Abstract
Background:
The number of patients with hepatocellular carcinoma (HCC) is showing a growing trend all over the world. The metabolic microenvironment has been shown to play a key role in the pathogenesis of HCC in recent studies. The expression of metabolites and metabolic processes in tumor cells can be regulated by gene regulation mediated by long non-coding RNAs (lncRNAs), the abnormal expression of which is closely related to tumor occurrence and metastasis. However, the fundamental mechanism of applying metabolism-related lncRNAs to predicting HCC is still unclear.
Methods:
With the complete RNA sequence data and clinical data obtained from The Cancer Genome Atlas database and metabolism-related genes downloaded from the Kyoto Encyclopedia of Genes and Genomes database, with false discovery rate < 0.001, log fold change > 1.5 selected as the screening criteria for lncRNA, the relationship between the expression level of metabolism-related LncRNAs (MRLs) and the overall survival rate was determined by the Univariate Cox regression analyses with the establishment of the metabolic prognosis model by the application of Multivariate Cox regression analyses, revealing the different biological processes and signaling pathways in both high-risk groups and low-risk groups by Gene Ontology, Kyoto Encyclopedia of Genes and Genomes enrichment analysis, and gene set enrichment analysis, leading the expression of lncRNA to be assessed by the reverse transcription-polymerase chain reaction results.
Results:
The overall survival rate of HCC patients is significantly correlated with signature of 5-MRLs. The prognosis characteristics of lncRNA reveal the relatively high death rate of patients in the high-risk groups, with the predicted signals by functional and pathway enrichment analysis related to biosynthesis, metabolic process, and metabolic pathway, with the prognostic characteristics of 5-MRLs by the combined analysis showing that it is an independent factor of HCC superior to the traditional clinical indicators in predicting the prognosis. A trend of high-expression was shown in MRLs in tumors by reverse transcription-polymerase chain reaction.
Conclusion:
The new 5-MRLs as potential biomarkers provide more powerful prognostic information for HCC patients. In the future clinical treatment of HCC, it will provide doctors with more methods.
Keywords: hepatocellular carcinoma, metabolism-related long non-coding RNA, prognostic signature, The Cancer Genome Atlas
1. Introduction
Ranking in the sixth of the most common cancers, primary liver cancer comes in the fourth leading cause of cancer-related death, leading to, according to the 2020 statistic, about 906,000 new cases and 830,000 deaths annually,[1] with the hepatocellular carcinoma (HCC) accounting for 75% to 85% the confirmed liver cancer cases, thus making advancement in the diagnosis, treatment, and prognosis of HCC in light of the limitedness of the current ones.[2–4] Hepatitis A virus (HBV), hepatitis C virus (HCV) infection, obesity, and type 2 diabetes are the main causes of high risk of HCC.[5] Cancer cells receive, by generating cumulative changes in metabolism, regular and unconventional nutrition, with these nutrients used to create new biomass to maintain uncontrolled proliferation and metabolites selected to affect the survival of cancer cells themselves and various normal cell types in the tumor microenvironment. As the result of the selective evolution of tumor growth, proliferation and survival, support for the fundamental needs of cancer cells, such as increasing energy production, macromolecular biosynthesis, and maintaining redox balance is provided by metabolic changes,[6] pathway of which is favorable for tumor proliferation and growth with ensured energy and substrate, thus creating essential conditions for accelerating tumor growth in the tumor microenvironment.[7] These processes are reprogrammed in cancer metabolism, resulting in the hepatocarcinogenesis and development with the presence of abnormal cell metabolism. Innumerable studies have shown that lipid metabolism, glucose metabolism, purine metabolism, and inositol phosphate metabolism play an active role in the carcinogenesis of HCC which usually manifests a variety of metabolic changes such as increased aerobic glycolysis, enhanced synthesis of new lipids, depletion of glutamine, and imbalance of oxidative metabolism, through which energy and biological macromolecular substances are provided for the rapid growth and proliferation of tumor cells.[8] Also, tumor metabolism, with its correlation between HCC disclosed by these studies, is regulated by a variety of factors, such as changes in metabolic enzyme activity, aberrant gene expression, disturbance of signal pathways.
With more cancer data published by The Cancer Genome Atlas (TCGA) through which the required shared data sets can be accessed and assessed by the researchers at any time,[9] a powerful platform was provided by the development of high-throughput sequencing technology and TCGA for the discovery of novel biomarkers and metabolic targets of liver cancer, leading to the results that it is of great significance to obtain the characteristic metabolic genes and their regulatory mechanisms by bioinformatics in predicting the survival rate of HCC patients. This study was designed to identify metabolic-related lncRNAs associated with HCC prognosis on the basis of TCGA and Kyoto Encyclopedia of Genes and Genomes (KEGG) database, with the prognostic risk model established based on univariate and multivariate Cox regression analysis, prognostic characteristics analyzed according to the expression of lncRNA, and revealed the different biological processes and signaling pathways in both high-risk groups and low-risk groups by Gene Ontology (GO), KEGG enrichment analysis, and gene set enrichment analysis (GSEA). The consistency of these results of the samples verified by utilizing reverse transcription-polymerase chain reaction. Our goal is to use the expression profile of lncRNA to identify metabolism-related lncRNA, which will help us find potential prognostic indicators and promising treatment targets of HCC.
2. Materials and methods
2.1. Dataset source
From the TCGA database (https://xenabrowser.net/, accessed October 21, 2019) were the tumor samples and normal samples of HCC patients obtained with gene expression profiles, clinicopathological information, and from KEGG database (https://www.genome.jp/kegg/, accessed October 21, 2019) were the metabolism-related genes obtained with the sample collecting conforming to the TCGA guidelines and data access policies, and from TCGA was all the available clinical information downloaded, making it not necessary to gain the approval of the ethics committee.
2.2. Differential expression lncRNA screening
RNA-seq data were obtained from the whole genome expression profile including mRNA and lncRNA expression data, with false discovery rate (FDR) < 0.001, log (fold change) > 1.5 selected as the screening criteria for lncRNA with differential expression using the “limma” package on R (version 3.6.1). With the gene expression data obtained from TCGA intersecting with metabolism-related genes downloaded from KEGG and the selected genes used as metabolism-related genes (MRGs) in expression data, metabolism-related lncRNAs (MRLs) were identified according to the Pearson correlation coefficient between MRGs (|r| > 0.5, P < .001) and extracted lncRNAs, which were adopted in the following studies.
2.3. Construction of prediction features based on MRL expression
The expression of MRLs and their predictive roles by the “survival” package were determined with the performance of univariate Cox regression analyses by identifying P value < .01 as prognosis MRLs, revealing significant correlation between the expression level and overall survival (OS) of HCC patients. The prognosis of MRL was estimated with the “step” function in survival analyses by selecting the most suitable combination for predicting prognosis. With the incorporation of MRL identified by “step” function into multivariate Cox regression analyses by taking OS as a dependent variable, the multivariate Cox regression coefficient expression and the weight based on multivariate Cox regression coefficient (β) were brought together to construct a risk score model expressing prognosis,[10] thus forming the risk score formula: Risk Score = expression of MRL 1 × β 1 MRL 1 + expression of MRL 2 × β 2 MRL 2 + expression of MRL n × β n MRL n,[11] leading the accuracy of predicting the prognostic characteristics of HCC to be determined by receiver operating characteristic (ROC) curve of “survival” packages on R.[12]
2.4. Evaluation of MRL-based prognostic signature
With the prognosis model assessed by prediction features based on MRL expression, the survival status of the high and low response groups compared, and the predictive value of MRL on the prognostic characteristics of HCC patients evaluated, a nomogram was built on the basis of risk score and clinicopathological features to predict the prognosis survival rate, with the prognostic value of risk score and the impact of the risk score on the survival rate assessed according to stratified analysis and combined effect analysis, thus rendering the distribution pattern of high and low-risk cases determined by the principal component analysis (PCA).
2.5. Functional assessment and enrichment analysis
The “cluster Profiler” R package was used to analyze the co-expressed MRLs for GO terminology and Kyoto gene and genome Encyclopedia (KEGG) database path analysis, with the GO analysis revealing the MRL function in the biology process, cell component, and molecular function, and the KEGG analysis showing the pathway enrichment of MRLs. As P-value < .05 is of statistical significance in GO and KEGG enrichment analysis, the different functional phenotypes between the high and low-risk groups were explored for GSEA (version 4.0.1, http://www.broadinstitute.org/GSEA/index.jsp), and the Molecular Signatures Database (MSigDB) of c2 (c2.cp.kegg.v7.0.symbols.gmt) and c5 (c5.all.v7.0.symbols.gmt) was used to GESA, in which P < .05 and FDR < 0.05 were considered to be of statistical significance.
2.6. RNA extraction and real time polymerase chain reaction assay
With the ethical approval, approval number: 2020 (KY-E-172), granted by the First Affiliated Hospital of Guangxi Medical University of the Ethics Committee, and written informed consent obtained from all patients. Inclusion criteria: age from 18 to 75 years old, no gender limit; cases undergoing hepatectomy, and postoperative histopathological diagnosis of HCC. Six tumor tissues and the adjacent tissues from hepatectomy patients from 2020 to 2021 were collected from the First Affiliated Hospital of Guangxi Medical University of the Hepatobiliary Surgery Department, with subjects identity information accessible during or after data collection. The specimens were collected during the operations and frozen in liquid nitrogen immediately afterward, with total RNA extracted by using RNAiso Plus (TaKaRa, Japan) complying with the manufacturer's protocol and reversely transcribed into complementary DNA (cDNA) by using a PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, Japan) in the condition that real-time PCR in the ABI7500 real-time PCR system was carried out by using the TB Green Premix Ex Taq II (Tli RNaseH Plus) (2 × conc) (TaKaRa, Japan).
The primers pairs were:
MIR210HG Forward CCCTTTCTCTGGAGCACAC
MIR210HG Reverse TTCCCTGTTCCCTGCCT
AL365203.2 Forward ACACTTCATGGGCTTACTGCT
AL365203.2 Reverse GCTGCCTTTCACCTCTTACAC
AL031985.3 Forward CACCTATTCAACTTCCCCATT
AL031985.3 Reverse CCAAGGATTCCCCTAAACATC
AC099850.3 Forward GAAAATATGGAAACAGGAACAGGAC
AC099850.3 Reverse GGAAATCTCAAAACCCAAAGG
LUCAT1 Forward GGTGCCAAGGTCCCATA
LUCAT1 Reverse AAGCTCGGATTGCCTTAGA
2.7. Statistical analysis
The measurement data conforming to the normal distribution are expressed by the mean ± standard deviation, the comparison between groups is by t test; the count data is expressed by percentage, and the comparison between groups is by Chi-squared test; survival status comparisons between different subgroups were carried out via Kaplan–Meier survival analysis by log-rank test, with the clinical characteristics and survival time analyzed by the Cox hazard model and all statistical analyses implemented by SPSS version 22.0 (IBM Corporation, Armonk, NY) and R 3.6.1.
3. Results
3.1. Identification of based MRLs
lncRNA (FDR < 0.001, |log fold change| > 1.5) in 374 HCC samples and 50 normal samples were identified by using the “limma” package on R, with 370 HCC samples with complete survival information further analyzed, under the condition that a total number of 937 MRGs were obtained from KEGG database and the expression data of these genes in the liver cancer cohort of the TCGA database were extracted (Table S1, Supplemental Digital Content). Meanwhile, a total number of 2052 different lncRNAs of cancer tissues and the adjacent tissues were screened by 14,141 lncRNAs, with the different analysis results shown in Fig. 1A and B, indicating the result of 166 MRLs being identified between 937 MRGs and 14,141 lncRNAs by Pearson correlation analysis (|r| > 0.5 and P < .001).
Figure 1.
Differential expression LncRNA screening (A). Volcano plot of MRLs (B). Heatmap of MRLs in 374 HCC samples and 50 normal samples. HCC = hepatocellular carcinoma, MRL = metabolism-related LncRNA.
3.2. Construction of the MRLs prognostic signature predicts survival
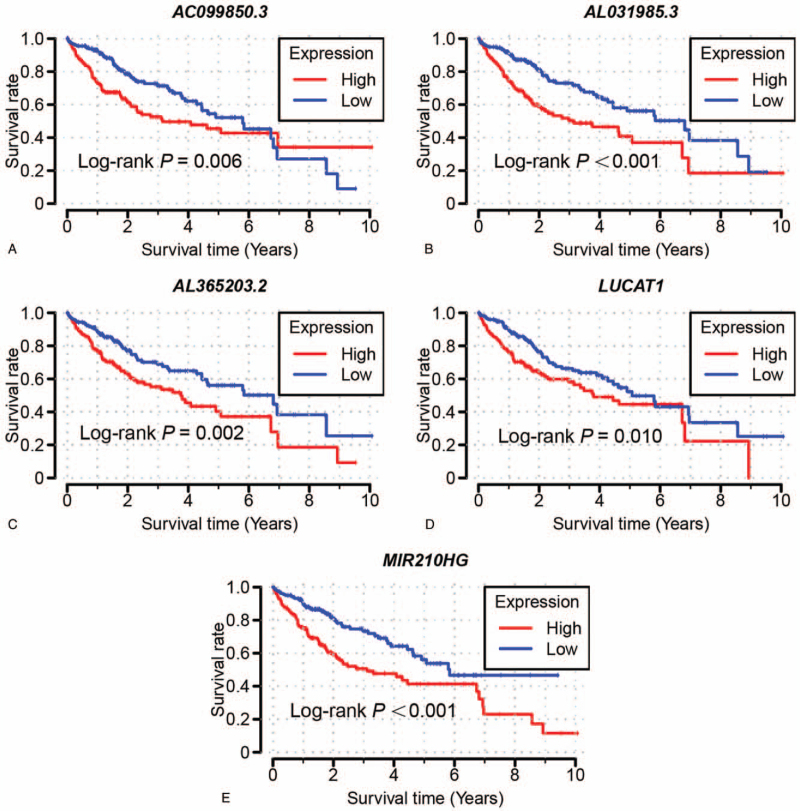
Univariate Cox analysis was used to analyze the expression level and survival time of 166 MRLs with “survival” package, with 20 of them significantly correlated with the survival time (P < .01, Table S2, Supplemental Digital Content). The best combination (5-MRLs: AC099850.3, AL031985.3, AL365203.2, LUCAT1, MIR210HG) was selected by the “step” function (Table 1). for the purpose of constructing a multivariate COX analysis model of MRLs related to prognosis, with prognostic analysis of 5-MRLs in the MRLs prognostic model falling into high and low-risk groups (Fig. 2), thus leading the contribution of prognostic MRLs to prognosis prediction to be determined by the multivariate Cox analysis. The risk score grades, survival outcomes, and prognosis of the 5-MRLs expression in different groups are shown in Fig. 3A–D, with the prognostic analysis of clinicopathological features and risk score showing that tumor stage and radical resection score were of statistical significance and the 2 factors were significantly related to the prognosis and survival of HCC (Table 2). Based on the median risk score of MRL expression, HCC patients were subdivided into high-risk groups and low-risk groups. Results show that, despite significant improvement made in the risk of death with high-risk score (P < .001; hazard ratio = 2.206; 95% confidence intervals = 1.539–3.163; Table 2, Fig. 3A–D), the median survival time (MST) of high-risk groups was still significantly shorter than that of low-risk groups (high risk vs low risk: 1149 days vs 2131 days). As displayed in ROC analysis using the survival package, the 1, 2, 3, and 5-year survival rates were respectively 0.769, 0.729, 0.703, and 0.648 (Fig. 3E), result in the conclusion that this prognostic feature based on the expression of LncRNA also is well-performed in predicting the survival rate of HCC, with the expression levels distribution of 5 MRLs in different subgroups shown in Fig. 4A and B.
Table 1.
The 5 MRLs of prognostic signature identified from Cox regression analysis.
| LncRNA | Ensemble ID | Hazard ratio∗ | Coefficient† | P value∗ |
| AC099850.3 | ENSG00000265303 | 1.523 | 0.188 | .001 |
| AL031985.3 | ENSG00000260920 | 3.205 | 0.597 | .001 |
| AL365203.2 | ENSG00000273038 | 1.654 | 0.225 | .001 |
| LUCAT1 | ENSG00000248323 | 1.692 | 0.367 | .001 |
| MIR210HG | ENSG00000247095 | 1.519 | 0.196 | .001 |
HCC = hepatocellular carcinoma, MRLs = metabolism-related long noncoding RNAs.
Data from the univariate Cox regression analysis in HCC cohort.
Data from the multivariate Cox regression analysis in HCC cohort.
Figure 2.
The Kaplan–Meier curve of 5-MRLs divided into high and low expression group. 5-MRLs selected through univariate cox regression analysis and multivariate cox risk model: A AC099850.3; B AL031985.3; C AL365203.2; D LUCAT1; E MIR210HG. MRL = metabolism-related LncRNA.
Figure 3.
Analysis of risk prognostic models for 5-MRLs. (A) The risk scores of the 5-MRLs of the low- and high-risk groups, (B) the distribution of the patient's survival status, and (C) the expression heat map. (D) Kaplan–Meier curves for low-risk and high-risk groups. (E) ROC curve analysis predicts survival of patients with HCC. HCC = hepatocellular carcinoma, MRL = metabolism-related LncRNA, ROC = receiver operating characteristic.
Table 2.
Clinical and pathologic characteristics of HCC patients and prognostic analysis.
| Variables | Count of events/total (n = 142) | MST, d | HR (95% CI) | P value |
| Age, y | .143 | |||
| ≤60 | 52/177 | 2532 | 1 | |
| >60 | 74/193 | 1622 | 1.303 (0.913–1.860) | |
| Gender | .362 | |||
| Female | 48/121 | 1560 | 1 | |
| Male | 78/249 | 2486 | 0.845 (0.588–1.214) | |
| Serum AFP, ng/mL∗ | .852 | |||
| ≤400 | 60/213 | 2456 | 1 | |
| >400 | 21/64 | 2486 | 1.049 (0.633–1.738) | |
| Child-Pugh grade† | .077 | |||
| A | 57/216 | 2542 | 1 | |
| B/C | 9/22 | 1005 | 1.872 (0.924–3.795) | |
| Ishak fibrosis score‡ | .847 | |||
| 0 | 29/74 | 2456 | 1 | |
| 1/2 | 7/31 | 1791 | 0.757 (0.325–1.762) | |
| 3/4 | 6/28 | NA | 0.686 (0.281–1.675) | |
| 5 | 2/9 | 1386 | 0.720 (0.170–3.056) | |
| 6 | 17/69 | NA | 0.750 (0.408–1.380) | |
| Tumor stage§ | <.001 | |||
| I | 41/171 | 2532 | 1 | |
| II | 25/85 | 1852 | 1.436 (0.871–2.369) | |
| III/IV | 47/90 | 770 | 2.751 (1.803–4.198) | |
| Histologic grade|| | .786 | |||
| G1 | 18/55 | 2116 | 1 | |
| G2 | 58/177 | 1685 | 1.148 (0.676–1.950) | |
| G3 | 41/121 | 1622 | 1.180 (0.676–2.060) | |
| G4 | 5/12 | NA | 1.825 (0.648–5.140) | |
| MVI¶ | .185 | |||
| No | 59/206 | 2131 | 1 | |
| Yes | 34/108 | 2486 | 1.331 (0.870–2.034) | |
| Radical resection# | .003 | |||
| R0 | 106/323 | 2116 | 1 | |
| R1/R2/RX | 17/40 | 837 | 2.137 (1.276–3.581) | |
| Risk index | <.001 | |||
| Low | 51/185 | 2131 | 1 | |
| High | 75/185 | 1149 | 2.206 (1.539–3.163) |
HCC = hepatocellular carcinoma, HR = hazard ratio, MST = media survival time.
Ninety three patients’ data were unavailable.
One hundred thirty two patient's data were unavailable.
One hundred fifty nine patient's data were unavailable.
Twenty four patients’ data were unavailable.
Five patients’ data were unavailable.
Fifty six patients’ data were unavailable.
Seven patients’ data were unavailable.
Figure 4.
Comparison of the expression levels of 5-MRLs. (A) Expression of 5-MRLs between cancer and para-cancerous, (B) expression of 5-MRLs between high and low-risk group. ∗∗∗P < .001. MRL = metabolism-related LncRNA.
3.3. Combined analysis of the prognostic characteristics of 5-MRLs
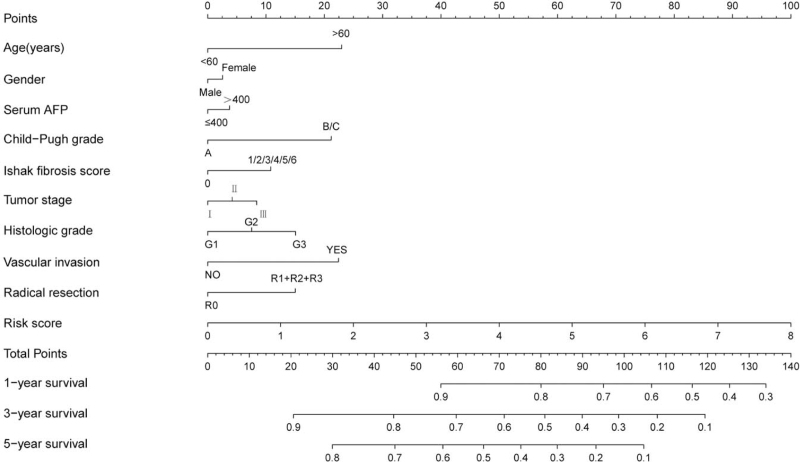
According to the combined effect analysis, the prognostic characteristics of 5-MRLs combined with the clinicopathological characteristics have a good performance in predicting the survival rate of HCC patients (Table 3, Fig. 5A–I), making it reasonable for the risk scores and clinical information to be integrated to construct a prognostic nomogram for predicting 1 to 0.3 to 0.5-year survival rates (Fig. 6). In this research, PCA analysis was carried out to describe the distribution pattern of prognosis in high-risk and low-risk populations, leading to the conclusion that, compared with traditional prognostic indicators, the risk score of nomogram has a more significant impact on the prognosis of HCC. Significantly different prognosis distributions of the high-risk and low-risk groups were obtained base on the 5 MRL prognostic models (Fig. 7A), while there was no significant difference in prognostic distributions of all genes, 166 MRLs, and 20 prognostic-related MRLs (Fig. 7B–D).
Table 3.
Combined effects analysis of clinicopathologic characteristics and the MRLs risk score in HCC patients.
| Groups | Risk | Variables | Count of events/total (n = 370) | MST, d | HR (95% CI) | P value |
| Age, y | ||||||
| A | Low risk | ≤60 | 20/87 | 2532 | 1 | |
| B | Low risk | >60 | 31/98 | 1791 | 1.224 (0.692–2.166) | .487 |
| C | High risk | ≤60 | 32/90 | 2542 | 2.211 (1.260–3.880) | .006 |
| D | High risk | >60 | 43/95 | 1005 | 3.046 (1.765–5.258) | <.001 |
| Gender | ||||||
| a | Low risk | Female | 24/59 | 2116 | 1 | |
| b | Low risk | Male | 27/126 | NA | 0.628 (0.356–1.108) | .108 |
| c | High risk | Female | 24/62 | 1135 | 1.595 (0.875–2.908) | .128 |
| d | High risk | Male | 51/123 | 1149 | 1.598 (0.971–2.630) | .065 |
| Serum AFP, ng/mL∗ | ||||||
| 1 | Low risk | ≤400 | 30/118 | 2456 | 1 | |
| 2 | Low risk | >400 | 7/27 | NA | 0.791 (0.341–1.834) | .584 |
| 3 | High risk | ≤400 | 30/95 | 2542 | 1.828 (1.090–3.065) | .022 |
| 4 | High risk | >400 | 14/37 | 2486 | 1.911 (1.004–3.638) | .049 |
| Child-Pugh grade† | ||||||
| (A) | Low risk | A | 29/121 | 3125 | 1 | |
| (B) | Low risk | B/C | 4/12 | 1624 | 1.668 (0.582–4.777) | .340 |
| (C) | High risk | A | 28/95 | 2542 | 1.765 (1.034–3.012) | .037 |
| (D) | High risk | B/C | 5/10 | 601 | 5.323 (1.623–11.511) | .003 |
| Ishak fibrosis score‡ | ||||||
| (a) | Low risk | 0 | 16/45 | 2456 | 1 | |
| (b) | Low risk | 1/2/3/4/5/6 | 17/75 | NA | 0.820 (0.401–1.676) | .587 |
| (c) | High risk | 0 | 13/29 | 1005 | 2.069 (0.964–4.443) | .062 |
| (d) | High risk | 1/2/3/4/5/6 | 15/62 | NA | 1.245 (0.584–2.657) | .570 |
| Tumor stage§ | ||||||
| (1) | Low risk | I/II | 28/136 | 2532 | 1 | |
| (2) | Low risk | III/IV | 15/35 | 931 | 2.852 (1.507–5.397) | .001 |
| (3) | High risk | I/II | 38/120 | 1852 | 2.448 (1.469–4.079) | <.001 |
| (4) | High risk | III/IV | 32/55 | 412 | 4.410 (2.637–7.374) | <.001 |
| Histologic grade|| | ||||||
| I | Low risk | G1/G2 | 39/139 | 2131 | 1 | |
| II | Low risk | G3/ G4 | 11/44 | NA | 0.855 (0.437–1.671) | .646 |
| III | High risk | G1/G2 | 37/93 | 1271 | 2.128 (1.350–3.354) | .001 |
| IV | High risk | G3/G4 | 35/89 | 1149 | 2.262 (1.408–3.633) | <.001 |
| MVI¶ | ||||||
| AA | Low risk | No | 29/123 | 2456 | 1 | |
| AB | Low risk | Yes | 12/41 | 3258 | 1.614 (0.818–3.184) | .168 |
| AC | High risk | No | 30/83 | 1372 | 2.591 (1.522–4.412) | <.001 |
| AD | High risk | Yes | 22/67 | 2486 | 2.253 (1.279–3.970) | .005 |
| Radical resection# | ||||||
| i | Low risk | R0 | 45/167 | 2456 | 1 | |
| ii | Low risk | R1 + R2 + RX | 5/14 | 837 | 4.402 (1.656–11.704) | .003 |
| iii | High risk | R0 | 61/156 | 1271 | 2.232 (1.510–3.300) | <.001 |
| iv | High risk | R1 + R2 + RX | 12/26 | 1135 | 3.387 (1.775–6.465) | <.001 |
Abbreviations: HCC = hepatocellular carcinoma, HR = hazard ratio, MST = media survival time, MVI = microvascular invasion.
Ninety three patients’ data were unavailable.
One hundred thirty two patient's data were unavailable.
One hundred fifty nine patient's data were unavailable.
Twenty four patients’ data were unavailable.
Five patients’ data were unavailable.
Fifty six patients’ data were unavailable.
Seven patients’ data were unavailable.
Figure 5.
Combined analysis of survival time by risk score and clinicopathological characteristics. (A) Age, (B) gender, (C) serum AFP, (D) Child-Pugh grade, (E) Ishak fibrosis score, (F) tumor stage, (G) histologic grade, (H) microvascular invasion, (I) radical resection.
Figure 6.
A nomogram between risk score and clinical data that predict 1-, 3-, and 5-year survival rates.
Figure 7.
PCA between the high and low-risk group of the genome expression set. (A) Prognostic model set; (B) all genes set; (C) 166 MRLs set; (D) 20 prognostic-related MRLs set. MRL = metabolism-related LncRNA.
3.4. Function and pathway enrichment analysis
Previous researches have established that proteins could not be encoded by the biological function of lncRNA, with many of them acting as CIS regulators and their functions linked to co-expressed mRNAs with the protein-coding ability.[13,14] Pearson correlation analysis showed that MRLs were highly correlated with metabolism and biosynthesis in function and pathway enrichment analysis, by the result of which these lncRNAs were mainly concentrated in 30 GO items and 18 KEGG pathways (P < .001), with GO enrichment analysis showing the obvious enrichment effects of the co-expressed MRLs on nucleoside phosphate, nucleotide, purine-containing compounds, nucleoside phosphate biosynthesis, and glycoside compound metabolism (Fig. 8A), and KEGG analysis disclosing the close connection between most co-expressed MRLs and purine metabolism, inositol phosphate metabolism, and DNA replication (Fig. 8B). The results of GSEA analysis on C2, which is enriched with polysaccharide biosynthesis, purine metabolism, pyrimidine metabolism, RNA degradation, and ubiquitin-mediated proteolysis (Fig. 9A–F), and C5, which is rich ATPase activity, DNA catalytic activity, and cell cycle regulation, in genomes of diverse risk populations were consistent with those of GO and KEGG of MRGs (Fig. 9G–I).
Figure 8.
Enrichment analysis of functions and pathways in 5-MRLs. (A) GO results reveal the MRL function in the biology process, cell component, and molecular function. (B) KEGG result shows the enrichment pathway of MRLs. KEGG = Kyoto Encyclopedia of Genes and Genomes, MRL = metabolism-related LncRNA.
Figure 9.
Gene set enrichment analysis results of 5-MRLS between high and low-risk groups. C2 (A–C): and C5 (D–I) gene sets. MRL = metabolism-related LncRNA.
3.5. Clinical validation of lncRNA levels of 5-MRLs
Six pairs of tumor and para-cancerous controls were analyzed to validate the lncRNA levels of 5-MRLs, revealing that the expressions of AC099850.3, AL031985.3, AL365203.2, LUCAT1, and MIR210HG were relatively highly expressed in tumors, indicating that the trend of our experimental results was almost consistent with data analysis (Fig. 10). The flow-process diagram of this experiment is shown in Fig. 11.
Figure 10.
RT-PCR tests on 5-MRLs in HCC tissues and para-cancerous tissues. (A) AC099850.3; (B) AL031985.3; (C) AL365203.2; (D) LUCAT1; (E) MIR210HG. ∗∗∗P ≤ .001. HCC = hepatocellular carcinoma, MRL = metabolism-related LncRNA, RT-PCR = reverse transcription-polymerase chain reaction.
Figure 11.
Research flow-process diagram.
4. Discussion
Most cases of hepatocellular carcinoma, a highly lethal tumor, are already at an advanced stage when diagnosed, with its morbidity close to its mortality, along with the continuous change of the metabolic environment resulting in the rapid development of HCC and the incessant changes of metabolic processes and metabolites leading to the rapid development of liver cancer, thus posing a major challenge to global public health and demonstrating the importance of comprehensively studying the relationship between metabolism-related genes and the prognosis of liver cancer. In this study, with the advancement in science and technology, the cancer prediction researches were carried out by utilizing TCGA in the search for metabolism-related genes that can predict the survival rate of HCC, with 5-MRLs proved by the bioinformatics methods to play an important role in predicting the prognosis of HCC and verified by the real-time PCR to be highly expressed in HCC tumors.
Long non-coding RNAs (LncRNAs), a common type of non-coding RNAs, are >200 nucleotides in length, with a great volume of published literature describing their biological processes, including gene activation, autophagy, metabolism, inflammation, and immune response,[15,16] and out-of-control lncRNA usually inducing cancer cells related to the occurrence, development, and metastasis of cancer as well,[17,18] proved by such examples as lncRNA-HEIH promoting the progression of liver cancer through the enhancer of Zete homolog 2,[19] lncRNA-UFC1 promoting tumor growth by targeting microRNA-34a,[20] lncRNA-PRAL being the key stimulator to the development of liver cancer,[21] lncRNA FAL1 being positive in HCC tissues and playing a role as an oncogene,[22] LINC00460 being a prognostic marker for lung cancer and has also been reported in recent literature.[23] And, in prostate cancer, the up-regulated lncRNA-HULC being associated with low overall survival of patients with prostate cancer.[24] In the latest study, Elhendawy et al[25] detected expression profiles of miRNAs in serum samples of 20 HCC patients and 10 healthy controls were detected and they have detected a panel of serum miRNAs that can be used as a reliable noninvasive screening biomarker of HCC. Despite the fact that, in previous studies, many abnormal lncRNAs have been found in various cancers, it is rare to see the reports of metabolic biomarkers on risk assessment and prognosis prediction in the early diagnosis and treatment of HCC.
To better understand the biological status and clinical status of cancer patients and help them to choose better treatment options, a diagnostic screening platform using metabolomics was established by the researchers in previous researches,[26] with the focus of identifying metabolism-related biomarkers of HCC,[27] as identifying the changes of HCC cells in every step of metabolism is essential for the tumor staging and treatment progress,[28] which was proved by the examples like significant reduction of, in the metabolomic analysis of liver cancer, both the metabolism related to the glycolysis process and the metabolism of malic acid, alanine, and linoleic acid.[29] Blood lipid measurement in patients with hepatitis C virus (HCV) and HCV-HCC showed that cholesterol, bile acid synthesis, and fatty acid oxidation were down-regulated compared with normal subjects,[13] with the increase of glycolysis, glycine, serine, threonine, aromatic amino acid metabolism, nucleotide metabolism, and succinate 5 secretion during HCC recurrence, leading a variety of metabolites and abnormal subtle changes in metabolic processes to be found before the occurrence and development of HCC owing to the above mentioned advantages of metabolomics.[14] It is a known fact that there are 3 obvious metabolic disorders found in tumor and hepatoma cell lines in the occurrence of liver cancer[30]: molecular processes related to hepatocyte function involving lipid/fatty acid/bile synthesis, inflammatory processes related to cytokine, sphingomyelin, and chondroitin sulfate metabolism, and nucleotide rich metabolic processes involving purine/pyrimidine and glucose-mediated, and glucose-mediated catabolism. With these results in agreement with the results of this paper. Lipid metabolism has been investigated as an energy source, microenvironment adaptation, and cell signal transduction of tumor cells, revealing the fact of the occurrence and development of liver cancer accelerated Fatty acid synthesis, β-oxidation, and changes in cell lipid composition.[31]
A poor prognosis was caused by high expression of adipose triglyceride lipase (ATGL) in HCC, the expression of which was regulated by LncRNA-NEAT1 which destroys the lipolysis of hepatoma cells and promotes the proliferation of hepatoma cells.[32] With the deregulation of lipid metabolism in hepatocellular carcinoma was regulated by LncRNA-HULC activating of the acyl CoA synthase subunit ACSL1, HULC mRNA level was shown, by real-time PCR detection, to be positively correlated with ACSL1 level in 60 HCC patients. About 77% (180/233) of HCC tissues were revealed, by immunohistochemical analysis of tissue microarray, to be positive for ACSL1, and the proliferation of HCC cells was promoted by the overexpression of cholesterol producing ACSL1, thus leaving the growth of HCC accelerated by the abnormal lipid metabolism enhanced by HULC.[33] Considered to be a marker of cancer is reprogrammed glucose metabolism (or Warburg effect) of aerobic glycolysis,[34] in which LINC01554 promotes the degradation of PKM2 proteasome, inhibits Akt/mTOR signaling pathway, and reduces the level of aerobic glycolysis in HCC cells, indicating the association between the down-regulation of LINC01554 and poor prognosis of HCC patients.[35] A rate-limiting enzyme implicated in purine metabolism is xanthine dehydrogenase, a low mRNA level of which is associated with a higher tumor stage and poor prognosis of HCC, with the migration and invasion of HCC cells promoted and the expression of xanthine dehydrogenase down-regulated by means of blocking or inhibiting xanthine dehydrogenase, which is conducive to the occurrence and development of HCC.[36]
In this study, it was investigated when 5-MRLs were used as an important prognostic signal to predict the survival rate of HCC patients, and 937 MRLs by “limma” package were identified to clarify the effect of 5-MRLs on HCC OS, with the construction of a multivariate prognostic analysis model based on MRLs. Meanwhile, the best combination of “step” function were further evaluated and screened, leaving 5-MRLs (AC099850.3, AL031985.3, AL365203.2, LUCAT1, MIR210HG) identified suitable for constructing prognosis characteristics, leading to the ROC analysis determination that the area under the curve values for 1-, 2-, 3-, and 5-year survival was 0.769, 0.729, 0.703, and 0.648, respectively, leaving the prognostic characteristics of 5-MRLs shown to be significantly correlated with HCC OS and the prognostic characteristics shown to be having a high accuracy in predicting the survival of HCC, by the survival analysis.
Stronger than the traditional clinical prediction index, prognostic characteristics obtained through the final analysis were confirmed by the stratified analysis to be independent factors of HCC, causing the combination of risk score and clinicopathological characteristics to make the prediction effect more obvious with combined effect analysis. The significant impact of the risk score on the survival rate of HCC was supported by the prognostic indicators of nomogram, with 5-MRLs shown to be divided into 2 different subgroups by PCA, indicating the significant difference between prognosis of HCC patients in high-risk group and that in low-risk group.
The 5-MRLs in biological process analysis by GO analysis were shown mainly to be enrichment in nucleoside phosphate, nucleotide biosynthesis, and glycosyl compounds metabolism, with the genes analyzed by cellular component which is rich in transferase complex, phosphorous group transfer, DNA, and nuclear DNA oriented RNA polymerase complex and the genes analyzed by molecular function which is abundant in nucleotide transferase activity and oxidoreductase activity. Most of the co-expressed MRGs were shown by KEGG analysis to be closely linked to DNA replication, purine metabolism, and inositol phosphate metabolism. Besides, according to GSEA analysis, C2 genes were significantly rich in polysaccharide biosynthesis, purine metabolism, pyrimidine metabolism, RNA degradation, and ubiquitin-mediated proteolysis, while C5 genes were abundant in ATPase activity, DNA catalytic activity, and cell cycle regulation. From a variety of studies, these 5-MRLs, which can serve as potential independent risk factors and prognostic indicators for HCC, have profound potential value in the diagnosis and prognosis of HCC patients.
The expression of 5-MRLs in HCC was finally confirmed by PCR, with the expressions of AC099850.3, AL031985.3, AL365203.2, LUCAT1, and MIR210HG relatively highly expressed in tumors, revealing the experimental results were generally consistent with the results from the TCGA, thus confirming the reliability of the model designed in this research.
AC099850.3, AL031985.3, AL365203.2, LUCAT1, and MIR210HG were included in the 5-MRLs, among which MIR210HG overexpression was found to be one of the independent factors of hepatocarcinogenesis by Wang et al,[37] with MIR210HG serving as a carcinogenic lncRNA in liver cancer. MIR210HG has proved to be the strongest candidate lncRNA for predicting the prognosis of colon cancer, with the high expression of MIR210HG confirmed, by Ruan et al,[38] to be related to the shortening of overall survival time in colon cancer in an investigation into MIR210HG which was assumed to be an important biomarker for glioma diagnosis by Min et al,[39] LUCAT1 expression proved to be an independent predictor of poor prognosis and assumed to be a useful biomarker for patients with liver cancer by Jiao et al,[40] and the lncRNA-miRNA-mRNA ceRNA network constructed to investigate lncRNA by Zhou et al,[41] revealing a new lncRNA (AC099850.3) related to ceRNA, which is conductive to the diagnosis and treatment of squamous cell carcinoma of the tongue. On the basis of RNA sequencing, including AL031985.3, a 9-lncRNAs prognosis model was established by Deng et al.[42] In those study, showing the predictability of the prognosis of HCC, achieving the results similar to results for MIR210HG, AL031985.3, AC099850.3, LUCAT1 in this study. However, there are no other relevant literature about the effects of lncRNA-AL365203.2 on cancer occurrence, development, and prognosis, leading the functions of these lncRNAs to be supported by future experiments and in-depth studies.
Previous works with different focuses on lncRNA analysis of HCC prognosis by TCGA have been found in literature, while in this study, a prognostic risk score model for 5-MRLs, with better performance than traditional prognostic indicators, was established to foresee HCC through comprehensive survival analysis of prognostic characteristics. However, in spite of a good application prospect in predicting the prognosis of liver cancer, there are still some limitations in the current research: first, a comprehensive survival analysis based on lncRNA expression is unable to be carried out due to such incomplete clinical parameters in TCGA as the unavailable details of postoperative treatment; second, results in this study may be biased owing to the relative small number of patient samples; third, a lncRNA was rarely reported in the literature, thus leaving the research work to be further experimented and verified for the purpose of remedying the limitations listed.
5. Conclusion
Identified by genome-wide screening analysis of the HCC cohort from TCGA database the 5 MRLs were consistent with the characteristics of MRLs and had independent prognostic significance in predicting the clinical prognosis of HCC patients, with the potential significance, as the result of constructing MRL based prognostic markers, in the clinical application of potential prognostic biomarkers and therapeutic targets for HCC and other malignant tumors.
Acknowledgments
The author thanks Junxin He, Jian Li, Jue Wang, Qilin Yi, Zhujing Lan, and Keyu Huang for their help in surgery and follow-up data collection. The authors would like to thank Editage for English language editing.
Author contributions
Conceptualization: Wei Wang, Zhenfeng Deng, Zongrui Jin, Guolin Wu, Jilong Wang, Hai Zhu, Banghao Xu, Zhang Wen, Ya Guo.
Data curation: Wei Wang, Zhenfeng Deng, Guolin Wu.
Formal analysis: Wei Wang, Zhenfeng Deng, Zongrui Jin, Guolin Wu.
Funding acquisition: Wei Wang, Zhenfeng Deng, Zhang Wen, Ya Guo.
Investigation: Wei Wang, Zhenfeng Deng, Zongrui Jin, Jilong Wang, Hai Zhu, Banghao Xu, Zhang Wen, Ya Guo.
Methodology: Wei Wang, Zhenfeng Deng, Zongrui Jin, Guolin Wu, Jilong Wang, Hai Zhu, Banghao Xu, Zhang Wen, Ya Guo.
Project administration: Wei Wang, Zhenfeng Deng, Jilong Wang, Hai Zhu, Banghao Xu, Zhang Wen, Ya Guo.
Resources: Wei Wang, Zhenfeng Deng.
Software: Wei Wang, Zhenfeng Deng.
Supervision: Jilong Wang, Hai Zhu, Zhang Wen, Ya Guo.
Validation: Wei Wang, Zhenfeng Deng, Guolin Wu.
Visualization: Wei Wang, Zhenfeng Deng, Guolin Wu.
Writing – original draft: Wei Wang, Zhenfeng Deng.
Writing – review & editing: Wei Wang, Zhenfeng Deng, Zongrui Jin, Guolin Wu, Jilong Wang, Hai Zhu, Banghao Xu, Zhang Wen, Ya Guo.
Supplementary Material
Supplementary Material
Footnotes
Abbreviations: FDR = false discovery rate, GO = Gene Ontology, GSEA = gene set enrichment analysis, HCC = hepatocellular carcinoma, KEGG = Kyoto Encyclopedia of Genes and Genomes, MRG = metabolism-related gene, MRL = metabolism-related LncRNA, OS = overall survival, PCA = principal components analysis, ROC = receiver operating characteristic, TCGA = The Cancer Genome Atlas.
How to cite this article: Wang W, Deng Z, Jin Z, Wu G, Wang J, Zhu H, Xu B, Wen Z, Guo Y. Bioinformatics analysis and experimental verification of five metabolism-related lncRNAs as prognostic models for hepatocellular carcinoma. Medicine. 2022;101:4(e28694).
WW, ZD, YG, and ZW contributed equally to this work.
This work was supported by the National Natural Science Foundation of China (grant no. 81560387, 81902983), the Guangxi Natural Science Foundation of China (grant no. 2018JJB140382), “Medical Excellence Award” funded by the creative research development grant from the first affiliated hospital of Guangxi Medical University (grant no.180327), the Guangxi medical and health technology development and application project (grant no. S2019097, S2018100). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials: The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate: All patients provided written informed consent, and the study was approved by the Ethics Committee of The First Affiliated Hospital of Guangxi Medical University (Nanning, China).
Patient consent for publication: Not applicable.
The authors declare that they have no competing interests.
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
Supplemental digital content is available for this article.
References
- [1].Sung H, Ferlay J, Siegel R, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- [2].Liver EAftSot. EASL Clinical Practice Guidelines: management of hepatocellular carcinoma. J Hepatol 2018;69:182–236. [DOI] [PubMed] [Google Scholar]
- [3].Yang J, Hainaut P, Gores G, Amadou A, Plymoth A, Roberts L. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol 2019;16:589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301–14. [DOI] [PubMed] [Google Scholar]
- [5].Miller K, Ortiz A, Pinheiro P, et al. Cancer statistics for the US Hispanic/Latino population, 2021. CA Cancer J Clin 2021;71:466–87. [DOI] [PubMed] [Google Scholar]
- [6].De Matteis S, Ragusa A, Marisi G, et al. Aberrant metabolism in hepatocellular carcinoma provides diagnostic and therapeutic opportunities. Oxid Med Cell Longev 2018;2018:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mossenta M, Busato D, Dal Bo M, Toffoli G. Glucose metabolism and oxidative stress in hepatocellular carcinoma: role and possible implications in novel therapeutic strategies. Cancers (Basel) 2020;12: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ning Z, Tan G. Cancer metabolism: a novel perspective on precision diagnosis and treatment for liver cancer. Zhonghua Wai Ke Za Zhi 2020;58:31–6. [DOI] [PubMed] [Google Scholar]
- [9].Wang M, Wen T, He L. A six-microRNA set as prognostic indicators for bile duct cancer. Int J Clin Exp Med 2015;8:17261–70. [PMC free article] [PubMed] [Google Scholar]
- [10].Lossos IS, Czerwinski DK, Alizadeh AA, et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med 2004;350:1828–37. [DOI] [PubMed] [Google Scholar]
- [11].Deng Z, Wang J, Xu B, et al. Mining TCGA Database for tumor microenvironment-related genes of prognostic value in hepatocellular carcinoma. Biomed Res Int 2019;2019:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Combescure C, Perneger T, Weber D. Prognostic ROC curves: a method for representing the overall discriminative capacity of binary markers with right-censored time-to-event endpoints. Epidemiology 2014;25:103–9. [DOI] [PubMed] [Google Scholar]
- [13].Wu J, Skill N, Maluccio M. Evidence of aberrant lipid metabolism in hepatitis C and hepatocellular carcinoma. HPB 2010;12:625–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ye G, Zhu B, Yao Z, et al. Analysis of urinary metabolic signatures of early hepatocellular carcinoma recurrence after surgical removal using gas chromatography-mass spectrometry. J Proteome Res 2012;11:4361–72. [DOI] [PubMed] [Google Scholar]
- [15].Susan C, Fitzgerald KA. Cytokines and long noncoding RNAs. Cold Spring Harb Perspect Biol 2017;10:a028589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mathy NW, Chen X. LncRNAs and their transcriptional control of inflammatory responses. J Biol Chem 2017;292: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yuan J, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014;25:666–81. [DOI] [PubMed] [Google Scholar]
- [18].Tang J, Jiang R, Deng L, Zhang X, Wang K, Sun B. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget 2015;6:4505–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yang F, Zhang L, Huo X, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology 2011;54:1679–89. [DOI] [PubMed] [Google Scholar]
- [20].Cao C, Sun J, Zhang D, et al. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of β-Catenin in HCC cells. Gastroenterology 2015;148:415.e18–26.e18. [DOI] [PubMed] [Google Scholar]
- [21].Zhou C, Yang F, Yuan S, et al. Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. Hepatology 2016;63:850–63. [DOI] [PubMed] [Google Scholar]
- [22].Li B, Mao R, Liu C, Zhang W, Tang Y, Guo Z. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci 2018;197:122–9. [DOI] [PubMed] [Google Scholar]
- [23].Ye J, Cheng Y, Deng J, Tao W, Wu L. LncRNA LINC00460 promotes tumor growth of human lung adenocarcinoma by targeting miR-302c-5p/FOXA1 axis. Gene 2019;685:76–84. [DOI] [PubMed] [Google Scholar]
- [24].Zheng P, Li H, Xu P. High lncRNA HULC expression is associated with poor prognosis and promotes tumor progression by regulating epithelial-mesenchymal transition in prostate cancer. Arch Med Sci 2018;14:679–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Elhendawy M, Abdul-Baki E, Abd-Elsalam S, et al. MicroRNA signature in hepatocellular carcinoma patients: identification of potential markers. Mol Biol Rep 2020;47:4945–53. [DOI] [PubMed] [Google Scholar]
- [26].Wei J, Xie G, Zhou Z, et al. Salivary metabolite signatures of oral cancer and leukoplakia. Int J Cancer 2011;129:2207–17. [DOI] [PubMed] [Google Scholar]
- [27].Xue R, Lin Z, Deng C, et al. A serum metabolomic investigation on hepatocellular carcinoma patients by chemical derivatization followed by gas chromatography/mass spectrometry. Rapid Commun Mass Spectrom 2008;22:3061–8. [DOI] [PubMed] [Google Scholar]
- [28].Wang X, Zhang A, Sun H. Power of metabolomics in diagnosis and biomarker discovery of hepatocellular carcinoma. Hepatology 2013;57:2072–7. [DOI] [PubMed] [Google Scholar]
- [29].Beyoğlu D, Imbeaud S, Maurhofer O, et al. Tissue metabolomics of hepatocellular carcinoma: tumor energy metabolism and the role of transcriptomic classification. Hepatology 2013;58:229–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ramesh V, Ganesan K. Integrative functional genomic analysis unveils the differing dysregulated metabolic processes across hepatocellular carcinoma stages. Gene 2016;588:19–29. [DOI] [PubMed] [Google Scholar]
- [31].Sangineto M, Villani R, Cavallone F, Romano A, Loizzi D, Serviddio G. Lipid metabolism in development and progression of hepatocellular carcinoma. Cancers (Basel) 2020;126: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liu X, Liang Y, Song R, et al. Long non-coding RNA NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular carcinoma proliferation. Mol Cancer 2018;17:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cui M, Xiao Z, Wang Y, et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res 2015;75:846–57. [DOI] [PubMed] [Google Scholar]
- [34].Lunt S, Vander Heiden M. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol 2011;27:441–64. [DOI] [PubMed] [Google Scholar]
- [35].Zheng Y, Li L, Jia Y, et al. LINC01554-mediated glucose metabolism reprogramming suppresses tumorigenicity in hepatocellular carcinoma via downregulating PKM2 expression and inhibiting Akt/mTOR Signaling pathway. Theranostics 2019;9:796–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chen G, Ye T, Chen H, et al. Xanthine dehydrogenase downregulation promotes TGFβ signaling and cancer stem cell-related gene expression in hepatocellular carcinoma. Oncogenesis 2017;6:e382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang Y, Li W, Chen X, Li Y, Wen P, Xu F. MIR210HG predicts poor prognosis and functions as an oncogenic lncRNA in hepatocellular carcinoma. Biomed Pharmacother 2019;111:1297–301. [DOI] [PubMed] [Google Scholar]
- [38].Ruan Z, Xu Z, Li Z, Lv Y. Integral analyses of survival-related long non-coding RNA MIR210HG and its prognostic role in colon cancer. Oncol Lett 2019;18:1107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Min W, Dai D, Wang J, et al. Long Noncoding RNA miR210HG as a potential biomarker for the diagnosis of glioma. PLoS One 2016;11:e0160451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jiao Y, Li Y, Ji B, Cai H, Liu Y. Clinical value of lncRNA LUCAT1 expression in liver cancer and its potential pathways. J Gastrointestin Liver Dis 2019;28:439–47. [DOI] [PubMed] [Google Scholar]
- [41].Zhou R, Zhang E, Sun Q, et al. Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue. BMC Cancer 2019;19:779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Deng B, Yang M, Wang M, Liu Z. Development and validation of 9-long Non-coding RNA signature to predicting survival in hepatocellular carcinoma. Medicine (Baltimore) 2020;99:e20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.