Abstract
How multidrug-resistant tuberculosis (MDR-TB) spreads and expands in Wuhan population is not clear. The study aimed to determine the transmission patterns of MDR-TB in Wuhan city, China, including 149 patients with MDR-TB.
Tuberculosis isolates were genotyped by deletion-targeted multiplex polymerase chain reaction, mycobacterial interspersed repetitive unit-variable number tandem repeat typing, and sequencing of drug resistance-associated genes. The risk factors of genomic-clustering were analyzed with logistic regression. The genomic-clustering patients were deeply investigated.
The analysis identified 111 unique and 11 clustered genotypes (38 isolates). The clustering rate was 25.50% and the minimum estimate proportion of recent transmission was 18.12%. Two clusters (5 isolates) shared the same mutation, the remain 9 clusters (33 isolates) had different mutation. Logistic regression showed that older than 60 years (adjusted OR 2.360, 95% CI:1.052-5.292) was an independent factor associated with the genomic-clustering of MDR-TB. Among the 38 genomic-clustering cases, 14 cases had epidemiological transmission links. The most common type of transmission link was social contact.
The local transmission of MDR-TB in Wuhan was really an issue. The elderly population might be the high-risk groups for transmission of MDR-TB, and the community or public transportation might be the main transmission places.
Keywords: genotype, multidrug-resistant tuberculosis, Mycobacterium tuberculosis, transmission pattern
1. Introduction
According to the World Health Organization global tuberculosis report,[1] 3.3% of new tuberculosis (TB) cases and 17.7% of previously treated TB patients were MDR/rifampicin drug resistant tuberculosis (RR-TB) (multidrug resistant tuberculosis [MDR-TB]; drug resistant tuberculosis, RR-TB) in 2019. It is estimated that 465,000 cases were RR-TB and 78.0% of them were MDR-TB. China has the second largest proportion (14.0%) of drug-resistant TB, and MDR-TB brings great challenges to tuberculosis control in China.
Although previous studies have explored the genetic diversity of MDR-TB isolates in China,[2–5] few studies combined the genotyping and epidemiological investigation on the transmission of MDR-TB. Yang et al[6] explored the transmission dynamics of MDR-TB in Shanghai, China, and concluded that transmission of MDR-TB had an important role in the burden of MDR-TB. Jiang et al's study[7] found that local transmission of MDR-TB was a serious problem in Shenzhen, China, and suggested schools and factories are common places of transmission occurrence. Wuhan, where our study took place, is the largest land, water and air transport hub in China's inland, and known as “the thoroughfare of nine provinces”. Its unique geographical advantages also mean that this area is densely populated and mobile, which undoubtedly creates an ideal environment for the spread and epidemic of MDR-TB. How multidrug-resistant tuberculosis spreads and expands in Wuhan population is not clear.
In order to comprehensively understand the transmission level and pattern of MDR-TB in Wuhan, we combined 24-locus mycobacterial interspersed repetitive unit-variable number tandem repeat typing (MIRU-VNTR) and deletion-targeted multiplex polymerase chain reaction (DTM-PCR) to genotype the isolates. Rifampicin resistance determining region of rpoB gene and isoniazid resistance-related genes (katG, inhA, fabG gene) were also sequenced to cluster. Furthermore, the genomic-clustering subjects were interviewed to explore the transmission dynamic of MDR-TB.
2. Materials and methods
2.1. Ethics statement
This study was approved by the Wuhan Pulmonary Hospital Medical Ethics Committee (protocol number: 2018-12). All the patients signed an informed consent form before they participated in this study.
2.2. Subjects enrollment and drug susceptibility testing
In 2017, a total of 341 MDR Mycobacterium tuberculosis (MTB) were detected from 6072 active pulmonary tuberculosis cases registered in Wuhan TB reporting network, which belongs to the National Tuberculosis Program system. Among them, 151 cases were detected by Wuhan Pulmonary Hospital, which is 1 of the 2 designated hospitals for tuberculosis in Wuhan. After excluding 2 lost isolates, 149 isolates were included in this study.
MDR-TB was defined as being resistant to at least 2 primary first-line anti-TB drugs (rifampicin and isoniazid) in vitro.[8] The standard strain H37Rv was provided by the National Tuberculosis Reference Laboratory as the control strain in this experiment. MDR-TB isolates were cultured according to the national standardized rules for tuberculosis bacteriological test and stored in the strain bank at −80 °C. MDR-TB isolates were confirmed by proportional drug susceptibility test with the following concentration: isoniazid (0.2 μg/mL), rifampin (40.0 μg/mL), streptomycin (4.0 μg/mL), ethambutol (2.0 μg/mL).
2.3. DNA extraction
Genomic DNA of the isolate was extracted according to the bacterial genomic DNA extraction kit provided by Daan Gene Biology Co., Ltd.
2.4. MIRU-VNTR typing
MIRU-VNTR typing was performed according to the international standard 24 loci method.[9] The primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd. Each locus amplification reaction was performed in a total volume of 20 μL consisting of 10 μL 2 × Taq PCR MasterMix (CWBIO), 2 μL 2 μmol/L each primer, 1 μL DNA lysate and 5 μL ddH2O. The PCR products were analyzed in a 1.5%∼2.0% agarose gel using a 100 bp DNA ladder as the molecular weight standard. H37Rv and H2O were used as positive and negative controls, respectively, in each PCR reaction. The number of tandem repeats was calculated based on the length of the repeat and flank sequences for each locus.
2.5. Deletion-targeted multiplex polymerase chain reaction
DTM-PCR was used to detect the deletion of RDl05 fragment to differentiate Beijing family strains. The modern or ancient sublineage of Beijing family was identified by detecting the IS6110 in the NTF region. The detail of this method was described in Khosravi AD's[10] and Wada T's study.[11]
2.6. Drug resistance-associated genes sequencing
Drug resistance-associated genes sequencing was used for secondary genotyping of after the isolates were clustered by MIRU-VNTR and DTM-PCR. The following major resistance-related gene regions were directly sequenced: rifampicin resistance determining region of rpoB gene and isoniazid resistance-related genes (katG, inhA, and fabG).
2.7. Interview and epidemiological transmission links analysis
The clinical variables and demographic information of each MDR-TB case were derived from electronic medical records. Patients whose isolates were identified as genomic-clustered were invited for a questionnaire survey. The social relations, medical history, contacts with active TB patients and their commonly acting places was included in the survey. The epidemiological transmission links were defined as the following: confirmed epidemiological transmission link (patients knew each other or shared the same address or complex), probable transmission link (patients did not know each other but the distance of residence less than 2 km, or shared the same bus line in similar time period, or visited the same food market), and without transmission link (patients did not know each other and lacked a common neighborhood, setting or bus line).
2.8. Data analysis
The gel images of different MIRU-VNTR loci and their background information of each isolate were input into the computer, and the PCR amplification fingerprints of the tested isolates were compared with H37Rv. The molecular weights and repetitions of different MIRU locus of TB were calculated based on the repeated H37Rv sequences.
The online tool at website http://www.MIRU-VNTRplus.org was used to perform the clustering analysis.[12] We constructed minimum spanning tree by unweighted pair group method with arithmetic mean. The Hunter-Gaston discriminatory index (HGDI) values of 24 loci was calculated by the equation of Hunter and Gaston.[13] The discriminatory power of each locus was judged as poor if HGDI<0.3, moderate if 0.3≤ HGDI≤0.6, or high if HGDI>0.6.
The MTB isolates were considered to be clustered if they had identical MIRU-VNTR and DTM-PCR genotype.[14] The minimum estimate of the recent infection rate was calculated as follows: (nc – c)/n, where nc represents the clustered isolates, c represents the number of clusters, and n represents all samples of isolates.
SPSS 21.0 (SPSS Inc., Chicago, IL) was used for statistical analysis. The demographic and clinical features of patients were compared between modern-Beijing and ancient-Beijing genotypes, and between clustered and non-clustered groups were assessed with univariate logistic regression. To analyze the influencing factors associated with the genomic-clustering, the multivariable logistics regression model was used. A forward, step-wise approach was used to select the variables.
3. Results
3.1. Demographic and clinical features
The relationships of age, sex, sputum smear results, and treatment history with Beijing sublineage (modern and ancient sublineage) of 149 patients were shown in Table 1. The relationship between the variables and Beijing sublineage did not show statistical significance (P > .05).
Table 1.
Demographic and clinical features of patients infected with Modern-Beijing and Ancient-Beijing genotype isolates.
| Number of Ancient Beijing family (n = 17) | Number of Modern Beijing family (n = 125) | OR | 95% CI | P value | |
| Sex | |||||
| Male | 10 | 93 | 1.000 | ||
| Female | 7 | 32 | 2.034 | 0.715-5.791 | .18 |
| Age (yrs) | |||||
| 14-60 | 14 | 97 | 1.000 | ||
| ≥60 | 3 | 28 | 1.347 | 0.361-5.022 | .66 |
| Treatment history | |||||
| New case | 11 | 80 | 1.000 | ||
| Retreated | 6 | 45 | 0.970 | 0.336-2.798 | .96 |
| Positive sputum smear result | |||||
| No | 6 | 31 | 1.000 | ||
| Yes | 11 | 94 | 1.654 | 0.565-4.843 | .36 |
3.2. DTM-PCR
All 149 strains were divided into Beijing strain (142 strains) and non-Beijing strain (7 strains). Among Beijing strains, 125 strains were identified as modern-Beijing sublineage and 17 strains were ancient-Beijing sublineage (Table 1).
3.3. Mycobacterial interspersed repetitive unit-variable number tandem repeat typing
Allelic diversity of each MIRU-VNTR locus are displayed in Table 2. Two out of 24 loci (QUB11b, MIRU26) had the high discriminatory power, 6 loci (Mtub21, Mtub04, QUB26, MIRU10, Mtub39, MIRU24) had the moderate discriminatory power, and the remain loci had the low discriminatory power.
Table 2.
Allelic diversity of each MIRU-VNTR locus for MDR-MTB isolates in Wuhan, China (n = 149).
| Allele number | ||||||||||||||
| VNTR alias | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | HGDI | Conclusion∗ |
| QUB11b | 1 | 3 | 3 | 5 | 6 | 46 | 70 | 14 | 1 | 0 | 0 | 0 | 0.6738 | High |
| MIRU26 | 0 | 3 | 1 | 6 | 6 | 44 | 77 | 10 | 1 | 1 | 0 | 0 | 0.6393 | High |
| Mtub21 | 0 | 3 | 1 | 6 | 33 | 93 | 10 | 2 | 1 | 0 | 0 | 0 | 0.5553 | Moderate |
| Mtub04 | 0 | 0 | 5 | 52 | 88 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0.5279 | Moderate |
| QUB26 | 0 | 1 | 3 | 3 | 1 | 2 | 8 | 13 | 105 | 11 | 1 | 1 | 0.4861 | Moderate |
| MIRU10 | 0 | 5 | 36 | 106 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.4333 | Moderate |
| Mtub39 | 5 | 5 | 119 | 17 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.3444 | Moderate |
| MIRU24 | 0 | 118 | 0 | 0 | 0 | 31 | 0 | 0 | 0 | 0 | 0 | 0 | 0.3272 | Moderate |
| ETRA | 0 | 0 | 2 | 18 | 124 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0.2887 | Poor |
| QUB4156 | 0 | 0 | 0 | 128 | 9 | 11 | 0 | 1 | 0 | 0 | 0 | 0 | 0.2495 | Poor |
| MIRU31 | 0 | 0 | 3 | 8 | 131 | 5 | 1 | 1 | 0 | 0 | 0 | 0 | 0.2188 | Poor |
| MIRU39 | 0 | 2 | 12 | 132 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.2042 | Poor |
| MIRU40 | 0 | 1 | 8 | 134 | 2 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0.1834 | Poor |
| Mtub30 | 0 | 0 | 10 | 2 | 137 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.1452 | Poor |
| ETRC | 0 | 3 | 1 | 139 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0.1231 | Poor |
| MIRU16 | 0 | 2 | 5 | 141 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0978 | Poor |
| MIRU04 | 0 | 1 | 3 | 142 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0856 | Poor |
| MIRU27 | 0 | 0 | 4 | 144 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0593 | Poor |
| ETRB | 0 | 2 | 145 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0466 | Poor |
| Mtub34 | 0 | 0 | 2 | 145 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0466 | Poor |
| MIRU02 | 0 | 3 | 146 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0332 | Poor |
| MIRU23 | 0 | 0 | 0 | 0 | 0 | 147 | 2 | 0 | 0 | 0 | 0 | 0 | 0.0201 | Poor |
| MIRU20 | 0 | 1 | 148 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0068 | Poor |
| Mtub29 | 0 | 0 | 0 | 149 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0000 | Poor |
HGDI = Hunter-Gaston discriminatory index, MDR-MTB = multidrug-resistant Mycobacterium tuberculosis, MIRU-VNTR = mycobacterial interspersed repetitive unit-variable number tandem repeat typing.
HGDI>0.6, highly discriminatory; 0.3≤ HGDI≤0.6, moderately discriminatory; HGDI<0.3, poorly discriminatory.
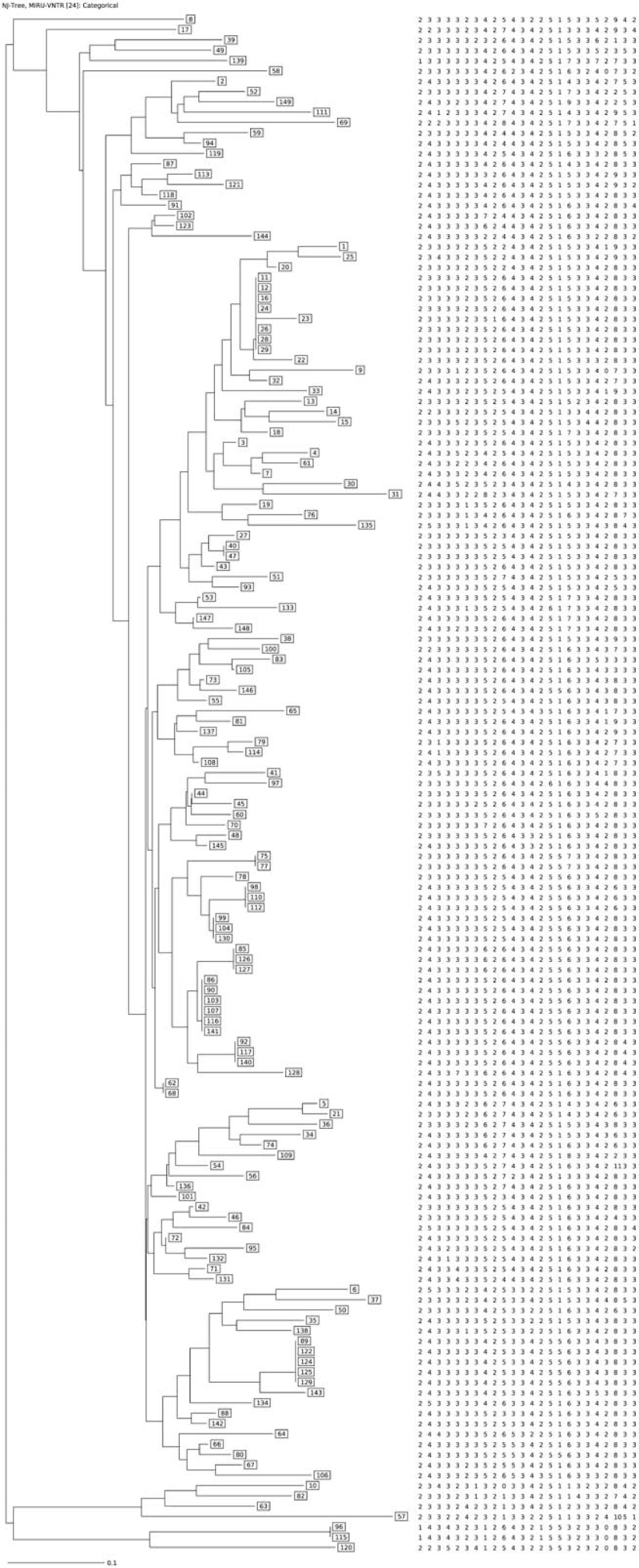
By 24 loci MIRU-VNTR, 111 unique genotypes and 11 clustered genotypes (38 isolates) from 149 isolates were identified, and the cumulative HGDI was 0.9943 (Table 3, Fig. 1). The clustering rate was 25.50% and the minimum estimate proportion of recent transmission was 18.12%. The largest cluster contained 7 isolates and the smallest cluster contained 2 isolates. Except the isolates from the 2 cases in cluster 11 were non-Beijing lineage, the others were Beijing lineage (Table 4). The genotyping results from MIRU-VNTR and DTM-PCR were consistent.
Table 3.
Cumulative HGDI of each MIRU-VNTR locus (n = 149).
| VNTR alias | Individual HGDI | Number of patterns | Number of clusters | Number of clustered isolates | Clustering rate (%) | Number of isolates in each cluster | Cumulative HGDI |
| QUB11b | 0.6738 | ||||||
| MIRU26 | 0.6393 | 26 | 19 | 142 | 95.30 | 2-39 | 0.8637 |
| Mtub21 | 0.5553 | 43 | 21 | 127 | 85.23 | 2-30 | 0.9221 |
| Mtub04 | 0.5279 | 62 | 22 | 109 | 73.15 | 2-22 | 0.9521 |
| QUB26 | 0.4861 | 81 | 19 | 87 | 58.39 | 2-16 | 0.9711 |
| MIRU10 | 0.4333 | 89 | 19 | 79 | 53.02 | 2-15 | 0.9769 |
| Mtub39 | 0.3444 | 96 | 18 | 71 | 47.65 | 2-13 | 0.9817 |
| MIRU24 | 0.3272 | 102 | 18 | 65 | 43.62 | 2-9 | 0.9881 |
| ETRA | 0.2887 | 107 | 17 | 59 | 39.60 | 2-9 | 0.9896 |
| QUB4156 | 0.2495 | 108 | 18 | 59 | 39.60 | 2-9 | 0.9912 |
| MIRU31 | 0.2188 | 111 | 17 | 55 | 36.91 | 2-8 | 0.9923 |
| MIRU39 | 0.2042 | 112 | 17 | 54 | 36.24 | 2-8 | 0.9926 |
| MIRU40 | 0.1834 | 116 | 14 | 47 | 31.54 | 2-8 | 0.9931 |
| Mtub30 | 0.1452 | 118 | 14 | 45 | 30.20 | 2-8 | 0.9934 |
| ETRC | 0.1231 | 119 | 13 | 43 | 28.86 | 2-8 | 0.9935 |
| MIRU16 | 0.0978 | 120 | 12 | 41 | 27.52 | 2-8 | 0.9936 |
| MIRU04 | 0.0856 | 121 | 11 | 39 | 26.17 | 2-8 | 0.9937 |
| MIRU27 | 0.0593 | 121 | 11 | 39 | 26.17 | 2-8 | 0.9937 |
| ETRB | 0.0466 | 121 | 11 | 39 | 26.17 | 2-8 | 0.9937 |
| Mtub34 | 0.0466 | 121 | 11 | 39 | 26.17 | 2-8 | 0.9937 |
| MIRU02 | 0.0332 | 121 | 11 | 39 | 26.17 | 2-8 | 0.9937 |
| MIRU23 | 0.0201 | 121 | 11 | 39 | 26.17 | 2-8 | 0.9937 |
| MIRU20 | 0.0068 | 122 | 11 | 38 | 25.50 | 2-7 | 0.9943 |
| Mtub29 | 0.0000 | 122 | 11 | 38 | 25.50 | 2-7 | 0.9943 |
HGDI = Hunter-Gaston discriminatory index, MIRU-VNTR = mycobacterial interspersed repetitive unit-variable number tandem repeat typing.
Figure 1.
Minimum spanning tree of 149 MDR Mycobacterium tuberculosis isolates from Wuhan.
Table 4.
Demographic and clinic characteristics of patients and genetic information of 38 MDR-MTB clustered isolates.
| Drug resistance gene mutation | |||||||||
| ID of cluster | Age | Sex | Previous treatment | MIRU-VNTR | Beijing | rpoB | katG | inhA | fabG |
| 1 | 45 | M | New | 243333352543425563342833 | Yes | Ser450Leu | – | – | −15C>T |
| 1 | 40 | M | New | 243333352543425563342833 | Yes | His445Tyr | Ser315Thr | – | – |
| 1 | 22 | M | Retreatment | 243333352543425563342833 | Yes | Ser450Leu | Ser315Thr | – | – |
| 2 | 60 | M | Retreatment | 243333352543425563342633 | Yes | Ser450Leu | Ser315Thr | – | – |
| 2 | 60 | M | New | 243333352543425563342633 | Yes | Ser450Leu | Ser315Thr | – | – |
| 2 | 78 | M | New | 243333352543425563342633 | Yes | Ser450Leu | Ser315Thr | – | – |
| 3 | 68 | M | Retreatment | 243333362643425563342833 | Yes | His445Leu | Ser315Thr | – | – |
| 3 | 59 | M | Retreatment | 243333362643425563342833 | Yes | Ser450Leu | Ser315Thr | – | – |
| 3 | 14 | F | New | 243333362643425563342833 | Yes | Ser450Leu | – | – | – |
| 4 | 61 | M | New | 243333352643425563342833 | Yes | Asp435Val | Ser315Thr | – | −15C>T |
| 4 | 48 | M | New | 243333352643425563342833 | Yes | Ser450Leu | Trp191Arg | – | −15C>T |
| 4 | 32 | F | Retreatment | 243333352643425563342833 | Yes | Ser450Leu | – | – | −15C>T |
| 4 | 61 | M | Retreatment | 243333352643425563342833 | Yes | Asp435Val | Ser315Thr | – | – |
| 4 | 22 | F | New | 243333352643425563342833 | Yes | Ser450Leu | Ser315Thr | – | – |
| 4 | 47 | M | New | 243333352643425563342833 | Yes | Ser450Leu | Ser315Thr | – | – |
| 5 | 28 | F | Retreatment | 243333352643425563342843 | Yes | Ser450Leu | Ser315Thr | – | – |
| 5 | 51 | M | Retreatment | 243333352643425563342843 | Yes | Ser450Leu | Ser315Thr | – | – |
| 5 | 88 | F | New | 243333352643425563342843 | Yes | Ser450Leu | – | – | – |
| 6 | 51 | M | Retreatment | 243333352643425163342833 | Yes | Ser450Leu | – | – | – |
| 6 | 61 | M | New | 243333352643425163342833 | Yes | His445Leu | Ser315Thr | – | – |
| 7 | 61 | M | Retreatment | 233332352643425153342833 | Yes | Ser450Leu | – | – | – |
| 7 | 63 | M | Retreatment | 233332352643425153342833 | Yes | Ser450Leu | – | – | −15C>T |
| 7 | 45 | M | New | 233332352643425153342833 | Yes | Ser450Leu | Ser315Thr | – | – |
| 7 | 37 | M | Retreatment | 233332352643425153342833 | Yes | Asp435Val | Ser315Thr | – | – |
| 7 | 22 | M | New | 233332352643425153342833 | Yes | Ser450Leu | Ser315Thr | – | – |
| 7 | 59 | M | New | 233332352643425153342833 | Yes | Asp435Tyr | – | – | – |
| 7 | 41 | F | New | 233332352643425153342833 | Yes | Asn437Asp, Asp435Val | Ser315Thr | – | −15C>T |
| 8 | 29 | M | New | 233333352543425153342833 | Yes | Ser450Leu | – | – | −15C>T |
| 8 | 77 | M | Retreatment | 233333352543425153342833 | Yes | Ser450Leu | – | – | – |
| 9 | 36 | F | Retreatment | 233333352643425573342833 | Yes | Ser450Leu | Ser315Thr | – | −8C>T |
| 9 | 30 | F | New | 233333352643425573342833 | Yes | Ser450Leu | Ser315Thr | – | – |
| 10 | 26 | M | New | 243333342533425563343833 | Yes | Ser450Leu | Ser315Thr | – | – |
| 10 | 35 | F | New | 243333342533425563343833 | Yes | His445Tyr | – | – | −15C>T |
| 10 | 73 | M | Retreatment | 243333342533425563343833 | Yes | Ser450Leu | Ser315Asn | – | – |
| 10 | 72 | M | New | 243333342533425563343833 | Yes | Ser450Leu | Ser315Thr | – | – |
| 10 | 35 | M | Retreatment | 243333342533425563343833 | Yes | His445Tyr | Ser315Thr | – | – |
| 11 | 78 | M | New | 143432312643215532330832 | No | Ser450Leu | Ser315Thr | – | – |
| 11 | 55 | M | New | 143432312643215532330832 | No | Ser450Leu | Ser315Thr | – | – |
MDR-MTB = multidrug-resistant Mycobacterium tuberculosis, MIRU-VNTR = mycobacterial interspersed repetitive unit-variable number tandem repeat typing.
3.4. Sequencing results of drug resistance-associated gene
RpoB, katG, inhA, and fabG genes were sequenced in genomic-clustered isolates, and their mutation characteristics are shown in Table 4. Among the rpoB gene mutations, 28 cases had a well-known mutation of Ser450Leu substitution, 1 case had Asn437Asp substitution, 1 case had Asp435Tyr substitution, 4 cases had Asp435Val substitution, 2 cases had His445Leu substitution, and 3 cases had His445Tyr substitution. One out of the 38 isolates had double mutations of rpoB gene. Among the katG gene mutations, 25 cases had the most common Ser315Thr substitution, 1 case had Trp191Arg substitution, 1 case had Ser315Asn substitution, and 11 cases had no mutation. There was no mutation in inhA gene of all 149 isolates. Among all 149 isolates, 8 isolates had fabG gene mutation of C. –15C >T, only 1 isolate had C. –8C > T, and the other isolates had no mutation.
Among the 11 clusters, 5 clusters shared the same rpoB gene mutations, all of which were the most common Ser450Leu substitution. In the largest cluster that contained 7 cases, 2 MDR-TB cases whose isolates had the same rare substitution of Asp435Val, and the distance between their residence was within 5 km. In the cluster 4, 2 cases had isolates with the same rare substitution of Asp435Val, and the 2 cases shared same bus line in 1 year before the diagnosis of MDR-TB. Among the 11 clusters, 4 clusters had the same mutation in katG gene, of which 3 clusters had the most common Ser315Thr, and the other cluster had the wild-type gene. In the largest cluster that contained 7 cases, 4 cases had isolates with the substitution of Ser315Thr, and the other 3 cases had isolates with wild-type gene. In addition, all isolates showed wild-type inhA gene. Five out of 11 clusters had identical fabG gene mutations, all of which were wild-type gene. One out of 11 clusters had wild-type inhA gene and –8C>T, and the remain 5 clusters had wild-type inhA gene and –15C>T.
Besides, the gene mutation of isolates in 2 clusters (5 isolates) were totally same, and the other clusters contained at least 1 isolate with different mutation.
3.5. Epidemiological transmission links of clustered cases and risk factors of genomic-clustering
According to the epidemiological survey, 8 out of the 38 MDR-TB cases had confirmed epidemiological transmission links, and 6 cases had probable epidemiological transmission links, and the remaining had no epidemiological transmission links (Table 5). The most common type of epidemiological transmission link was social contact, such as visiting the same food market, taking the same bus line, and residence proximity. Only 2 cases had close family contact and 2 cases had nosocomial infection.
Table 5.
Characteristics of multidrug-resistant tuberculosis genomic clusters (n = 38).
| Number of cases in cluster | Number of cases living in the same district | Number of cases with positive sputum smear result | Number of new cases | Known risk factors (number of cases) | Epidemiologically linked | Nature of epidemiological link [number of cases] | |
| Cluster 1 | 3 | 2 | 2 | 2 | Retreatment (1) | No | |
| Cluster 2 | 3 | 2 | 3 | 2 | Retreatment (1), old (1)∗ | Yes | Household [2] |
| Cluster 3 | 3 | 0 | 3 | 1 | Retreatment (2), hospitalization (2) | Yes | Nosocomial (health center [2]) |
| Cluster 4 | 6 | 2,2 | 3 | 4 | Retreatment (2) | Probable | Social (bus [2], residence proximity [2]) |
| Cluster 5 | 3 | 3 | 2 | 1 | Retreatment (2), old (1)∗ | No | |
| Cluster 6 | 2 | 0 | 2 | 1 | Retreatment (1) | No | |
| Cluster 7 | 7 | 2,2 | 7 | 4 | Retreatment (3) | Yes | Social (community, food market [2]) |
| Cluster 8 | 2 | 0 | 2 | 1 | Retreatment (1), old (1)∗ | No | |
| Cluster 9 | 2 | 0 | 2 | 1 | Retreatment (1) | No | |
| Cluster 10 | 5 | 0 | 2 | 3 | Retreatment (2), Cook in shopping plaza (1) | Probable | Social (bus [2]) |
| Cluster 11 | 2 | 0 | 1 | 2 | Old (1)∗ | Yes | Social (former residence place [2]) |
Older than 75 yrs old.
Table 6 shows the results of risk factors associated with genomic-clustering of MDR-TB. Using the multivariate analysis, we found that older than 60 years was the independent factors associating with the genomic-clustering (adjusted OR 2.360, 95% CI:1.052-5.292).
Table 6.
Univariable analysis of risk factors associated with multidrug-resistance in genomic clusters.
| Non-cluster (n = 111) | Cluster (n = 38) | OR | 95% CI | P value | |
| Sex | |||||
| Male | 80 | 29 | 1.000 | ||
| Female | 31 | 9 | 1.249 | 0.531-2.936 | .61 |
| Age (yrs) | |||||
| 14-60 | 89 | 24 | 1.000 | ||
| ≥60 | 22 | 14 | 2.360 | 1.052-5.292 | .04 |
| Treatment history | |||||
| New case | 73 | 22 | 1.000 | ||
| Retreated | 38 | 16 | 0.716 | 0.337-1.521 | .39 |
| Positive sputum smear result | |||||
| No | 29 | 9 | 1.000 | ||
| Yes | 82 | 29 | 1.140 | 0.483-2.691 | .77 |
| Treatment outcome | |||||
| Cured or treatment completed | 58 | 16 | 1.000 | ||
| Treatment failed | 5 | 0 | 0.000 | 0.000 | .99 |
| Default, moved, or lost to follow-up | 24 | 11 | 1.661 | 0.673-4.099 | .27 |
| Died | 4 | 0 | 0.000 | 0.000 | .99 |
| Stop treatment due to drug-adverse-event | 4 | 4 | 3.625 | 0.815-16.122 | .09 |
| unknown | 16 | 7 | 1.586 | 0.557-4.516 | .39 |
| Beijing strains | |||||
| No | 5 | 2 | 1.000 | ||
| Yes | 106 | 36 | 1.178 | 0.219-6.337 | .85 |
4. Discussion
The MDR-TB is still a serious public health issue in China. According to the National Baseline Survey of Drug-resistant Tuberculosis conducted in 2007 to 2008, the overall prevalence of MDR-TB was 8.3% in smear-positive tuberculosis patients (5.7% among new cases; 25.6% among retreatment cases).[15] Wuhan city is the transportation hub of the central part and the fourth largest city in China. Based on the retrospective population-based study in Wuhan, we combined the genotyping method and traditional epidemiological investigation to explore the transmission patterns of MDR-TB. The genotyping method included MIRU-VNTR, DTM-PCR and drug resistance-associated genes sequencing.
The genotyping of MTB plays an important role in tracing the source of infection in contact investigation,[16,17] distinguishing exogenous infection and endogenous relapse,[18] and outbreak investigation,[19,20] which can make up for the shortcomings of traditional epidemiological survey. Identification of Beijing family genotype is often the first step of genotyping MTB. The most widely used methods to detect Beijing family were spoligotyping[21,22] and DTM-PCR.[2] This study adopted DTM-PCR which was simpler and quickly. The second step of genotyping was RFLP[23] or MIRU-VNTR,[22] and MIRU-VNTR was more operational. The third step was whole-genome sequencing (WGS). However, because of the high cost and complex data analysis, sequencing of the drug-resistant gene was conducted as a substitute of WGS. In this study, we found that the HGDI of standard 24-locus MIRU-VNTR was relatively high, and the result of DTM-PCR genotyping remained unchanged. The further genotyping by sequencing of drug resistance-associated genes showed that several isolates in the same cluster shared the same rare mutation, indicated more chance to have the same transmission source.
National Baseline Survey of Drug-resistant Tuberculosis in China demonstrated that the proportion of drug-resistant patients who did not have treatment history was more than 70.0%. Shao's[18] study reported that 47.8% of treated patients with TB was due to reinfection rather than relapse. Several studies that combined WAS and traditional epidemiological methods showed the evidence of recent transmission of MDR-TB in China.[6,7] In the present study, we found that the clustering rate was 25.50%, the minimum estimate proportion of recent transmission was 18.12%, and the proportion of patients who had epidemiological transmission link accounting for 9.40%. The recent transmission rate observed in the present study may be underestimated. First, the MDR-TB patients in this study was the total patients in Wuhan Pulmonary Hospital, not included the patients in another designated hospital (Jin-yin-tan hospital), thus only representing about 40.0% of patients in Wuhan. Second, the proportion of culture-positive tuberculosis was only 40.6% (data in Wuhan TB reporting network) in Wuhan in 2017. Such a low proportion of culture-positive TB cases may reduce the effectiveness of molecular epidemiological studies. Third, WGS was more efficient in detecting epidemiological association than MIRU-VNTR.[24–26] Due to the high cost and complex analysis of WGS, we used MIRU-VNTR combing sequencing of common drug-resistant gene. Thus, the method rendered a suboptimal discrimination among different genotypes.
In this study, we found that the rate of epidemiological transmission link among genomic-clustered patients was lower than that observed in Yang et al's[6] study in Shanghai, whereas the result was similar to that in Jiang et al's[7] study in Shenzhen. Maybe the reason is that as well as in the present study, the subjects in the study in Shenzhen only represented about 40.0% of the whole MDR-TB cases. We also found the places where transmission most frequently occurred were community and public transportation. This was similar as Shanghai,[6] whereas it was different from Shenzhen[7] in which school and factory were the most common place transmission occurring. Maybe this difference is due to the proportion of inter-migrant in Shenzhen being much higher than Wuhan.
In addition, Yang et al[6] also investigated the risk factors associated with genomic-clustering. Consistent with their results, our observation also showed that older than 60 years was an independent risk factor of genomic-clustering. The elderly are more likely to lack knowledge about tuberculosis, which leads to serious delay in diagnosis or treatment.[27,28] This maybe a possible reason for high chance of transmission in the elderly. Therefore, elderly population deserved increased vigilance in MDR-TB control.
This study had several strengths. The subjects were the total MDR-TB cases detected by Wuhan Pulmonary Hospital, representing about half of the registered cases in Wuhan TB reporting network, thus this study is population-based, not hospital-based.
What is more, we combined molecular with traditional epidemiological method to explore the transmission pattern of MDR-TB in Wuhan city. The DTM-PCR, MIRU-VNTR, and sequencing of drug-resistance-associated genes were used for genotyping of MTB. The traditional epidemiological investigation was meanwhile conducted. Combining these 2 methods, we can better explore the population transmission dynamic of MDR-TB.
But this study had some limitations. MDR-TB patients in this study represented only about 40.0% patients in Wuhan. The sputum culture positivity rate of confirmed TB cases was relatively low (40.6%) in Wuhan in 2017, and the genotyping methods by this study had a suboptimal genotyping discrimination. Thus, the transmission level of MDR-TB in Wuhan might be underestimated.
Due to the retrospective study design, information such as the social relations, medical history, contacts with active TB patients and their commonly acting places among patients was based on the recall of the subjects, the recall bias cannot be avoided.
In summary, we concluded that the local transmission of MDR-TB in Wuhan was really an issue. The elderly population might be the high-risk groups for transmission of MDR-TB, and the community or public transportation might be the main transmission places. The findings of this study remind related department to take actions to prevent the transmission of MDR-TB.
Author contributions
Conceptualization: Duan Qionghong, Zhou Melian, Li Yuehua, Chen Jun.
Data Curation: Duan Qionghong, Chen Jun.
Formal analysis: Duan Qionghong, Zhou Melian.
Funding acquisition: Duan Qionghong, Zhou Melian.
Investigation: Zhang Zhengbin, Tian Dan, Hu Yanjie, Wu Jun, Wang Tiantian,
Methodology: Li Yuehua, Chen Jun.
Project administration: Chen Jun.
Supervision: Chen Jun.
Validation: Duan Qionghong, Zhang Zhengbin, Tian Dan.
Visualization: Duan Qionghong.
Writing – original draft: Duan Qionghong, Zhang Zhengbin, Tian Dan, Zhou Melian.
Writing – review & editing: Duan Qionghong, Zhang Zhengbin, Tian Dan, Zhou Melian, Li Yuehua, Chen Jun.
Footnotes
Abbreviations: DTM-PCR = deletion-targeted multiplex polymerase chain reaction, HGDI = Hunter-Gaston discriminatory index, MDR-TB = multidrug-resistant tuberculosis, MIRU-VNTR = mycobacterial interspersed repetitive unit-variable number tandem repeat typing, MTB = Mycobacterium tuberculosis, RR-TB = rifampicin drug resistant tuberculosis, TB = tuberculosis, WGS = whole-genome sequencing.
How to cite this article: Duan Q, Zhang Z, Tian D, Zhou M, Hu Y, Wu J, Wang T, Li Y, Chen J. Transmission of multidrug-resistant Mycobacterium tuberculosis in Wuhan, China: a retrospective molecular epidemiological study. Medicine. 2022;101:4(e28751).
This work was supported by Wuhan Municipal Health Commission Health [grant number: WG20A03 and WG15A04]; and Hubei Municipal Health Commission Health [grant number: WJ2021M026].
The data that support the findings of this study are available from Wuhan pulmonary hospital but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of Wuhan pulmonary hospital.
The authors have no conflicts of interest to disclose.
The data that support the findings of this study are available from a third party, but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are available from the authors upon reasonable request and with permission of the third party.
References
- [1].WHO. Global tuberculosis report 2020. 2020. Available at: https://apps.who.int/iris/handle/10665/336069. Accessed August 28, 2020. [Google Scholar]
- [2].Luo D, Chen Q, Xiong G, et al. Prevalence and molecular characterization of multidrug-resistant M. tuberculosis in Jiangxi province, China. Sci Rep 2019;9:7315. 10.1038/s41598-019-43547-2 10.1038/s41598-019-43547-2, PMID:31086215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lin S, Wei S, Zhao Y, et al. Genetic diversity and drug susceptibility profiles of multidrug-resistant tuberculosis strains in Southeast China. Infect Drug Resist 2021;14:3979–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Li D, Song Y, Yang P, et al. Genetic diversity and drug resistance of Mycobacterium tuberculosis in Yunnan, China. J Clin Lab Anal 2019;33:e22884. 10.1002/jcla.22884 10.1002/jcla.22884, PMID:30896073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jia QJ, Zeng MC, Li QC, et al. Recent transmission analysis of 66 MDR-TB strains using the 15-locus MIRU-VNTR method. Biomed Environ Sci 2020;33:791–5. [DOI] [PubMed] [Google Scholar]
- [6].Yang C, Luo T, Shen X, et al. Transmission of multidrug-resistant Mycobacterium tuberculosis in Shanghai, China: a retrospective observational study using whole-genome sequencing and epidemiological investigation. Lancet Infect Dis 2017;17:275–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jiang Q, Liu Q, Ji L, et al. Citywide transmission of multidrug-resistant tuberculosis under China's rapid urbanization: a retrospective population-based genomic spatial epidemiological study. Clin Infect Dis 2020;71:142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yao C, Guo H, Li Q, et al. Prevalence of extensively drug-resistant tuberculosis in a Chinese multidrug-resistant TB cohort after redefinition. Antimicrob Resist Infect Control 2021;10:126. 10.1186/s13756-021-00995-8 10.1186/s13756-021-00995-8, PMID:34446095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chizimu JY, Solo ES, Bwalya P, et al. Genetic diversity and transmission of multidrug-resistant Mycobacterium tuberculosis strains in Lusaka, Zambia. Int J Infect Dis 2021;114:142–50. [DOI] [PubMed] [Google Scholar]
- [10].Khosravi AD, Goodarzi H, Alavi SM, Akhond MR. Application of deletion- targeted multiplex PCR technique for detection of Mycobacterium tuberculosis Beijing strains in samples from tuberculosis patients. Iran J Microbiol 2014;6:330–4. [PMC free article] [PubMed] [Google Scholar]
- [11].Wada T, Iwamoto T, Maeda S. Genetic diversity of the Mycobacterium tuberculosis Beijing family in East Asia revealed through refined population structure analysis. FEMS Microbiol Lett 2009;291:35–43. [DOI] [PubMed] [Google Scholar]
- [12].Fang Y, Ma Y, Lu Q, Sun J, Pei Y. An outbreak of pulmonary tuberculosis and a follow-up investigation of latent tuberculosis in a high school in an eastern city in China, 2016–2019. PLoS One 2021;16:e0247564. 10.1371/journal.pone.0247564 10.1371/journal.pone.0247564, PMID:33626108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ordaz-Vazquez A, Torres-Gonzalez P, Cruz-Hervert P, et al. Genetic diversity and primary drug resistance transmission in Mycobacterium tuberculosis in southern Mexico. Infect Genet Evol 2021;93:104994. 10.1016/j.meegid.2021.104994 10.1016/j.meegid.2021.104994, PMID:34245908. [DOI] [PubMed] [Google Scholar]
- [14].Conceicao EC, Salvato RS, Gomes KM, et al. Molecular epidemiology of Mycobacterium tuberculosis in Brazil before the whole genome sequencing era: a literature review. Mem Inst Oswaldo Cruz 2021;116:e200517. 10.1590/0074-02760200517 10.1590/0074-02760200517, PMID:33729319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Xiao DL. Chinese Ministry of Health. Summary of the national baseline survey of TB resistance. National Baseline Survey of Drug-resistant Tuberculosis (2007–2008). Beijing: People's Medical Publishing House; 2010. 02. [Google Scholar]
- [16].Pan D, Palittapongarnpim P, Chaiprasert A, et al. Infectivity of Mycobacterium tuberculosis genotypes and outcome of contact investigation in classroom in Guangxi, China. Biomed Res Int 2019;2019:3980658. 10.1155/2019/3980658 10.1155/2019/3980658, PMID:31111051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chamie G, Kato-Maeda M, Emperador DM, et al. Spatial overlap links seemingly unconnected genotype-matched TB cases in rural Uganda. PLoS One 2018;13:e0192666. 10.1371/journal.pone.0192666 10.1371/journal.pone.0192666, PMID:29438413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shao Y, Song H, Li G, et al. Relapse or re-infection, the situation of recurrent tuberculosis in Eastern China. Front Cell Infect Microbiol 2021;11:638990. 10.3389/fcimb.2021.638990 10.3389/fcimb.2021.638990, PMID:33816342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].You NN, Zhu LM, Li GL, et al. A tuberculosis school outbreak in China, 2018: reaching an often overlooked adolescent population. Epidemiol Infect 2019;147:e303. 10.1017/S0950268819001882 10.1017/S0950268819001882, PMID:31736459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rudeeaneksin J, Phetsuksiri B, Nakajima C, et al. Drug-resistant Mycobacterium tuberculosis and its genotypes isolated from an outbreak in western Thailand. Trans R Soc Trop Med Hyg 2021;115:886–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shi J, Zheng D, Zhu Y, et al. Role of MIRU-VNTR and spoligotyping in assessing the genetic diversity of Mycobacterium tuberculosis in Henan Province, China. BMC Infect Dis 2018;18:447. 10.1186/s12879-018-3351-y 10.1186/s12879-018-3351-y, PMID:30176820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Moe Sann WW, Namwat W, Faksri K, et al. Genetic diversity of Mycobacterium tuberculosis using 24-locus MIRU-VNTR typing and Spoligotyping in Upper Myanmar. J Infect Dev Ctries 2020;14:1296–305. [DOI] [PubMed] [Google Scholar]
- [23].Ansarin K, Sahebi L, Aftabi Y, Khalili M, Seyyedi M. Comparing IS6110-RFLP, PGRS-RFLP and IS6110-Mtb1/Mtb2 PCR methods for genotyping of Mycobacterium tuberculosis isolates. J Appl Microbiol 2020;129:1062–70. [DOI] [PubMed] [Google Scholar]
- [24].Alaridah N, Hallback ET, Tangrot J, et al. Transmission dynamics study of tuberculosis isolates with whole genome sequencing in southern Sweden. Sci Rep 2019;9:4931. 10.1038/s41598-019-39971-z 10.1038/s41598-019-39971-z, PMID:30894568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chee CBE, Lim LKY, Ong RTH, et al. Whole genome sequencing analysis of multidrug-resistant tuberculosis in Singapore, 2006–2018. Eur J Clin Microbiol Infect Dis 2021;40:1079–83. [DOI] [PubMed] [Google Scholar]
- [26].Meehan CJ, Moris P, Kohl TA, et al. The relationship between transmission time and clustering methods in Mycobacterium tuberculosis epidemiology. EBioMedicine 2018;37:410–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kwak SH, Choi JS, Lee EH, et al. Characteristics and risk factors associated with missed diagnosis in patients with smear-negative pulmonary tuberculosis. Korean J Intern Med 2021;36:S151–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yazdani-Charati J, Rezai MS, Fendereski A, Mohammadi S, Alipour N. Treatment delay and total delay among pulmonary tuberculosis patients in the North of Iran: application survival data analysis. Tanaffos 2017;16:13–21. 28638420. [PMC free article] [PubMed] [Google Scholar]