remillard D, Shi J, et al. Targeted degradation of BRD9 reverses oncogenic gene expression in synovial sarcoma. Elife. 2018;7:e41305. 10.7554/eLife.41305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Theodoulou NH, Bamborough P, Bannister AJ, et al. Discovery of I-BRD9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition. J Med Chem 2016;59:1425-39. 10.1021/acs.jmedchem.5b00256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bewersdorf JP, Shallis R, Stahl M, et al. Epigenetic therapy combinations in acute myeloid leukemia: what are the options?. Ther Adv Hematol 2019;10:2040620718816698. 10.1177/2040620718816698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu XL, Liu HQ, Li J, et al. Role of epigenetic in leukemia: From mechanism to therapy. Chem Biol Interact 2020;317:108963. 10.1016/j.cbi.2020.108963 [DOI] [PubMed] [Google Scholar]
- 20.Alqahtani A, Choucair K, Ashraf M, et al. Bromodomain and extra-terminal motif inhibitors: a review of preclinical and clinical advances in cancer therapy. Future Sci OA 2019;5:FSO372. 10.4155/fsoa-2018-0115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mou Y, Wang J, Wu J, et al. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J Hematol Oncol 2019;12:34. 10.1186/s13045-019-0720-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang C, Zhang X, Yang M, et al. Recent Progress in Ferroptosis Inducers for Cancer Therapy. Adv Mater 2019;31:e1904197. 10.1002/adma.201904197 [DOI] [PubMed] [Google Scholar]